Abstract
Macrophages tightly scale their core metabolism upon activation, but the precise regulation of the mitochondrial electron transport chain (ETC) and its functional implications are currently unknown. Here we show that recognition of live bacteria by macrophages transiently decreased the assembly of ETC complex I (CI) and CI-containing supercomplexes and switched the relative contribution of CI and CII to mitochondrial respiration. This was mediated by the phagosomal NADPH-oxidase and the reactive oxygen species (ROS)-dependent tyrosine-kinase Fgr. It required Toll-like receptor signalling and the NLRP3 inflammasome, which were both connected to bacterial viability-specific immune responses. Inhibition of CII during E. coli infection normalized serum levels of interleukin 1β (IL-1β) and IL-10 to levels found in mice treated with dead bacteria, and impaired control of bacteria. We thus identified ETC adaptations as an early immune-metabolic checkpoint that adjusts innate immune responses to bacterial infection.
Macrophages are phagocytic immune cells that reside in most tissues thereby constituting the first line of host-defence to invading microorganisms1. They express a wide variety of innate immune receptors allowing them to precisely determine microbial features such as pathogenicity, invasiveness or viability in order to fine-tune their differentiation program2,3. To meet their immunological functions, macrophages metabolize a variety of carbon substrates including glucose, fatty acids, ketone bodies and amino acids. Adaptations of cellular metabolism thus appeared to be critically associated with macrophage activation4,5,6,7 and may contribute to macrophage functions requirements8,9,10. Activation of macrophages by Gram-negative bacterial cell wall component lipolysaccharide (LPS) through the Toll-like receptor 4 (TLR4) induces profound metabolic reprogramming that culminates in enhanced glycolysis, while glutamine replenishes the tricarboxylic acid (TCA) cycle through glutaminolysis5,7. In turn, both the accumulation of the TCA intermediate succinate and the induction of the glycolytic enzyme hexokinase-1 contribute to the production of the pro-inflammatory cytokine interleukin-1β (IL-1β) by regulating the transcription of pro-IL-1β7 and activating the NLRP3 inflammasome11, respectively. Accumulation of TCA cycle intermediates may also directly contribute to macrophage antimicrobial functions. For example, LPS-activated macrophages accumulate citrate5 that can be metabolized to itaconic acid, which exerts direct antibacterial properties on various pathogens including Salmonella enterica Typhimurium, Mycobacterium tuberculosis or Legionella pneumophila 12,13.
At the core of the metabolic pathways is the mitochondrion, a bioenergetic organelle that not only contributes to energy supply, biosynthesis or cellular redox maintenance, but also serves as a signalling platform for various innate immune signalling pathways9,10,14. Mitochondria balance their contribution to anabolism and catabolism in response to fuel and oxygen availability as well as extracellular signals including danger signals and cytokines8,9. All catabolic processes converge on the mitochondrial electron transport chain (ETC) by supplying electrons in the form of the reductive equivalents NADH and FADH2. The ETC comprises two electron carriers (coenzyme Q [CoQ] and cytochrome c) and four respiratory complexes (complex I to IV [CI to CIV]), which, except for CII, can dynamically assemble as larger molecular supercomplexes (SCs) in the mitochondrial inner membrane15,16. The dynamic assembly of respiratory complexes into SCs has been proposed to confer functional advantages to the cells by potentiating electron flux within the ETC, preventing the generation of ROS by sequestering reactive intermediates or stabilizing individual respiratory complexes15. Whether super-assembly of ETC respiratory complexes can contribute to immune function remains to be determined. Nevertheless, several recent studies highlighted the potential importance for ETC respiratory complexes in macrophage activation. Chemical inhibition of CI impairs the production of the pro-inflammatory cytokine IL-1β while inducing secretion of the anti-inflammatory cytokine IL-10 in activated macrophage17. In contrast, the genetic ablation of the CI subunit NDUFS4 promotes an inflammatory macrophage phenotype18, suggesting that CI activity rather dampens macrophage activation.
Here, we investigated if ETC adaptations contribute to the metabolic switch of myeloid cells upon activation via innate immune receptors5,7, and would therefore impact antimicrobial responses. We report that recognition of viable Gram-negative bacteria through TLR- and NLRP3-dependent pathways transiently decreased abundance of SCs within the macrophage mitochondria due to destabilization of CI. The resulting dampening in CI activity was accompanied by an increase in the activity of CII. The induction of CII activity was not observed upon phagocytosis of dead bacteria, but could be restored with bacterial RNA, a viability-associated pathogen-associated molecular pattern (vita-PAMP) that signals microbial life to the immune system19. Conversely, inhibition of CII during viable bacteria challenge normalized the amount of IL-1β and IL-10 to levels found upon dead bacteria encounter. These findings uncover a critical role for mitochondrial ETC in innate immune responses to bacterial infections, and highlight the potential therapeutic interest of manipulating the ETC.
Results
Sensing of bacteria affects mitochondrial ETC architecture
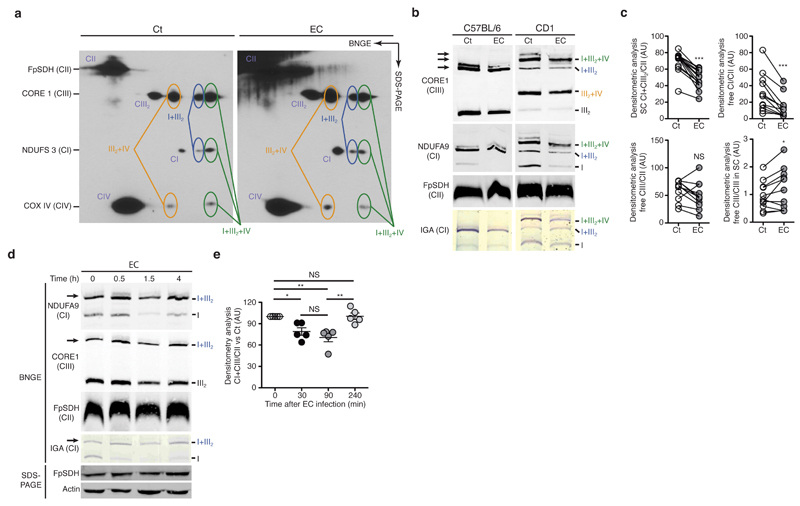
To determine whether innate immune cell activation impacts the mitochondria respiratory chain, we first analysed the ETC organization of mouse CD1 bone-marrow-derived macrophages (BMDMs) challenged or not with viable Escherichia coli K12, strain DH5α (hereafter defined as E. coli) for 1.5h. Unlike C57BL/6 mice, CD1 mice express the long isoform of supercomplex assembly factor I (SCAFI, Cox7a2l) and consequently assemble CIV-containing SC reflecting the situation found in human cells20. Two-dimensional gel analysis of mitochondria isolated from CD1 BMDMs revealed several respiratory SCs including SC I+III2+IV, SC I+III2 and SC III2+IV20,21 in both resting and stimulated macrophages (Fig. 1a and Supplementary Fig. 1a). We then performed blue-native (BN)-PAGE analysis of whole BMDMs cell lysates. We observed a decrease in the abundance of CI and CI-containing SC (SC I+III2+IV and SC I+III2) in CD1 BMDMs that have phagocytosed live E. coli compared with untreated BMDMs (Fig. 1b). Protein amount of various ETC subunits was unaffected by E. coli challenge, despite mild transcriptional variations of some ETC-subunit-encoding nuclear genes (Supplementary Fig. 1b-f), suggesting the changes in SC abundance were due to alteration of SC assembly, rather than changes in protein expression. BMDMs from C57BL/6 mice (here called wild-type, WT) also exhibited a transient reduction in the amounts of CI- and CIII-containing SCs when incubated with E. coli (Fig. 1b-e and Supplementary Fig. 2a,b). Therefore, the disassembly of SCs upon detection of viable bacteria affects all CI containing SCs. ETC alteration was also observed in mouse peritoneal macrophages incubated with E. coli, (Supplementary Fig. 2c), and was also evident upon recognition of Salmonella enterica serovar Typhimurium SL1344 (S. enterica Typhimurium) (Supplementary Fig. 2d-f) by BMDMs. Thus, our data indicated that the recognition of Gram-negative bacteria by macrophages induced modifications of the ETC architecture that were characterized by a decrease abundance of CI-associated structures.
Figure 1.
Detection of bacteria decreases the abundance of mitochondrial electron transport chain complex I (CI) and CI-containing supercomplexes. (a) Two-dimensional gel immunoblot analysis of mitochondria isolated from CD1 BMDMs stimulated with medium (Ct) or viable E. coli (EC) for 1.5h. Circles indicate localization of the different complexes. (b, d) Blue-native gel electrophoresis (BN-PAGE) immunoblot and in-gel activity (IGA) assay of CI in mitochondria isolated from CD1 BMDMs (b) and C57BL/6J BMDMs (b, d) stimulated with EC for the indicated time. (c) Densitometric analysis of ‘CI+CIII2/CII’, ‘free CI/CII’, ‘free CIII/CII’ and ‘free CIII/CIII in SC’ signal ratio as observed by BNGE immunoblot of C57BL/6J BMDMs treated as in (b). (e) Densitometric analysis of BN-PAGE showing CI+CIII2 SC proportion vs. CII as observed by BNGE immunoblot of C57BL/6J BMDMs treated as in (d). NS, not significant (P > 0.05); *P < 0.05; ***P < 0.001 (paired t-test analysis (c) or one-way ANOVA followed by Tukey post-test analysis (e)). Data are from three (e, mean and s.e.m.) or ten (c) independent experiments, one representative (a, b, d) of three (a, d) or ten (b) experiments with similar results. Immunoblot membranes were probed with the indicated mETC-subunit-specific antibodies (a, b, d). Arrows indicate the main SCs affected (b, d).
Detection of bacteria modulates ETC complex activities
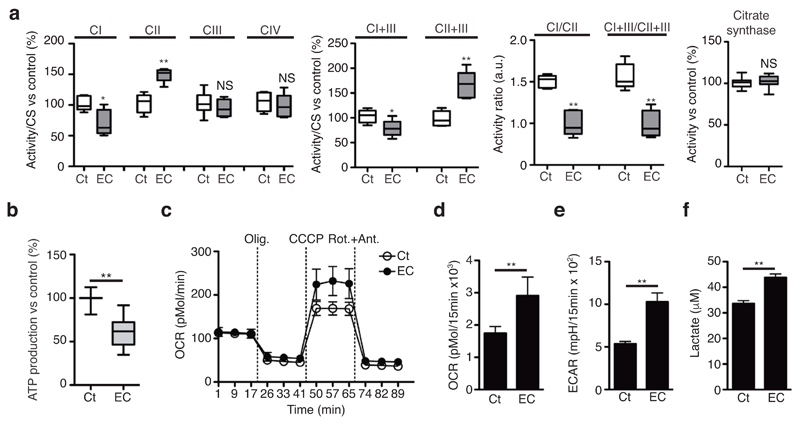
Macrophages activated through innate immune receptors use glutamine rather than glucose or fatty acid as the carbon source for oxidative phosphorylation4,5,6,7. This adaptation results in a shift in the proportion of NADH-FADH2 electrons feeding the ETC22, which requires the modulation of relative SC proportions20,23. Stimulation of BMDMs with E. coli or S. enterica Typhimurium transiently impaired the in-gel NADH dehydrogenase activity of CI within SC I+III2+IV and SC I+III2 (Fig. 1b,d and Supplementary Fig. 2f) indicating a decreased CI abundance within SCs. Quantitative spectrophotometric assessment of CI and CI+CIII activities in mitochondria isolated from BMDMs also indicated a decrease in CI-mediated respiration in response to E. coli compared with resting condition (Fig. 2a). However, we found that CII and CII+CIII activities were increased and total CIII and CIV activities remained unaltered in this context (Fig. 2a). To determine whether the decreased CI activity translated into a drop in energy production, we measured mitochondrial-ATP production rate in permeabilized BMDMs in the presence of glutamate plus malate, which generate intramitochondrial NADH that feeds electrons to CI20. CI-dependent ATP production was diminished in BMDMs incubated with E. coli in contrast to resting cells (Fig. 2b). Thus, macrophage responses to bacteria are characterized by reduced CI assembly into SCs and decreased overall CI activity, consistent with an anti-inflammatory role of CI in macrophages18. In line with the effect of LPS on macrophages4,5,7, E. coli-stimulated but not resting BMDMs markedly lowered their O2 consumption on glucose substrate, while exerting a high glycolytic activity that increased the extracellular acidification rate (ECAR) and lactate production at 18h after infection (Supplementary Fig. 3a-c). However, we found that the maximal respiration rate (MRR) was increased in E. coli-treated BMDMs at 2h post-challenge compared untreated cells (Fig. 2c and Supplementary Fig. 3d). This created a situation in which spare respiratory capacity (SRC) (Fig. 2d), ECAR (Fig. 2e) and lactate production (Fig. 2f) were increased simultaneously in BMDMs upon E. coli recognition. Taken together, these data indicated that E. coli detection by macrophage decreased ETC CI activity thereby modulating mitochondrial respiratory parameters.
Figure 2.
Detection of bacteria induces changes in mitochondrial ETC complex activities and influences mitochondrial respiration and glycolysis. (a) Spectrophotometric activities of the indicated mitochondrial respiratory complexes, normalized to citrate synthase (CS) activity in mitochondria isolated from BMDMs treated or not with E. coli for 1.5h. Specific activities (in IU/mg protein) corresponding to 100% activity are 0.105±0.013, 0.040±0.008, 0.055±0.01, 0.1±0.03, 0.23±0.01, 0.02±0.003, 0.3±0.01 for CI, CII, CIII, CIV, CI+III, CII+III and CS, respectively. (b) Effect of EC-stimulation on WT BMDMs glutamate+malate-driven ATP synthesis. 100% activity corresponds to a rate of 153±24.9 nmol ATP/min/mg protein. (c-f) Oxygen consumption rate (OCR) upon sequential treatment of oligomycin (olig.), CCCP, and rotenone+antimycin (Rot.+Ant.) (c) spare respiratory capacity (SRC) (d), basal extracellular acidification rate (ECAR) (e) and extracellular lactate concentration (f) in WT BMDMs treated or not with EC for 2h. NS, not significant; *P < 0.01; **P < 0.001 (two-tailed unpaired Student’s t-test). Data (mean and s.e.m. (a, b, d-f) and mean and s.d. (c)) are from three (a, d-f) or eight (b) independent experiments, or one representative of three independent experiments with similar results (c).
Recognition of bacteria stimulates mitochondrial CII
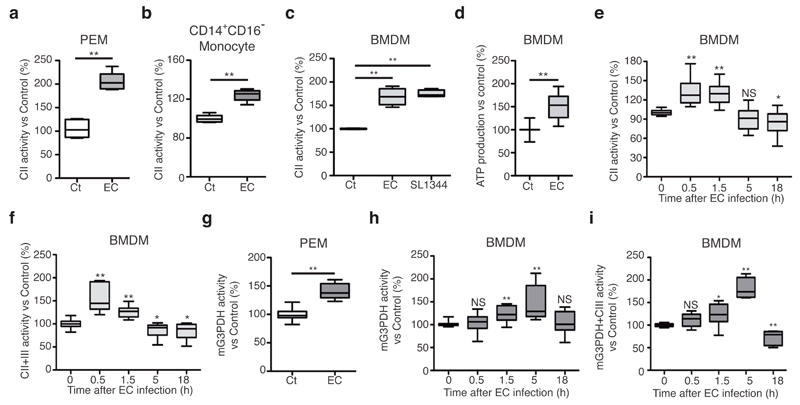
The concomitant reduction in CI+CIII activity and the increased MRR of E. coli-stimulated macrophages on glucose substrate appeared contradictory and suggested that an alternative electron source would compensate the expected reduction in NADH (CI)-dependent electron flux. We tested whether bacterial infection-driven alterations of macrophage ETC and the resulting reduced CI activity would favour the use of electrons from FADH2-oxidizing enzymes, as shown previously15,20,24. We found a robust increase in CII enzymatic activity in peritoneal macrophages (Fig. 3a) or human CD14+CD16- primary monocytes (Fig. 3b) challenged with E. coli and BMDMs challenged with S. enterica Typhimurium (Fig. 3c) compared with untreated cells. In addition, BMDMs incubated with E. coli increased their CII-mediated ATP production rate compared to resting cells (Fig. 3d). The increased activities of CII (Fig. 3e) and CII+III (Fig. 3f) were transient. The use of glucose catabolism to increase the maximum respiration rate despite a decrease in CI function would necessitate the delivery of NADH electrons generated by glycolysis to the mitochondrial ETC without the utilization of CI. This can be achieved by shuttling cytoplasmic NADH electrons to CoQ by mitochondrial bound glycerol-3-phosphate dehydrogenase (mG3PDH)25. Similar to CII, mG3PDH (Fig. 3g,h) and mG3PDH+CIII (Fig. 3i) activities were increased in BMDMs challenged with E. coli compared with resting condition. Nevertheless, increased CII activity reached a maximum at 0.5h to 1.5h after E. coli challenge (Fig. 3e) whereas the induction of mG3PDH was maintained for longer (Fig. 3h). Taken together, these data indicate that E. coli recognition by macrophages induces a transient increase in the activity of mitochondrial FADH2-dependent enzymes that feeds electron to CIII.
Figure 3.
Sensing of bacteria transiently induces mitochondrial complex II and glycerol-3-phosphate dehydrogenase activities. (a, b, c, e, f) CII activity (a, b, c, e) and CII+CIII activity (f) in thioglycollate-elicited WT macrophages (a), CD14+CD16- human monocytes (b), or WT BMDMs (c, e, f) stimulated with EC or S. enterica Typhimurium (SL1344) (c) for 1.5h or for the indicated time. (d) Succinate (Succ)-driven ATP synthesis in WT BMDMs stimulated with EC for 1.5h. 100% activity corresponds to a rate of 70.4±18.1 nmol ATP/min/mg protein. (g-i) Mitochondrial glycerol-3-phosphate dehydrogenase (mG3PDH) (g, h) and G3PDH+CIII (i) in WT thioglycollate-elicited macrophages (g) or BMDMs (h, i) stimulated with EC for 1.5h or for the indicated time. NS, not significant; *P < 0.05, **P < 0.001 (two-tailed unpaired Student’s t-test). Data are means ± s.e.m. of three (a-c, h, i), four (f), six (e), or eight (d) independent experiments performed in three to five technical replicates, or one experiment with five mice per group (a) or three donors (b) and performed in two technical replicates.
Phagosomal NADPH oxidase triggers ETC adaptations
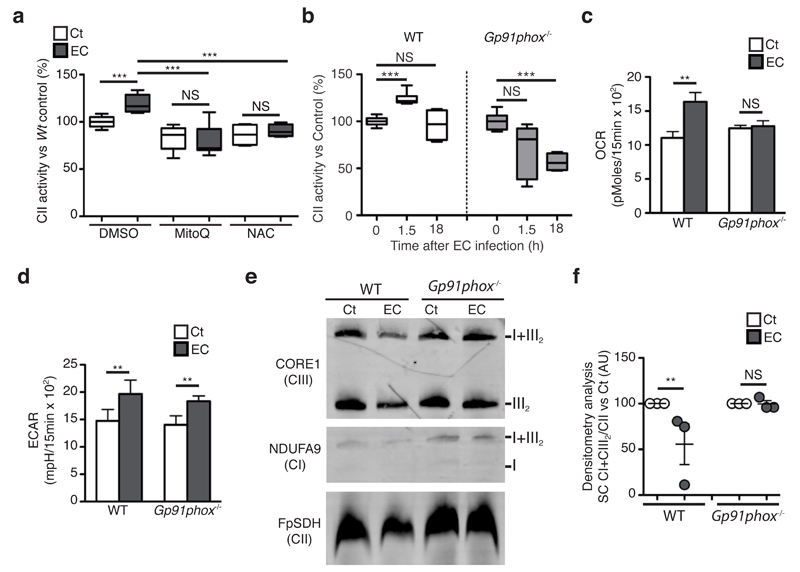
Mitochondrial ROS (mROS), which increases within a few hours upon E. coli sensing26, are inherent to ETC function, making them potential regulators of CII activity in E. coli-stimulated macrophages. We were unable to detect substantial mROS production enhancement in BMDMs at 1.5h after E. coli challenge compared with BMDMs left untreated or treated with the mROS-inducer rotenone (Supplementary Fig. 4a,b). However, we found that both the mROS-specific inhibitor mitoQ27 and the broad antioxidant N-acetyl cysteine (NAC) decreased CII enzymatic activity in untreated or E. coli-stimulated BMDMs (Fig. 4a), suggesting that mROS was required for proper CII functions in macrophages. We next investigated if an alternative source of ROS, namely phagosomal NADPH-oxidase28, was a regulator of CII activity. In BMDMs, a 15min challenge with E. coli strongly induced ROS production compared with untreated cells (Supplementary Fig. 4c). BMDMs deficient in the NADPH oxidase subunit gp91phox (Cybb-/-, called ‘Gp91phox-/-’ here) did not produce ROS in response to E. coli as opposed to WT cells BMDMs (Supplementary Fig. 4d-e), demonstrating that the NADPH oxidase mediates ROS production in this context. Contrary to WT cells, Gp91phox-/- BMDMs did not increase CII enzymatic activity in response to E. coli (Fig. 4b). Consequently, the E. coli-mediated increase in SRC observed in WT was absent in Gp91phox-/- BMDMs (Fig. 4c and Supplementary Fig. 4f,g) while ECAR induction was similar in WT and KO cells (Fig. 4d). The absence of gp91phox also prevented the decrease in abundance of SCs in mitochondria isolated from E. coli-stimulated BMDMs that was observed in WT cells (Fig. 4e,f). Taken together, these results indicate that phagosomal ROS is an early inducer of ETC adaptations during bacterial sensing in macrophages and cooperates with mROS to complete those adaptations.
Figure 4.
Induction of mitochondrial complex II activity and decrease in supercomplex abundance upon detection of E. coli relies on phagosomal reactive oxygen species (ROS). (a, b) CII activity in WT and Gp91phox-/- BMDMs stimulated with EC for 1.5h and treated with the ROS inhibitors N-acetylcysteine (NAC) or mitoQ (a). (c, d) spare respiratory capacity (SRC) (c) and extracellular acidification rate (ECAR) (d) in Gp91phox-/- and WT BMDMs stimulated with EC for 2h. (e) BNGE immunoblot of mitochondria isolated from Gp91phox-/- and WT BMDMs stimulated with EC for 2h. Representative of 3 independent experiments. (f) Densitometric analysis of BN-PAGE showing CI+CIII SC proportion vs. CII in 3 independent experiments. NS, not significant; *P < 0.01; **P < 0.001 (two-tailed unpaired Student’s t-test). Data (mean and s.e.m. (a-d, f)) are from three (b, c, d, f) or four (a) independent experiments performed in two (b, f) or five (c, d) technical replicates, one representative of three independent experiments with similar results (e).
Fgr is required for E. coli-mediated induction of CII activity
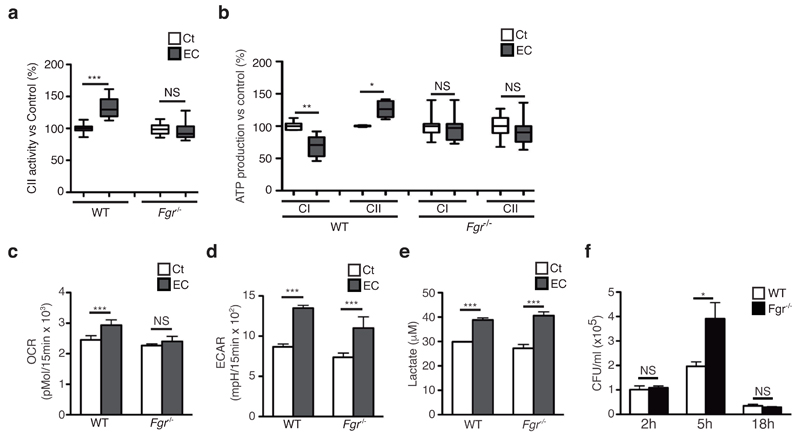
CII activity adapts to fuel use through H2O2-mediated activation via phosphorylation of the CII subunit SDHA by the Src-family tyrosine kinase Fgr29, the partner kinase of the phosphatase PTPMT130. Fgr deficiency prevented the decrease in abundance of SC induced by E. coli challenge in permeabilized BMDMs (Supplementary Fig. 5a-e) or mitochondria isolated from BMDMs (Supplementary Fig. 5f-i) when compared with WT situation. BMDMs from Fgr-/- mice did not show the decrease in CI activity and the increase in CII activity observed in WT cells stimulated by E. coli (Fig 5a and Supplementary Fig. 5j). Similarly, Fgr-/- BMDMs preserved CI-dependent ATP production and did not enhance the CII-mediated ATP production upon E. coli detection as opposed to WT BMDMs (Fig. 5b). As a consequence, Fgr-/- BMDMs challenged with E. coli did not show an increase in MRR (Supplementary Fig. 5k) and SRC (Fig. 5c) compared to resting condition, but did show an increase in ECAR (Fig. 5d) and lactate production (Fig. 5e) similar to that of WT BMDMs. These results indicate that Fgr is an important regulator of macrophage ETC adaptations during bacterial infection.
Figure 5.
Fgr kinase is required form induction of complex II activity upon detection of E. coli requires Fgr kinase. (a-e) Effect of E. coli-stimulation on CII activity (a), substrate-driven CI- and CII-mediated ATP synthesis (b), SRC (c), ECAR (d) and extracellular lactate concentration (e) of Fgr-/- and WT BMDMs. In (b), 100% activity corresponds to a rate of 72.1, 66.3, 21.2, 17.8 nmol ATP/min/mg protein for CI and CII-mediated synthesis in WT and Fgr-/-, respectively. (f) Intracellular colony-forming units (CFU) after E. coli infection of Fgr-/- and WT BMDMs for the indicated times at MOI = 5. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data (mean and s.e.m.) are from two (f), three (b-e) or seven (a) independent experiments performed in two (a), three (b, f) to five (c-e) technical replicates.
Metabolic reprogramming towards glycolysis and accumulation of succinate7, together with an increase in mitochondrial ROS production26 contribute to antimicrobial functions of macrophages. In line with this, Fgr-/- BMDMs showed increased intracellular E. coli upon infection in vitro compared with WT cells (Fig. 5f), indicating enhanced survival of the bacteria within BMDMs. However, Fgr-deficient and WT mice showed equal number of colony-forming units (CFU) in the spleen after intraperitoneal E. coli infection (Supplementary Fig. 5l,m). In line with this, the induction of the inflammatory genes Il1b, Ifnb and Tnf in response to E. coli challenge was similar between WT and Fgr-/- BMDMs (Supplementary Fig. 5n), despite a slight reduction in IL-1β secretion by Fgr-/- BMDMs compared with WT (Supplementary Fig. 5o). Taken together, this indicated that other Src-family kinases might compensate for Fgr loss for inflammatory cytokine expression31.
CII activity contributes to anti-microbial responses
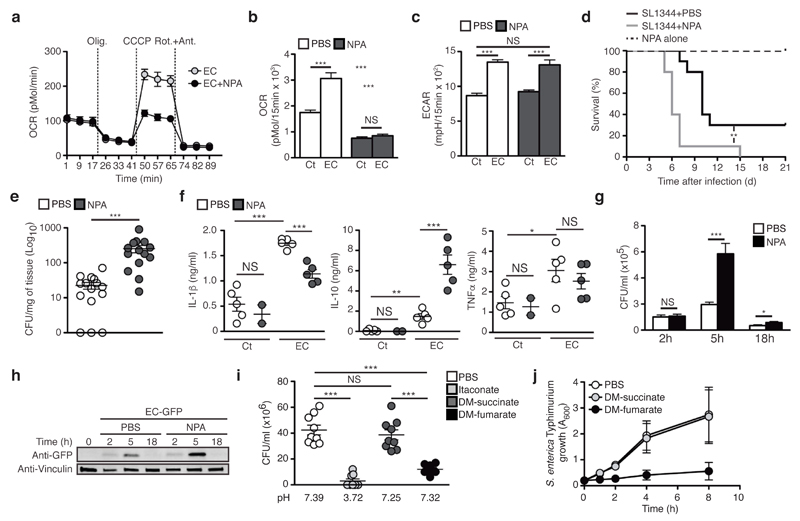
Because Fgr has other targets besides CII, we used the SDH-specific inhibitor 3-nitropropionic acid (NPA)32 to block CII activity. At the concentrations tested, NPA did not affect WT BMDMs survival or phagocytic activity (Supplementary Fig. 6a,b), but strongly repressed CII activity in resting and E. coli-stimulated BMDMs compared with untreated cells (Supplementary Fig. 6c) without affecting bacterial SDH activity or growth (Supplementary Fig. 6d,e). Treatment of WT BMDMs with NPA did not prevent SC disassembly (Supplementary Fig. 6f) or the drop in CI-mediated ATP production, but efficiently prevented the increase in CII-mediated ATP production induced by E. coli challenge (Supplementary Fig. 6g) in contrast to untreated BMDMs. NPA treatment also strongly reduced the MRR induced by E. coli (Fig. 6a and Supplementary Fig. 6h) and ablated the SRC (Fig. 6b) without affecting ECAR induction (Fig. 6c and Supplementary Fig. 6i) or lactate production (Supplementary Fig. 6j) in WT BMDMs. Similar results were obtained using the CII competitive inhibitors dimethyl-malonate (DM)33 or thenoyltrifluoroacetone (TTFA) (Supplementary Fig. 6k-o).
Figure 6.
Mitochondrial complex II contributes to macrophage bactericidal capacity. (a-c) OCR upon sequential treatment with oligomycin (olig.), CCCP, and rotenone+antimycin (Rot.+Ant.) (a), SRC (b), and ECAR (c) in BMDMs stimulated for 2h with E. coli ± NPA. (d) Survival of WT mice infected with 1x109 S. enterica Typhimurium by gavage and treated or not with 50mg/kg NPA. (e, f) Splenic bacterial burdens at 72h (e) and cytokine serum levels at 2h (f) after intra-peritoneal injection of 1 x 108 (e) or 1 x 109 (f) of E. coli (f) into WT mice treated or not NPA. (g, h) Intracellular CFU (g) and anti-GFP immunoblot of SDS-solubilized extracts (h) from WT BMDMs treated or not with NPA and infected with E. coli (g) or GFP-expressing E. coli (h) at MOI = 5 for the indicated times. (i) CFU of 1x105 E. coli incubated 3h with itaconic acid, dimethyl (DM)-succinate or DM-fumarate. (j) S. enterica Typhimurium growth in presence of 10mM of the indicated reagent. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t-test (b, c, e-g, i, j) or Log-rank (Mantel-Cox) test (d)). Data are from three independent experiments performed in two to five technical replicates (b-g, i, j). Data (mean and s.e.m. in (b,c, e-g, i, j); mean and s.d. in (a)) are from two (d, f, g) and three (b, c, e, i, j) independent experiments performed in three (j) or five (b, c) technical replicates or with two to five mice per group (e, f), one representative of at three independent experiments with similar results (a, h).
We next investigated whether CII activity contributed to anti-microbial functions. When compared with mice treated with vehicle, mice treated with NPA were more susceptible to S. enterica Typhimurium infection (Fig. 6d), and presented an increased splenic bacterial burden at 72h after intra-peritoneal infection with viable E. coli (Fig. 6e) despite normal recruitment of inflammatory cells to the peritoneal cavity (Supplementary Fig. 7a). The higher bacterial in NPA-treated mice load correlated with decreased serum amounts of the pro-inflammatory cytokine IL-1β and increased amounts of anti-inflammatory cytokine IL-10, while TNF amount was not affected compared with control mice (Fig 6f). We also found increased numbers of viable E.coli in NPA-treated BMDMs compared with WT (Fig. 6g,h), indicating that inhibition of CII impaired macrophage bactericidal activity in vitro. In addition, NPA-treated BMDMs had reduced expression of E. coli-induced IL-1β protein and Il1b and Ifnb mRNA, but normal TNF expression compared with untreated BMDMs (Supplementary Fig. 7b,c), consistent with the relative insensitivity of TNF production to macrophage-metabolic fluctuations and oxygen availability7,34. Itaconic acid, which results from decarboxylation of the tricarboxylic acid (TCA) cycle intermediate cis-aconitate, shows anti-bacterial properties12. We found that the TCA cycle intermediate fumarate, which is produced by the oxidation of succinate by CII, strongly inhibited bacterial growth (Supplementary Fig. 7d) and induced bacterial death, while succinate had negligible effects when compared with PBS-treated bacteria (Supplementary Fig. 7e,f). To exclude that mere lowering of the pH accounted for the observed effects of fumarate on bacteria viability, we used ester forms of these TCA intermediates. Dimethyl-fumarate, but not dimethyl-succinate, impaired E. coli (Fig. 6i) and S. enterica Typhimurium growth in vitro (Fig. 6j). These data identify CII as an important contributor to the macrophage mitochondrial respiratory functions needed for anti-microbial responses.
Microbial viability drives ETC adaptations
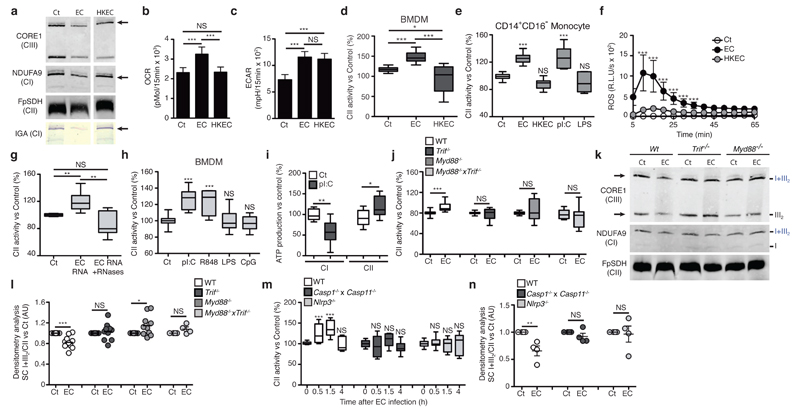
In contrast to viable E. coli, heat-killed E. coli did not affect the assembly of the abundance of CI and SC I+III2 in BMDMs (Fig. 7a and Supplementary Fig. 8a,b). In contrast to viable E. coli, heat-killed E. coli did not impair CI in-gel activity within SC I+III2 (Fig. 7a) and failed to increase mitochondrial MRR (Supplementary Fig. 8c) or SRC in BMDMs (Fig. 7b). However, viable and heat-killed E. coli both efficiently induced ECAR (Fig. 7c), lactate release (Supplementary Fig. 8d) and mG3PDH activity (Supplementary Fig. 8e) in BMDMs, likely reflecting the capacity of LPS to trigger a glycolytic switch in macrophages5,7. Unlike viable E. coli, heat-killed E. coli did not induce CII activity in BMDMs (Fig. 7d), human CD14+CD16- monocytes (Fig. 7e) or peritoneal macrophages (Supplementary Fig. 8f). Importantly, live E. coli were more efficient than heat-killed E. coli at inducing phagosomal ROS production by WT BMDMs (Fig. 7f and Supplementary Fig. 8g), which was required for CII and SRC induction in macrophages. These findings indicate that stimuli associated with viable bacteria trigger ETC adaptations in macrophages.
Figure 7.
Sensing of bacterial viability induces MyD88/TRIF- and NLRP3 inflammasome-dependent decrease in supercomplex abundance and increase in CII activity. (a) BN-PAGE immunoblot analysis and CI-in gel activity (IGA) assay in BMDMs treated as indicated. (b-e) SRC (b), ECAR (c) and CII activity (d, e) in BMDMs (b-d) and CD14+CD16- human monocytes (e) stimulated with EC or HKEC are shown. (f) ROS production by BMDMs stimulated as in (b). (g, h) CII activity in WT BMDMs stimulated as indicated for 1.5h. (i) Substrate-driven ATP synthesis assay in WT BMDMs stimulated with pI:C. 100% activity corresponds to a rate of 25.1 and 19.8 nmol ATP/min/mg protein for CI- and CII-mediated synthesis. (j) CII activity in WT, Trif-/-, Myd88-/- and Trif-/-Myd88-/- BMDMs treated as indicated. (k) BN-PAGE immunoblot analysis of resting and E. coli-stimulated WT, Trif-/- and Myd88-/- BMDMs. Arrow indicate the main SCs affected. (l, n) Densitometric analysis of CI+CIII2/CII signal ratio as observed by BNGE immunoblot of BMDMs of the indicated genotype stimulated with EC for 1.5h. (m) CII activity in WT, Nlrp3-/- and Caspase1-/- Caspase11-/- BMDMs stimulated with EC for the indicated time point. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data (mean and s.e.m. (b-j, l-n)) are from three (b, c, e, i), four (d, g, h, m, n) or five (f, j, l) independent experiments performed in two (d, e, g, h, j), three (f, i) or five (b, c) technical replicates, one representative of four independent experiments (a, k).
An association with bacterial viability has been suggested for several bacterial molecules2,35, including bacterial RNA19. We found that CII activity in BMDMs was enhanced in response to RNA purified from E. coli and unaffected when the RNA preparations were pre-treated with RNases (Fig. 7g and Supplementary Fig. 8h). The double-stranded RNA-mimic polyinosinic:polycytidylic acid (poly(I:C)) and the single-stranded RNA-mimic R848, which respectively trigger TLR3 and TLR7, also induced CII activity in BMDMs (Fig. 7h), whereas the TLR4 agonist LPS or the TLR9 agonist CpG oligodeoxynucleotide did not (Fig. 7h and Supplementary Fig. 8i-j). This was also evident in human CD14+CD16- monocytes (Fig. 7e). In addition, poly(I:C) treatment decreased CI-mediated ATP production and promoted CII-mediated ATP production in permeabilized BMDMs (Fig. 7i). These results indicated that recognition of microbial RNA controls CII activity.
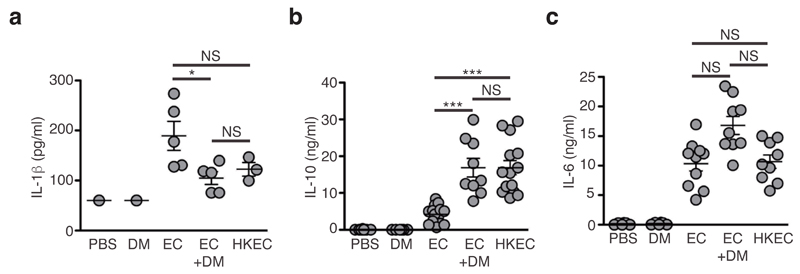
Because bacterial RNA triggers viability-specific immune responses through the adaptor TRIF19, we evaluated the contribution of TLR adaptors to ETC changes induced by E. coli. In contrast to WT cells, BMDMs generated from Trif-/-, Myd88-/- and Trif-/- Myd88-/- mice did not induce CII activity when exposed to bacteria (Fig. 7j). In contrast, induction of CII activity upon E. coli challenge was unaffected in STING-deficient (Tmem173gt, here called GT-Sting) or MAVS-deficient (Mavs-/-) BMDMs (Supplementary Fig. 8k), two adaptors that use mitochondria to mediate nucleic acids and poly(I:C) sensing by cytosolic receptors14. In addition, both TRIF and MyD88 were required to initiate changes in ETC composition upon E. coli sensing (Fig. 7k,l and Supplementary Fig. 8l). Thus, adjustments of the CII function in response to bacteria were likely regulated by phagosomal RNA-sensing TLRs, rather than cytosolic nucleic acid-sensing innate immune receptors2. Viability-specific immune responses and bacteria RNA sensing also involve the NLRP3 inflammasome2,19,36, the activation of which has been linked to mitochondria, phagosomal NADPH oxidase and ROS release2,14,19,36,37. Both CII induction (Fig. 7m) and ETC superassembly decrease (Fig. 7n and Supplementary Fig. 8m) were markedly impaired in Nlrp3-/- or Casp1-/-Casp11-/- BMDMs compared with WT, thereby placing CII activation in the centre of viability-specific immune responses to bacteria. Consistently, treatment of E. coli-infected WT mice with the CII inhibitor dimethyl-malonate reduced the serum amounts of IL-1β and increased the serum amounts of IL-10 to levels measured in mice treated with heat-killed E. coli, without influencing serum amount of the IL-6 (Fig. 8). Taken together these data suggested that CII activity regulates cytokine production by macrophages during bacterial infection (Supplementary Fig. 8n).
Figure 8.
Inhibition of CII in vivo modulates cytokine production upon viable E. coli challenge. (a-c) Serum levels of IL-1β (a), IL-10 (b) and IL-6 (c) at the indicated time point after intra-peritoneal injection of 1x109 viable EC or 1x1010 HKEC in WT mice treated or not with 600mg/kg dimethyl-malonate (DM). Poly(I:C), (pI:C); LPS, lipopolysaccharide; CpG, CpG oligodeoxynucleotide. NS, not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed unpaired Student’s t-test). Data (means and s.e.m.) are from one (a) or two (b, c) independent experiments performed with two to five mice per group. Each symbol represents one mouse.
Discussion
Our study describes a mechanism by which macrophages adjust their metabolism in response to viable bacteria by coupling TLR engagement, the NLRP3 inflammasome and ROS signalling to mitochondrial electron transport chain. This metabolic adjustment in turn contributes to pathogen-specific immune responses. The ETC architecture was altered in response to detection of viable bacteria because of a decrease in the abundance of assembled CI. Several explanations can account for this. CI is an unstable ETC multiprotein complex composed of 44 different protein subunits, the assembly of which is influenced by the oxidative environment within the ETC15 and/or by previous assembly of other ETC components including CIII38 or cytochrome c oxidase39. A switch in substrate that fuels the TCA cycle modifies the NADH to FADH2 ratio, which can saturate the oxidation capacity of the CoQ pool thereby inducing a reverse electron transfer (RET) towards CI. This in turn increases superoxides that oxidize specific CI proteins thereby leading to disassembly of the complex23. Upon surface TLR engagement ROS expression within the ETC is increased26, and thus constitute a potential source of CI-destabilizing ROS. In addition, a RET-inducing modulation of the NADH/FADH2 ratio is likely to occur here because innate immune receptor activation diverts pyruvate from entering the mitochondria and limit fatty acid oxidation while glutaminolysis ensures the replenishment of the TCA cycle5,40.
Unless a yet to be determined bacteria viability-specific metabolic flux adaptation further modulates the NADH/FADH2 ratio, CI destabilization should occur in response to both heat-killed and viable bacteria. However, we only observed ETC alterations in response to the later. In addition, the inhibition of CII in macrophage challenged with viable E. coli did not impair the decrease in CI and CI-containing SC abundance, nor it prevented the decrease in CI-mediated ATP synthesis. Phagosomal ROS may also account for the oxidative destabilization of CI. Indeed, we found that gp91phox-deficient macrophage had preserved ETC architecture in response to viable E. coli, and that phagosomal ROS levels were substantially lower in response to heat-killed E. coli. However, gp91phox deficiency also impaired CII activity induction, and as a consequence could also prevent CI-destabilizing RET. Therefore, whether changes in ETC architecture upon viable bacterial challenge are a consequence of the rapid increase in oxidative burst within the ETC due innate immune signalling or result from fuel switch remains to be determined. Nevertheless, our data indicate that E. coli recognition induces a transient alteration of SCs in macrophages, which could make CIII available for electrons provided by FADH2 dependent enzymes and thereby allowing mitochondria to re-oxidize cytoplasmic NADH without the use of CI as shown in other models15,20.
Whether metabolic reprogramming directly contributes to macrophage effector function remains unclear. Itaconate, a nonamino organic acid, can exert antimicrobial functions in concentration range about 10mM12,13. Consistent with this notion, we found that CII-enzymatic product fumarate, but not its precursor succinate, prevented bacteria growth in vitro and decreased bacterial viability at a concentration of 10mM. At such concentration pH is merely decreased and should therefore impair macrophage normal functions. However, upon bacteria encounter by macrophages, mitochondria are juxtaposed to microbe-containing phagosomes26,41. This is mediated by the formation of a complex between the TLR signalling adaptor TRAF6 and the mitochondrial complex I-assembly factor ECSIT26, the interaction of which is regulated by the Mst1-Mst2-Rac signalling axis41. Such proximity between mitochondria and bacteria-containing phagosomes might permit the delivery of mROS or mitochondrial metabolites to contribute to bacteria killing14. Such model presents some advantages because it would allow reaching sufficient concentration of those ‘antimicrobial metabolites’ locally (i.e. within the phagosome) while sparing host cellular metabolism. Future work will likely provide additional insights on this issue.
The use of LPS to activate macrophages has generated a considerable amount of information on the metabolic pathways and reprogramming engaged during inflammation8. We found that challenge of macrophage with heat-killed bacteria recapitulates many aspects of LPS-mediated TLR4 engagement on mitochondrial respiration, including induction of glycolytic flux (increased ECAR and lactate release) and decreased oxygen consumption in the mitochondria. However, the induction of CII activity and destabilization of the ETC was observed only in response to viable bacteria. Therefore, while the use of a single PAMP certainly presents advantages, it only offers a partial view of the complexity of the innate immune signals that may regulate metabolic adjustments upon the encounter of a whole viable microorganism. This is not trivial because macrophages tightly scale their response to many features of bacteria including viability-specific signals2,35. Most importantly, we found that the inhibition of CII during viable bacteria challenge in mice increased IL-10 and decreased Il-1β in the serum to those observed when heat-killed bacteria were used. Thus, the establishment of a functional link between pattern recognition receptors, ETC organization and subsequent inflammatory immune responses may offer substantial benefits for vaccine design and provide valuable new targets for pharmacological intervention both during infection and in metabolic inflammatory disorders.
Methods
Mouse strains
C57BL/6J and CD1 mice were purchased from Harlan Laboratories. Myd88-/- and Trif-/- mice were originally generated by S. Akira and bred to homozygosity to generate Trif-/-Myd88-/- mice by R. Medzhitov. Ips1-/- mice were originally obtained from S. Akira and backcrossed with Trif-/- mice to obtain Ips1-/-Trif-/- mice. C57BL/6J-Tmem173gt/J mice (here called GT-Sting mice) and Gp91phox-/- (B6.129S6-Cybbtm1Din/J) mice were obtained from the Jackson Laboratory. Nlrp3-/- and Casp1-/-Casp11-/- mice have been described previously42, 43. We used 8- to 10-week-old animals (males or females) for all experiments. Experiments were repeated 3 times and 3-5 animals per group were used to reach statistical significance. No blinding or randomization strategy was used and no animal was excluded from analysis. All experimental procedures were approved by institutional care and use committees and performed in agreement with EU directive 86/609/EEC and recommendation 2007/526/EC regarding the protection of laboratory animals and enforced under Spanish law by Royal decree 1201/2005.
Reagents
Lipopolysaccharide, polyinosinic:polycytidylic acid (polyI:C), CpG ODN were purchased from Invivogen. 3-nitropropionic acid (NPA), succinate, succinate hexahydrate, glutamate, malate disodium-salt, fumarate, dimethyl-fumarate, dimethyl succinate, dimethyl malonate (DM), itaconic acid, thenoyltrifluoroacetone (TTFA), carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP), CCCP, oligomycin, rotenone, antimycin A, ubiquinone, sn-glycerol 3-phosphate, oxidized cytochrome c, adenosine tri-phosphate (ATP), adenosine di-phosphate (ADP), phenazine methosulfate (PMS) and digitonin were all from Sigma. Luciferin and luciferase were from Promega and Roche, respectively.
Bacteria
Escherichia coli K12, strain DH5α, were purchased from Invitrogen. Salmonella enterica serovar Thyphimurium strain SL1344 were provided by F. Norel-Bozouklian. SL1344 were grown in LB broth supplemented with 50μg/ml streptomycin (Sigma). For phagocytosis experiments, bacteria were grown overnight in Luria-Bertani (LB) broth with shaking, diluted 1/50, and grown until log-phase [optical density at 600 nm (OD600) of 0.8-1.2] without shaking. Bacteria were washed with phosphate buffer saline (PBS) to remove LB salts before addition to cells. For heat-killing, E. coli (HKEC) were grown to log phase, washed, re-suspended in PBS and subsequently incubated at 60°C for 60-90 min. Aliquots of heat-killed bacteria were stored at -80°C until use. Efficient killing was confirmed by overnight plating on LB-agar plates. Total RNA was isolated from E. coli using the e.z.n.a. Bacterial RNA kit (Omega Bio-Tek). E. coli-GFP were generated by transformation of BL21pLysS bacteria (Invitrogen) with a pET-28 vector encoding the GFP-OT fusion protein44. E. coli-GFP were grown in the presence of 50 μg/ml kanamycin and 50 μg/ml chloramphenicol. To induce GFP expression, overnight-grown bacteria were diluted to an OD600 of 0.8 and incubated for 4h in the presence of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG).
Antimicrobial assay
The following bacteria strains were used: Salmonella enterica serovar Thyphimurium strain SL1344, Escherichia coli K12, strain DH5α (Invitrogen). Bacteria were cultured at 37 °C overnight in lysogeny broth (LB). Bacterial concentrations were measured by spectrophotometry at 600 nm and diluted to a concentration of 105 CFU/ml in PBS supplemented with 0.1, 1, or 10mM of itaconate, fumarate, succinate, dimethyl-fumarate or dimethyl-succinate and incubate for 6h at room temperature. Serial dilutions were then plated on LB-agar plates supplemented with 50mM streptomycin (for SL1344) and grown overnight at 37°C. Photographs were taken using a scanner. Alternatively, bacteria were diluted to a final OD600=0.2 in LB supplemented as above and growth was measured every 2h by spectrophotometry. For CFU enumeration, bacteria were diluted to a concentration of 105 CFU/ml in PBS supplemented as above, incubated for 6h and serial dilution were plated on LB-agar plates. The number of colonies formed after overnight incubation was counted. For analysis of cell death, bacteria were stained with 5μM propidium iodide (to stain nucleic acids) and 5μM of cell SYTO red dye (Life Technology) for 15min, and analyzed by flow cytometry.
Macrophage preparation and treatment with Escherichia coli
Murine bone marrow-derived macrophages (BMDMs) were generated from C57BL/6J, CD1, Fgr-/-, Myd88-/-, Trif-/-, Trif-/-Myd88-/-, GT-sting, Ips-1-/-, Trif-/-Ips-1-/-, Gp91phox-/-, Nlrp3-/- and Casp1-/- Casp11-/- mice, as described previously45, in RPMI 1640 supplemented with M-CSF (30% mycoplasma-free L929 cell supernatant, NCBI Biosample accession # SAMN00155972) and 10% FBS, plus 100 μg/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 1 nM sodium pyruvate and 50 mM 2-mercaptoethanol (all from Gibco). Peritoneal macrophages were harvested 72h after intraperitoneal injection of 1 ml 3% thioglycollate medium (BD Bioscience). Human CD14+CD16- monocytes were obtained from buffy coats using the EasySep Human Monocyte Enrichment Immunomagnetic kit (Stemcell Technologies). For treatment with viable E. coli and HKEC, cells were plated at 1.5x106 cells/well in non-treated 6-well cell culture plates (BD Bioscience) and left to adhere for at least 4h. BMDMs were challenged with E. coli or HKEC at a multiplicity of infection (MOI) of 20 and plates were spun at 400 g for 5min. Cells were incubated for 1.5h unless otherwise indicated. For longer time points, 50 μg/ml gentamicin sulfate (Gibco) was added after 1h incubation. Alternatively, cells were stimulated with soluble ligands as follows: 200 ng/ml LPS, 20 μg/ml poly(I:C), 5 μg/ml CpG ODN. For treatment with metabolic inhibitors, 0.5 mM 3-nitropropionic acid (NPA), 0.5mM dimethyl-fumarate or 0.5mM TTFA were added to the cells 30 min-to-1 h prior challenge. For stimulation of human, CD14+ CD16- monocytes were isolated from buffy coats. Cells were plated at 1x106 cells/ml in non-treated 12-well cell culture plates (BD Bioscience) and left to adhere for at least 2h. Cells were challenged with E. coli and HKEC at a multiplicity of infection (MOI) of 10 and plates were spin at 2500rpm for 1min. Cells were incubated for 1.5h. Alternatively, cells were stimulated with soluble ligands as follows: 250 ng/ml Ultrapure EK-LPS and 10 μg/ml LMW poly(I:C). For supernatant collection, cells were plated at 3x105 cells/well in a 48-well plate and stimulated as described above.
Oxygen consumption rate and glycolytic flux evaluation
Real-time oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in BMDMs were determined with an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience); 1x105 cells/well in 5-6 wells were used for each condition. The assay was performed in DMEM supplemented with 2mM glutamine, 100 μg/ml penicillin, 100 μg/ml streptomycin, phenol red and 25 mM glucose + 1mM pyruvate or 5 mM L-carnitine + 50 μM palmitoyl-CoA. The pH was adjusted to 7.4 with KOH (herein called seahorse medium). Three consecutive measurements were performed under basal conditions and after the sequential addition of the following ETC inhibitors: 1μM oligomycin, 1 μM CCCP, 1 μM rotenone and 1 μM antimycin. Basal respiration rate (BRR) was defined as OCR in the absence of any inhibitor. Maximal respiration rate (MRR) was defined as the OCR after addition of oligomycin and FCCP. Spare respiration capacity (SRC) was defined as the difference between MRR and BRR. ECAR was measured in the absence of drug. Where indicated, cells were treated with 0.5 mM NPA for 30 min prior to stimulation. For lactate production measurement, cells (1x105/well) were plated on a 96-well plate and stimulated as indicated. Cells were washed 5 times with PBS and 100μl of seahorse medium was added. Plates were incubated at 37°C without CO2 for 1h and supernatants were harvested. 25μl of 5-time-diluted supernatant was used to measure lactate production using a Lactate assay kit II (Sigma) according to manufacturer’s instructions.
Isolation of mitochondria and BMDMs permeabilization
Mitochondria were isolated as described by Schägger et al.46 with some modifications. 1x108 BMDMs were collected in PBS supplemented with 5 mM EDTA and washed with PBS. Cell pellets were frozen at −80°C to increase cell breakage and were homogenized in a tightly fitting glass-teflon homogenizer with 10 volumes of buffer A (83mM sucrose, 10 mM MOPS, pH 7.2). An equal volume of buffer B (250 mM sucrose, 30 mM MOPS, pH 7.2) was added and nuclei and unbroken cells were removed by centrifugation at 1000 g for 5 minutes. Supernatants were collected and centrifuged at 12 000 g for 2 min. Mitochondria pellets were washed once with buffer C (320 mM sucrose, EDTA 1 mM, 10 mM Tris-HCl, pH 7.4). Mitochondria were then suspended in an appropriate volume of PBS for storage at −80°C.
Blue-native polyacrylamide gel electrophoresis (BN-PAGE), two-dimensional gel analysis, and in gel activity assay
For BMDMs permeabilization, 3x106 macrophages were resuspended in 100 μl of PBS. 32.5 μl of digitonin (8 mg/ml) was added and cells incubated on ice for 10 min. Cold PBS (1 ml) was added and cells were centrifuged for 5 min at 10 000 g. The pellet was suspended in 100 μl of AA buffer (500 mM 6-aminohexanoic acid, 50 mM immidazole, 1 mM EDTA, pH 7) and 10 μl of a 10% digitonin solution was added. Cells were centrifuged for 30 min at 18 000 g. Supernatant was harvested and 10 μl of sample buffer (5% Blue G-250, 5% glycerol in AA Buffer) was added. Samples were stored at −80°C until use. BN-PAGE was performed as described by Wittig et al.47. For 2-dimensional gel analysis, 50 to 75 μg of mitochondria were digitonin-permeabilized with 4 μg digitonin per μg of protein and loaded on a BN polyacrylamide gel. Each individual band on the BN-PAGE was cut out and incubated for 1 hour in buffer containing 1% SDS and 1% 2-mercaptoethanol. The buffer was replaced with a 1% SDS solution and incubation was continued for 30 min. BN-PAGE bands were loaded on an SDS polyacrylamide gel composed of a 10% acrylamide/Bis-acrylamide (AB) stacking gel and a 16% AB resolving gel. Electrophoresis was performed overnight at 12-to-15 mA, and proteins were transferred to a PVDF membrane using a Trans-Blot Semi-Dry Transfer Cell (Bio-Rad). For in-gel OXPHOS complex activity assays, CI was revealed by incubating the BN gel for 1h-to-3h in 0.1M Tris-HCl, pH 7.5, containing 1mg/ml NBT and 0.14mM NADH. For densitometry analysis, the ImageJ64 software was used. Band limits were determined using low exposure images to efficiently distinguish the different bands. Background correction was applied for each analysis.
OXPHOS function and enzyme activities
CII activity was measured spectrophotometrically from the reduction of 2,6-diclorophenol-indophenol (DCPIP) by tracking the absorbance at 600 nm over 3min as described48 with some modifications. Briefly, 3x106 cells were suspended in 100μl PBS on ice. Protein concentration was determined and the volume was adjusted to the lowest concentrated sample. Sample (20 μl) was suspended in 950 μl of buffer C1/C2 (25 mM potassium phosphate (K2HPO4) pH=7.2, 5 mM MgCl2, 3 mM KCN, 2.5 mg/ml BSA) supplemented with 100 mM succinate and 0.1% Triton X-100, and incubated 10 min at room temperature in a cuvette. After addition of 6 μl 5 mM DCPIP, 2 μl 1 mg/ml of antimycin A and 2 μl 1 mM rotenone, samples were incubated for 2 min. Then, 6 μl of 10 mM UQ1 was added and absorbance was measured. CII activity was extrapolated using the following formula: [CII Activity = ((rate/min)/19.1) / sample volume x 1000 x dilution factor], where 19.1 is the molar extinction coefficient at 30°C (mM-1cm-1). For CII+CIII, UQ1 was replaced with 1 mM oxidized cytochrome c (Sigma). For SDH activity, UQ1 was replaced with phenazine methosulfate (PMS). For mitochondrial glycerol 3-phosphate dehydrogenase (mG3PDH) activity, succinate was replaced with 1 M glycerol-3-phosphate. For OXPHOS enzymatic activities in isolated mitochondria, individual and combined complex activities of isolated mitochondria were measured spectrophotometrically as described48. The CII, CII+III and mG3PDH specific activities corresponding to 100% activity measured in permeabilized untreated BMDMs are 0.0165 ± 0.005 IU/mg protein (N=25), 0.020 ± 0.008 IU/mg protein (N=4) and 0.0180 ± 0.006 IU/mg protein (N=18), respectively.
ATP synthesis assay
ATP synthesis was measured in permeabilized cells by kinetic luminescence assay49. Cells (2x106) were suspended in 160 μl of buffer A (150mM KCl, 25mM Tris-HCl, 2mM EDTA, 0.1% BSA FA, 10mM K-phosphate, 0.1mM MgCl2, pH 7.4) at room temperature (RT) and 50 μg/ml digitonin was added. Samples were mixed gently for 1 min, and the reaction was stopped by addition of 1ml of buffer A. Cells were centrifuged at 3000 rpm for 2 min at RT, and pellets were suspended in 160 μl of buffer A and dispensed into the wells of a 96-well luminescence reading plate (Costar). Substrate cocktail (50 μl) and 20 μl of buffer B (0.5M Tris-acetate, pH 7.75, 0.8mM luciferine, 20 μg/ml luciferase) were added, and luminescence was measured over 1 min. Substrate cocktails were composed of 6 mM diadenosin pentaphosphate and 6 mM ADP supplemented with 1 M glutamate + 1 M malate for determination of CI activity or with 1 M succinate for CII activity. ATP production rate is expressed as ‘nmol of ATP/min/mg of protein.’ All measurements were performed in triplicate.
Phagocytosis assay
Macrophages (3x105) were seeded in triplicate on a non-treated 24-well plate (BD Biosciences). Cells were challenged with 6x106 3μm Fluoresbrite®-microspheres (Polysciences) or 3x106 E. coli-GFP and centrifuged for 5 min at 400 g. After 20 min incubation, cells were washed with PBS, harvested in PBS containing 5 mM EDTA, and analyzed by flow cytometry.
Cytokine enzyme-linked immunosorbent assay (ELISA)
IL-1β, IL-10 and TNF-α ELISA kits were from BD Biosciences. Capture/detection antibodies for IL-6 antibodies were from BD Biosciences. Supernatants from BMDMs were collected at 24h after stimulation. ELISA kits are used according to the manufacturer’s instructions. Detection antibodies were biotinylated and labeled with streptavidin-conjugated horseradish peroxidase (HRP, from invitrogen) and visualized by incubation with 5,5’-tetramethylbenzidine solution (TMB, KPL). Colour development was stopped with TMB-stop solution (KPL). Recombinant cytokines served as standards and were purchased from Peprotech. Absorbances at 450 nm were measured on a microplate reader (Benchmark Plus, Bio-Rad)
In vitro and in vivo infections and bactericidal activity experiments
For in vitro experiments, BMDMs were plated at 2x105 cells/well in triplicate on a 24-well plate in an antibiotic-free complete medium. BMDMs were infected with DH5α or SL1344 at a multiplicity of infection of 5 and centrifuged for 5 min at 400 g. After 30 min incubation, cells were washed and complete medium supplemented with 50 μg/ml gentamycin was added. At the indicated time point after infection, cells were washed with PBS and 1 ml of PBS containing 1% Triton X-100 was added. Plates were incubated at room temperature for 15 min and serial dilutions (1/10, 1/100, 1/1000) were plated on an LB-agar plate, which in the case of SL1344 contained 50 μg/ml streptomycin. Plates were incubated at 37°C and bacterial colonies were counted. When needed, cells were pretreated with 3-nitropropionic acid (NPA) 30 min before infection; the inhibitor concentration was maintained throughout the experiment. For in vivo experiments, mice were injected intraperitoneally (i.p.) with 50mg/kg NPA 1 hour before infection. Injection of inhibitor was repeated every second day over the course of the experiment. For E. coli infection, mice were injected i.p. with 1x108 DH5α and sacrificed at 72h post-infection. Spleens were harvested and homogenized in 5 ml PBS, and serial dilutions were plated on LB-agar plates for colony counting. For peritoneal cell analysis, mice were injected i.p. with 1x108 DH5α. Twelve hours later, mice were sacrificed and peritoneal cells were collected in 8 ml ice-cold PBS. Each experiment included 4-5 mice per group and was repeated 3 times with similar results. No specific blinding or randomization strategy was used. No animal was excluded from analysis.
Immunoblot
For protein extract collection, 1.5x106 cells were lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktails (both from Roche) and subsequently sonicated and boiled for 5 min at 95°C. Protein lysates were separated on 4-12% SDS-gradient gels (Bio-Rad). Proteins were transferred to PVDF membranes (Millipore). Membranes were blocked with 5% bovine serum albumin (BSA) in PBS and probed with antibodies sourced as follows: anti-CORE1 (1/5000 dilution, ab110252, Abcam), -NDUSF3 (1/5000 dilution, ab110246, Abcam), -ATP-B (1/2000 dilution, Abcam), and -NDUFA9 (1/5000 dilution, ab14713, Abcam), -FpSDH (1/5000 dilution, #459200, Invitrogen), -Cox5b (1/5000 dilution, Proteintech Europe), anti-vinculin (1/10000 dilution, Sigma); and anti-β-actin (1/10000 dilution, Santa Cruz Biotechnology).
Real-time PCR
Total RNA was isolated from macrophages using the RNeasy kit (Qiagen). Contaminating genomic DNA was removed by DNase digestion (Qiagen). Reverse transcription was performed using the High Capacity cDNA Reverse Transcriptase kit (Applied Biosystem), and cDNA was used for subsequent real-time PCR reactions. Quantitative real-time PCR was conducted on an 7900 HT Fast Real-Time PCR system (Lifetechnologies) using SYBR green qPCR Master Mix (Promega) with the following primer pairs: β-Actin, FW 5’- GAAGTCCCTCACCCTCCCAA-3’, RV 5’-GGCATGGACGCGACCA-3’; Il1b, FW 5’- AAAGACGGCACACCCACCCTGC-3’, RV 5’-TGTCCTGACCACTGTTGTTTCCCAG-3’; Ifnb, FW 5’- TCAGAATGAGTGGTGGTTGC ; RV 3’- GACCTTTCAAATGCAGTAGATTCA; Tnf FW 5’-CCCCAAAGGGATGAGAAGTT, RV 3’-TGGGCTACAGGCTTGTCACT.
Flow cytometry
Cells were stained with the appropriate antibody cocktails in ice-cold PBS supplemented with 2 mM EDTA, 1% fetal calf serum and 0.2% sodium azide for 15min. Samples were processed by FACS canto-3L or LSR-Fortessa analyzers (BD Biosciences) and data were analyzed with FlowJo software. Antibodies used: CD11b-APC-Cy7 (1/200 dilution, M1/70, #557657, BD Bioscience), Ly6G-PE-Cy7 (1/200 dilution, 1A8, #560601, BD Bioscience), F4/80-PE (1/400 dilution, BM8, eBioscience).
Gene microarray analysis
Affymetrix Microarray data from BMDMs were previously deposited with the NCBI Gene Expression Omnibus under accession number GSE27960 by Sander et al.19. Data for genes encoding ETC subunits were analyzed and plotted using Genesis software from Graz university of Technology (http://genome.turgraz.at).
Statistical analysis
Statistical differences were analysed with Prism software (version 5, GraphPad Software Inc.). Comparisons of two groups were calculated with two-tailed unpaired Student’s t-test and, where indicated, with paired Student’s t-test or one-way ANOVA followed by Tukey post-test analysis. For survival experiments, statistical significance was tested by a Log-Rank (Mantel-Cox) test. A P value of less than 0.05 was considered statistically significant. No randomization or exclusion of data points was used. Pilot in vivo studies were used for estimation of the sample size required to ensure adequate power.
Supplementary Material
Acknowledgments
We are grateful to A. Hidalgo for critical reading of the manuscript, J. Magarian Blander (Mount Sinai School of Medicine) for Myd88-/-, Trif-/- and Trif-/-Myd88-/- mice, F. Norel-Bozouklian (Institut Pasteur) for Salmonella thyphimurium SL1344, S. Trombetta (New York University) for the plasmid encoding GFP-OT and S. Bartlett for English editing. We thank M. Fernández-Monreal, M. Villalba, F. Ruperez-Pascualena and Sancho and Enríquez lab members for insightful discussions and support. This work was supported by the European Community (FP7-Marie Curie-CIG#332881 to J.G., ERC-2010-StG 260414 and 635122-PROCROP H2020 to D.S.; UE0/MCA1108 and UE0/MCA1201 to J.A.E.), the French association ‘La Ligue Contre le Cancer-comité du Gard’ (CG/59-2013 to J.G.), the Spanish Ministry of Economy and Competitiveness (SAF-2013-42920R to D.S.; SAF2012-1207 to J.A.E., RyC2011-07826 to R. A.-P.), the Comunidad de Madrid (CAM/API1009 to J.A.E.), the German research council (DFG grant SA1940/2-1 and SFB-TR84 TP-C08 to L.E.S.). CNIC is supported by the MINECO and the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Footnotes
Author Contributions
J.G and R.A.-P. designed and performed all experiments. S.M.-C. and M.E. performed experiments measuring OXPHOS enzymatic activities, prepared samples for BN-PAGE and western blot and helped with experiments in mice. M.U. and L.E.S. performed experiments with human monocytes. E.N.-V. and S.H.-S. provided bone marrow progenitor cells from GT-Sting and Mavs-/- mice and generated Mavs-/-Trif-/- mice. P.P. provided Nlrp3-/- and Casp1-/-Casp11-/- mice. J.G, R.A.-P., J.A.E. and D.S directed the study, analysed the data and wrote the manuscript.
Competiting Financial Interests
The authors declare no competing financial interests
References
- 1.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14(6):392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 2.Blander JM, Sander LE. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol. 2012;12(3):215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- 3.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annual review of immunology. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 4.Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature immunology. 2014;15(9):846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity. 2015;42(3):419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185(1):605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 7.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. The Journal of experimental medicine. 2016;213(1):15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley IA, Ribeiro SM, Gimenez-Cassina A, Norberg E, Danial NN. Changing appetites: the adaptive advantages of fuel choice. Trends Cell Biol. 2014;24(2):118–127. doi: 10.1016/j.tcb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon JS, Hisata S, Park MA, DeNicola GM, Ryter SW, Nakahira K, et al. mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell reports. 2015;12(1):102–115. doi: 10.1016/j.celrep.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, et al. IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRG1-Derived Itaconic Acid. PLoS Pathog. 2016;12(2):e1005408. doi: 10.1371/journal.ppat.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enriquez JA. Supramolecular Organization of Respiratory Complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 16.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. The EMBO journal. 2000;19(8):1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly B, Tannahill GM, Murphy MP, O'Neill LA. Metformin Inhibits the Production of Reactive Oxygen Species from NADH:Ubiquinone Oxidoreductase to Limit Induction of Interleukin-1beta (IL-1beta) and Boosts Interleukin-10 (IL-10) in Lipopolysaccharide (LPS)-activated Macrophages. The Journal of biological chemistry. 2015;290(33):20348–20359. doi: 10.1074/jbc.M115.662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z, Wei W, Yang M, Du Y, Wan Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell metabolism. 2014;20(3):483–498. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 21.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32(4):529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Speijer D. Oxygen radicals shaping evolution: why fatty acid catabolism leads to peroxisomes while neurons do without it: FADH(2)/NADH flux ratios determining mitochondrial radical formation were crucial for the eukaryotic invention of peroxisomes and catabolic tissue differentiation. BioEssays : news and reviews in molecular, cellular and developmental biology. 2011;33(2):88–94. doi: 10.1002/bies.201000097. [DOI] [PubMed] [Google Scholar]
- 23.Guaras A, Perales-Clemente E, Calvo E, Acin-Perez R, Loureiro-Lopez M, Pujol C, et al. The CoQH2/CoQ Ratio Serves as a Sensor of Respiratory Chain Efficiency. Cell reports. 2016;15(1):197–209. doi: 10.1016/j.celrep.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Benard G, Faustin B, Galinier A, Rocher C, Bellance N, Smolkova K, et al. Functional dynamic compartmentalization of respiratory chain intermediate substrates: implications for the control of energy production and mitochondrial diseases. The international journal of biochemistry & cell biology. 2008;40(8):1543–1554. doi: 10.1016/j.biocel.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Mracek T, Drahota Z, Houstek J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochimica et biophysica acta. 2013;1827(3):401–410. doi: 10.1016/j.bbabio.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 26.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, et al. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. The Journal of biological chemistry. 2001;276(7):4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 28.Nunes P, Demaurex N, Dinauer MC. Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic. 2013;14(11):1118–1131. doi: 10.1111/tra.12115. [DOI] [PubMed] [Google Scholar]
- 29.Acin-Perez R, Carrascoso I, Baixauli F, Roche-Molina M, Latorre-Pellicer A, Fernandez-Silva P, et al. ROS-Triggered Phosphorylation of Complex II by Fgr Kinase Regulates Cellular Adaptation to Fuel Use. Cell metabolism. 2014 doi: 10.1016/j.cmet.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nath AK, Ryu JH, Jin YN, Roberts LD, Dejam A, Gerszten RE, et al. PTPMT1 Inhibition Lowers Glucose through Succinate Dehydrogenase Phosphorylation. Cell reports. 2015 doi: 10.1016/j.celrep.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowell CA. Src-family kinases: rheostats of immune cell signaling. Molecular immunology. 2004;41(6–7):631–643. doi: 10.1016/j.molimm.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Alston TA, Mela L, Bright HJ. 3-Nitropropionate, the toxic substance of Indigofera, is a suicide inactivator of succinate dehydrogenase. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(9):3767–3771. doi: 10.1073/pnas.74.9.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutman M. Modulation of mitochondrial succinate dehydrogenase activity, mechanism and function. Mol Cell Biochem. 1978;20(1):41–60. doi: 10.1007/BF00229453. [DOI] [PubMed] [Google Scholar]
- 34.Pan H, Wu X. Hypoxia attenuates inflammatory mediators production induced by Acanthamoeba via Toll-like receptor 4 signaling in human corneal epithelial cells. Biochemical and biophysical research communications. 2012;420(3):685–691. doi: 10.1016/j.bbrc.2012.03.069. [DOI] [PubMed] [Google Scholar]
- 35.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell host & microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440(7081):233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 37.Sokolovska A, Becker CE, Ip WK, Rathinam VA, Brudner M, Paquette N, et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nature immunology. 2013;14(6):543–553. doi: 10.1038/ni.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell. 2004;13(6):805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol. 2006;26(13):4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang YL, Morales-Rosado J, Ray J, Myers TG, Kho T, Lu M, et al. Toll-like receptor agonists promote prolonged triglyceride storage in macrophages. The Journal of biological chemistry. 2014;289(5):3001–3012. doi: 10.1074/jbc.M113.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng J, Sun X, Wang P, Zhang S, Wang X, Wu H, et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nature immunology. 2015;16(11):1142–1152. doi: 10.1038/ni.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267(5206):2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 43.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440(7081):237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 44.Drutman SB, Trombetta ES. Dendritic cells continue to capture and present antigens after maturation in vivo. J Immunol. 2010;185(4):2140–2146. doi: 10.4049/jimmunol.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304(5673):1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 46.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Analytical biochemistry. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 47.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nature protocols. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 48.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods in cell biology. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 49.Vives-Bauza C, Yang L, Manfredi G. Assay of mitochondrial ATP synthesis in animal cells and tissues. Methods in cell biology. 2007;80:155–171. doi: 10.1016/S0091-679X(06)80007-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.