Abstract
RNA editing is a finely tuned, dynamic mechanism for post-transcriptional gene regulation that has been thoroughly investigated in the last decade. Nevertheless, RNA editing in non-coding RNA, such as microRNA (miRNA), have caused great debate and have called for deeper investigation. Until recently, in fact, inadequate methodologies and experimental contexts have been unable to provide detailed insights for further elucidation of RNA editing affecting miRNAs, especially in cancer.
In this work, we leverage on recent innovative bioinformatics approaches applied to a more informative experimental context in order to analyze the variations in miRNA seed region editing activity during a time course of a hypoxia-exposed breast cancer cell line. By investigating its behavior in a dynamic context, we found that miRNA editing events in the seed region are not depended on miRNA expression, unprecedentedly providing insights on the targetome shifts derived from these modifications. This reveals that miRNA editing acts under the influence of environmentally induced stimuli.
Our results show a miRNA editing activity trend aligning with cellular pathways closely associated to hypoxia, such as the VEGF and PI3K/Akt pathways, providing important novel insights on this poorly elucidated phenomenon.
INTRODUCTION
Low O2 tension (hypoxia) is a characteristic feature of pathophysiological conditions such as cancer. The rapid and uncontrolled growth of a tumor outgrows its blood supply, leaving certain regions of the cancer mass greatly deprived of the necessary oxygen intake, causing a substantial alteration of their metabolism. Hypoxic microenvironements in solid tumors lead, in fact, to the activation of several cellular pathways, such as AKT and VEGF (1), altering the activity not only of several coding transcripts but also of non-coding genes, such as microRNAs (Kulshreshtha et al., 2007).
MicroRNAs (miRNAs) are endogenous, non-coding RNA molecules, ∼22 nt long, found in eukaryotes and capable of negatively regulating gene expression at the post-transcriptional level. They represent a dominant class of small RNAs in most somatic tissues and play important regulatory roles in most biological pathways (2).
miRNAs originate from longer precursor transcripts called primary miRNAs (pri-miRNA) (3), processed in the nucleus into a transitional hairpin-shaped form (pre-miRNA) (4). Once exported into the cytoplasm, pre-miRNAs are cleaved into mature miRNAs (5). Specifically, each arm of a pre-miRNA (−5p and −3p) encodes a potential mature sequence, nevertheless only one is predominantly loaded into the RNA-induced silencing complex (RISC) (6). In the RISC, the mature miRNA sequence allows interaction with the 3′-untranslated region (3′-UTR) of mRNA targets through canonical binding, namely, a partial sequence complementarity mediated by a 6–8 nt long region at the 5′ end of the miRNA called the ‘seed’, thus exerting its regulatory function via inhibition of protein translation or degradation of the mRNA.
A single mRNA can be targeted by several miRNAs, and a single miRNA might target hundreds of mRNAs, thus producing so-called miRNA networks (7) that modulate the translation of a large fraction of the transcriptome (8). In fact, it has been shown that miRNAs are involved in a plethora of biological processes, including development and differentiation (9), cell cycle control (10), metabolism (11) and apoptosis (12). It is thus not surprising that the perturbation of miRNA networks can lead to the onset of diseases such as metabolic disorders (13), neurodegenerative diseases (14), as well as cancer (15). In the last decade, the emergence of a large number of studies making use of platforms for the global assessment of miRNA expression has enabled the identification of specific miRNA signatures characteristic of specific cancer types and subtypes (16), along with their specific oncogenic aberrations (17).
In the last few years, the advent of high-throughput sequencing (HTS) technology has led to a radical improvement in the accuracy and sensitivity of cancer specifications in comparison with previous expression profiling techniques, especially in relation to the detection of non-coding RNAs (18). HTS technology is gradually becoming essential, especially in the genome-wide identification and investigation of polymorphisms occurring in miRNA seed regions (MSRs) as well as within target 3′-UTRs, phenomenons that can disrupt miRNA function in many human diseases, including cancers (19–23) (http://compbio.uthsc.edu/miRSNP/) (24).
Though previously primary emphasis was on genetic variants to elucidate biological pathways perturbed in human cancers, recent focus has shifted toward post-transcriptional modifications, such as RNA editing. RNA editing is a post-transcriptional mechanism that alters the sequence of primary RNA transcripts. A-to-I (Adenosine-to-Inosine) RNA editing is the most prevalent type of RNA editing in mammals, and is mediated by members of the family of adenosine deaminases acting on RNA (ADAR) (25,26), which bind double-stranded RNAs (dsRNA) (27,28), deaminating adenosine (A) to inosine (I), which in turn is interpreted both by the splicing and translation machineries as guanosine (G) (29). A-to-I RNA editing events can occur in both coding and non-coding RNA molecules, such as miRNAs (30,31), with 10–20% of unique sequences potentially able to undergo A-to-I RNA editing (30,32) at the pri-miRNA level (33). While pri- or pre-miRNA modifications, such as A-to-I editing, outside the mature sequence may change both the maturation (34) and the expression of miRNAs (33). Editing by ADAR1 in pri-miR-455 at the +2 and +17 positions were reported to reduce the ability to bind to Drosha and then be processed into mature miR-455-5p in human melanocytes (35). Negative regulation of ADAR1 expression mediated by CREB in metastatic melanoma cell lines leads to an increase in expression for miR-455-5p and, consequently, to the suppression of CPEB1, which in turn enhances melanoma growth and metastasis in vivo (35). ADAR1 regulates the expression of several miRNAs essential for differentiation and neural induction in human embryonic stem cells by acting as an RNA-binding protein (36). Another editing-independent activity of ADAR1, namely, the suppression of miR-222 with consequent immune resistance of melanoma cells, has also been discovered (37). On the other hand, through a global study of ADAR1 binding, it was observed that there may be a possible competition with DGCR8 in pri-miRNA binding in the nucleus (38). Moreover, modulation of miRNA editing and expression by ADAR2, as well as its tumor-promoting function, were reported (39). Finally, modification events occurring in the mature sequence, particularly in the seed region, could affect target recognition and modify miRNA function (24,40). Indeed, a single editing site in a MSR could drastically alter the set of mRNA targets (41).
While RNA editing has been associated with many biological processes (25,26), including hypoxia (42–44), this has not yet been established for miRNA editing. To investigate potential variation in miRNA editing activity in relation to environmental stimuli, we have analyzed small RNA-seq (sRNA-seq) data obtained from an experiment examining over time a hypoxia-treated breast cancer cell line, focusing specifically on miRNA post-transcriptional modifications occurring in the seed region, with particular attention to A-to-I editing events. Results have indicated, for the first time, how the hypoxic state is accompanied by a significant change over time in miRNA editing activity. Finally, by integrating variations in gene expression along with target prediction, we investigated the potential new roles which environmental factors play on the editing of the seed region in miRNAs, thus resulting in the alteration of their respective targetomes. We have thus detected an association between miRNA editing and fundamental biological pathways linked to hypoxia.
MATERIALS AND METHODS
Data sets used
We considered the sRNAseq data sets from Camps et al. (45) deposited in GEO (GSE47602), comprising a time-course (16, 32, 48 h) small RNA expression profiling of breast adenocarcinoma cell line MCF-7 exposed to hypoxia (1% Oxygen), along with small RNA expression profiling of cells from the same cell line maintained in normoxic conditions (21% Oxygen). We generated all data from RNA samples obtained from two biological replicates for each experimental condition (normoxia, 16 h hypoxia, 32 h hypoxia, 48 h hypoxia).
We also considered normalized data on mRNA expression from three biological replicate samples for each of the same experimental conditions as deposited by Camps et al. (45) in GEO (GSE47533).
Detection and analysis of miRNA editing events
We detected miRNA editing events from the sRNAseq data by employing the computational approach implemented by Alon and Eisenberg (46).
Briefly, we filtered all reads according to a read quality ≥20 in more than three positions. In addition, we removed sequences identified as 5′ or 3′ adaptors. Subsequently, we also removed reads whose length did not fall within the typical length range for a mature miRNA (16–27 bases).
We applied the Bowtie software (v1.1.2) (47) to align the filtered and trimmed reads against the human genome (UCSC hg19/GRCh37), allowing one mismatch at most, while trimming the last two bases of the read (48) (Bowtie parameters: -n 1 -e 70 -a -m 1 -best -strata -trim3 2). We then mapped the total resulting number of reads against the known pre-miRNA sequences (miRBase, release 21) (49). All nucleotide positions in each mature or miRNA* were screened for mismatches that were overrepresented considering the expected sequencing error rate of 0.1% (as consequence of applying a Phred score filter of >30) (41). We accomplished this by applying the binomial cumulative distribution on the counts of each sequenced nucleotide. We applied a very permissive expression filter, as we took into consideration any miRNA against which more than five reads were aligned. Then, we applied a Benjamini–Hochberg false-discovery rate of 5% (50) (Benjamini and Hochberg 1995), to detect statistically significant modifications, which were subsequently filtered of known single nucleotide polymorphisms (dbSNP build 142).
The editing motifs in the bases flanking the A-to-I editing sites detected, as well as sequence preference in the bases opposing the A-to-I editing sites, were created using the WebLogo tool (51).
For each of these modification events, we calculated the average modification levels (AMLs) in each condition as the average of the modification levels of each replicate.
We calculated a Pearson's correlation for the series of AMLs (as in each condition) and the series of linear fold-changes (at each time point relative to normoxia control (45)) for each modification event/miRNA pair, by using the function cor.test in the stats R package.
The significance of the differences between editing levels of different time points during hypoxia progression was assessed through a Wilcoxon signed rank test by employing the function wilcox.test in the stats R package, and considering all putative A-to-I editing sites for each time point.
Prediction of WT and ED miRNA targetomes
We predicted binding sites for WT and ED miRNA on the whole 3′UTR-ome (UCSC.hg19) through a consensus of four miRNA target prediction tools: miRanda (v3.3a) (52), TargetScan (v7.0) (53), PITA (v6.0) (54) and miRiam (55), our in-house tool, enhanced with the scoring function described in (56). Standard parameters were employed for the tools miRanda, TargetScan and PITA. miRiam's parameters were set to detect canonical binding sites only (6mer, 7mer-A1, 7mer-m8 and 8mer), allowing no mismatches in the seed (e.g. wobble pairs). miRiam's scoring function is based on the combination of the tree-based multiple linear regression learning system M5P with CTree and takes into account six different features of miRNA/target interactions: type of seed match, miRNA nucleotide composition, pairing of the 3′ region of the miRNA, AU content of the binding site and its flanking regions, structural accessibility of the binding site and presence of AU Rich Element (ARE) and Cytoplasmic Polyadenylation Element (CPE) motifs upstream of the binding sites.
We created the Venn diagrams that represent the intersection between the sets of predicted mRNA targets for the WT and ED miRNAs by using the R package VennDiagram.
mRNA differential expression and WT\ED miRNA targets functional enrichment analyses
We performed differential expression analysis of microarray gene expression data from publicly available data set GSE47533, in each time-point by using the R/Bioconductor package Limma (57). P-values were used to rank all genes, retaining those under a significant threshold of 0.01 adjusted P-value (BH) (50) and included in at least one of the targetomes of the mature miRNAs (WT and ED) affected by the 4 statistically significant A-to-I editing events analyzed above, for further functional enrichment analysis. Furthermore, as control for the enrichment analysis, we considered the remaining DE genes not included in the predicted targetome of the edited or unedited miRNA in question.
We performed functional enrichment analysis on the retained set of differentially expressed (DE) genes in each time-point by employing the software integrated pathway analysis (IPA) by Ingenuity. Settings used included experimentally observed data for human species, specifically, pathways exclusively associated to the MCF-7 cell line.
Cell culture, transfection and chemicals
Cells were seeded and grown in RPMI (A549) or DMEM (HeLa) with 10% fetal bovine serum (FBS), L-glutamine and antibiotics (Invitrogen, Carlsbad, CA, USA). All the transfections were performed by using Lipofectamine 2000 (Invitrogen) as suggested by the manufacturer. A549 and HeLa cells for Western blot assay were cultured to 80% confluence in six-well plates and transfected with 100 nmol of miRNAs or negative control in a serum-free medium without antibiotics, after 6 h the medium was replaced with a complete grown media. The cells were harvested after 48 h. HeLa cells transfection for luciferase assay is described below.
Luciferase assay
We used the luciferase reporter constructs described in another work (58). We introduced mutations in hsa-miR-27a-3p-ED binding sites on the MET construct by using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA, USA). We seeded HeLa cells in 12 well plate and after 24 h transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), 1.2 µg of pGL3control containing EGFR, MET or MET mutants, 200 ng of Renilla luciferase expression construct. After 24 h from transfection, we lysed and assayed cells with Dual Luciferase Assay (Promega) according to the manufacturer's instructions.
The mutagenesis primers used are:
Met27ed mutFw 5′-gaccaatggcctgcagcaacactcctgtcata-3′
Met27ed mutRv 5′-tatgacaggagtgttgctgcaggccattggtc-3′
Western-blot analysis
A549 and HeLa cells were seeded and grown in appropriate media with 10% FBS in six-well plates for 24 h before the transfection, 48 h after which we washed cells with cold phosphate buffered saline and subjected them to lysis in lysis buffer (50 mM Tris-HCl, 1 mM EDTA, 20 g/l SDS, 5 mM dithiothreitol, 10 mM phenylmethylsulfonyl fluoride). Equal amounts of protein lysates (50 µg each) and rainbow molecular weight marker (Bio-Rad Laboratories, Hercules, CA, USA) were separated by 4–20% SDS–PAGE and then electrotransferred to nitrocellulose membranes. The membranes were blocked with a buffer containing 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20 for 2 h and incubated overnight with antibodies at 4°C. After a second wash with Tris-buffered saline with 0.1% Tween 20, the membranes were incubated with peroxidase-conjugated secondary antibodies (GE Healthcare, Amersham, Pittsburg, PA, USA) and developed with an enhanced chemiluminescence detection kit (Pierce, Rockford, IL, USA).
Antibody used for Western-blot analysis and RNA extraction
MET was obtained from Cell signaling (#4560), EGFR from Santa Cruz (#Sc-03), Tubulin from Sigma (#T6199). Total RNA was extracted with TRIzol solution (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions.
Q-real-time PCR (Q-rt PCR)
For the detection of single-target a-miRs, we performed quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) by using a standard TaqMan PCR Kit protocol on an Applied Biosystem 7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). For the TaqMan qRT, the 10 µl PCRreaction included 0.67 ml RT product, 1 ml TaqMan Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA), 0.2 mM Taqman probe, 1.5 mM forward primer and 0.7 mM reverse primer. The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions ran in triplicate. The threshold cycle (Ct) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The comparative Ct method for relative quantization of gene expression (Applied Biosystems, Carlsbad, CA, USA) was used to determine miRs expression levels. The y-axis represents the relative expression of the different miRs expression was calculated relative to U44 rRNA. We carried out experiments in triplicate for each data point, and performed data analysis by using software tools (Bio-Rad Laboratories, Hercules, CA, USA).
RESULTS
Systematic identification of miRNA-sequence modifications in human breast cancer cells during a time-course of exposure to hypoxic conditions
The advent of high-throughput sequencing technology has considerably improved the exploration of the transcriptome. HTS has, indeed, allowed a more precise analysis of differential expression under different conditions and the detection of different types of sequence modifications in transcripts, such as those caused by RNA editing. Recently, Camps et al. studied in depth the regulation of miRNA expression in the human breast cancer cell line MCF-7 during hypoxia (45), identifying 41 and 28 miRNAs significantly up- and down-regulated, respectively. Leveraging on their published data (GEO reference: GSE47534; miRNA-seq: GSE47602), including sequences of a small RNA library for HTS originating from biological duplicate RNA samples for each experimental condition (normoxia, 16, 32 and 48 h hypoxia), to investigate the presence of post-transcriptional modification events in miRNA mature sequences and evaluate such changes as they occur in the hypoxic cellular context. To systematically identify such events, we have employed the Alon–Eisenberg pipeline (41,46) (see Supplementary Figure S1 and Material and Methods section), currently the only method for accurate detection and quantification of canonical and non-canonical editing sites in mature miRNAs from HTS data (59). We identified a total of 31 statistically significant modification sites in 21 different miRNAs, with 7 sites previously known (see Supplementary Figure S2). Interestingly, 83% of A-to-G events (10 of 12) occurred in seed regions compared to 37% of non A-to-G events (7 of 19), as shown in Supplementary Table S1 and Supplementary Figure S3. Since post-transcriptional A-to-G modifications are mostly expected to be the result of A-to-I editing (29), it is of relevant importance to observe a significant incidence of such modifications in the miRNA seed region. This reveals how the A-to-I editing phenomenon may preferentially occur in the miRNA seed region under hypoxic cellular conditions, thus causing significant shifts in the miRNA targetome (24,40).
The discovery of these A-to-I editing sites in pre-miRNAs (Supplementary Figure S4) was further supported by the presence of specific editing motifs near the sites, detected by neighborhood profiling, in line with previous studies (60,61). As shown in Supplementary Figure S5A, changes to nucleotides C and U are over-represented upstream of the edited site, while G is under-represented upstream and over-represented downstream of the edited site, as previous studies have established (62,63). The nucleotide opposing the editing site is U (see Supplementary Figure S5B), as previously documented (41).
Levels of miRNA modifications in the seed region change during hypoxia
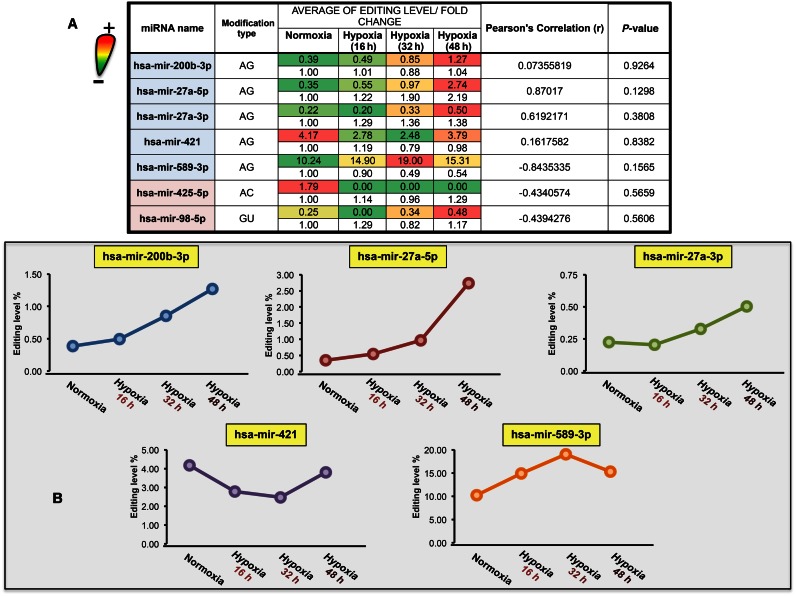
Given the lack of substantial knowledge on the matter, we sought to determine whether the level of miRNA modification events changes during a dynamic cellular context, such as in a hypoxia time course, and whether such changes are proportionally associated to miRNA expression or not. To accomplish this, we considered only those miRNA modification events occurring in the seed region in both biological replicates to assure robustness to our analysis. Such was the case for 7 of the statistically significant modification events, 5 of which are A-to-G (all in MSRs) (Figure 1).
Figure 1.
Average modification levels have been calculated for each replicate in normoxia and in each hypoxia time point. (A) The colors used for the rows are summarized in the ‘volume’ icon on the left of the table in the figure, with green representing low levels and red representing high levels. Fold-changes (in linear scale) for each microRNA at each time point relative to normoxia control have been reported as obtained by Camps et al. (45) (clear rows). Pearson's correlation (r), with relative P-value, was calculated between the average modification level and the linear fold-change for each miRNA. (B) Plotting of observed average modification levels over time-course for each selected A-to-I edited miRNA.
The AMLs of the majority of miRNAs affected (5 out of 7) generally increased with time of exposure to hypoxia (Figure 1A and B), a result which is in line with relatively recent findings where higher post-trascriptional modification rates in coding genes are detected under increased exposure to hypoxia (42,43,64). Nevertheless, we also observed decreased (e.g. A-to-G modifications in miR-421) and null levels (e.g. non A-to-G modifications in miR-425-5p) of the editing activity. To relate the level of modifications with differential expression under hypoxia, we calculated Pearson's correlation (Figure 1 and Material and Methods section) for the AML in each condition and the linear fold-change obtained at each time point relative to normoxia control (45). Interestingly, there is no significant correlation among the AMLs and the linear fold changes shows that miRNA seed sequence modifications are at least not positively correlated with miRNA expression (Figure 1). This suggests that the trend of the miRNA seed sequence modification phenomenon does not follow miRNA expression during dynamic cellular changes, such as those occurring during progressive hypoxia. Additionally, in order to specifically assess the significance of the differences between editing levels of different time points during hypoxia progression, we performed a Wilcoxon signed rank test considering all putative A-to-I editing sites for each time point, as presented in Figure 1A. Results have shown a significant increase in editing levels only between the 16 h and 48 h hypoxia time points (P-value < 0.05).
miRNA targetome changes due to A-to-I seed region editing during hypoxia
To estimate the impact of editing on miRNA function during hypoxia progression, we selected to investigate A-to-G miRNA modifications, as these are expected to be the result of A-to-I editing. Specifically, we decided to consider all A-to-I editing events (Figure 1A, Supplementary Figure S3) occurring within MSRs (6-mers comprising nucleotide positions 2–7 corresponding to the seed region, thus excluding hsa-miR-27a-5p whose putative A-to-I editing site occurs in position 1), extended by nucleotide position 8 (in order to also take into account offset 6-mer sites comprising nucleotide positions 3–8), as the fundamental role of the 6-mer seed sequence in the miRNA–mRNA interaction has been widely documented (65). A single editing site in the MSR could determine important changes, as shown in Supplementary Figure S6. In fact, by comparing edited miRNA seed regions with their un-edited references (49), there are two possible scenarios for an edited miRNA: either it is modified into a new miRNA (acquiring an entirely unprecedented seed sequence) or its seed sequence is modified into that of another known human miRNA (Supplementary Figure S6).
Subsequently, in order to compare the target sets of the wild type (WT) and edited (ED) versions of each miRNA, we performed a robust binding site prediction analysis on gene 3′-UTRs with a consensus of four miRNA target prediction tools, namely, miRanda (52), TargetScan (53), PITA (54) and miRiam, our in-house tool, enhanced with our recently developed scoring function which takes into account both sequence and structure features (55,56). The prediction tools we employed consider seeds when evaluating target genes, starting from a 6-mer MSRs within nucleotide positions 2–8 (as stated above and as the established norm for miRNA target prediction tools). Any modification occurring outside such MSRs, under such paradigm, would thus not imply any change in the predicted targetome, contributing, at best, to a strengthening or weakening of an already established potential binding site.
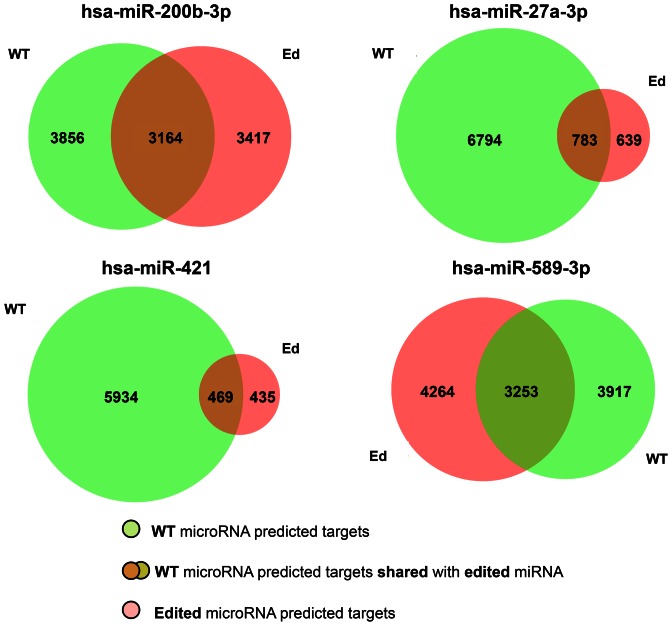
In line with previous studies, we performed target predictions for A-to-I edited miRNAs by considering inosine as guanosine (41,66). In fact, inosine can bind cytidine with approximately the same energy as guanosine, while the binding of inosine to uridine is weaker than that of guanosine. Moreover, unlike guanosine, inosine can bind weakly to adenosine (67). As shown in Figure 2, the sets of predicted mRNA targets of the WT and A-to-I ED miRNAs respectively, overlap with an average of 19%, a surprisingly larger percentage compared to the 3% known so far (41). These results are consistent with a very recent study (68), in which Hill et al. have demonstrated that a single nucleotide substitution in positions 2–8 of a mature miRNA can determine a targetome shift >60–70%. In fact, the considered editing events may lead to an average loss >60% of mRNA targets for WT miRNAs when edited. In particular, we predicted drastic changes for miR-27a-3p and miR-421, suggesting alteration of their functions.
Figure 2.
Venn diagrams of predicted mRNA target sets for the WT and A-to-I ED miRNAs.
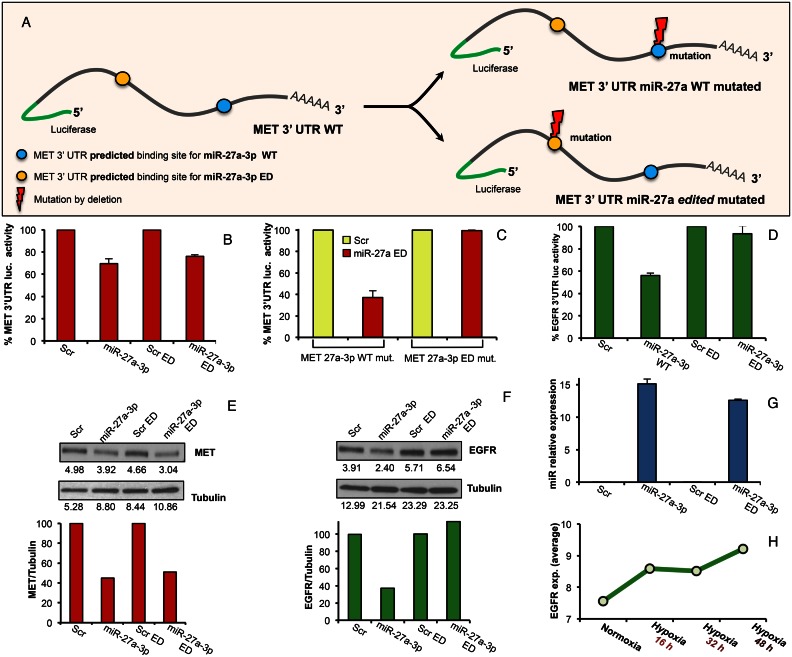
In light of previous reports which proved that the seed of miR-27a-3p possesses binding matches in the 3′-UTR of human MET (nucleotide 1564–1571; NM_000245) and human EGFR (nucleotide 200–207 and nucleotide 430–436 NM_005228) (58), we decided to employ this knowledge to validate our target prediction. Moreover, the choice of miR-27a-3p has allowed us to validate our prediction results in relation to two specific and confirming contexts: that of complete loss of targeting (as with miR-27a-3p ED on EGFR) as well as that of replacement of an old binding site with a new one (as with miR-27a-3p ED and MET) (Supplementary Figure S7).
To verify that MET was still a direct target of miR-27a-3p ED, we cloned the MET 3′ UTR containing the predicted WT and ED binding sites for miR-27a-3p, into the pGL3 control vector, downstream of the luciferase open reading frame (ORF) (Figure 3A). Transfecting HeLa cells with WT and ED miR-27a-3p, respectively, together with the MET 3′-UTR luciferase construct, resulted in a significant inhibition of luciferase activity in both cases as compared to the negative control, confirming our prediction (Figure 3B). To determine that miR-27a-3p ED was not affecting the MET 3′-UTR by nonspecific binding to the miR-27a-3p WT binding site, we performed deletion of this predicted binding site within the MET 3′-UTR (Figure 3A). As shown in Figure 3C, miR-27a-3p ED could still regulate the luciferase expression of MET 3′-UTR, despite the mutation of miR-27a-3p WT binding site. Subsequently, we mutated by deletion the predicted binding site for miR-27a-3p ED on the MET 3′UTR (Figure 3A). As shown in Figure 3C, miR-27a-3p ED was no longer able to repress the luciferase expression of MET 3′-UTR with miR-27a-3p ED binding site mutated. That proves the binding site of the ED version of the miRNA to be the only functional one, confirming our prediction. We also predicted that miR-27a-3p ED would no longer be able to directly target the EGFR 3′-UTR, as shown in Supplementary Figure S7. To validate this prediction, we co-transfected HeLa cells with both miR-27a-3p ED and the EGFR 3′-UTR containing both the WT and ED miR-27a-3p binding sites, previously cloned into the pGL3 control vector downstream of the luciferase ORF. As a result, we noted that miR-27a-3p ED could not inhibit the luciferase activity of the EGFR 3′ UTR compared to negative control (Figure 3D). In addition, the over-expression of miR-27a-3p ED reduced the endogenous MET level but could no longer reduce the endogenous EGFR protein level in A549 and HeLa cells compared to control (Figure 3E, F and Supplementary Figure S8). We confirmed the increased expression of WT and ED miR-27a-3p in transfected cells by RT-qPCR (Figure 3G). We also confirmed the EGFR expression trend in the hypoxic samples in which the above miRNA editing analysis was performed. The EGFR expression shown in Figure 3H shows its increase during the hypoxia time course. These results thus confirm our target predictions as displayed in Figure 2, while also specifically showing that the increase in A-to-I editing of miR-27a-3p is in accord with increased EGFR expression during hypoxia.
Figure 3.
Effects of A-to-I miRNA seed editing on targeting. (A) c-MET 3′ UTR binding sites for miR-27a-3p, ED (yellow spot) and WT (blue spot), along with corresponding deletions (lightening bolt). (B) Luciferase assay for pGL3-MET 3′ UTR WT construct co-transfected with miR-27a-3p WT, miR-27a-3p ED or negative scramble miRNA control (Scr) in HeLa cells (error bars: ±). (C) Luciferase assay for pGL3-MET 3′ UTR WT/mut and pGL3-MET 3′ UTR ED/mut constructs co-transfected with miR-27a3p ED or negative control (Scr) in HeLa cells. (D) Luciferase assay for pGL3-EGFR 3′ UTR WT construct co-transfected with miR-27a-3p WT, miR-27a-3p ED or negative controls (Scr) in HeLa cells. (E and F) c-MET and EGFR expression by western blot in A549 cells transfected with miR-27a-3p ED, miR-27a-3p WT or negative control (Scr) and harvested after 48 h, with graphs for c-Met/Tubulin or EGFR/Tubulin ratio quantification. (G) qRT-PCR of WT and ED miR-27a-3p respectively after miR transfection in A549 cells as control for Figure 3E and F. (H) EGFR expression as in (45).
miRNA editing is in line with dynamic phenotype alteration
The hypoxia-Inducible Factor 1 (HIF-1), a key factor in cellular hypoxia response, is known to regulate specific genes and influence several cellular pathways (69). For instance, the VEGF and PI3K/Akt pathways are closely associated with the hypoxic condition and are activated during this cellular process (70–72).
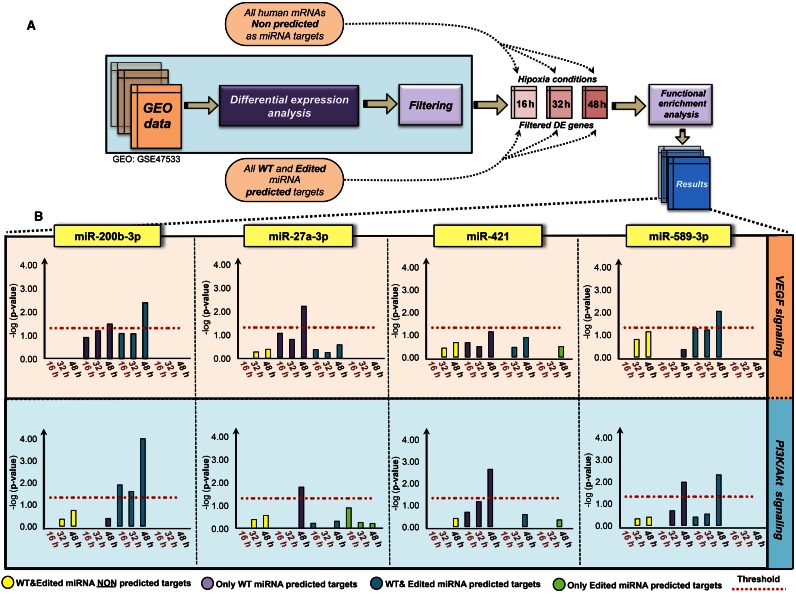
After having analyzed the RNA-seq data, we proceeded to perform differential gene expression analysis on microarray data also provided by Camps et al. (GEO reference: GSE47534; mRNA: GSE47533) (45), originating from the same hypoxia time-course samples. We then applied statistical significance filtering, by considering only genes which were differentially expressed with an adjusted P-value below 0.01. Subsequently, at each time point, for each miRNA with A-to-I editing events occurring in their MSRs (2–8 nt seed region) we isolated their predicted targets as present in the DE gene set (namely, the set of targets exclusive to the WT and ED versions, respectively, as well as shared targets); on the other hand, we also considered the DE genes which were not predicted to be targets of either version of the considered miRNA, as depicted in Figure 4A. We thus performed an MCF7-specific functional enrichment of both subsets of genes by Ingenuity Pathway Analyzer software. Finally, we focused on hypoxia-related pathways, such as VEGF and PI3K, to evaluate how the miRNA editing phenomenon globally behaves in relation to these pathways during the hypoxic time course, by confronting the set of targets of the WT miRNA with the set of targets of its ED version, using as control all those DE transcripts which were not predicted to be targets of either version. Our data clearly shows how A-to-I editing of the MSR of these miRNAs translates into a diminished ability to target key genes involved in these two important pathways throughout the full time course of the observed hypoxia process (Figure 4B, Supplementary Figure S9 and Supplementary Table S2). Our results unprecedentedly show that A-to-I miRNA editing is not a merely random phenomenon, but rather a molecular mechanism that, specifically through miRNA seed mutation events, is in line with important biological processes.
Figure 4.
Functional enrichment of differentially expressed targets. (A) General scheme representing the functional enrichment workflow: differentially expressed genes with a significant adjusted P-value (BH < 0.01) in each hypoxia time point relative to normoxia were separated according to whether or not belonging to predicted miRNA targetome (WT or ED) for each miRNA, followed by MCF7-specific functional enrichment by Ingenuity Pathway Analyzer (IPA) software. (B) IPA analysis on VEGF and PI3K/Akt pathways in MCF7 cell line. Graphs show -log (P-value) over time course for both WT (purple) and A-to-I ED (green) predicted miRNA targets. Bars represent the level of significance for the indirect involvement in the considered pathways for WT and A-to-I ED miRNAs, respectively. P-values represent the significance of the set of target genes (which and how many) involved in a given pathway for each miRNA (WT and ED, respectively). The red dotted line represents the significance threshold level (-log(p), where P = 0.05).
DISCUSSION
Until recently, miRNA editing has been at the center of a debate concerning its purpose and even its very occurrence. After several reports attempted to shed light on the matter, discordant opinions and widespread scepticism on the topic seem to have started to fade in favor of affirming the existence of such biological phenomenon (41,73). Additionally, all studies conducted on the matter have investigated miRNA editing under static cellular states, performing a comparison analysis between fixed pathological (i.e. cancerous) and normal conditions (39). This type of study provides insights on on what changes without elucidating how miRNA editing behaves.
Recently, the advent of innovative bioinformatics approaches have allowed for an unprecedentedly precise evaluation of miRNA editing events from deep sequencing data (41,46). Combining such novel methodologies with the nature of HTS technology, has, for instance, presented the opportunity to investigate the phenomenon under specific cellular states over time, providing a dynamic view which is necessary for a more informative analysis and understanding of the phenomenon of RNA editing.
In the present study, we have indeed applied the recently published bioinformatics pipeline devised by Alon and Eisenberg to time course sRNA-seq data originating from MCF-7 breast cancer cells cultured under hypoxic conditions, to globally analyze the miRNA editing phenomenon in a dynamic cellular context. Indeed, the main core of our work focuses on evaluating how the miRNA editing activity varies in relation to hypoxia progression, and, secondly, to elucidate how this phenomenon globally relates to cellular changes. As we have observed, the general miRNA seed region modification level varied significantly as well as proved to be significantly independent from miRNA expression during the hypoxic time course.
Our research thus sought to specifically investigate three possibilities regarding the phenomenon: miRNA editing could have indeed displayed a collective behavior which resulted in line with, contrary to, or independent of the cellular response to external factors. In order to accomplish this, we focused on editing events occurring in the MSR as it is fundamental for targeting effectiveness and specificity (65). Target prediction for both WT and ED miRNAs was followed by experimental validation. Interestingly, functional enrichment analysis of four putative A-to-I edited miRNAs revealed a common trend during hypoxia. In fact, according to our observations as presented in Figure 4, we have detected a collective alignment of the targetome shifts for all four MSR-edited miRNAs which were predicted to no longer target key genes of hypoxia-related pathways such as VEGF and PI3K/AKT, as summarized in Supplementary Figure S9 and Table S2. This shows a global disinterest of miRNA editing in affecting hypoxia-related pathways. This does not necessarily signify that miRNA editing, due to very low editing levels, plays an active role, rather that there is a non-random trend of disinterest. Future studies on other tissue and cellular contexts more quantitatively informative for the editing phenomenon (i.e. glioblastoma) could provide further confirmation on the matter. Nonetheless, our goal is to establish a novel and more appropriate approach of investigation of this largely unexplored phenomenon, with regards to its behavior in relation to functional cellular changes. This allows to elucidate how the fine-tuning miRNA editing activity locates itself within the life of the cell, especially in light of external stimuli.
The current study is also the first to investigate the biological behavior of miRNA editing within a global and dynamic context, with the hope of providing new elements to further elucidate the cellular role of this biological phenomenon in the near future. The dynamic dimension of our study is, in our view, essential to better assess the nature of the molecular phenomenon. In addition to clarifying its purposeful existence, we were able to successfully relate miRNA editing to a biological condition such as hypoxia, along a time frame. It should be noted that hypoxia represents but a limited instance of a much wider panorama, in which miRNA editing could be considered as a component of a response mechanism employed by the cell to rapidly shift the miRNA targetome according to contextual needs, especially in reaction to stressful external factors. The evidence that emerges from our functional data shows, indeed, how the transformation (editing) of certain miRNAs is associated with the cellular response to hypoxic stimulus. Additionally, despite the low editing levels, we do not intend to focus on the editing of a single miRNA, rather consider the phenomenon globally so to evaluate its collective effects and behavior. Adding a temporal dimension to the study of this phenomenon provides a quantity and quality of information on how the cell responds to cellular changes. Specifically, miRNA editing has thus shown to align itself to cellular needs and thus, potentially contribute to cell economy. In fact, instead of transcribing novel miRNAs, the cell can more easily leverage on the already existing population.
Unfortunately, lack of proper investigation of editing events in the seed region of miRNA binding sites on mRNAs, and of editing affecting pre-miRNA maturation due to limitations in current technology, do not yet allow for a more comprehensive and detailed evaluation of the phenomenon. Nevertheless, our results provide a foundation for further elucidation of miRNA editing, giving a direction to future innovative investigation of its biological importance and potential involvement in physiological and pathological cellular changes.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. F. Kay Hubner for her useful support during the drafting process and Dr Shahar Alon for his precious suggestions during the application of the computational approach for RNA editing detection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [U01-CA152758 to M.A. and C.M.C]; Italian Foundation for Cancer Research (FIRC) [15046 to G.N.]; Italian Funding for Cancer Research (FIRC) [16572 to D.V.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Namiki A., Brogi E., Kearney M., Kim E.A., Wu T., Couffinhal T., Varticovski L., Isner J.M. Hypoxia induces vascular endothelial growth factor in cultured human endothelial cells. J. Biol. Chem. 1995;270:31189–31195. doi: 10.1074/jbc.270.52.31189. [DOI] [PubMed] [Google Scholar]
- 2.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Gregory R.I., Chendrimada T.P., Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 5.Lund E., Dahlberg J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz D.S., Zamore P.D. Why do miRNAs live in the miRNP? Genes Dev. 2002;16:1025–1031. doi: 10.1101/gad.992502. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S., Galasso M., Costinean S., Tagliavini L., Gamberoni G., Drusco A., Marchesini J., Mascellani N., Sana M.E., Abu Jarour R., et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harfe B.D. MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev. 2005;15:410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Carleton M., Cleary M.A., Linsley P.S. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2137. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 11.Boehm M., Slack F.J. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5:837–840. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 12.Jovanovic M., Hengartner M.O. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 13.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 14.Jin P., Alisch R.S., Warren S.T. RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 15.Iorio M.V., Croce C.M. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio M.V., Croce C.M. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veneziano D., Nigita G., Ferro A. Computational approaches for the analysis of ncRNA through deep sequencing techniques. Front. Bioeng. Biotechnol. 2015;3:1–6. doi: 10.3389/fbioe.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y., He Y., Ding J., Wu K., Hu B., Liu Y., Wu Y., Guo B., Shen Y., Landi D., et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30:2064–2069. doi: 10.1093/carcin/bgp283. [DOI] [PubMed] [Google Scholar]
- 21.Yue C., Wang M., Ding B., Wang W., Fu S., Zhou D., Zhang Z., Han S. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol. Oncol. 2011;122:33–37. doi: 10.1016/j.ygyno.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Nicoloso M.S., Sun H., Spizzo R., Kim H., Wickramasinghe P., Shimizu M., Wojcik S.E., Ferdin J., Kunej T., Xiao L., et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brewster B.L., Rossiello F., French J.D., Edwards S.L., Wong M., Wronski A., Whiley P., Waddell N., Chen X., Bove B., et al. Identification of fifteen novel germline variants in the BRCA1 3'UTR reveals a variant in a breast cancer case that introduces a functional miR-103 target site. Hum. Mutat. 2012;33:1665–1675. doi: 10.1002/humu.22159. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya A., Ziebarth J.D., Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2015;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jepson J.E.C., Reenan R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 30.Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara Y. Quantification of adenosine-to-inosine editing of microRNAs using a conventional method. Nat Protoc. 2012;7:1426–1437. doi: 10.1038/nprot.2012.073. [DOI] [PubMed] [Google Scholar]
- 32.Blow M.J., Grocock R.J., Van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chawla G., Sokol N.S. ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 2014;42:5245–5255. doi: 10.1093/nar/gku145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoshan E., Mobley A.K., Braeuer R.R., Kamiya T., Huang L., Vasquez M.E., Salameh A., Lee H.J., Kim S.J., Ivan C., et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T., Xiang J.-F., Zhu S., Chen S., Yin Q.-F., Zhang X.-O., Zhang J., Feng H., Dong R., Li X.-J., et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Research. 2015;25:459–476. doi: 10.1038/cr.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galore-Haskel G., Nemlich Y., Greenberg E., Ashkenazi S., Hakim M., Itzhaki O., Shoshani N., Shapira-Fromer R., Ben-Ami E., Ofek E., et al. A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme. Oncotarget. 2015;6:28999–29015. doi: 10.18632/oncotarget.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahn J.H., Ahn J., Lin X., Zhang Q., Lee J.-H., Civelek M., Xiao X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:1–13. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomaselli S., Galeano F., Alon S., Raho S., Galardi S., Polito V., Presutti C., Vincenti S., Eisenberg E., Locatelli F., et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16:5. doi: 10.1186/s13059-014-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawahara Y., Zinshteyn B., Chendrimada T.P., Shiekhattar R., Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alon S., Mor E., Vigneault F., Church G.M., Locatelli F., Galeano F., Gallo A., Shomron N., Eisenberg E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevo-Caspi Y., Amariglio N., Rechavi G., Paret G. A-to-I RNA editing is induced upon hypoxia. Shock. 2011;35:585–589. doi: 10.1097/SHK.0b013e31820fe4b7. [DOI] [PubMed] [Google Scholar]
- 43.Borik S., Simon A.J., Nevo-Caspi Y., Mishali D., Amariglio N., Rechavi G., Paret G. Increased RNA editing in children with cyanotic congenital heart disease. Intensive Care Med. 2011;37:1664–1671. doi: 10.1007/s00134-011-2296-z. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Zvi M., Amariglio N., Paret G., Nevo-Caspi Y. F11R expression upon hypoxia is regulated by RNA editing. PLoS One. 2013;8:e77702. doi: 10.1371/journal.pone.0077702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camps C., Saini H.K., Mole D.R., Choudhry H., Reczko M., Guerra-Assunção J.A., Tian Y.-M., Buffa F.M., Harris A.L., Hatzigeorgiou A.G., et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer. 2014;13:1–21. doi: 10.1186/1476-4598-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alon S., Eisenberg E. Identifying RNA editing sites in miRNAs by deep sequencing. Methods Mol. Biol. 2013;1038:159–170. doi: 10.1007/978-1-62703-514-9_9. [DOI] [PubMed] [Google Scholar]
- 47.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burroughs A.M., Ando Y., de Hoon M.J.L., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y., et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 51.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Correction: Human MicroRNA Targets. PLoS Biol. 2005;3:e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;12:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 55.Laganà A., Forte S., Russo F., Giugno R., Pulvirenti A., Ferro A. Prediction of human targets for viral-encoded microRNAs by thermodynamics and empirical constraints. J. RNAi Gene Silencing. 2010;6:379–385. [PMC free article] [PubMed] [Google Scholar]
- 56.Laganà A., Acunzo M., Romano G., Pulvirenti A., Veneziano D., Cascione L., Giugno R., Gasparini P., Shasha D., Ferro A., et al. miR-Synth: a computational resource for the design of multi-site multi-target synthetic miRNAs. Nucleic Acids Res. 2014;42:5416–5425. doi: 10.1093/nar/gku202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth G.K. Limma: linear models for microarray data. In: Gentleman RVC, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. NY: Springer; 2005. [Google Scholar]
- 58.Acunzo M., Romano G., Palmieri D., Laganà A., Garofalo M., Balatti V., Drusco A., Chiariello M., Nana-Sinkam P., Croce C.M. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8573–8578. doi: 10.1073/pnas.1302107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nigita G., Veneziano D., Ferro A. A-to-I RNA editing: Current knowledge sources and computational approaches with special emphasis on non-coding RNA molecules. Front. Bioeng. Biotechnol. 2015;3:1–7. doi: 10.3389/fbioe.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleinberger Y., Eisenberg E. Large-scale analysis of structural, sequence and thermodynamic characteristics of A-to-I RNA editing sites in human Alu repeats. BMC Genomics. 2010;11:1–17. doi: 10.1186/1471-2164-11-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nigita G., Alaimo S., Ferro A., Giugno R., Pulvirenti A. Knowledge in the investigation of A-to-I RNA editing signals. Front. Bioeng. Biotechnol. 2015;3:1–8. doi: 10.3389/fbioe.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polson A.G., Bass B.L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehmann K.A., Bass B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities †. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 64.Fang X., Nevo E., Han L., Levanon E.Y., Zhao J., Avivi A., Larkin D., Jiang X., Feranchuk S., Zhu Y., et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 2014;5:1–9. doi: 10.1038/ncomms4966. [DOI] [PubMed] [Google Scholar]
- 65.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vendeix F.A.P., Munoz A.M., Agris P.F. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA. 2009;15:2278–2287. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hill C.G., Jabbari N., Matyunina L.V., McDonald J.F. Functional and evolutionary significance of human microRNA seed region mutations. PLoS One. 2014;9:e115241. doi: 10.1371/journal.pone.0115241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semenza G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirota K., Semenza G.L. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit. Rev. Oncol./Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 71.Liao D., Johnson R.S. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 72.Coleman M.L., Ratcliffe P.J. Angiogenesis: escape from hypoxia. Nat. Med. 2009;15:491–493. doi: 10.1038/nm0509-491. [DOI] [PubMed] [Google Scholar]
- 73.de Hoon M.J.L., Taft R.J., Hashimoto T., Kanamori-Katayama M., Kawaji H., Kawano M., Kishima M., Lassmann T., Faulkner G.J., Mattick J.S., et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.