Abstract
Cell cycle (CC) and TP53 regulatory networks are frequently deregulated in cancer. While numerous genome-wide studies of TP53 and CC-regulated genes have been performed, significant variation between studies has made it difficult to assess regulation of any given gene of interest. To overcome the limitation of individual studies, we developed a meta-analysis approach to identify high confidence target genes that reflect their frequency of identification in independent datasets. Gene regulatory networks were generated by comparing differential expression of TP53 and CC-regulated genes with chromatin immunoprecipitation studies for TP53, RB1, E2F, DREAM, B-MYB, FOXM1 and MuvB. RNA-seq data from p21-null cells revealed that gene downregulation by TP53 generally requires p21 (CDKN1A). Genes downregulated by TP53 were also identified as CC genes bound by the DREAM complex. The transcription factors RB, E2F1 and E2F7 bind to a subset of DREAM target genes that function in G1/S of the CC while B-MYB, FOXM1 and MuvB control G2/M gene expression. Our approach yields high confidence ranked target gene maps for TP53, DREAM, MMB-FOXM1 and RB-E2F and enables prediction and distinction of CC regulation. A web-based atlas at www.targetgenereg.org enables assessing the regulation of any human gene of interest.
INTRODUCTION
The tumor suppressors RB and TP53 serve central roles in regulation of cell cycle (CC) gene expression. TP53 mediates its tumor suppressor function as a transcription factor to activate a plethora of target genes (1,2). In recent years, several genome-wide analyses have been used to identify TP53 target genes and each has identified many shared as well as unique candidates (3–9). However, the increased number of available datasets has not led to a more complete picture of TP53 target genes since the overlap between any two expression profile studies is often quite small. The apparent discrepancies between studies have made it difficult to be confident in the regulation of a specific gene of interest across multiple studies. Furthermore, recent genome-wide analyses suggest that TP53 itself may function exclusively as a transcription activator and not as a direct repressor (10).
Similarly, starting with the study by Whitfield et al., CC-regulated genes have been identified in several expression profiling analyses of synchronized cells (11–15). However, there is limited overlap between studies. Furthermore, it has become apparent that there are several distinct transcription factors that regulate CC genes but it is not clear how they interact with each other. The repressive retinoblastoma protein (RB, encoded by the RB1 gene) and the activating E2F transcription factors E2F1, E2F2 and E2F3 are central to regulation of the CC genes (16). However, it is not clear how RB and the activating E2Fs contribute to regulation of CC-regulated genes late in the CC during G2 and mitosis. Instead, the DREAM (DP, RB-like, E2F4 and MuvB) complex that does not contain either RB or E2F1 functions as a master coordinator of CC transcription (17–19). DREAM consists of the RB-like pocket proteins p130 (RBL2) or p107 (RBL1), the repressor E2F transcription factor E2F4 or E2F5 together with DP1 and the MuvB core complex that contains LIN9, LIN37, LIN52, LIN53 (RBBP4) and LIN54. Similar to RB, the DREAM complex is important for repression of CC gene expression during quiescence and early G1. When cells exit quiescence and enter into the CC, the repressive components p130/p107, E2F4/5 and DP1 become inactivated and the MuvB core forms a new complex with B-MYB (MYBL2) and FOXM1 that drives expression of a distinct subset of CC genes (13,20–21). The activating MMB (B-MYB-MuvB)-FOXM1 complex binds to the promoters of G2/M CC genes via the CC genes homology region (CHR) motif in their promoters (21–23). Although RB-E2F, DREAM and MMB-FOXM1 form distinct CC regulatory complexes, their target genes are often grouped together and termed ‘RB-E2F targets’ or ‘E2F-responsive genes’. In this case, ‘RB’ refers to all three pocket proteins, RB, p107 and p130, and ‘E2F’ refers to all E2F transcription factors E2F1-8 making it challenging to evaluate the specific regulation of any given CC gene.
Crosstalk between the TP53 and CC gene regulatory networks is well-known. CC genes are often found to be regulated in a TP53-dependent manner (24) and this regulation is mediated at least in part by the TP53 target gene p21 (CDKN1A) (25,26) in concert with RB (27–29) and DREAM (30–32). However, it is not fully understood how TP53, RB and DREAM coordinate their efforts to regulate the CC and how they cooperate with additional CC regulators such as E2F7, B-MYB and FOXM1.
To take advantage of many large genome-wide datasets focused on TP53 and the CC, we developed a meta-analysis approach that integrates expression profiling datasets and chromatin binding sites to identify gene regulatory mechanisms. We show that integration of data from a variety of cell types and perturbations can provide high confidence maps of TP53 regulated genes ranked by the number of datasets that agree on their regulation. A similar approach was used to interrogate DREAM, MMB-FOXM1, RB-E2F studies and identify 1408 high confidence CC-regulated genes to provide an overview of how these factors function to regulate CC gene expression. This resource is made available as a web-based atlas on www.targetgenereg.org providing a useful tool to identify the TP53- and CC-dependent transcriptional regulation of any gene of interest.
MATERIALS AND METHODS
Meta-analysis approach
In an effort to generate a more complete picture of the gene regulatory networks governed by TP53 and the CC, we combined expression profiling and transcription factor binding datasets and ranked individual genes by the number of datasets that found them to be differentially expressed or bound by a transcription factor.
Publicly available datasets involving TP53 and CC gene regulation were curated. In many cases microarray data was available at a pre-processed stage that included normalization. In these cases GEO2R (33,34) was used to obtain fold expression changes and P-values, which were adjusted for multiple testing using Benjamini-Hochberg correction. For the remaining microarray datasets and for the RNA-seq datasets fully pre-analyzed data presenting genes with their fold expression changes and adjusted P-values were made available by the respective authors. Common thresholds for absolute log2 (fold-change expression) ≥ 0.5 and adj. P-value ≤ 0.05 were employed to identify significantly differentially expressed genes. In some cases, deviating thresholds were used to conform to settings used in the original study. For CC expression profiling studies, the datasets were available pre-analyzed providing significantly differentially expressed genes with the CC phase that displays peak expression.
Genes were ranked by p53 Expression Scores reflecting the number of datasets finding the gene to be significantly upregulated minus the number of datasets that find the gene to be downregulated upon TP53 activation. Genes were ranked by the number of CC datasets that identify the gene as CC regulated. In addition, genes were ranked by a CC Expression Score reflecting the number of datasets finding the gene to display peak expression during ‘G2’ or ‘G2/M’ minus the number of datasets finding the gene to be a ‘G1/S’ or ‘S-phase’ expressed gene.
Chromatin immunoprecipitation (ChIP) peak datasets were publicly available and intersections of binding peaks and promoter regions were calculated using BETA-minus in Cistrome (35,36). Protein binding was required to occur within 1000 bp around the transcriptional start site (TSS) except for TP53, where binding was required to occur within 25 000 or 2500 bp of the TSS. Similar to expression profiling datasets, genes were ranked by the number of ChIP datasets that identify a binding peak near the gene's TSS.
Additional information is provided in the Supplementary Methods.
RESULTS
Meta-analysis of TP53-regulated genes across cell types and treatments
Identification of TP53 responsive genes has been a research focus for decades with many genome-wide gene expression datasets becoming available recently. We aimed to combine multiple such datasets to generate a more complete picture of the TP53 gene regulatory network. Because it is generally agreed that gene expression data from different experimental platforms are not directly comparable, we instead used a unique meta-analysis approach that ranks genes by the number of datasets that find them significantly differentially regulated. For this meta-analysis, we assessed 18 845 protein-coding genes and asked how many individual datasets found them to be significantly differentially regulated upon TP53 activation (see ‘Materials and Methods’ section).
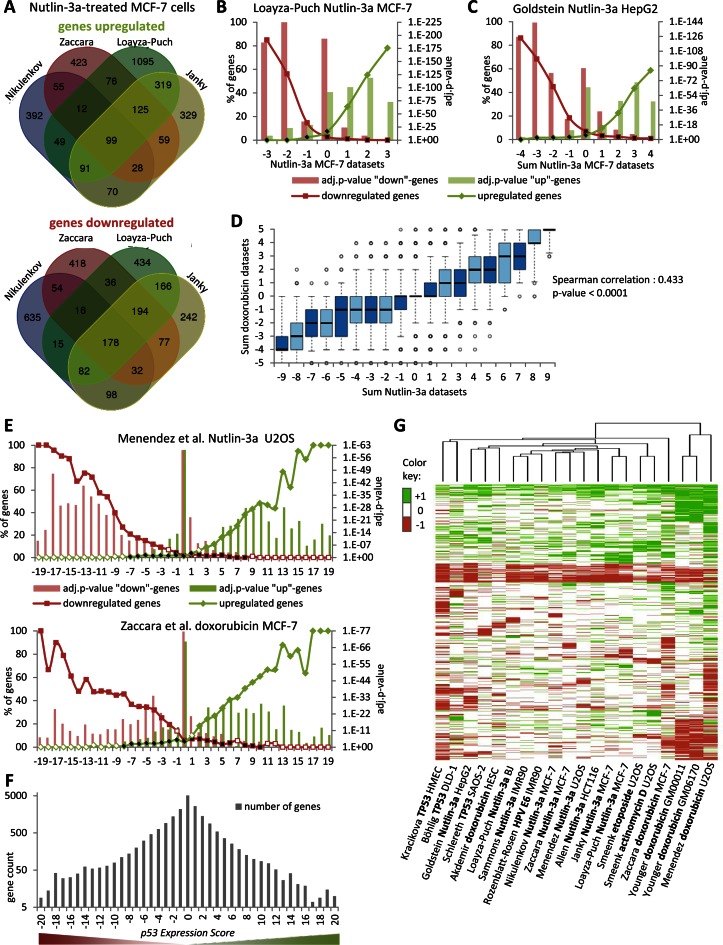
Initially, we compared four different datasets using Nutlin-3a treated MCF-7 cells (5,37–39). Even though these experiments were performed in a similar manner, the overlap between any two datasets was small. For example, 10.4–39.5% of responsive genes were identified between any two datasets and 60.5–89.6% of genes were unique to one study (Figure 1A and Supplementary Figure S1A–D). While there were minor differences in the protocols to isolate mRNA, different platforms and software were used to quantify gene expression. We reasoned that differential gene expression that was not reproducible across platforms was more likely to represent noise. We asked whether integration of these four datasets would result in a more robust joint dataset comprising genes that were repeatedly identified to be regulated. In each dataset, a gene was classified as upregulated ‘+1’, downregulated ‘−1’ or not regulated ‘0’. We plotted the subset of genes that were considered significantly up- or downregulated in one dataset against the sum of the remaining three datasets (Figure 1B). We observed that when several datasets agreed on a gene being significantly regulated the more likely that it was also identified by the remaining dataset. Thus, the number of datasets that agree on a gene's regulation reflects a confidence score. We refer to this as ‘step-wise meta-analysis’.
Figure 1.
Meta-analysis of TP53-dependent gene expression. (A) Venn diagram displaying the overlap of genes that were detected as upregulated or downregulated by TP53 activation in datasets from Nikulenkov et al., Zaccara et al., Loayza-Puch et al. and Janky et al. (B) In each dataset on TP53-dependent gene regulation, a gene can be found as upregulated ‘+1’, downregulated ‘−1’ or not regulated ‘0’. The number of genes identified in a Nutlin-3a MCF-7 dataset as either upregulated or downregulated is compared to the sum of the remaining three datasets from Nutlin-3a treated MCF-7 cells (see Supplementary Figure S1A–D for more). (C) The number of genes identified in datasets from other cell types treated with Nutlin-3a compared to the sum of the four Nutlin-3a MCF-7 datasets (see Supplementary Figure S1E–I for more) (D) Boxplot displaying the sum of the five doxorubicin datasets compared to the sum of the nine Nutlin-3a datasets. Correlation coefficient and two-tailed P-value was calculated using GraphPad Prism version 6.00. (E) Integration of 20 datasets on TP53-dependent gene regulation from multiple cell types and treatments. The number of genes identified in each dataset as either upregulated or downregulated is compared to the sum of the remaining 19 datasets (see Supplementary Figures S3 and 4). (B, C and E) A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). White data points are not significantly different. (F) Hierarchical clustering of the 10 000 most variant genes across the 20 TP53 datasets. (G) The number of genes is displayed that is found in each of the 41 p53 Expression Score groups.
Next, we asked whether datasets using other cell types treated with Nutlin-3a could be integrated with the MCF-7 datasets. We compared the genes affected by Nutlin-3a treatment in HepG2 (40), U2OS (6), IMR90 (41), BJ (39) and HCT116 (8) cells against the sum of the four MCF-7 datasets (Figure 1C and Supplementary Figure S1E–I). We found that when more Nutlin-3a MCF-7 datasets agreed a gene was regulated by TP53, the more likely it was regulated in other cell lines.
In a similar manner, we integrated five datasets assessing TP53-dependent genes responsive to doxorubicin treatment (6,37,42–43) (Supplementary Figure S2). Comparing the sum of the five doxorubicin and nine Nutlin-3a datasets revealed a strong correlation (Figure 1D) with a common set of genes up- or downregulated by TP53 across multiple cell types and treatments. Consequently, we integrated datasets using alternative methods to induce TP53: etoposide or actinomycin D treatment (4), overexpression of TP53 cDNA (7,44–45) and inactivation of TP53 with human papilloma virus (HPV) E6 (46). Each of these datasets detected genes that were also identified by many of the remaining 19 datasets (Figure 1E; Supplementary Figures S3 and 4). We based our meta-analysis on these 20 datasets that displayed the least amount of data heterogeneity. In contrast, a dataset from RITA treated HCT116 cells (47) and a meta-analysis of IR treated cells (48) failed to find a substantial number of genes that was identified by most of the other 20 datasets (Supplementary Figure S5) and were excluded from this analysis (see below).
We performed unsupervised hierarchical clustering of the regulation profile for the 10 000 most variant genes to test whether cell types and treatments could be distinguished. We found that all Nutlin-3a studies formed a cluster with cells expressing HPV E6 or treated with etoposide or actinomycin D (Figure 1F). Smaller clusters of Nutlin-3a treated fibroblasts (IMR90 and BJ) and etoposide or actinomycin D treated U2OS cells were also identified. Studies using cells overexpressing TP53 clustered separately, while studies using doxorubicin treated cells shared the least similarity with other studies. Although data heterogeneity could account for substantial differences between any two datasets derived from the same cell type and treatment, the clustering analysis indicates that gene expression profiles of similar TP53 activation mechanism are similar; this also holds for similar cell type, however to a smaller extent.
Integration of the 20 datasets on TP53-dependent gene regulation resulted in 41 gene groups ranging from −20 to +20 in ‘p53 Expression Score’ (Supplementary Table S1). In general, the number of genes declines substantially with the number of datasets that agree on a gene's regulation (Figure 1G). P-values were dependent in part on group size, accounting for the smallest P-values found in the p53 Expression Score group ‘0’. Together, integration of the 20 datasets revealed that many genes were commonly regulated across cell types and treatments.
Proximal TP53 binding is associated with activation of 311 target genes
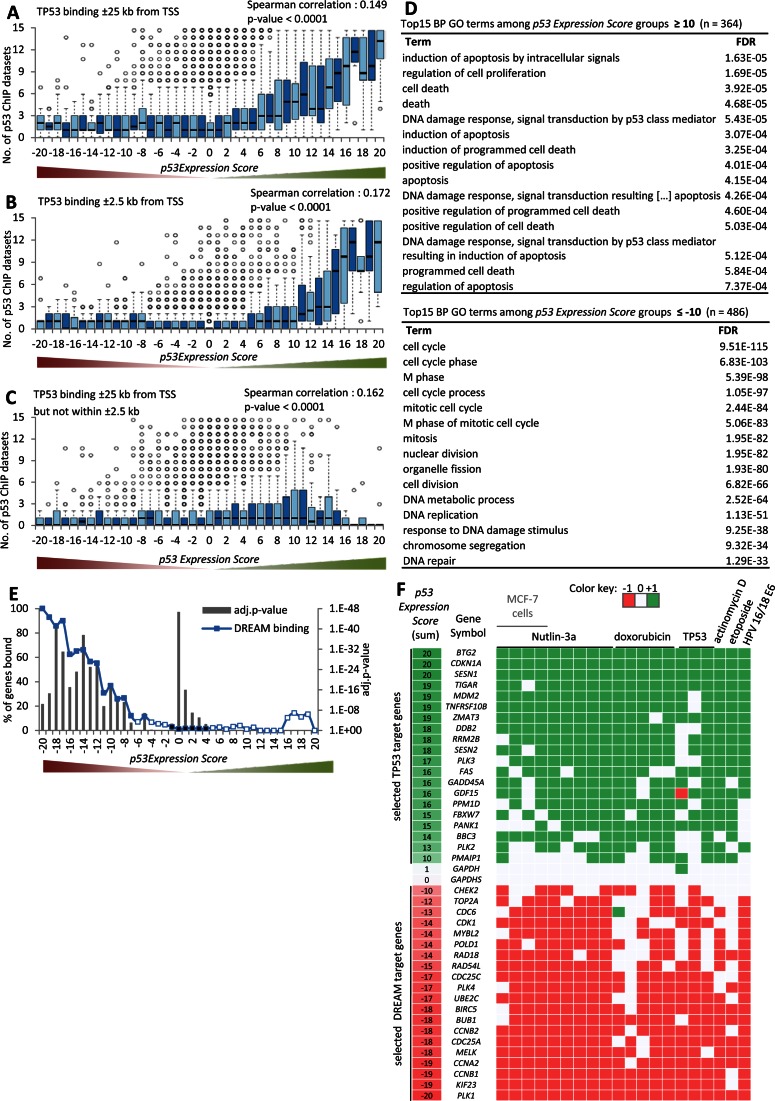
TP53 itself is the central transcription factor mediating TP53-dependent gene regulation (1,2). To assess the role of TP53 in regulating gene expression, we used the step-wise meta-analysis to integrate 15 genome-wide ChIP datasets of TP53 binding derived from several cell lines with the 20 expression profiling datasets (4–6,41–43,49–50). Using the stepwise meta-analysis approach, we ranked genes by the number of datasets that detected a TP53 binding peak near their TSS. Initially, we used TP53 binding within ±25 kb of the TSS for our analysis (Supplementary Table S2). We observed that genes upregulated were enriched for TP53 binding (Figure 2A; Supplementary Figures S6 and 7). A similar pattern was also observed using the conservative threshold of ±2.5 kb from the TSS (Figure 2B). In contrast, genes downregulated upon TP53 activation were not enriched for TP53 binding (Figure 2A and B).
Figure 2.
Proximal TP53 binding correlates with transcriptional activation. Boxplot displaying the number of ChIP datasets that find a gene to be bound by TP53 within (A) ±25 kb, (B) ±2.5 kb and (C) ±25 kb but not within ±2.5 kb of their TSS across the 41 p53 Expression Score groups. Correlation coefficient and two-tailed P-value was calculated using GraphPad Prism version 6.00. (D) Top 15 BP GO terms with their FDR value as identified using the DAVID Functional Annotation Tool (53) enriched at genes that are found down- (left) or upregulated (right) in at least half of the 20 datasets. The number of (E) DREAM (17) bound genes across the 41 p53 Expression Score groups. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). White data points are not significantly different. (F) A heatmap displaying the regulation of 20 well-established TP53 or DREAM target genes across the 20 datasets on TP53-dependent gene regulation. GAPDH and GAPDHS serve as negative controls.
A genome-wide analysis of gene regulation by TP53 in mouse embryonic stem cells suggested that distal binding of TP53 to the TSS was more likely to control TP53-dependent downregulation compared to upregulation by more proximal TP53 binding (51). We tested this hypothesis by examining genes that were distally bound by TP53 at ±25 kb but not within ±2.5 kb of the TSS. We observed that distal binding of TP53 was weakly correlated with gene upregulation, but not with downregulation (Figure 2C). Our finding is in agreement with reported distal TP53 binding in enhancer regions that may convey long-distance upregulation (52). The previous finding of gene downregulation through distal TP53 binding might stem from the use of mouse stem cells (51). Thus, we conclude that proximal TP53 binding to a gene promoter contributes to transcriptional activation but not repression, while distal TP53 binding appears to have a relatively minor but positive influence on transcription.
By integrating the TP53 binding and TP53-dependent expression profiling datasets, we generated a list of TP53 target genes with a threshold for p53 Expression Score of ≥ 5 and TP53 binding within +/− 2.5 kb of the TSS in at least four ChIP datasets. These thresholds ensure that TP53 regulation and binding was observed in at least two different cell lines or treatments. These criteria were passed by 311 genes including many well-known TP53 target genes such as CDKN1A (p21; p53 Expression Score = 20), BTG2 (= 20), TIGAR (also known as C12orf5; = 19), MDM2 (= 19), SUSD6 (KIAA0247; = 19), PLK3 (= 17), FAS (= 16), GADD45A (= 16) and BBC3 (PUMA; = 14) (Supplementary Table S3). We found that 223 of these 311 genes (71.7%) were identified in previous genome-wide TP53 target maps (3–9). Another five genes were captured by the literature collection from Riley et al. including KRT8, PTEN, RNF144B, TNFRSF10A and TP53 (1). Our approach complements information that dropped below the thresholds of individual studies and dismisses information that was unique to a comparably small number of studies. Based on our analysis, we identify 83 potential direct TP53 target genes that to our knowledge have not been previously described as TP53 targets. Examples include several highly-studied genes such as IER3, POU2F2, POU3F1, RPS19, SMAD3 and STAT3.
To identify biological processes for TP53-regulated genes, we performed an enrichment analysis for gene ontology (GO) terms (53) among genes up- or downregulated upon TP53 activation in at least half of the datasets (Figure 2D). As expected, the genes upregulated by TP53 were enriched for regulation of cell proliferation, induction of apoptosis and DNA damage response. In contrast, the genes downregulated by TP53 were significantly enriched for CC, mitosis and DNA replication.
CC genes have often been found to be downregulated upon TP53 activation (24). The DREAM complex was shown to regulate many CC genes in response to TP53 activation (30–32). To address the potential connection between DREAM target genes and TP53 mediated downregulation, we integrated binding data on DREAM (17) with TP53 expression. We found that genes downregulated by TP53 are highly enriched for binding by DREAM (Figure 2E).
To illustrate the utility of the meta-analysis approach, we selected 20 direct TP53 target genes (Supplementary Table S3) and 20 previously published DREAM targets (17,22–23,32,54–56) and examined their regulation across the 20 TP53 expression profiling datasets (Figure 2F and Supplementary Figure S8). The meta-analysis approach identifies target genes that were missed in some datasets but identified in several others.
Downregulation by TP53 requires p21
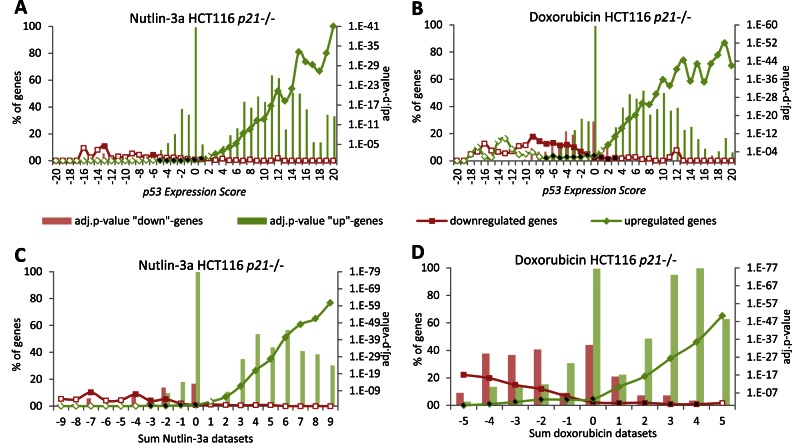
The CDKN1A gene, encoding the cyclin dependent kinase (CDK) inhibitor p21, was identified as a TP53 target by all 20 expression datasets as well as all 15 TP53 ChIP datasets. Inhibition of CDK activity by p21 was shown to be important for TP53-mediated downregulation of genes targeted by DREAM (30,31), the DREAM component E2F4 (57,58) and RB (27–29). To evaluate the global requirement for p21 in TP53-dependent downregulation, we performed RNA-seq using HCT116 p21-/- cells treated with Nutlin-3a or doxorubicin compared to control DMSO (Supplementary Table S4). We plotted the number of genes that were significantly up- or downregulated by Nutlin-3a or doxorubicin treatment against the p53 Expression Score. For the most part, genes upregulated by TP53 in cells containing p21 were also upregulated by Nutlin-3a and doxorubicin in HCT116 p21-/- cells. In contrast, the vast majority of genes downregulated by TP53 in our meta-analysis were not downregulated in HCT116 p21-/- cells (Figure 3A and B; Supplementary Figure S9). These results indicate that p21 is required for downregulation of gene expression upon TP53 activation.
Figure 3.
Downregulation upon TP53 induction requires p21. The number of genes identified in HCT116 p21−/− cells treated with (A) Nutlin-3a or (B) doxorubicin as either upregulated or downregulated is compared to the 41 p53 Expression Score groups. (C) The number of genes identified as regulated in Nutlin-3a treated HCT116 p21−/− cells is compared to the nine Nutlin-3a datasets. (D) The number of genes identified as regulated in doxorubicin treated HCT116 p21−/− cells is compared to the five doxorubicin datasets. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). White data points are not significantly different.
We compared the p21-/- gene sets with the nine Nutlin-3a and five doxorubicin datasets to identify potential treatment-specific effects. Few genes that were downregulated in the nine Nutlin-3a datasets were also downregulated by Nutlin-3a in HCT116 p21-/- cells (Figure 3C). However, we observed that a significant number of genes were downregulated by doxorubicin treatment in the absence of p21 (Figure 3D). This subset was not downregulated by Nutlin-3a treatment in p21-/- cells (Supplementary Figure S10A and B). A direct involvement of TP53 is unlikely since TP53 ChIP binding was not enriched among genes downregulated by doxorubicin (Supplementary Figure S10C). This indicates that a subset of genes was significantly downregulated by doxorubicin treatment in a p21 and TP53 independent manner.
A major distinction between doxorubicin and Nutlin-3a treatment is induction of TP53 activity through DNA damage response (59) and MDM2 inhibition (60), respectively. Recently, it was reported that DNA damage response also contributes to the cellular response to RITA relative to Nutlin-3a (61). Furthermore, RITA may impair p21 function (47), which could account for the low number of genes downregulated by RITA treatment in our meta-analysis (Supplementary Figure S5A). Interestingly, we found that genes downregulated in doxorubicin treated HCT116 p21-/- cells were also downregulated by RITA treatment of HCT116 cells including ATF2, GSK3B, NRF1, WRN and ZEB1 (Supplementary Figure S10D–F). This indicates that DNA damaging agents such as doxorubicin and RITA downregulate a specific subset of genes independent of p21 and TP53 activity.
Genes commonly downregulated by TP53 are cell cycle genes
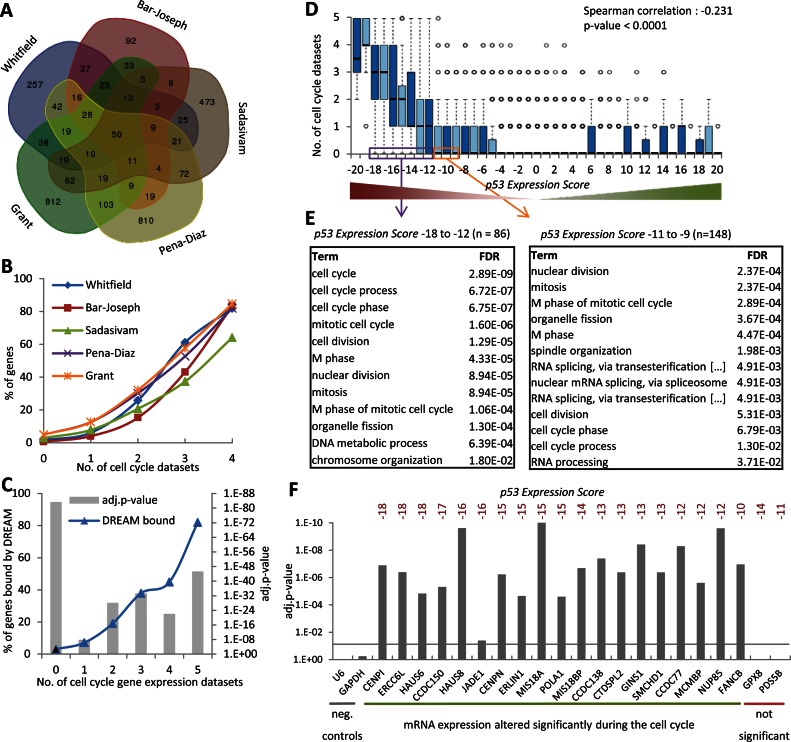
Genes downregulated by TP53 were enriched for GO terms associated with CC (Figure 2D) and DREAM binding (Figure 2E). To examine the effect of TP53 on CC genes in more detail, we analyzed five genome-wide expression profiles of CC-dependent gene regulation (11–15) (Supplementary Table S5). Similar to what was observed with the TP53 analysis, the overlap of CC-regulated genes between any two datasets was small ranging from 11.7 to 22.7% (Figure 4A and Supplementary Figure S11), whereas grouping of the five datasets using the step-wise meta-analysis approach led to a robust joint dataset where the number of datasets in agreement led to a measure of confidence that a given gene had differential expression during the CC (Figure 4B). Strikingly, when an increasing number of expression profile datasets agreed on a gene being regulated by the CC, it was more likely to also be bound by DREAM (Figure 4C). After performing this step-wise meta-analysis of CC genes, we plotted the number of CC datasets against the p53 Expression Score. We observed that genes downregulated by TP53 were also CC-regulated (Figure 4D). Genes in the p53 Expression Score groups of ‘−20’ and ‘−19’ were each found to be a CC-regulated gene in at least one dataset.
Figure 4.
Cell cycle (CC) genes are downregulated by TP53 and bind the DREAM complex. (A) Venn diagram displaying the overlap between the five CC datasets. (B) The number of genes identified in each of the five CC datasets is compared to the number of the remaining datasets that identify these genes. (C)The number of DREAM bound genes (Litovchick et al., (17)) is compared to the number of datasets that identify a gene as CC gene. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. (D) Boxplot displaying the number of datasets that find a gene to be a CC gene across the 41 p53 Expression Score groups. Correlation coefficient and two-tailed P-value was calculated using GraphPad Prism version 6.00. (E) Top 15 BP GO terms with their FDR value as identified using the DAVID Functional Annotation Tool (53) enriched at genes that display a p53 Expression Score ≤ −12 (left) or −9 to −11 (right) that were not identified as CC gene. (F) Significant CC regulation was tested for 21 TP53 repressed genes and the negative controls U6 and GAPDH using an unpaired Student's t-test for data from time points 0 to 10 h and 16 to 30 h and P-values were adjusted for multiple testing using Bonferroni correction (see Supplemental Figure S10).
Given these results, we asked whether any other gene sets were commonly downregulated by TP53. To this end, we performed a GO term enrichment analysis of the genes in p53 Expression Score groups −18 to −12 and −11 to −9 that were not found in the five CC datasets. CC terms were primarily enriched among these genes although they were not previously identified in the five CC studies (Figure 4E). Several highly-validated CC genes such as B-MYB (MYBL2) and POLD1 (23) were found in this group, revealing that the five genome-wide CC datasets together had failed to identify a substantial number of CC genes and that identifying genes consistently downregulated by p53 could be an alternative approach to identifying CC-regulated genes.
Consequently, we asked whether a negative p53 Expression Score could accurately predict CC genes that were not previously detected in the five CC datasets. We tested 21 genes for CC-dependent regulation with a p53 Expression Score ≤ −10 that were not previously identified as CC-regulated gene in any of the five CC datasets. We observed that 19 of the 21 genes tested display significant CC-dependent gene expression, while negative controls U6 and GAPDH did not (Figure 4F and Supplementary Figure S10). These results indicate that the majority of genes downregulated by TP53 are CC genes and that a low p53 Expression Score ≤ −10 can predict previously unidentified CC genes.
A comprehensive map of 971 candidate DREAM target genes
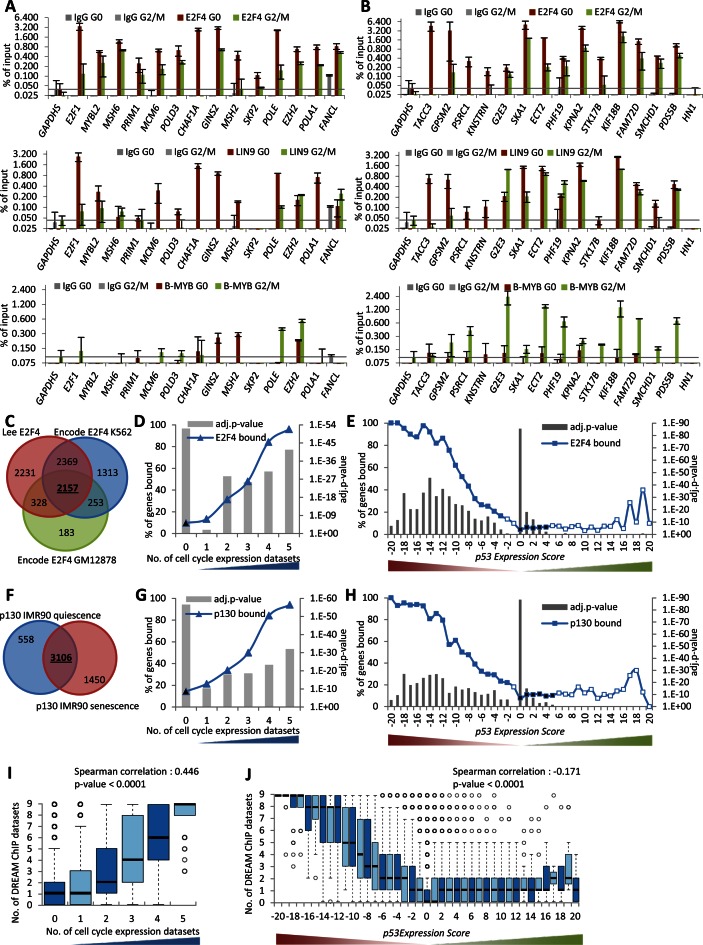
The step-wise meta-analysis approach was able to generate a more complete assessment of TP53 target genes by integrating a large number of expression profiling and ChIP-seq datasets. In a similar manner, we wanted to identify a more complete assessment of DREAM target genes. Several DREAM target genes such as E2F1 (17), GAS2L3 (62) and MYBL2 (32) were not identified as DREAM targets in the original ChIP-chip dataset by Litovchick et al. (17). Notably, 284 of 486 (58.4%) genes that have a p53 Expression Score ≤ −10 and 209 of 698 (29.9%) genes that were reported as CC genes in at least two datasets were identified as DREAM targets based on the dataset by Litovchick et al. (17). Considering that downregulation by TP53 and CC-dependent regulation are features of DREAM targets, we searched for genes with p53 Expression Scores ≤ −7 or identified as CC regulated in at least three of the five datasets that were not previously described as DREAM targets. In addition, we required the genes to harbor a phylogenetically conserved CHR or E2F element to support specific DREAM binding (17,22–23). From this group of 374 genes, we selected a total of 27 to test for binding of DREAM components E2F4 and LIN9 and the MMB component B-MYB in serum-starved (G0) and re-stimulated (22 h; G2/M) T98G cells (Figure 5A and B). E2F1 and MYBL2 were selected to serve as positive controls for DREAM binding as they have been validated earlier (17,32) while GAPDHS served as negative control. We found 26 of 27 (96.3%) genes bound to DREAM and MMB components above background. The only gene that tested negative was HN1 and it displayed the weakest p53 Expression Score ( = −4). These results indicate CC-dependent expression of mRNA combined with a low p53 Expression Score is predictive for DREAM target genes.
Figure 5.
Prediction and validation of candidate DREAM target genes. (A and B) Protein binding to promoters of the indicated genes was tested by ChIP in serum starved (G0) and 22 h re-stimulated (G2/M) T98G cells. The E2F1 and MYBL2 promoters served as a positive control for DREAM binding; the GAPDHS promoter was used as a negative control. One representative experiment with three technical replicates (n = 3) is displayed. Venn diagram of overlaps between genes identified as bound in the (C) E2F4 or (F) p130 ChIP-seq datasets. The number of common (D) E2F4 or (G) p130 bound genes is compared to the number of datasets that identify a gene as being a CC gene. The number of common (E) E2F4 or (H) p130 bound genes in the 41 p53 Expression Score groups. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). White data points are not significantly different. (I) Boxplot displaying the number of datasets that find a gene to be targeted by DREAM compared to the number of datasets that identify a gene as CC regulated. (J) Boxplot displaying the number of datasets that find a gene to be targeted by DREAM in the 41 p53 Expression Score groups. Correlation coefficient and two-tailed P-value was calculated using GraphPad Prism version 6.00.
The 26 newly identified DREAM target genes form two groups of genes expressed early (G1/S) or late (G2/M) in the CC. The early CC genes, including E2F1, MYBL2, MSH6, PRIM1, MCM6, POLD3, CHAF1A, GINS2, MSH2, SKP2, POLE, EZH2, POLA1 and FANCL, possess E2F elements in their promoters and bind DREAM but not MMB (Figure 5A). In contrast, the late CC genes, TACC3, GPSM2, PSRC1, KNSTRN, G2E3, SKA1, ECT2, PHF19, KPNA2, STK17B, KIF18B, FAM72D and SMCHD1, possess CHR elements in their promoters and bind MMB in addition to DREAM (Figure 5B). These results are in concordance with an earlier genome-wide classification showing that DREAM targets encompass both early and late CC genes, while MMB-FOXM1 targets late CC genes (23).
To generate a comprehensive genome-wide map of strong candidate DREAM target genes, we combined additional genome-wide ChIP datasets for the DREAM components E2F4 and p130 (Figure 5C and F). We examined the E2F4 and p130 binding data by comparing the 2157 common E2F4 bound genes and 3106 common p130 bound genes to the number of CC datasets a gene was identified in (Figure 5D and G) and the p53 Expression Score (Figure 5E and H). We found that both E2F4 and p130 were increasingly enriched for gene binding and reflected the confidence a gene was found to be either a CC gene or downregulated by TP53. These results are similar to findings we observed with the DREAM binding data based in part on E2F4 and p130 binding (Figures 2E and 4C).
We combined all three E2F4 ChIP-seq datasets, two p130 ChIP-seq datasets and the original four DREAM (E2F4, p130, LIN9 and LIN54) ChIP-chip datasets from Litovchick et al. The sum of these nine datasets was enriched for genes regulated within the CC and downregulated by TP53 (Figure 5I and J). To be considered a strong candidate DREAM target gene, we required a gene to be detected in at least four of the nine datasets to ensure binding of at least two DREAM components. Additionally, a gene was required to either be reported as being a CC gene in at least two of the five CC datasets or have a p53 Expression Score ≤ −5. These criteria were met by 968 genes (Supplementary Table S7). The finding that all 20 DREAM target genes displayed in Figure 2F were identified in this list demonstrates the ability of this approach to identify bona fide candidates. The list of strong candidate DREAM targets includes E2F1, GAS2L3, MYBL2 as well as 23 (88.5%) of 26 novel DREAM target genes we identified above (Figure 5A and B). We manually included the three false negative targets EZH2, FAM72D and STK17B that did not meet the stringent thresholds to be considered DREAM targets in our analysis but were instead validated experimentally (Figure 5A and B). Together, our screening approach identifies several hundred novel potential targets expanding the number of DREAM target genes to 971 strong candidates.
Identification of specialized subgroups of DREAM and TP53 target genes
The stepwise meta-analysis approach used to identify DREAM target genes combining CC expression, DREAM component binding and TP53 repression studies also uncovered several exceptions. For example, the gene encoding the apoptosis enhancing nuclease, AEN, was identified in the meta-analysis data as a CC gene but was also found to be upregulated by TP53 (p53 Expression Score 11). AEN was upregulated upon TP53 activation in HCT116 p21−/− cells as well (Supplementary Figure S13A and B). TP53 and DREAM were both recruited to the AEN promoter upon doxorubicin treatment. However, binding of TP53 but not DREAM was present in doxorubicin treated HCT116 p21−/− cells (Supplementary Figure S13C and D). TP53 binding to AEN was identified in all 15 ChIP datasets (Supplementary Table S2) and AEN has been confirmed in the literature to be a TP53 regulated gene (63). Additional examples of DREAM-bound but TP53 activated genes include E2F7 and PCNA, both previously reported to be E2F-regulated CC genes (64,65) and upregulated by TP53 binding (66,67). While AEN, E2F7 and PCNA represent early CC genes, late DREAM bound CC genes such as BTG1 and RAD51C were also bound and upregulated by TP53 (Supplementary Table S6). These results indicate that a small number of DREAM target genes are also direct TP53 target genes. Moreover, it suggests that TP53 activation can oppose the repressive DREAM complex upon DNA damage.
A different type of exception is represented by the PDS5B gene, which is a direct target of DREAM and not TP53. According to the meta-analysis data, PDS5B was downregulated by TP53 (p53 Expression Score −11) across multiple cell types and treatments (Supplementary Table S1). We confirmed this in HCT116 cells and found that the down-regulation was dependent on p21, as expected for a DREAM target gene (Supplementary Figure S13A and B). However, we also found that the levels of PDS5B mRNA were not CC regulated (Supplementary Figure S12). Thus, a small subgroup of DREAM target genes appears not to be regulated within the CC although it is downregulated by the TP53-p21-DREAM pathway. Taken together, the stepwise meta-analysis approach enabled identification of specialized subgroups of DREAM target genes.
DREAM target genes comprise two distinct subgroups: G1/S and G2/M cell cycle genes
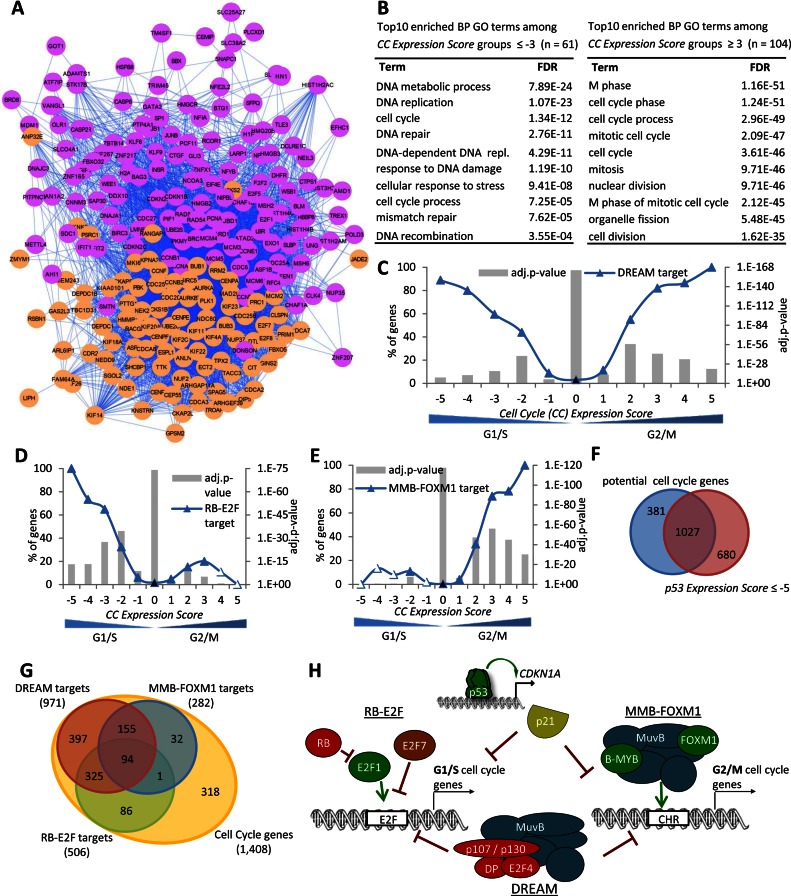
Temporal separation of gene transcription likely influences interactions of the encoded proteins. To address whether CC genes diverged into distinct subgroups, we analyzed the network of protein–protein interactions between 259 high confidence CC genes identified in at least three out of the five CC datasets. The protein–protein interaction links were extracted from the STRING database (68). We used a message-passing approach to detect significant communities in the network of CC genes. A community is a group of nodes that has more connections amongst themselves than expected by random chance (see ‘Materials and Methods’ section). We detected two robust communities in the network (Figure 6A). The magenta subgroup contained proteins with known functions during the G1 and S phases of the CC, such as E2F transcription factors, members of the minichromosome maintenance complex and histones. In contrast, the orange subgroup contained proteins with well-known functions during mitosis, such as kinesins and centromere proteins.
Figure 6.
DREAM target genes comprise distinct subgroups regulated by RB-E2F and MMB-FOXM1. (A) Message passing clustering of 259 high confidence CC genes based on their protein–protein interaction network obtained from string-db. (B) Top 10 BP GO terms with their FDR value as identified using the DAVID Functional Annotation Tool (53) enriched at genes that are found to be G1/S (left) or G2/M (right) CC genes in at least three of the five CC datasets. The number of (C) DREAM targets, (D) RB-E2F target genes and (E) MMB-FOXM1 targets in the 11 CC Expression Score groups. A two-sided Fisher's exact test was employed to test for significant over- and under-representation of gene sets and P-values were adjusted for multiple testing using Bonferroni correction. Colored and black data points are significantly over- and under-represented, respectively (adj. P-value ≤ 0.05). Venn diagram of predicted CC genes and (F) genes downregulated by TP53 (p53 Expression Score ≤ −5) or (G) DREAM, MMB-FOXM1 and RB-E2F targets. (H) The TP53 target p21 (CDKN1A) is required for downregulation by TP53. CC genes are downregulated by TP53 and bound by the DREAM complex. RB-E2F and MMB-FOXM1 bind discrete subsets of DREAM targets reflecting G1/S and G2/M CC genes.
To explore the characteristics of the two major CC gene subgroups in more detail, we took advantage of information on peak expression provided in the five CC datasets. We introduced the CC Expression Score based on the same logic as the p53 Expression Score. We calculated how many datasets agree on a gene displaying peak expression in ‘G2’ or ‘G2/M’ phase of the CC minus the number of datasets that find a gene expressed in ‘G1/S’ or ‘S phase’. To verify this approach, we examined the most confident G2/M and G1/S groups, CC Expression Score +5 and −5, respectively. The +5 group contained several well-known and validated late CC genes including CENPA, CENPE, KIF11, KIF23 and UBE2C (Supplementary Table S6). In contrast, the −5 group contained known early CC genes, such as CCNE2, CDC6, E2F1, MCM5 and PCNA. The group of G1/S CC genes was enriched, as expected, for functions in DNA replication, DNA metabolic process, DNA repair and DNA recombination (Figure 6B). In contrast, the group of G2/M CC genes was strongly enriched for mitotic functions.
The distribution of the 971 strong candidate DREAM target genes (Supplementary Table S7) across the CC Expression Score demonstrated the presence of both G1/S and G2/M CC genes (Figure 6C). This is consistent with DREAM binding to G1/S and G2/M CC genes harboring E2F and CHR elements, respectively (Figure 5A and B).
Identification of 282 MMB-FOXM1 targets reveals G2/M gene signature
Considering that DREAM binds G1/S and G2/M CC genes, we asked if specific transcription factors could also distinguish these subgroups. MMB and FOXM1 have been reported to bind specifically to G2/M genes (13,21,23). To test whether MMB-FOXM1 is a signature of G2/M genes, we integrated ChIP-seq datasets on B-MYB and LIN9, major components of the MMB complex (13) and FOXM1, a known MMB binding partner (13,20–21). MMB binding was significantly enriched for G2/M but not G1/S CC genes (Supplementary Figure S14A and B). The MMB interaction partner FOXM1 displays a similar binding pattern (Supplementary Figure S14C and D). In addition, phylogenetically conserved CHR elements that mediate binding of MMB and FOXM1 (21,22) were also significantly more abundant among G2/M but not G1/S CC genes (Supplementary Figure S14E).
Using the meta-analysis approach, we established a genome-wide map of MMB-FOXM1 target genes. If a gene was found in at least four of the seven MMB-FOXM1 datasets (Supplementary Figure S14F) and was either detected as a CC gene in at least two out of five CC datasets or had a p53 Expression Score ≤ −5, we considered it to be a potential MMB-FOXM1 target. A negative p53 Expression Score was included because a switch from MMB to DREAM complex binding was reported to be critical for TP53-dependent downregulation of several CC genes (30,31). These criteria were met by 276 genes (Supplementary Table S8) including known MMB and FOXM1 targets CCNB1, CCNB2 and PLK1 (13,21). This approach identified several novel MMB target genes including ECT2, EZH2, GPSM2, KIF18B, KPNA2, PHF19, PSRC1 and STK17B (Figure 5A and B). Identification of these MMB and FOXM1 target genes illustrates the utility of this approach to identify bona fide candidate genes. While the list of MMB-FOXM1 targets is not exhaustive due to the stringent criteria, loosening these criteria would result in an unfavorable increase in false positives. Genes such as FAM72D, G2E3, PDS5B, POLE, SKA1 and SMCHD1 are false negatives that did not meet the stringent thresholds to be considered MMB-FOXM1 targets in our analysis and were added manually as they were validated experimentally (Figure 5A and B). Together, these 282 strong candidate MMB-FOXM1 target genes identify many G2/M CC genes (Figure 6E).
A genome-wide map of 506 candidate RB-E2F targets includes G1/S cell cycle genes
Given that most, if not all, G2/M CC genes are bound by MMB-FOXM1 as well as the DREAM complex, we asked whether we also could establish a distinct signature for G1/S CC genes. In contrast to G2/M CC genes, G1/S CC genes are directly regulated by RB, the activating E2F1, E2F2 and E2F3 and repressive E2F6, E2F7 and E2F8 transcription factors, in addition to the repressive E2F4 and p130/p107-containing DREAM complex (16,19). We integrated ChIP-seq datasets for these transcription factors (Supplementary Figure S15). Similar to a previous analysis of the E2f1, E2f2, E2f4 and E2f6 genome-wide binding data from mouse (69), we found only a small overlap between the human E2F1, E2F4, E2F6 and E2F7 binding data.
We examined the distribution of genes binding RB, E2F1, E2F6 and E2F7 across the CC Expression Score. We found that binding of RB and E2F7 was enriched at G1/S but not at G2/M CC genes (Supplementary Figure S15A, B and F). Similarly, E2F1 was more significantly enriched at G1/S than G2/M CC genes (Figure 14C). Although E2F6 was reported to be specific for G1/S genes (70), we found that it was bound to both CC groups (Supplementary Figure S15D and E).
The E2F DNA element recruits RB and E2F transcription factors to gene promoters and is enriched among RB and E2F7 bound genes (65,71). We found that phylogenetically conserved E2F elements were significantly overrepresented in the promoters of G1/S but not G2/M CC genes (Supplementary Figure S15G). Together, the five datasets consisting of RB, E2F1 and E2F7 binding and E2F promoter elements function as a signature for G1/S CC genes (Supplementary Figure S15H). We classify this group as RB-E2F target genes. Genes found in at least three of these five datasets were included in a genome-wide map of RB-E2F targets if they had either a p53 Expression Score ≤ −5 or were present as a CC gene in at least two out of five CC datasets. The 506 potential RB-E2F targets (Supplementary Table S9) were significantly enriched for G1/S CC genes and contrast with the distribution of MMB-FOXM1 targets (Figure 6D and E).
Given that transcription factor binding profiles can predict CC-regulated genes (72), we combined all candidate target genes of DREAM, MMB-FOXM1 and RB-E2F with genes that were found in at least two of the five CC datasets to establish a genome-wide map of CC genes. This expanded dataset identifies 1406 potential CC genes including 24 (92.3%) of the 26 cell experimentally validated cycle-regulated genes that were missed in all five CC datasets (Supplementary Figure S12). We manually added the two false negative genes CENPI and CCDC138 that did not meet the stringent thresholds to be considered CC regulated in our analysis but were instead identified experimentally (Supplementary Figure S12). Together, this approach identifies a total of 1408 CC-regulated genes (Supplementary Table S10).
Many of the potential CC genes overlap with genes downregulated by TP53 (Figure 6F). DREAM, RB-E2F and MMB-FOXM1 targets account for 983 (57.6%) of 1707 genes having a p53 Expression Score ≤ −5 (Supplementary Figure S16A), indicating that these are the primary genes downregulated by TP53. Consistent with an earlier study reporting rRNA processing genes downregulated by TP53 in addition to CC genes (39), we found the remaining 724 genes were enriched for GO terms representing RNA processing and metabolism (Supplementary Figure S16B).
Most of the 1408 potential CC genes bind DREAM, RB-E2F or MMB-FOXM1 (Figure 6G). Overall, RB-E2F and MMB-FOXM1 targets appear to be distinct subgroups of DREAM target genes reflecting the G1/S and G2/M groups of CC genes (Figures 6H and 7).
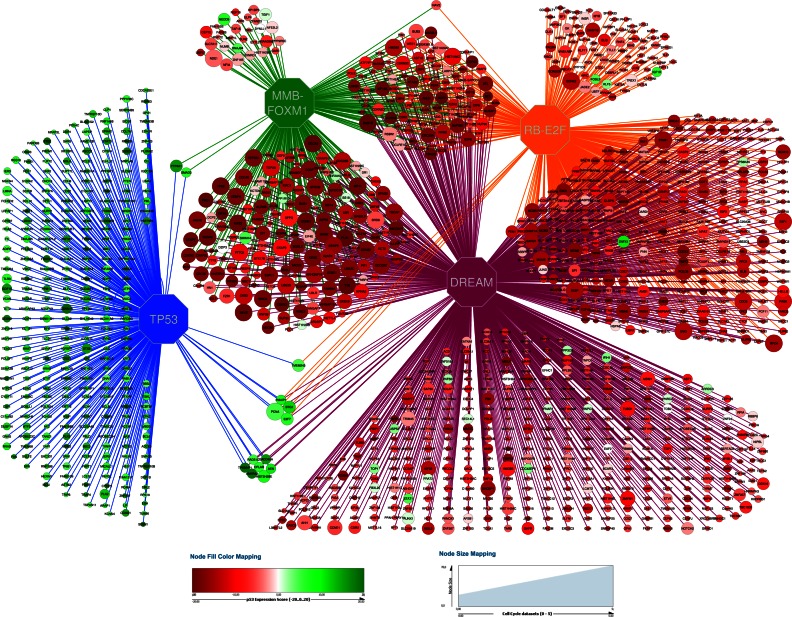
Figure 7.
Predicted direct target gene network governed by TP53, DREAM, MMB-FOXM1 and RB-E2F. Octagons represent the transcriptional regulators. All other nodes represent target genes. Edges represent predicted direct regulation. The size of the nodes reflects the number of CC datasets that identify the gene as CC regulated. The node color reflects the gene's p53 Expression Score.
DISCUSSION
Genome-wide approaches have increasingly shaped our understanding of TP53 and CC gene regulatory networks. However, due to the nature of these genome-wide datasets, the overlap between any two can be small even when the underlying data were derived from the same cell line undergoing identical treatments. We have developed an approach that captures the information from many of the recently reported datasets to gain a more complete overview of the TP53 and CC regulatory landscape.
To avoid including highly heterogenic datasets, we tested each TP53 expression dataset against the sum of the remaining datasets and consequently included 20 out of 22 datasets tested (Figure 1E; Supplementary Figures S3 and 4). Since most studies included stringent thresholds to identify genes that are significantly differentially expressed, only a few genes were identified in all datasets. Despite the stringent thresholds, many genes were found differentially expressed in only one or two datasets displaying little reproducibility across datasets. These genes may represent either false negatives or, alternatively, genes that are regulated uniquely through certain treatment-cell type combinations.
Considering that the findings of our meta-analysis approach are based on the data provided by the underlying datasets, there is a bias toward genes regulated by Nutlin-3a or doxorubicin treatment as these were applied in most studies. Treatment-cell type combinations that are present solely once have little influence on the whole meta-analysis. Although we find that many TP53 responsive genes are regulated by TP53 robustly across multiple cell types and treatments, our findings also support that there is cell type and treatment specific gene regulation by TP53. Thus, when investigating treatment-cell type combinations that are not represented, insights from our meta-analysis may be limited.
Integration of these various gene expression profiling datasets results in a robust joint dataset comprised of genes that were repeatedly identified to be regulated. By integrating publically available datasets, high confidence lists of TP53 and CC regulated genes were generated. This meta-analysis approach complemented incomplete information in individual studies with data from other studies with noise lowered using stringent thresholds. For example, the ChIP–chip dataset by Litovchick et al. identified 865 genes bound by DREAM based on the stringent criteria that a binding peak for p130, E2F4 and LIN9 had to be present within 1000 bp from the TSS (Supplementary Table S5) (17). Only 473 of these genes passed our stringent criteria to also be considered regulated by DREAM. In contrast, by including five additional binding studies of DREAM components, our meta-analysis approach identified 2,897 potential DREAM bound genes that display a binding peak in at least four out of the nine binding profiles. Out of these 2897 genes 971 also met the criteria of being regulated across multiple conditions and were consequently predicted to be high confidence targets (Supplementary Table S5). Our scoring system is based on the number of datasets that agree on transcription factor binding sites or on a gene's regulation. The scoring system can be used as a measure of confidence and enables visual evaluation of the impact of additional studies in the meta-analysis. Furthermore, our approach visualizes thresholds and provides ranked maps of regulated genes.
The stepwise meta-analysis approach refines our understanding of the TP53 and CC gene regulatory networks. It enabled us to integrate a variety of genome-wide gene expression datasets with transcription factor binding profiles from multiple treatments and cell types. Combining various types of studies revealed a more comprehensive insight into how gene expression is regulated by TP53 and the CC. We used this method to build high confidence ranked target gene maps for TP53, DREAM, MMB-FOXM1 and RB-E2F that predicted potential CC regulation. This integrated view of CC regulation highlights the distinct role for RB-E2F and MMB-FOXM1 binding in DREAM target genes reflecting the G1/S and G2/M groups of CC genes (Figures 6H and 7).
Our meta-analysis approach also provided clear insight into the role of TP53 as an activator through proximal promoter binding (Figure 2A–C). In agreement with the latest model on TP53-dependent transcriptional regulation (10), a direct repressor function of TP53 appears to be absent. The most striking finding was that the TP53 target gene and CDK inhibitor p21 is critical to TP53-mediated transcriptional downregulation in general (Figure 3). These results challenge gene regulatory models that do not incorporate p21 in mediating TP53-dependent transcriptional downregulation, such as E2F7 (7), multiple microRNAs (73) and long non-coding RNAs (74). Our results suggest that the group of CC genes is primarily and generally regulated through the TP53-p21-DREAM (32) and TP53-p21-RB/E2F (28,29) pathways across cell types and treatments (Figure 6H).
The meta-analysis approach also enabled identification of several interesting exceptions. One group of genes appears to be downregulated independent of both p21 and TP53 by the DNA damaging agents doxorubicin and RITA. Another outlier group contains a small subset of DREAM bound CC genes that are also bound by TP53. Genes in this group are transcriptionally activated by TP53 and include AEN, BTG1, E2F7, PCNA and RAD51C (Figure 7).
Significantly, our target gene resource can help to identify co-expression signatures identified in other cell types and treatments. For example, high levels of EZH2, a member of the PRC2 complex, and its co-expression signature comprising 116 genes enabled identification of high risk non-small-cell lung cancer patients (75). Here, we identified EZH2 as novel DREAM target gene (Figure 5B). Examining the 116 gene EZH2 co-expression signature revealed that 99 genes were also CC-regulated and DREAM targets (Supplementary Table S5). CC gene expression signatures help to stratify cancer patients into risk groups (76). Our meta-analysis approach provides an extensive resource of CC genes and their regulatory networks that can help to identify co-expression signatures.
Taken together, our results showcase the power of integrating gene expression and transcription factor binding profiles across cell types and treatments to gain insight into the gene regulatory networks and to establish high confidence ranked maps of transcription factor target genes. Our work provides an extensive and integrated resource on TP53 and CC-regulated genes available in the Supplementary Data and as a web-based atlas on www.targetgenereg.org. Our meta-analysis provides an opportunity to discover unique features and novel mechanisms when compared to genome-wide datasets from new treatments or cell lines. The information provided on many genes of interests may help guiding research resources to the most promising targets.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein for the kind gift of HCT116 cell lines, Galina Selivanova, Thorsten Stiewe, Ran Elkon, Reuven Agami, Annelien Verfaillie, Stein Aerts, Scott Younger and Axel Wintsche for providing pre-analyzed datasets, Gerd A. Müller for the kind gift of RNA samples, Lydia Steiner for help on dataset integration and motif search, Zach Herbert from the Dana-Farber Cancer Institute Molecular Biology Core Facility for help with RNA-seq, Marianne Quaas and Kurt Engeland for support during early stages of the project and Timothy Branigan for critical reading of the manuscript.
Author contributions: M.F. initiated and conceived the study. M.F. integrated the datasets and performed the meta-analysis. P.G. performed the hierarchical clustering analysis. M.F. performed and analyzed the experiments. M.P. and M.F. analyzed the protein–protein interaction network. M.F. and J.A.D. interpreted the data. P.G. and M.F. designed and P.G. created the website. M.F. and J.A.D. wrote the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Postdoctoral fellowship provided by the Fritz Thyssen Foundation; German National Academy of Sciences Leopoldina Postdoctoral Fellowship; Add-On Fellowship for Interdisciplinary Science by the Joachim Herz Foundation [to M.F.]; NIH [K25 HG006031 to M.P.]; US Public Health Service grants [R01CA63113, R01CA173023 to J.A.D., in part]. Funding for page charges: Fritz Thyssen Foundation [through M.F.]. Funding for open access charge: German Research Foundation (DFG) and University of Leipzig within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1.Riley T., Sontag E., Chen P., Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 2.Beckerman R., Prives C. Transcriptional regulation by p53. Cold Spring Harb.Perspect.Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 4.Smeenk L., van Heeringen S.J., Koeppel M., Gilbert B., Janssen-Megens E., Stunnenberg H.G., Lohrum M. Role of p53 Serine 46 in p53 Target Gene Regulation. PLoS One. 2011;6:e17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikulenkov F., Spinnler C., Li H., Tonelli C., Shi Y., Turunen M., Kivioja T., Ignatiev I., Kel A., Taipale J., et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez D., Nguyen T.A., Freudenberg J.M., Mathew V.J., Anderson C.W., Jothi R., Resnick M.A. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–7301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlereth K., Heyl C., Krampitz A.-M.M., Mernberger M., Finkernagel F., Scharfe M., Jarek M., Leich E., Rosenwald A., Stiewe T. Characterization of the p53 Cistrome—DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen M.A., Andrysik Z., Dengler V.L., Mellert H.S., Guarnieri A., Freeman J.A., Sullivan K.D., Galbraith M.D., Luo X., Kraus W.L., et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang G.S., Chen X.A., Park B., Rhee H.S., Li P., Han K.H., Mishra T., Chan-Salis K.Y., Li Y., Hardison R.C., et al. A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep. 2014;8:514–527. doi: 10.1016/j.celrep.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer M., Steiner L., Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Joseph Z., Siegfried Z., Brandeis M., Brors B., Lu Y., Eils R., Dynlacht B.D., Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadasivam S., Duan S., DeCaprio J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant G.D., Brooks L., Zhang X., Mahoney J.M., Martyanov V., Wood T.A., Sherlock G., Cheng C., Whitfield M.L. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell. 2013;24:3634–3650. doi: 10.1091/mbc.E13-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peña-Diaz J., Hegre S.A., Anderssen E., Aas P.A., Mjelle R., Gilfillan G.D., Lyle R., Drabløs F., Krokan H.E., Sætrom P. Transcription profiling during the cell cycle shows that a subset of Polycomb-targeted genes is upregulated during DNA replication. Nucleic Acids Res. 2013;41:2846–2856. doi: 10.1093/nar/gks1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertoli C., Skotheim J.M., de Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., von Eyss B., Gagrica S., Hanel F., Brehm A., Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 19.Sadasivam S., DeCaprio J.A. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Down C.F., Millour J., Lam E.W., Watson R.J. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Müller G.A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A.D., Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 2013;33:227–236. doi: 10.1128/MCB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller G.A., Quaas M., Schümann M., Krause E., Padi M., Fischer M., Litovchick L., DeCaprio J.A., Engeland K., Schümann M. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40:1561–1578. doi: 10.1093/nar/gkr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller G.A., Wintsche A., Stangner K., Prohaska S.J., Stadler P.F., Engeland K. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42:10331–10350. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spurgers K.B., Gold D.L., Coombes K.R., Bohnenstiehl N.L., Mullins B., Meyn R.E., Logothetis C.J., McDonnell T.J. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J. Biol. Chem. 2006;281:25134–25142. doi: 10.1074/jbc.M513901200. [DOI] [PubMed] [Google Scholar]
- 25.de Toledo S.M., Azzam E.I., Keng P., Laffrenier S., Little J.B. Regulation by ionizing radiation of CDC2, cyclin A, cyclin B, thymidine kinase, topoisomerase IIalpha, and RAD51 expression in normal human diploid fibroblasts is dependent on p53/p21Waf1. Cell Growth Differ. 1998;9:887–896. [PubMed] [Google Scholar]
- 26.Löhr K., Möritz C., Contente A., Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 2003;278:32507–32516. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 27.Flatt P.M., Tang L.J., Scatena C.D., Szak S.T., Pietenpol J.A. p53 regulation of G(2) checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 2000;20:4210–4223. doi: 10.1128/mcb.20.12.4210-4223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottifredi V., Karni-Schmidt O., Shieh S.S., Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol. Cell. Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schvartzman J.M., Duijf P.H., Sotillo R., Coker C., Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannefeld M., Klassen E., Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res. 2009;69:4073–4080. doi: 10.1158/0008-5472.CAN-08-4156. [DOI] [PubMed] [Google Scholar]
- 31.Quaas M., Müller G.A., Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012;11:4661–4672. doi: 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer M., Quaas M., Steiner L., Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44:164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis S., Meltzer P.S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 34.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y., et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaccara S., Tebaldi T., Pederiva C., Ciribilli Y., Bisio A., Inga A. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ. 2014;21:1522–1534. doi: 10.1038/cdd.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janky R., Verfaillie A., Imrichová H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., Svetlichnyy D., Atak Z. K., Fiers M., Marine J.-C., Aerts S. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loayza-Puch F., Drost J., Rooijers K., Lopes R., Elkon R., Agami R. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 2013;14:R32. doi: 10.1186/gb-2013-14-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein I., Ezra O., Rivlin N., Molchadsky A., Madar S., Goldfinger N., Rotter V. p53, a novel regulator of lipid metabolism pathways. J. Hepatol. 2012;56:656–662. doi: 10.1016/j.jhep.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 41.Sammons M.A., Zhu J., Drake A.M., Berger S.L. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015;25:179–188. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akdemir K.C., Jain A.K., Allton K., Aronow B., Xu X., Cooney A.J., Li W., Barton M.C. Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 2014;42:205–223. doi: 10.1093/nar/gkt866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Younger S.T., Kenzelmann-Broz D., Jung H., Attardi L.D., Rinn J.L. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res. 2015;43:4447–4462. doi: 10.1093/nar/gkv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Böhlig L., Friedrich M., Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res. 2011;39:440–453. doi: 10.1093/nar/gkq796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kracikova M., Akiri G., George A., Sachidanandam R., Aaronson S.A. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013;20:576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enge M., Bao W., Hedström E., Jackson S.P., Moumen A., Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–183. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 48.Rashi-Elkeles S., Elkon R., Shavit S., Lerenthal Y., Linhart C., Kupershtein A., Amariglio N., Rechavi G., Shamir R., Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol. Oncol. 2011;5:336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Botcheva K., McCorkle S.R., McCombie W.R., Dunn J.J., Anderson C.W. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle. 2011;10:4237–4249. doi: 10.4161/cc.10.24.18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smeenk L., van Heeringen S.J., Koeppel M., van Driel M.A., Bartels S.J., Akkers R.C., Denissov S., Stunnenberg H.G., Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M., He Y., Dubois W., Wu X., Shi J., Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melo C.A., Drost J., Wijchers P.J., van de Werken H., de Wit E., Oude Vrielink J.A.F., Elkon R., Melo S.A., Léveillé N., Kalluri R., et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Fischer M., Grundke I., Sohr S., Quaas M., Hoffmann S., Knörck A., Gumhold C., Rother K. p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes. PLoS One. 2013;8:e63187. doi: 10.1371/journal.pone.0063187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer M., Quaas M., Wintsche A., Müller G.A., Engeland K. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res. 2014;42:163–180. doi: 10.1093/nar/gkt849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer M., Quaas M., Nickel A., Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget. 2015;6:41402–41417. doi: 10.18632/oncotarget.6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor W.R., Schonthal A.H., Galante J., Stark G.R. p130/E2F4 binds to and represses the cdc2 promoter in response to p53. J. Biol. Chem. 2001;276:1998–2006. doi: 10.1074/jbc.M005101200. [DOI] [PubMed] [Google Scholar]
- 58.Benson E.K., Mungamuri S.K., Attie O., Kracikova M., Sachidanandam R., Manfredi J.J., Aaronson S.A. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene. 2014;33:3959–3969. doi: 10.1038/onc.2013.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tewey K.M., Rowe T.C., Yang L., Halligan B.D., Liu L.F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 60.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 61.Wanzel M., Vischedyk J.B., Gittler M.P., Gremke N., Seiz J.R., Hefter M., Noack M., Savai R., Mernberger M., Charles J.P., et al. CRISPR-Cas9–based target validation for p53-reactivating model compounds. Nat. Chem. Biol. 2016;12:22–28. doi: 10.1038/nchembio.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolter P., Schmitt K., Fackler M., Kremling H., Probst L., Hauser S., Gruss O.J., Gaubatz S. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J. Cell Sci. 2012;125:2393–2406. doi: 10.1242/jcs.097253. [DOI] [PubMed] [Google Scholar]
- 63.Kawase T., Ichikawa H., Ohta T., Nozaki N., Tashiro F., Ohki R., Taya Y. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene. 2008;27:3797–3810. doi: 10.1038/onc.2008.32. [DOI] [PubMed] [Google Scholar]
- 64.Li Y.-Y., Wang L., Lu C.-D. An E2F site in the 5′-promoter region contributes to serum-dependent up-regulation of the human proliferating cell nuclear antigen gene. FEBS Lett. 2003;544:112–118. doi: 10.1016/s0014-5793(03)00485-x. [DOI] [PubMed] [Google Scholar]
- 65.Westendorp B., Mokry M., Groot Koerkamp M.J., Holstege F.C., Cuppen E., de B.A. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 2012;40:3511–3523. doi: 10.1093/nar/gkr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shivakumar C. V, Brown D.R., Deb S., Deb S.P. Wild-type human P53 transactivates the human proliferating cell nuclear antigen promoter. Mol. Cell. Biol. 1995;15:6785–6793. doi: 10.1128/mcb.15.12.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carvajal L.A., Hamard P.J., Tonnessen C., Manfredi J.J. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012;26:1533–1545. doi: 10.1101/gad.184911.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laresgoiti U., Apraiz A., Olea M., Mitxelena J., Osinalde N., Rodriguez J.A., Fullaondo A., Zubiaga A.M. E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res. 2013;41:10185–10198. doi: 10.1093/nar/gkt821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giangrande P.H., Zhu W., Schlisio S., Sun X., Mori S., Gaubatz S., Nevins J.R. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R.A., Narita M., Zhang M., Lowe S.W. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng C., Ung M., Grant G.D., Whitfield M.L. Transcription factor binding profiles reveal cyclic expression of human protein-coding genes and non-coding RNAs. PLoS Comput. Biol. 2013;9:e1003132. doi: 10.1371/journal.pcbi.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat. Rev. Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 74.Grossi E., Sánchez Y., Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim. Biophys. Acta. 2016;1859:200–208. doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 75.Fillmore C.M., Xu C., Desai P.T., Berry J.M., Rowbotham S.P., Lin Y.-J., Zhang H., Marquez V.E., Hammerman P.S., Wong K.-K., et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitfield M.L., George L.K., Grant G.D., Perou C.M. Common markers of proliferation. Nat. Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.