Summary
Although connexin production is mainly regulated at the protein level, altered connexin gene expression has been identified as the underlying mechanism of several pathologies. When studying the latter, appropriate methods to quantify connexin mRNA levels are required. The present chapter describes a well-established reverse transcription quantitative real-time polymerase chain reaction procedure optimized for analysis of hepatic connexins. The method includes RNA extraction and subsequent quantification, generation of complementary DNA, quantitative real-time polymerase chain reaction and data analysis.
Keywords: connexins, RNA extraction, reverse transcription, Minimum Information for publication of Quantitative real-time PCR Experiments
1. Introduction
Connexin (Cx) signaling can be regulated by a plethora of mechanisms at the transcriptional, posttranscriptional, translational and posttranslational level (1, 2). Regarding the former, connexin expression is predominantly controlled by the conventional cis/trans machinery (1). A basal level of connexin gene transcription is maintained by general transcription factors, such as specificity protein 1 and activator protein 1, while tissue-specific expression depends on cell type-specific repressors and activators, such as hepatocyte nuclear factor 1α for Cx32 expression in the liver (3, 4). In addition, epigenetic mechanisms, including histone modifications, DNA methylation and microRNA-related control, are essential determinants of connexin gene transcription (5, 6).
Four methods are commonly used for studying individual target gene transcription, namely northern blotting (7), in situ hybridization (8), RNAse protection assays (9, 10) and reverse transcription polymerase chain reaction (RT-PCR) (11). The main limitation of the former 3 techniques is their relative low sensitivity, which is not the case for RT-PCR analysis (12). The latter has a wide dynamic range, as poor and abundant expressed genes can be detected with this technique. In contrast to the conventional RT-PCR procedure, typically followed by agarose gel electrophoresis, the read-out in reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is monitored throughout the PCR process as such and is characterized by the reaction time, during cycling, when amplification of the target is first detected. Being a quick, accurate, sensitive, specific and cost-effective method, RT-qPCR analysis has now become the benchmark assay for quantification of mRNA (13).
The liver was the first organ in which connexins have been described (14, 15). Hepatocytes, the main hepatic cellular population, express Cx32 and to a lesser extent Cx26. In contrast, most non-parenchymal liver cells harbor Cx43 (16, 17). In several liver diseases, such as chronic hepatitis, cirrhosis and hepatocellular carcinoma, connexin mRNA content is altered (18). When studying the latter, appropriate methods to quantify connexin mRNA levels are required. This chapter provides a 2-step RT-qPCR procedure optimized for analysis of hepatic connexins, specifically Cx26, Cx32 and Cx43. Compared to the 1-step RT-qPCR procedure, where the reverse transcription and the polymerase chain reaction take place in 1 buffer system, this is performed in 2 separate systems in the 2-step RT-qPCR procedure. In essence, the procedure implies RNA extraction and quantification, total RNA reverse transcription into complementary DNA (cDNA) followed by a separate amplification of the cDNA by PCR and data analysis. The protocol follows the recommendations provided in the Minimum Information for publication of Quantitative real-time PCR Experiment (MIQE) guidelines (19, 20), which is a state-of-the-art guide for all the necessary requirements for experimental set-up, analysis and publication.
2. Materials
2.1. RNA extraction
GenElute™ Mammalian Total RNA Miniprep Kit (Sigma, USA) (see Note 1): lysis solution, 2-mercaptoethanol, wash solution 1 (see Note 2), wash solution 2 concentrate, elution solution, GenElute™ filtration columns in tubes, GenElute™ binding columns in tubes and collection tubes of 2.0 mL. These reagents must be stored at room temperature.
Lysis solution/2-mercaptoethanol mixture: add 10 µL 2-mercaptoethanol for each 1 mL of lysis solution ex tempore (see Note 3).
≥ 99.5% anhydrous ethanol.
Wash solution 2: dilute 2.5 mL of the provided wash solution 2 concentrate to 10 mL with ≥ 99.5% anhydrous ethanol ex tempore.
70% ethanol solution.
Vortex.
RNase-free pipette tips: aerosol barrier recommended.
RNase-free microcentrifuge tubes.
Microcentrifuge.
On-Column DNase I Digestion Set (Sigma, USA): DNase digestion buffer, DNase I, binding column and wash solution 1. The set may be stored between 2 to 8°C for up to 6 months. For longer-term, storage at -20°C is recommended.
DNase I/digest buffer mixture: mix 10 µL of DNase I with 70 µL of DNase digest buffer for each preparation. Mix by inversion. Do not vortex the DNase I or the DNase I/digest buffer mixture. The mixture may be prepared up to 2 h in advance.
RNAlater™, RNA stabilization solution for tissue (Sigma, USA).
Rotor-stator homogenizer or RNA-free pellet mixer (VWR, USA).
2.2. RNA-DNA quantification and purity control
NanoDrop® ND-100 Spectrophotometer (Thermo Scientific, USA).
RNase-free pipette tips: aerosol barrier recommended.
2.3. cDNA synthesis
iCycler iQ™ (Bio-Rad, USA).
96-well thin wall plates.
Optical sealing tape.
iScript™ cDNA Synthesis Kit (Bio-Rad, USA): 5x iScript™ reaction mix (see Note 1), nuclease-free water and iScript™ reverse transcriptase. The reagents must be stored at -20°C, except for the nuclease-free water, which can be stored at room temperature. The reagents are stable for a minimum of 1 year.
0.2 mL nuclease-free tubes (Bio-Rad, USA).
2.4. cDNA purification
GenElute™ PCR Clean-Up Kit (Sigma, USA): column preparation solution, binding solution, wash solution concentrate, elution solution, GenElute™ plasmid mini spin column and collection tubes of 2.0 mL. The reagents should be stored at room temperature.
≥ 99.5% anhydrous ethanol.
Wash solution: dilute 12 mL of he provided concentrate with 48 mL of ≥ 99.5% anhydrous ethanol.
Microcentrifuge.
RNase-free pipette tips: aerosol barrier recommended.
RNase-free microcentrifuge tubes
Nuclease-free water.
2.5. Real-time qPCR
TaqMan® Universal PCR Master Mix (Applied Biosystems, USA): Taqman® probe, AmpliTaq Gold® DNA polymerase, AmpErase® uracil-N-glycosylase deoxynucleotides with 2’-deoxyuridine 5’-triphosphate, passive reference and optimized buffer components. The reagents must be stored at 2-8°C.
Taqman® primer (Table 1) (Applied Biosystems, USA).
StepOnePlus™ real-time PCR system (Applied Biosystems, USA).
RNase-free microcentrifuge tubes.
Microcentrifuge.
Assay-on-Demand™ Gene Expression Assay Mix (Applied Biosystems, USA).
Nuclease-free water.
Centrifuge suitable for cooling at 4°C with adapter for 96-well plate.
MicroAmp® optical 96-well reaction plates (Applied Biosystems, USA).
MicroAmp® optical caps (Applied Biosystems, USA).
MicroAmp® optical tubes (Applied Biosystems, USA).
Pipette tips: aerosol barrier recommended.
Vortex.
Table 1.
| Gene Symbol | Assay ID | Accession number | Assay location | Amplicon size (base pairs) | Exon boundary |
|---|---|---|---|---|---|
| Gjb2 | Mm00433643_s1 | NM_008125.3 | 603 | 72 | 2-2 |
| Gjb1 | Mm01950058_s1 | NM_008124.2 | 466 | 65 | 1-1 |
| Gja1 | Mm01179639_s1 | NM_010288.3 | 2937 | 168 | 2-2 |
| 18S | Hs99999901_s1 | X03205.1 | 604 | 187 | 1-1 |
| Actb | Mm00607939_s1 | NM_007393.3 | 1233 | 115 | 6-6 |
| B2m | Mm00437762_m1 | NM_009735.3 | 111 | 77 | 1-2 |
| Gapdh | Mm99999915_g1 | NM_008084.2 | 265 | 107 | 2-3 |
| Hmbs | Mm01143545_m1 | NM_013551.2 | 473 | 81 | 6-7 |
| Ubc | Mm02525934_g1 | NM_019639.4 | 370 | 176 | 2-2 |
2.6. Data processing
Reference gene validation systems, such as geNorm, NormFinder and BestKeeper. GeNorm is currently integrated in the qbase+ software (Biogazelle, Belgium). The BestKeeper and NormFinder software can be downloaded from http://www.gene-quantification.de/bestkeeper.html#download and http://moma.dk/normfinder-software, respectively.
Normalization and relative quantification of mRNA software, such as qbase+ and the relative expression software tool (REST) (Qiagen, USA).
3. Methods
3.1. Maintenance of a contamination-free workplace
Since RNases and DNases can rapidly degrade RNA and DNA, respectively, it is of utmost importance to take measures to avoid degradation. Sample acquisition constitutes the first potential source of experimental variability due to degradation. RNA yield and quality are easily perturbed by sample collection and processing methods (21).
The following steps should be taken into consideration throughout the complete RT-qPCR procedure in order to diminish the risk of contamination:
A strict separation between the pre-PCR and post-PCR procedures. If this is not possible, a separate enclosure for either operation should be considered. This separation also implies different sets of reagents and equipment.
A process flow in a unidirectional way must be constructed, implying that the PCR set-up should be performed in a template-free area with reagents that under no circumstances will come in contact with possible contamination sources.
Benchtop hoods with high-efficiency particulate arrestance filters may be useful.
Non-porous surfaces should be habitually cleaned with a 10% bleach solution.
A water container should not be used for long-term water storage, since some bacterial species may flourish.
When stock solutions of the PCR reaction mix are recurrently entered with pipettes that may be contaminated with nucleic acids, aliquots of this mix should be prepared. Thus, if contamination is suspected, the aliquots currently in use may be discarded and replaced.
Sample handling should be minimized and tubes opened very carefully, preferably with a tube opener that can be easily decontaminated.
Gloves should be changed frequently. Nothing should be touched with bare hands.
Aerosol-filtered pipette tips or positive displacement pipettors should be used. Tips should never be touched by anything but the pipettor.
Often used equipment, such as pipettors and work surfaces, should be regularly decontaminated.
3.2. RNA extraction
Different RNA extraction kits are commercially available (e.g. RNeasy Minikit, Qiagen, USA). In this protocol, the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma, USA) is used. This kit provides a simple and convenient procedure to isolate total RNA from mammalian cells and tissues, in casu liver tissue. For RT-qPCR purposes, an additional purification step is needed, as even minor DNA contamination can give false positive detections. Therefore, a digestion step of DNA has been introduced to the outlined procedure using the DNase digestion set (Sigma, USA). All steps in this section are carried out at room temperature.
3.2.1. Cultured hepatic cells
Cells grown on cell culture dishes can be lysed in situ or pelleted and stored at -80°C for several month prior to lyzation. For in situ lyzation, the following procedure should be followed:
Remove the cell culture medium.
Add 250 µL of the lysis solution/2-mercaptoethanol mixture for up to 5 x 106 cells or 500 µL for 5 x 106 to 107 cells to the cell culture dish.
Rock the culture dish while tapping the side for a few seconds to completely cover the cells with the mixture. Let the mixture react for 1 to 2 min.
Continue with step 4 from the procedure for pelleted cells.
For pelleted cells, vortex pellet to loosen cells and continue as follows:
Add 250 µL of lysis solution/2-mercaptoethanol mixture for up to 5 x 106 cells or 500 µL for 5 x 106 to 107 cells.
Vortex or pipette thoroughly until all clumps disappear.
Pipette the lysed cells into a GenElute™ filtration column, a blue insert with a 2.0 mL receiving tube (see Note 4).
Centrifuge at 12000xg to 16000xg (see Note 5) for 2 min.
Discard the filtration column.
Add an equal volume of 70% ethanol solution, 250 or 500 µL, to the filtered lysate.
Vortex or pipette thoroughly to mix.
Pipette up to 700 µL of lysate/ethanol mixture into a GenElute™ binding column, a colorless insert with a red o-ring seated in a 2.0 mL receiving tube. If the volume of lysate/ethanol mixture exceeds 700 µL, the RNA must be bound to the column in 2 steps.
Centrifuge at 12000xg to 16000xg for 15 s.
Retain the collection tube and discard the flow-through liquid.
Repeat steps 9 to 11 if any remaining lysate/ethanol mixture is left.
Continue the procedure using the On-Column DNase I Digestion Set by pipetting 250 µL of wash solution 1 into the binding column and centrifuge at 12000xg to 16000xg for 15 s.
Add 80 µL of the DNase I/digest buffer mixture directly onto the filter in the binding column.
Incubate at room temperature for 15 min.
Pipette 250 µL of wash solution 1 into the binding column and centrifuge at 12000xg to 16000xg for 15 s.
Transfer the binding column into a fresh 2.0 mL collection tube.
Pipette 500 µL of wash solution 1 into a binding column of the GenElute™ Mammalian Total RNA Miniprep Kit.
Centrifuge at 12000xg to 16000xg for 15 s.
Transfer the binding column into a fresh 2.0 mL collection tube.
Discard the flow-through liquid and the original collection tube.
Pipette 500 µL of the ethanol containing wash solution 2 into the column.
Centrifuge at 12000xg to 16000xg for 15 s.
Retain the collection tube and discard the flow-through liquid.
Pipette a 500 µL of wash solution 2 into the column.
Centrifuge at 12000xg to 16000xg for 2 min. If any residual wash solution 2 is seen on the surface of the binding column, centrifuge the column for an additional 1 min at 12000xg to 16000xg. Empty and re-use the collection tube if an additional centrifugation step is needed.
Transfer the binding column to a fresh 2.0 mL collection tube.
Pipette 50 µL of elution solution into the binding column.
Centrifuge at 12000xg to 16000xg for 1 min.
If more than 50 µg of RNA is expected, repeat steps 28 and 29, collecting both eluates in the same tube.
Purified RNA is ready for immediate use or storage at -80°C for several months.
3.2.2. Liver tissue
To yield intact RNA, the liver tissue must be harvested as quickly as possible. Tissue may be instantly flash-frozen in liquid nitrogen and stored at -80°C for several months prior to RNA extraction or directly harvested after collection. Alternatively, the tissue may be kept in RNAlater® stabilization solution and stored for 1 day at 37°C, 1 week at 25°C, 1 month at 4°C or at -20°C or colder temperatures for longer-term storage.
Quickly slice and weigh of a piece of fresh, frozen or RNAlater® stabilized liver tissue for up to 40 mg per preparation. Do not allow frozen tissue to thaw before disruption.
Add 500 µL of lysis solution/2-mercaptoethanol mixture (see Note 6).
Homogenize immediately until no visible pieces remain using a rotor-stator homogenizer or a disposable RNase-free pellet mixer.
Pipette the homogenized tissue into a GenElute™ filtration column, a blue insert with a 2.0 mL receiving tube.
Continue procedure from step 5 in section 3.2.1.
3.2. RNA-DNA quantification and purity control
Quantification of RNA is recommended, as the same amounts of RNA should be used when comparing different samples. Several quantification methods are routinely applied, including spectrophotometric assays (Nanodrop®, Thermo Scientific, USA), capillary gel electrophoresis (QIAxcel®, Qiagen, USA), microfluidic analysis (Experion™, Bio-Rad, USA) or fluorescent dye detection systems (RiboGreen®, Applied Biosystems, USA). In this protocol, a spectrophotometric assay is used.
Open the sampling arm of the NanoDrop® ND-1000 spectrophotometer and pipette 2 µL of purified RNA or blank control, elution solution, on the lower pedestal (see Note 7).
Close the sampling arm and measure the absorbance at 260 nm (A260) and 280 nm (A280) (see Note 8).
Optionally, the integrity of the RNA can be checked by gel electrophoresis and detection of ribosomal RNA (rRNA), which makes up > 80% of total RNA in mammalian cells. The intact total RNA run on a denaturing gel will have sharp 28S rRNA and 18S rRNA bands. rRNA forms 2 sharp bands in the gel. The 28S rRNA band should be approximately twice as intense as the 18S rRNA band. Alternatively, 2100 Bioanalyser (Agilent Technologies, USA) is an easy-to-use device to assess RNA integrity of 12 samples for 1 biochip. Its algorithm gives a RNA Integrity Number (RIN), which scales from 0 to 10, where 10 is the highest quality. The RIN must be between 8 and 10 for further RT-qPCR analysis.
3.3. cDNA synthesis
It is recommend that the reverse transcription step be carried out in duplicate or triplicate.
Thaw the 5x iScript™ reaction mix and the iScript™ reverse transcriptase at room temperature.
Prepare the complete reaction mix in a total volume of 40 µL (Table 2).
Incubate complete reaction mix (Table 3) in the iCycler iQ™.
Table 2.
| Reagent | Volume per reaction (µL) |
|---|---|
| 5x iScript™ reaction mix | 8 |
| iScript™ reverse transcriptase | 2 |
| Nuclease-free water | 30 - x |
| RNA template (200 fg to 2 µg total RNA) | x |
Table 3.
| Time (min) | Temperature (°C) |
|---|---|
| 5 | 25 |
| 30 | 42 |
| 5 | 85 |
| Hold (optional) | 4 |
3.4. cDNA purification
Although purification of cDNA is optional, it is highly recommended to include this step. It is designed for rapid purification of single-stranded or double-stranded PCR amplification products (i.e. 100 base pairs to 10 kilobase) from the other components in the reactions, such as excess primers, nucleotides, DNA polymerase, oil and salts. Different cDNA purification kits are commercially available (e.g. QIAquick PCR Purification Kit® from Qiagen, DNAclear™ kit from Applied Biosystems, Jetquick PCR Purification Kit from Genomed). The protocol here below is outlined using the GenElute™ PCR Clean-Up Kit (Sigma, USA).
Insert a GenElute™ plasmid mini spin column, with a blue o-ring, into a provided collection tube if not already assembled.
Add 0.5 mL of column preparation solution to each GenElute™ plasmid mini spin column.
Centrifuge at 12000xg for 30 s to 1 min.
Discard the eluate.
Add 5 volumes of binding solution to 1 volume of the PCR reaction and mix (i.e. add 500 µL of binding solution to 100 µL of the PCR reaction).
Transfer the solution into the binding column.
Centrifuge the column at 12000xg to 16000xg for 1 min.
Retain the collection tube and discard the eluate.
Replace the binding column into the collection tube.
Apply 0.5 mL of diluted ethanol-containing wash solution to the column.
Centrifuge at 12000xg to 16000xg for 1 min.
Retain the collection tube and discard the eluate.
Replace the binding column into the collection tube.
Centrifuge at 12000xg to 16000xg for 2 min without any additional wash solution.
Discard any residual eluate as well as the collection tube.
Transfer the binding column to a fresh 2.0 mL collection tube.
Apply 50 µL of elution solution or water (see Note 9) to the center of each column.
Incubate at room temperature for 1 min.
Centrifuge the binding column at 12000xg to 16000xg for 1 min.
The PCR amplification product is now present in the eluate and is ready for immediate use or storage at -20°C.
3.5. Real-time PCR
The first step of the real-time qPCR process is to gather information about the DNA sequence of the target gene (i.e. Cx26, Cx32 and Cx43) to be used for primer design (see Note 10). The real-time qPCR procedure, described in this chapter, is optimized for primers targeting Cx26, Cx32 and Cx43 (Table 1). Next to the target genes, appropriate reference genes should be selected as internal controls for normalization purposes. Reference gene mRNAs should be stably expressed in all samples of the study and their abundances should show strong correlation with the total amounts of mRNA present in the samples (19). Normalization against a single reference gene is not acceptable unless clear evidence is presented confirming its invariant expression under the experimental conditions described. Ideally, a pool of candidate reference genes (Table 1) should be used and the optimal number and choice of the reference genes must be experimentally determined during data analysis (see section 3.6.).
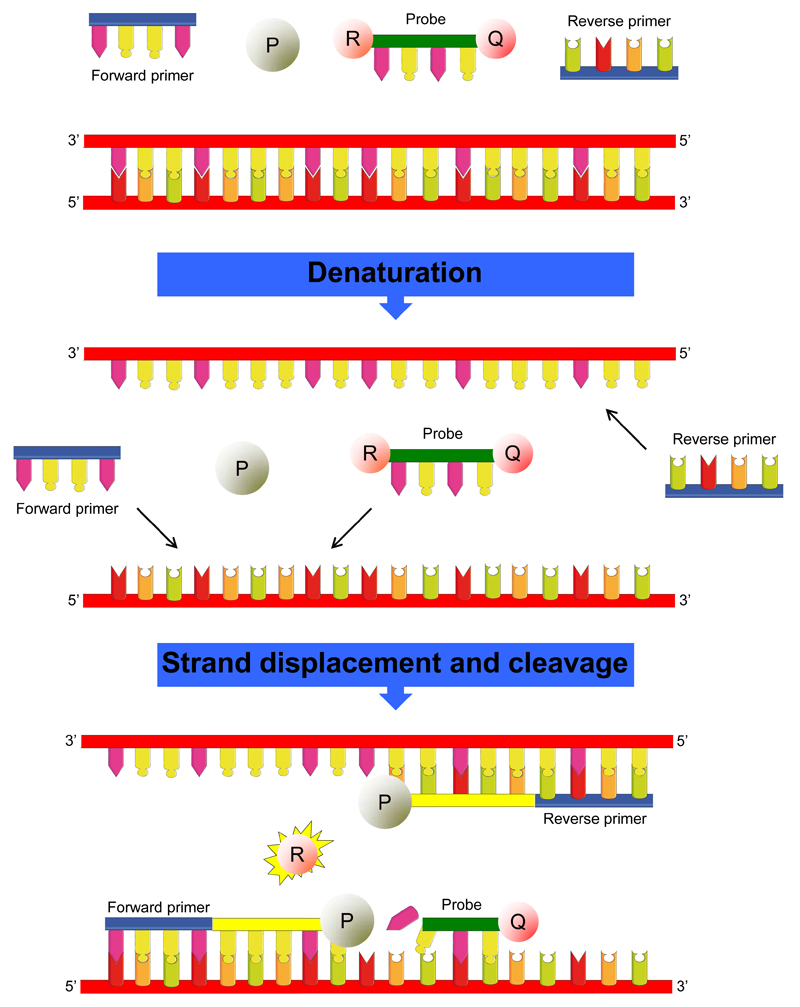
It is also strongly advised to use non-template controls (NTC), thus reaction mixes containing no DNA template as negative control (see Note 11), on each plate or batch of samples. The RT-qPCR system applied in this protocol is the StepOnePlus™ real-time PCR system (Applied Biosystems, USA) combined with the TaqMan® reagents (Applied Biosystems, USA). The TaqMan® reagents consist of 2 primers and a hydrolysis probe. The primers are designed to amplify the target, while the hydrolysis probe is designed to hybridize to the target and generate fluorescence when the target is amplified (Fig. 1). Alternatively, SYBR® Green reagents (Life Technologies, USA) can be used (see Note 12).
Figure 1.
Prepare the DNA standard 1 by pooling cDNA from the samples. A minimum of 7.5 µL cDNA should be pooled per target and reference gene if the standards will be analyzed in triplicate (i.e. prepare 67.5 µL when applying 3 target genes and 6 reference genes). Every sample should provide an equal volume for the preparation of DNA standard 1 (i.e. if 10 samples should be analyzed, 6.75 µL cDNA of every sample should be pooled to obtain standard 1).
Vortex thoroughly.
Prepare standard 2 by diluting standard 1 with a dilution factor 5 with nuclease-free water.
Repeat steps 2 and 3 to obtain a serial dilution of minimum 5 standards.
Prepare 20 µL reaction volume for each standard, sample and NTC (Table 4).
Each standard, NTC and sample is run in triplicate (i.e. 3 wells per sample). Therefore, the sample reaction mix volumes should be tripled.
Seal the 96-well plate and centrifuge at 1600xg for 1 min at 4°C (see Note 13).
Samples are placed in a MicroAmp® optical 96-well reaction plate and assay run in the StepOnePlus™ machine as instructed in the manufacturer’s guidelines (see Note 14).
Select the appropriate ramp speed for the instrument run (Table 5).
The associated StepOnePlus™ software will automatically calculate the quantification cycle (Cq) (see Note 15). Cq values exceeding 40 are questionable because they imply low efficiency (see Note 16) and generally should not be reported. However, the use of such arbitrary Cq cut-offs is not ideal, because they may be either too low, eliminating valid results, or too high, increasing false positive results (19).
Table 4.
| Reagent | Volume (µL) |
|---|---|
| Taqman® Universal PCR Master Mix (2x) | 10 |
| Assay-on-Demand™ Gene Expression Assay Mix (20x) | 1 |
| cDNA/Standards | 2 |
| Nuclease-free water | 7 |
Table 5.
| Function | Time | Temperature |
|---|---|---|
| Ramp time | 2 min | 50°C |
| Hold | 10 min | 95°C |
| Melt | 15 s | 95°C |
| Anneal/extension | 1 min | 60°C |
3.6. Data processing
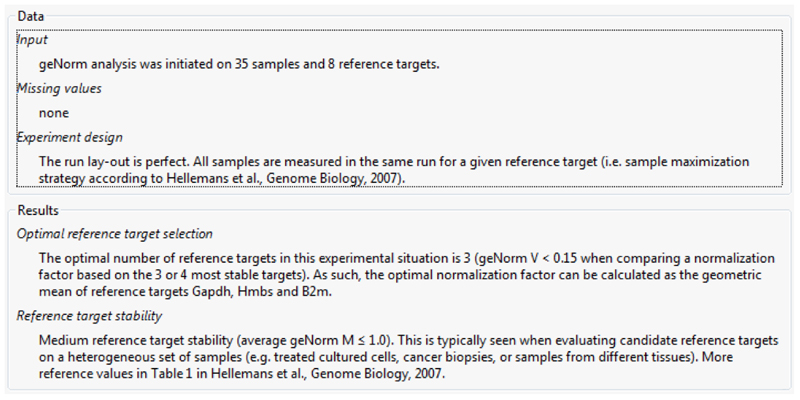
Many different methods have been proposed to normalize qPCR data (22). The use of reference genes is undoubtedly the most popular and adequate approach. However, accurate normalization can only be performed after validation of the candidate reference genes. Normalization against adequate reference genes correct for variable sample mass, nucleic acid extraction efficiency, reverse transcription efficiency and pipette calibration errors. Different software systems can be used for this purpose, including geNorm (23), NormFinder (24) and BestKeeper (25). These algorithms determine the most stable reference genes from a set of tested genes in a given sample panel. From this, a gene expression normalization factor can be calculated for each tissue sample based on the geometric mean of a user-defined number of reference genes. GeNorm calculates the gene expression stability measure M for a reference gene as the average pairwise variation V for that gene with all other tested candidate reference genes. Stepwise exclusion of the gene with the highest M value allows ranking of the tested genes according to their expression stability (23, 26). The geNorm is currently integrated in the qbase+ software, which additionally to the first version gives a fully automatic and expert result report, handles missing data and identifies single best reference genes (Fig. 2). Besides geNorm, the qbase+ software contains different normalization methods to accommodate a wide range of experiments, post-PCR quality control, interrun calibration and statistical analysis wizard. The qbase+ helps applying MIQE compliant procedures and guides the experimenter to highest quality results. Unlike geNorm, NormFinder adopts a model-based approach to give a score to the 2 most stable reference genes with the least intragroup and intergroup variation. Stability is expressed as a value in arbitrary units. Furthermore, NormFinder possesses the ability to discriminate between sample variability and bias among several groups (24). BestKeeper determines the variability in expression of a set of reference genes by analyzing Cq values and classifying variability by the coefficient of variance and the standard deviation. To define the most stable reference gene, the software generates an index, which finally is compared to each candidate reference gene. This comparison results in a value for the Pearson correlation coefficient and probability, which are then allocated to each candidate reference gene (25). Using this software systems, relative alterations (i.e. fold change) in mRNA levels can be calculated according to the Livak 2-ΔΔCq formula (27). Using the latter, data are presented as the fold change in gene expression normalized to the selected reference genes and relative to the untreated control.
Figure 2.
4. Notes
Reagents from the GenElute™ Mammalian Total RNA Miniprep Kit and the 5x iScript™ reaction mix may contain some precipitation upon thawing. It should be mixed thoroughly to resuspend the precipitation.
Lysis solution and wash solution 1 of the GenElute™ Mammalian Total RNA Miniprep Kit contain guanidine thiocyanate, which is a potent chaotripic agent and irritant. Wear gloves, safety glasses and suitable protective clothing when handling these solutions or any reagent provided with the kit.
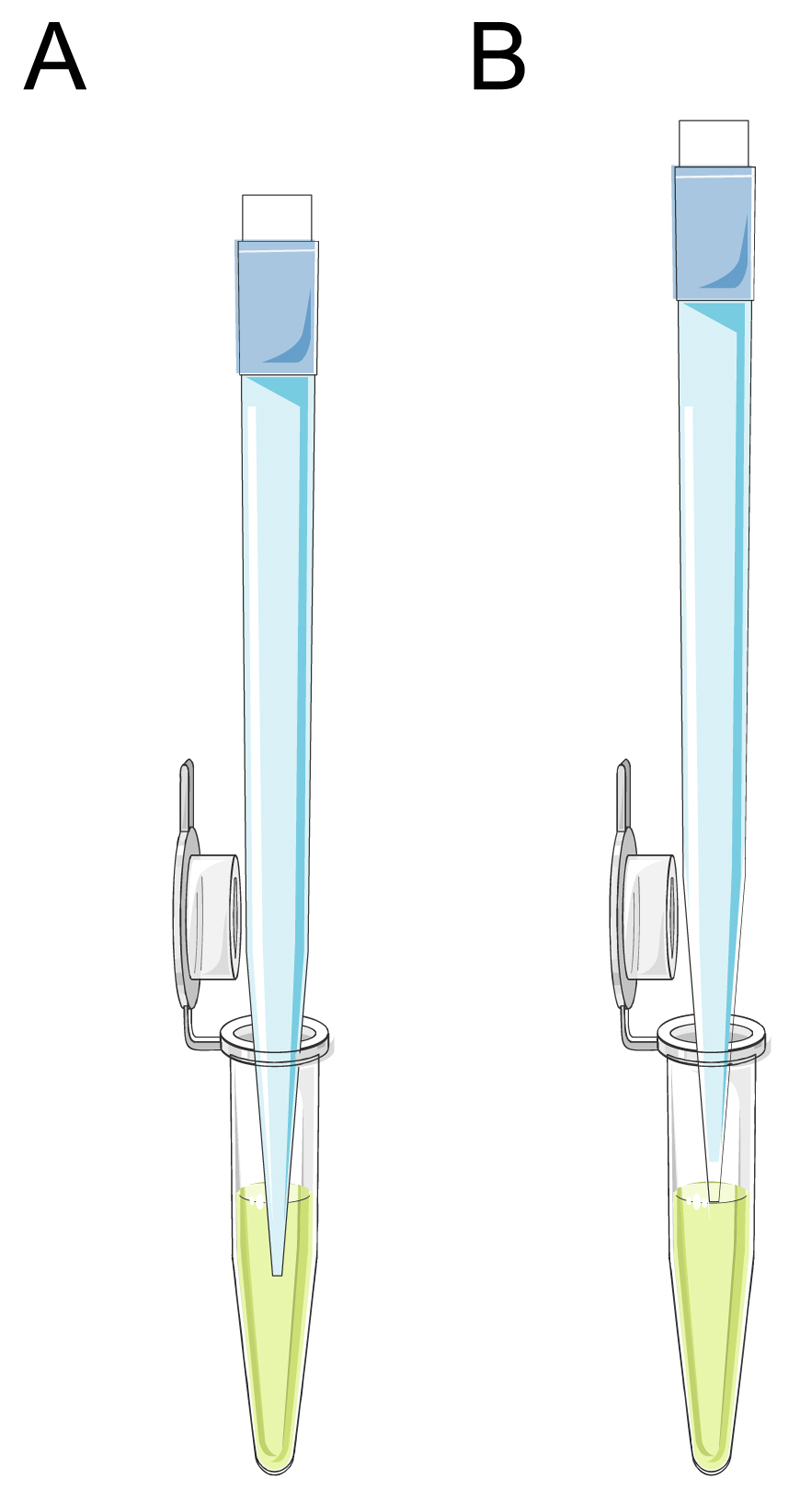
In general, RT-qPCR analysis requires working with small volumes and tubes, including handling volumes of less than1 µL. This necessitates correct standard operation procedures for pipetting small volumes. If the pipette tip is submerged into the sample or reagent, the delivered volume to the reaction will be larger than intended. Instead, the pipette tip should be touched to the surface of the sample or reagent (Fig. 3). To ensure reproducibility, pipettors should be calibrated on a regular basis (i.e. at least every 6 months). Alternatively, automated liquid handling systems (e.g. epMotion® 5070, Eppendorf, Germany) can be used for pipetting tasks in the RT-qPCR procedure. This automated systems help to eliminate manual pipetting errors and maximize the reproducibility.
The filtration step removes cellular debris and shears DNA. This step may be omitted with fewer than 1 x 106 cultured cells.
Centrifugation speeds are indicated in units of g. Convert the relative centrifugal force (RCF) to rotation per min (rpm) according to the following formula: RCF = 1.118 x 10-5 x radius (in cm) x rpm2.
For larger amounts, scale up the volume of the lysis solution/2-mercaptoethanol mixture proportionally. Divide the lysate into 500 to 700 µL aliquots and process through separate filtration and binding columns.
After each sample measurement the liquid on the upper and lower pedestals should be removed with a soft laboratory wipe to prevent sample carryover in successive measurements. After measuring a large number of samples it is recommended to clean the pedestals thoroughly using 2 µL water aliquots.
An A260 reading of 1.0 is equivalent to about 40 µg/mL of RNA. The A260/A280 ratio provides an indication of the RNA purity, in particular the presence of DNA or residual phenol. Pure RNA has an A260/A280 ratio between 1.8 and 2.1.
When eluting with water, make sure that the pH of the water is between 5.5 and 8.5. Elution may also be performed using the elution solution diluted 10-fold with water.
The most convenient way to gather information about the DNA sequence of the target gene is by consulting the GenBank™ database maintained at the National Center for Biotechnology Information (NCBI). Importantly, as one searches for DNA sequences for connexin genes, one should consider the correct designation. Indeed, 2 nomenclature systems are currently used to name the different connexin species (28). A first system is based on the predicted molecular weight in kilodaltons (i.e. Cx26 refers to a connexin with a molecular weight predicted by cDNA sequencing of 26 kilodaltons). A second system divides the connexin family into 4 subclasses according to genetic origin, overall sequence similarity and length of cytoplasmic domain. This classification system forms the basis for the current official nomenclature of connexin genes. Hence, the corresponding gene names for Cx26, Cx32 and Cx43 are GJB2, GJB1 and GJA1, respectively, for human, and Gjb2, Gjb1 and Gja1 for other species. The information about the DNA sequence can be used to carefully choose the oligonucleotide primers, which can be performed using software, such as Oligo® (29), Primer3 (30) and PRIMO (31).
Amplification in the NTC may indicate genomic contamination of samples or amplicon cross-contamination. RNA samples that contain DNA should be treated with DNase. When possible, probes should be designed spanning an exon-exon junction to avoid amplification of genomic DNA. It is essential to take precautions in order to clear samples from RNases and DNases. In case of contamination, all reagents (e.g. master mix) and stock buffers should be replaced, and all PCR areas should be cleaned meticulously.
Hydrolysis Taqman® reagents have the advantage to contain an increased specificity compared to traditional primer systems and to provide capability for use in multiplex systems. A limitation is that the system requires synthesis of a unique fluorogenic probe. The SYBR® Green reagents use a double-stranded DNA binding dye to detect PCR products, as they accumulate during PCR cycles. This system is financially more favorable and allows melt curve analysis. However, the dye binds non-specifically to all double-stranded DNA sequences. This could yield false positive results. To avoid false positive signals, formation of non-specific products should be identified using melt curve or gel analysis.
The reaction mix should be at the bottom of each well of the reaction plate. If not, centrifuge the reaction plate again for a longer period of time at a higher centrifugation speed. It is important not to allow the bottom of the reaction plate to become smudged. Fluids and other impurities that adhere to the bottom of the reaction plate can cause a contamination and create an abnormally high background signal.
The instrument needs monthly maintenance, including calibration. In addition, make sure that the instrument is set on the correct dye and filter settings.
The nomenclature describing the fractional PCR cycle used for quantification is inconsistent, with threshold cycle (Ct), crossing point (Cp) and take-off point (TPF) currently used in the literature. The MIQE guidelines therefore propose to use the term quantification cycle (Cq) (19).
The efficiency of each PCR assay needs to be taken into account for quality control purposes. If the efficiency is not between 90% and 110%, the PCR assay requires further optimization before results can be considered valuable and suitable for further purposes. The efficiencies for the target and reference genes should be approximately equal.
Figure 3.
Acknowledgements
This work was financially supported by the grants of Agency for Innovation by Science and Technology in Flanders (IWT), the University Hospital of the Vrije Universiteit Brussel-Belgium (Willy Gepts Fonds UZ-VUB), the Fund for Scientific Research-Flanders (FWO grants G009514N and G010214N), the European Research Council (ERC Starting Grant 335476), the University of São Paulo-Brazil and the Foundation for Research Support of the State of São Paulo (FAPESP SPEC grant 2013/50420-6).
References
- 1.Oyamada M, Oyamada Y, Takamatsu T. Regulation of connexin expression. Biochim Biophys Acta. 2005;1719:6–23. doi: 10.1016/j.bbamem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 2.D'hondt C, Iyyathurai J, Vinken M, et al. Regulation of connexin- and pannexin-based channels by post-translational modifications. Biol Cell. 2013;105:373–398. doi: 10.1111/boc.201200096. [DOI] [PubMed] [Google Scholar]
- 3.Piechocki MP, Toti RM, Fernstrom MJ, et al. Liver cell-specific transcriptional regulation of connexin32. Biochim Biophys Acta. 2000;1491:107–122. doi: 10.1016/s0167-4781(00)00036-1. [DOI] [PubMed] [Google Scholar]
- 4.Koffler LD, Fernstrom MJ, Akiyama TE, et al. Positive regulation of connexin32 transcription by hepatocyte nuclear factor-1alpha. Arch Biochem Biophys. 2002;407:160–167. doi: 10.1016/s0003-9861(02)00488-5. [DOI] [PubMed] [Google Scholar]
- 5.Vinken M, De Rop E, Decrock E, et al. Epigenetic regulation of gap junctional intercellular communication: more than a way to keep cells quiet? Biochim Biophys Acta. 2009;1795:53–61. doi: 10.1016/j.bbcan.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Oyamada M, Takebe K, Oyamada Y. Regulation of connexin expression by transcription factors and epigenetic mechanisms. Biochim Biophys Acta. 2013;1828:118–131. doi: 10.1016/j.bbamem.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 7.Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker RM, Barnes NM. mRNA: detection by in Situ and northern hybridization. Methods Mol Biol. 1999;106:247–283. doi: 10.1385/0-89603-530-1:247. [DOI] [PubMed] [Google Scholar]
- 9.Hod Y. A simplified ribonuclease protection assay. Biotechniques. 1992;13:852–854. [PubMed] [Google Scholar]
- 10.Saccomanno CF, Bordonaro M, Chen JS, et al. A faster ribonuclease protection assay. Biotechniques. 1992;13:846–850. [PubMed] [Google Scholar]
- 11.Weis JH, Tan SS, Martin BK, et al. Detection of rare mRNAs via quantitative RT-PCR. Trends Genet. 1992;8:263–264. doi: 10.1016/0168-9525(92)90242-v. [DOI] [PubMed] [Google Scholar]
- 12.Wang T, Brown MJ. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal Biochem. 1999;269:198–201. doi: 10.1006/abio.1999.4022. [DOI] [PubMed] [Google Scholar]
- 13.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Loewenstein WR, Kanno Y. Intercellular communication and tissue growth. I. cancerous growth. J Cell Biol. 1967;33:225–234. doi: 10.1083/jcb.33.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Revel JP, Karnovsky MJ. Hexagonal array of subunits in intercellular junctions of the mouse heart and liver. J Cell Biol. 1967;33:C7–C12. doi: 10.1083/jcb.33.3.c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maes M, Cogliati B, Crespo Yanguas S, et al. Roles of connexins and pannexins in digestive homeostasis. Cell Mol Life Sci. 2015;72:2809–2821. doi: 10.1007/s00018-015-1961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinken M, Henkens T, De Rop E, et al. Biology and pathobiology of gap junctional channels in hepatocytes. Hepatology. 2008;47:1077–1088. doi: 10.1002/hep.22049. [DOI] [PubMed] [Google Scholar]
- 18.Maes M, Crespo Yanguas S, Willebrords J, et al. Connexin and pannexin signaling in gastrointestinal and liver disease. Transl Res. 2015;166:332–343. doi: 10.1016/j.trsl.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 20.Bustin SA, Beaulieu JF, Huggett J, et al. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micke P, Ohshima M, Tahmasebpoor S, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86:202–211. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 22.Huggett J, Dheda K, Bustin S, et al. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 26.Hellemans J, Vandesompele J. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol. 2014;1160:19–26. doi: 10.1007/978-1-4939-0733-5_3. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Rychlik W, Rhoads RE. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 31.Li P, Kupfer KC, Davies CJ, et al. PRIMO: A primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics. 1997;40:476–485. doi: 10.1006/geno.1996.4560. [DOI] [PubMed] [Google Scholar]