Abstract
Lymph node (LN) stromal cells, particularly fibroblastic reticular cells (FRCs), provide critical structural support and regulate immunity, tolerance and transport properties of LNs. In many tumors, LN metastasis is predictive of poor prognosis. However, the stromal contribution to the evolving microenvironment of tumor draining LNs (TDLNs) remains poorly understood. Here we show that FRCs specifically of TDLNs proliferate in response to tumor-derived cues and that the network they form is remodeled. Comparative transcriptional analysis of non-draining and TDLN FRCs demonstrated reprogramming of key pathways including matrix remodeling, chemokine/cytokine signaling and immune functions including leukocyte recruitment, migration and activation. In particular, downregulation of FRC-derived CCL21 and IL-7 were accompanied by altered immune composition and aberrant localization. These data imply that stroma of TDLNs adapt on multiple levels, following exposure to tumor-derived factors, to exhibit features typically associated with immune suppression.
Introduction
LNs form an integral part of our immune system, acting as “filters” to survey potential lymph-borne pathogens, and as immunological hubs maintaining homeostasis or eliciting effective immune responses. To enable these functions, LNs are organized into discrete cellular compartments, which the supporting stromal cells are central to1–3.
Lymph draining from peripheral tissues enters via afferent lymphatic vessels and along the subcapsular sinus4, but small constituents such as chemokines and soluble antigen <70kDa cross the sinus floor and penetrate deeper into the LN, along narrow conduits formed by FRCs5–7. While this conduit network provides structural support4, FRCs have additional properties vital for LN function. They produce a number of cues that are critical for immune cell migration and localization such as CCL19 and CCL218, whose receptor CCR7 is present on naïve T cells, B cells and Dendritic Cells (DCs)9. Gradients of these chemokines direct intranodal migration during homeostasis and infection3, 10, 11, lymphocyte homing12 and mediate interactions between T cells and DCs8. FRCs are a major source of IL-7, essential for naïve T cell survival2. Recently, evidence indicates that FRCs also contribute to immunological tolerance via deletion of self-reactive T cells13–15.
LNs also feature in numerous pathologies. Many viruses specifically target FRCs for destruction to avoid detection during infection16. In cancer, LNs represent the first site of metastasis17, 18. Yet, despite our increasing understanding of LN metastasis19, mechanisms underlying the failure of effective anti-tumor immune responses in the TDLN remain poorly characterized. Tumor-derived factors drain to downstream LNs, bathing the cells encountered en route. Therefore, the potential exists for tumors to exploit similar communication to remotely control tissues such as the LN to its advantage. Given our current knowledge of stromal cells in LN function, we sought to determine responses of FRCs in TDLNs. Analysis of stromal populations demonstrated that FRCs, specifically within TDLNs, undergo structural remodeling and transcriptional modifications that correlate with gross alterations in cellular composition and localization. These observations imply that aberrant stromal cues impact the downstream structure and function of TDLNs.
Results
TDLN stromal networks expand and remodel
To study the role of stromal cells of TDLNs, prior to the onset of metastasis, we utilized a well-established B16.F10 melanoma model in which draining LNs were assessed over 14 days. Tumor-draining brachial LNs were significantly enlarged at pre-metastatic time points (Fig. 1a-c and Supplementary Fig. 1a). In contrast, LNs of PBS-injected control mice (non-draining lymph nodes; NDLNs) remained at a constant size (Fig. 1c). The absence of metastatic tumor cells was confirmed by qRT-PCR of Tyrp1 and Dct mRNA in total LNs (Supplementary Fig 1b). Increases in cellularity were confined to sentinel LNs, as adjacent LNs did not increase in size (Supplementary Fig. 1c).
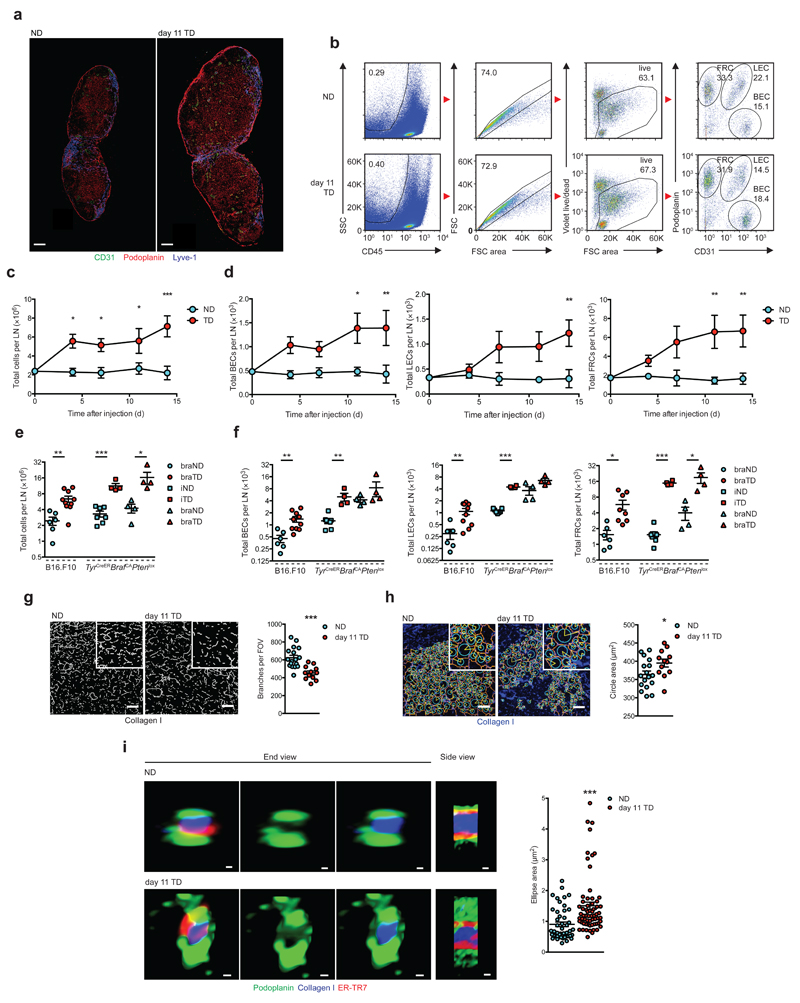
Figure 1.
LN expansion and FRC remodeling. (a) Confocal images of stromal populations in NDLNs and day 11 TDLNs. b) Flow cytometry gating strategy in NDLNs (top) and day 11 TDLNs (bottom). Stromal subtypes are gated on CD45-, live, singlets (numbers in box indicate percent of parent). Right panel; numbers in outlined areas denote percent of Podoplanin+CD31- FRCs (top left), Podoplanin+CD31+ LECs (top right) and Podoplanin-CD31+ BECs (bottom right). (c,d) Flow cytometry measurement of total LN cells (c), BECs, LECs, and FRCs (d). Total LN cells (e) and stromal cells (f) from B16.F10 (circles), TyrCreERBrafCAPtenlox brachial LNs (braLNs, squares) and inguinal LNs (iLNs, triangles). (g) Skeleton analysis, (h) Gap analysis of collagen I networks. (i) Confocal Airyscans of conduit end and side views (left) and quantification (right). Scale bars (a) 200 μm, (g,h) 50 μm, (i) 0.256 μm (NDLN side), 0.441 (NDLN end), 0.363 μm (TDLN side), 0.549 μm (TDLN end). Each symbol represents an individual LN (e,f); field of view (g,h); conduit cross section (i). Small horizontal lines indicate mean ± s.e.m. *P <0.05, **P <0.01 and ***P <0.001 (two-way ANOVA (c,d) or two-tailed unpaired t-test (e-i)). Data are representative of two independent experiments with n=3 NDLNs and n=5 TDLNs in C57BL/6 female mice (c,d; mean and s.e.m.), for B16.F10, n=6 NDLNs and n=10 TDLNs in female C57BL/6 mice and for TyrCreERBrafCAPtenlox mice n=7 iNDLNs, n=4 iTDLNs n=4 braNDLNs and n=4 braTDLNs (equal male:female ratio, e,f); or three independent experiments in female C57BL/6 mice, n=6 NDLNs and n=4 TDLNs (g,h) with 3 FOV analyzed/LN, or n=3 NDLNs and n=4 TDLNs (i) with 5 conduits/LN and 3 measurements/conduit.
Stromal populations within TDLNs were identified by differential expression of podoplanin and CD31 among CD45- cells (gating strategy Fig. 1b). Over the period examined, significant proliferation of BECs, LECs and FRCs was recorded specifically within TDLNs (Fig. 1d). Consistent with B16.F10 tumors, TDLNs from TyrCreERBrafCAPtenlox mice were also enlarged (Fig. 1e). Mice develop melanoma at multiple sites following activation of constitutively active mutant BRaf, BRaf(V600E), and deletion of the tumor suppressor Pten. Enlargement of TDLNs in this model was also supported by increased stromal populations (Fig. 1f and Supplementary Fig. 1d). Proliferation of FRCs was confirmed by in vivo EdU labeling, whereas turnover of LECs and BECs remained in-line with NDLN counterparts (Supplementary Fig. 1e). In contrast, a significant increase in the ratio of FRCs to whole node cell counts after 11 days was recorded (Supplementary Fig. 2a) leading us to examine FRC network changes in more detail. Networks remained intact in TDLNs, with collagen I cores surrounded by ER-TR7 matrix and podoplanin+ FRCs (Supplementary Fig. 2b-d). Skeleton analysis, however, revealed fewer branches per field of view in TDLNs (Fig. 1g), whereas branch length (Supplementary Fig. 2e) and FRC cell size (data not shown) were unchanged. Furthermore, gap analysis highlighted greater distances between adjacent FRC networks in TDLNs (Fig. 1h). Evaluation of podoplanin-lined conduits using high power Airyscans revealed that conduit thickness was significantly enlarged (Fig. 1i) and highlighted physical changes of individual conduits from TDLNs compared with NDLN counterparts (Fig. 1i and Supplementary Fig. 2d). Together, these data imply that conduits of TDLNs enlarge rather than increase in frequency, and hence increasing FRC numbers might be required to provide cellular coverage to support the growing conduit diameter.
FRCs of TDLNs are transcriptionally modified
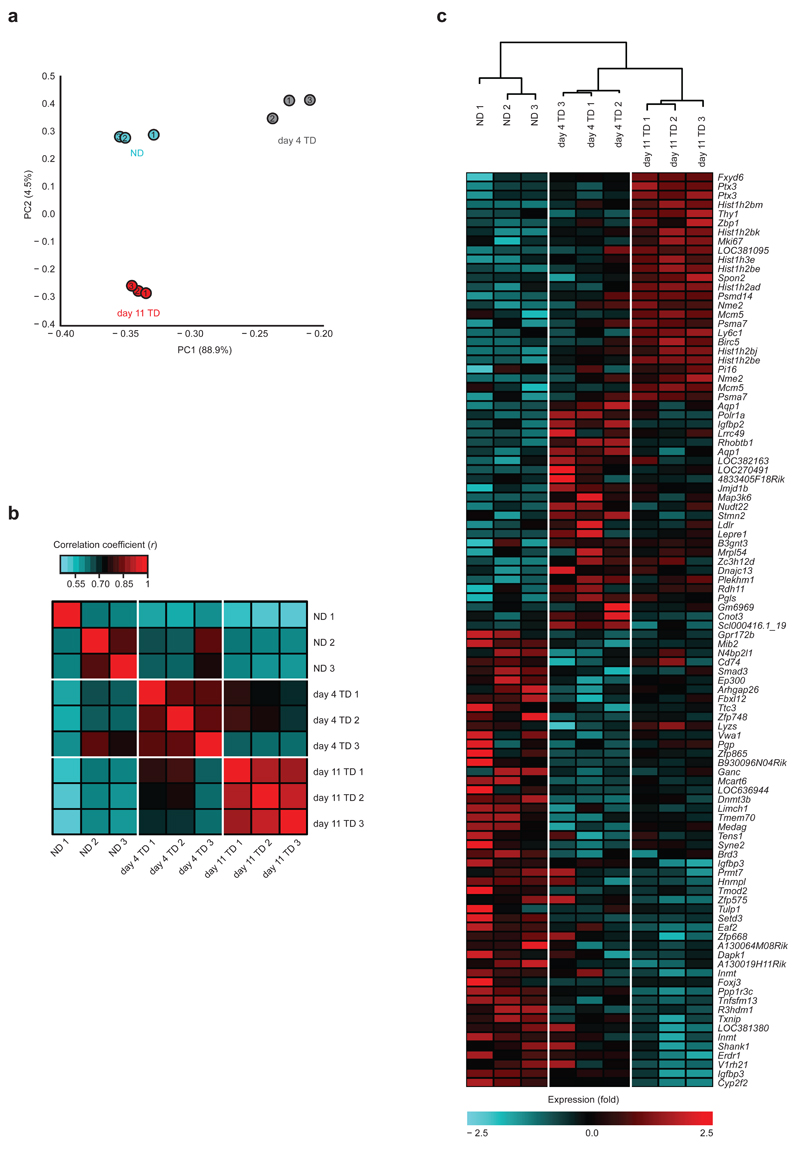
Considering these structural alterations, we next sought to identify how FRCs of TDLNs adapt to the evolving microenvironment, and the consequences of such changes. FRCs from LNs at days 4 and 11 post B16.F10 inoculation were sorted and subjected to whole genome transcriptional profiling. At day 4, tumors were barely palpable thus this time point was chosen to represent an early stage of development, where communication between lymphatics and TDLNs are likely at their earliest stages. In contrast, large day 11 tumors with established stroma and LN connections represent the late stage of LN transformation. Inter-replicate coefficients of variation confirmed consistency between samples, with means of 0.036, 0.037, and 0.035 for NDLNs, day 4 and day 11 TDLNs respectively. Applying a cutoff of a change in expression of 1.5-fold to the gene array data revealed distinct, significant changes between FRCs from TDLNs and NDLNs (Supplementary Fig. 2f-h). We initially calculated principal components (PC). When plotted, eigenvalues of these (Supplementary Fig. 2i) highlight that the majority of variance (88.9% and 4.5% respectively) is contained within the first two components, which partition into their respective sample types (Fig. 2a). PC1 separates NDLNs and day 4 TDLNs effectively, and PC2 separates NDLNs and day 11 TDLNs. Correlation matrix plots supported this relationship (Fig. 2b), showing strong association within all datasets. This was further confirmed by hierarchical clustering showing strong association within all datasets (Fig. 2c). The heat map, linked to hierarchical clusters (Fig. 2c, bottom) revealed that clusters of samples (NDLNs, day 4 TDLNs, and day 11 TDLNs) exhibit the same pattern of gene expression changes within their groups. These data demonstrate that gene expression profiles for day 4 TDLNs and day 11 TDLNs are distinct and replicable, and implies that FRCs undergo a gradual reprogramming response after exposure to tumor factors with day 4 representing a distinct and transitional state, rather than a weaker profile of day 11 TDLNs.
Figure 2.
Statistical analysis of microarray results. (a) Principle Component Analysis of NDLNs, day 4 TDLNs, and day 11 TDLNs. (b) Heat map of coefficients of correlation for most significantly deregulated genes. Red indicates the highest correlation. (c) Hierarchical clustering analysis of all samples with heat map of the top deregulated gene probes. All analyses were performed on genes applying a cutoff of a change in expression of 1.5-fold and a P value <0.05. Each symbol (a) or column (b,c) represent transcriptomes of FRCs from pooled from 2 brachial LNs per mouse. Data are representative of one experiment with n=3 C57BL/6 female mice per group.
Identification of pathways deregulated in FRCs
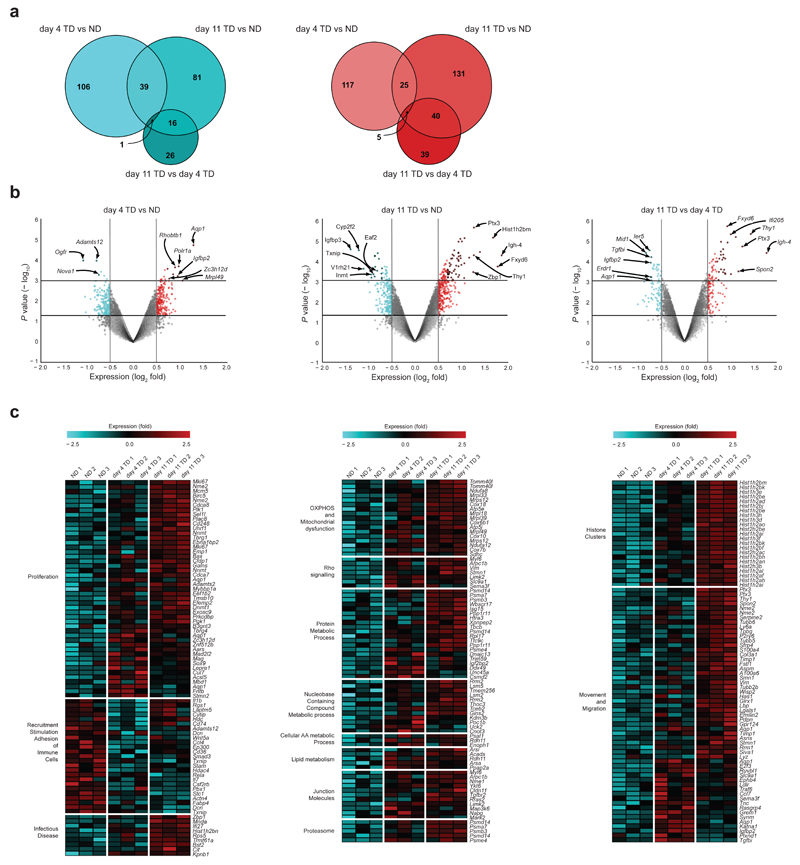
Significant overall changes in expression of 1.5-fold were observed between conditions (Supplementary Fig. 3a). Differential gene expression between day 4 and day 11 illustrated transient increases or decreases, which likely represent early activation or repression of FRC signaling pathways. These either return to control levels by 11 days or continue to be further up or downregulated. While expression levels within the array differ over time, the number of altered genes remains comparable. 106 genes were downregulated at day 4, and 81 at day 11 with an overlap (i.e. genes that similarly upregulated in both) of 39 (Fig. 3a), whereas 117 were upregulated at day 4, 131 at day 11 with an overlap of 25 genes (Fig. 3a). Volcano plots show the highest deregulated genes between day 4 and day 11 in TDLNs vs. NDLNs as ranked by their expression (Fig. 3b). Aqp1 is among the top upregulated genes after 4 days (Fig. 3b), but by 11 days returns to baseline (Fig. 3b). In contrast, Fxyd6, Igh4, Thy1 and Ptx3 are among the top upregulated genes at day 11 (Fig. 3b), indicating a unique late stage signature. Clustering of the top deregulated genes into functional groups highlighted differences in proliferation, metabolism, mitochondrial function, movement and migration, and cell junction pathways (Fig. 3c). Associated gene functions were collated using common pathways of deregulated genes in day 4 and day 11 TDLNs, generated by Gene Set Enrichment Analysis (GSEA) and ingenuity pathway analysis (IPA; also Supplementary Fig. 3). These data illustrate that as tumors develop, FRCs specifically within TDLNs respond via transcriptionally altering signaling pathways key to their behavior and function.
Figure 3.
Identification of specific genes and pathways deregulated in TDLN FRCs. (a) Venn diagrams displaying overlap between significantly downregulated and upregulated genes in NDLNs, day 4 TDLNs, and day 11 TDLNs. (b) Volcano plots of significantly deregulated genes for day 4 TDLNs vs. NDLNs (left), day 11 TDLNs vs. NDLNs (middle), and day 11 TDLNs vs. day 4 TDLNs (right) applying a cutoff of a change in expression of 1.5-fold. Downregulated genes (blue), upregulated genes (red). (c) Heatmaps of key pathways involving top deregulated genes compared between day 4 TDLNs vs. NDLNs and day 11 TDLNs vs. NDLNs. Functional groups were assigned with GSEA and IPA. Horizontal lines indicate P <0.05 or P <0.01 (b). Each data point represents transcriptomes of FRCs of 2 brachial LNs, pooled per female mouse.
FRC chemokine/cytokine signaling impacts immune composition
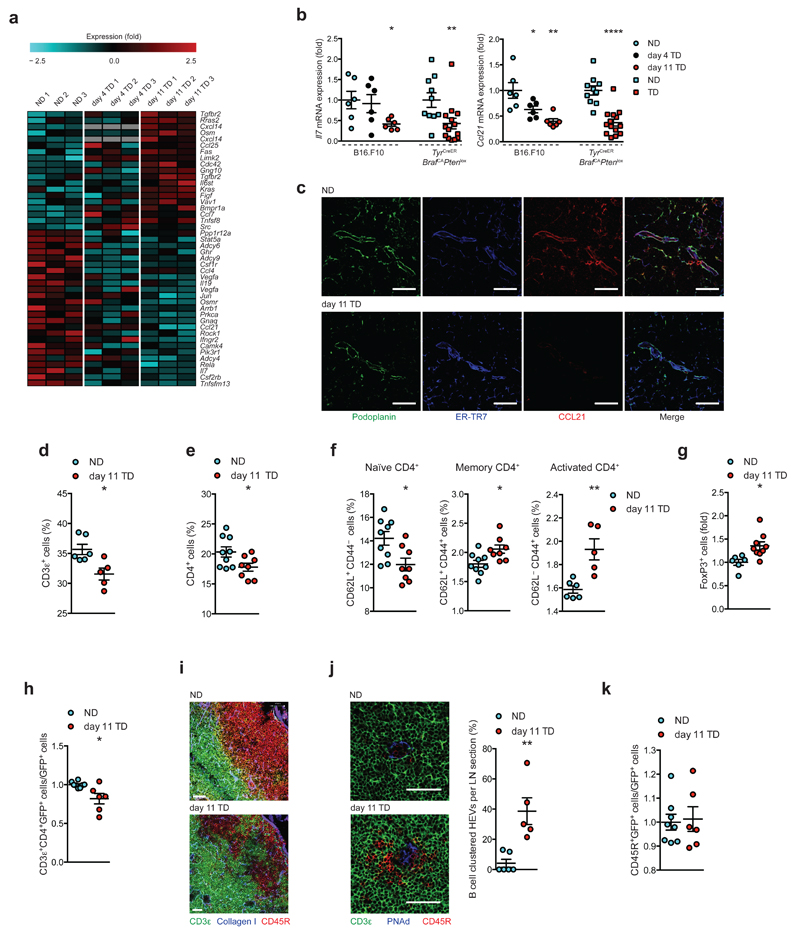
FRCs provide chemical cues necessary for immune homeostasis, leukocyte trafficking and survival within the LN20. Both GSEA and IPA analyses identified significant deregulation in chemokine and cytokine production by TDLN FRCs (Fig. 4a). While signaling molecules such as Limk2, Kras, Tgfbr2 and Src were upregulated, cytokines and chemokines including Il19, Il7, Ccl4 and Ccl21 were downregulated. As FRCs provide the bulk of CCL21 and IL-72, 8, 12, mRNA levels were verified in independent sample sets by qRT-PCR confirming significant downregulation in TDLN FRCs from both tumor models examined (Fig. 4b). Confocal imaging confirmed this at the protein level (Fig. 4c). Reduced T cell areas and concurrent increases in B cell follicle sizes per TDLN were measured (Supplementary Fig. 4a), and corresponding reductions in CD3ε+ cellularity were confirmed by flow cytometry (Fig. 4d). Although no change in CD8α+ T cells were measured (Supplementary Fig. 4b), a significant reduction in the percentage of CD4+ T cells was observed in TDLNs after 11 days (Fig. 4e). Within this population, the percentage of naïve CD4+CD62L+CD44- T cells decreased (Fig. 4f). This was accompanied by significant increases in memory (CD62L+CD44+), activated (CD62L-CD44+) CD4+ T cells (Fig. 4f) and CD4+FoxP3+ regulatory T cells (Fig. 4g and Supplementary Fig. 4c). Moreover, we observed impaired homing efficiency of CD4+ T cells into day 11 TDLNs (Fig. 4h). Upon examining the cellular architecture of TDLNs we observed mislocalization and disorganization of major immune cell populations. In contrast to NDLNs where T and B cell zones were clearly delineated (Supplementary Fig. 4d, left panel), TDLNs exhibited integration and loss of this margin (Supplementary Fig. 4d, right panel, and Fig. 4i). A transitional stage was observed in day 4 TDLNs (Supplementary Fig. 4d, middle panel). Furthermore, B cells clustered around high endothelial venules of TDLNs (HEV, Fig. 4j), but no differences in the homing capacity of B cells within adoptively transferred splenocytes were measured (Fig. 4k). The accumulation of B cells around HEVs specifically within TDLNs in the absence of homing defects suggests B cell migration and localization is disrupted once exited from HEVs. Moreover, consistent with the fall in IL-7 (Fig. 4b) in vivo EdU-labeling showed reduced proliferation of T and B cells in TDLNs (Supplementary Fig. 4e,f).
Figure 4.
Perturbation in chemokine/cytokine signaling alters immune cell composition and localization. (a) Heatmap of cytokine and chemokine signaling genes. (b) qRT-PCR validation of Il7 and Ccl21 expression in independent samples from B16.F10 (circles) and TyrCreERBrafCAPtenlox (squares). (c) Confocal images of NDLNs (top) and day 11 TDLNs (bottom) stained for Podoplanin (green), ER-TR7 (blue) and CCL21 (red). (d) Flow cytometric quantification of T cells (CD45+CD3ε+) as percentage of singlets (B16.F10). (e) Quantification of CD4+ T cells (CD45+CD3ε+CD4+), (f) naïve CD62L+CD44-, memory CD62L+CD44+, activated CD62L-CD44+, and (g) regulatory T cells (CD45+CD3ε+CD4+Foxp3+). (h) LN homing assay: flow cytometry quantification of homed CD4+GFP+ splenocytes in C57BL/6 recipients. (i) Confocal images of B cell follicles in NDLNs and day 11 TDLNs stained for CD3ε (green), CD45R (red) and collagen I (blue). (j) Confocal images of NDLNs and day 11 TDLNs stained for CD3ε (green) CD45R (red) and PNAd (blue; left), and quantification of clustering (right). (k) LN homed B cells: flow cytometry quantification of homed CD45R+GFP+ splenocytes. Scales bars (c and j) 50 μm, (i) 51 μm (NDLNs), 38 μm (TDLNs). Each symbol represents an individual LN (b,d,e-g,h-k). Small horizontal lines indicate mean ± s.e.m. *P <0.05, **P <0.01 and ****P <0.0001 (one-way ANOVA (b, B16.F10) or two-tailed unpaired t-test (b,d-h,j,k,)). Data represent one experiment in technical duplicates, n=3 per condition in C57BL/6 female mice for B16.F10, and n=5 NDLNs (3 female, 2 male) or n=7 TDLNs (4 female, 3 male) for TyrCreERBrafCAPtenlox (b); or two independent experiments in C57BL/6 female mice.n=6 NDLNs and n=5 TDLNs (d), n=9 NDLNs and n=8 TDLNs (e), n=9 NDLNs and n=8 TDLNs (naïve and memory), n=6 NDLNs and n=6 TDLNs (activated) (f), n=6 NDLNs and n=9 TDLNs (g), n=8 NDLNs and n=6 TDLNs (h,k), n=6 NDLNs and n=5 TDLNs (j).
Other deregulated factors at the mRNA level included Cxcl14, chemotactic to monocytes and DCs; Ccl25, chemotactic to DCs; and Ccl7, chemotactic to monocytes; all of which were upregulated in day 11 TDLNs. Consistent with this, and following a transient dip at day 4, CD11c+ DCs and CD11b+ Macrophages (MΦ, Supplementary Fig. 5a) were significantly increased in numbers (Supplementary Fig. 5b,c). Together, these data suggest that tumor-driven perturbation of FRC-derived guidance cues modifies both the composition but also the localization of key immune populations within TDLNs.
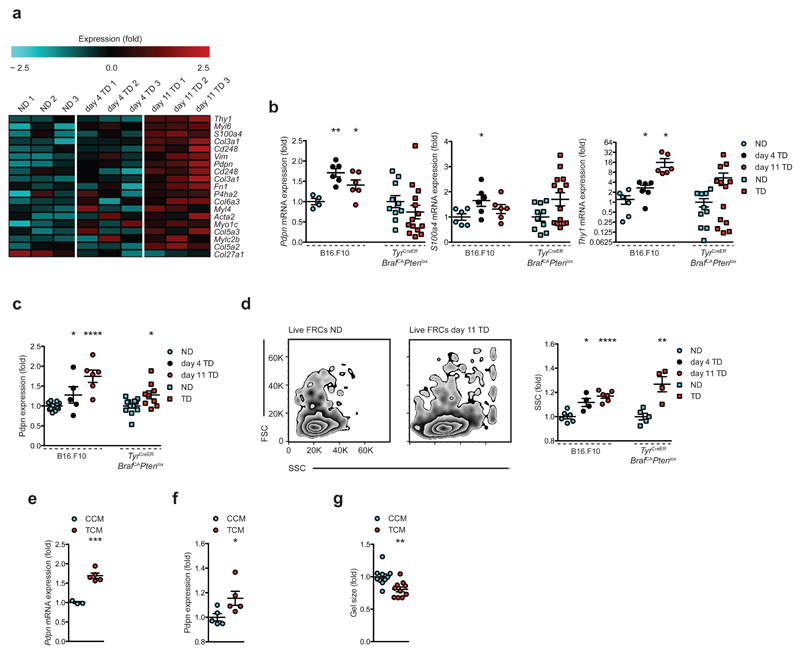
FRCs of TDLNs are more activated
Fibrosis at the primary tumor21, mediated by hyper-activated fibroblasts (cancer-associated fibroblasts, CAFs) recruited and educated in the local microenvironment provides pro-tumor support22. We therefore sought to determine if fibroblasts of pre-metastatic TDLNs undergo similar changes. Transcriptional profiling showed increased expression of genes encoding typical activation markers Pdpn, Fn1, Cd248, Acta2, S100a4, Vim, Myl and Col families (Fig. 5a) indicating increased activation of FRCs in TDLNs. Pdpn, S100a4, Thy1 and Cd248 were further verified on independent data sets (Fig. 5b and Supplementary Fig. 5d). These trends were largely supported in the genetic model, and protein expression of podoplanin was significantly increased in both systems (Fig. 5c). Moreover, flow cytometry showed tumor-draining FRCs increase in granularity, indicative of increased internal complexity and corresponding activation status (Fig. 5d). To investigate the activation status further in vitro, cultured FRCs treated with tumor conditioned medium (TCM) obtained from B16.F10 cells were compared to control-conditioned medium (CCM)-treated cells. Podoplanin was upregulated at both mRNA (Fig. 5e) and protein levels (Fig. 5f), and TCM treatment enhanced the capacity of FRCs to contract collagen gels (Fig. 5g and Supplementary Fig. 5e). Thus, exposure to tumor-derived factors at the TDLN may push FRCs towards an activated, pro-fibrotic ‘CAF-like’ state.
Figure 5.
FRCs in TDLNs are more activated. (a) Heatmap of genes related to fibroblast activation. (b) qRT-PCR validation of Pdpn, S100a4 and Thy1 in independent FRC samples from B16.F10 (circles) and TyrCreERBrafCAPtenlox (squares). (c) Flow cytometric analysis of podoplanin expression from B16.F10 and TyrCreERBrafCAPtenlox. (d) Flow cytometry scatter profiles of live FRCs from NDLNs vs. day 11 TDLNs (left), quantification of geometric mean of side scatter (right). (e) mRNA and (f) protein expression of podoplanin and (g) collagen gel contractile activity of FRCs treated with control conditioned medium (CCM) or tumor conditioned medium (TCM) in vitro. Each symbol represents an individual LN (b-d) or biological replicate (e-g). Small horizontal lines indicate mean ± s.e.m. *P <0.05, **P <0.01, ***P <0.001 and ****P <0.0001 (one-way ANOVA with Tukeys post-hoc (b-d B16.F10) or two-tailed unpaired t-test (b-g)). Data represent one experiment in technical duplicates, n=3 per condition in C57BL/6 female mice for B16.F10, and n=5 NDLNs (3 female, 2 male) or n=7 TDLNs (4 female, 3 male) for TyrCreERBrafCAPtenlox (b); or two independent experiments in C57BL/6 female mice n=14 NDLNs, n=5 day 4 and n=6 day 11 TDLNs for B16.F10, and n=12 NDLNs (6 female, 6 male) and n=9 TDLNs (5 female, 4 male) for TyrCreERBrafCAPtenlox (c), n=6 NDLNs, n=4 day 4 and n=6 day 11 TDLNs for B16.F10, and n=5 NDLNs (3 female, 2 male) and n=4 TDLNs (2 female, 2 male) for TyrCreERBrafCAPtenlox (d); or three independent experiments n=3 CCM, n=5 TCM(e), n=5 each (f), n=10 each (g).
TDLN conduits allow enhanced solute transport
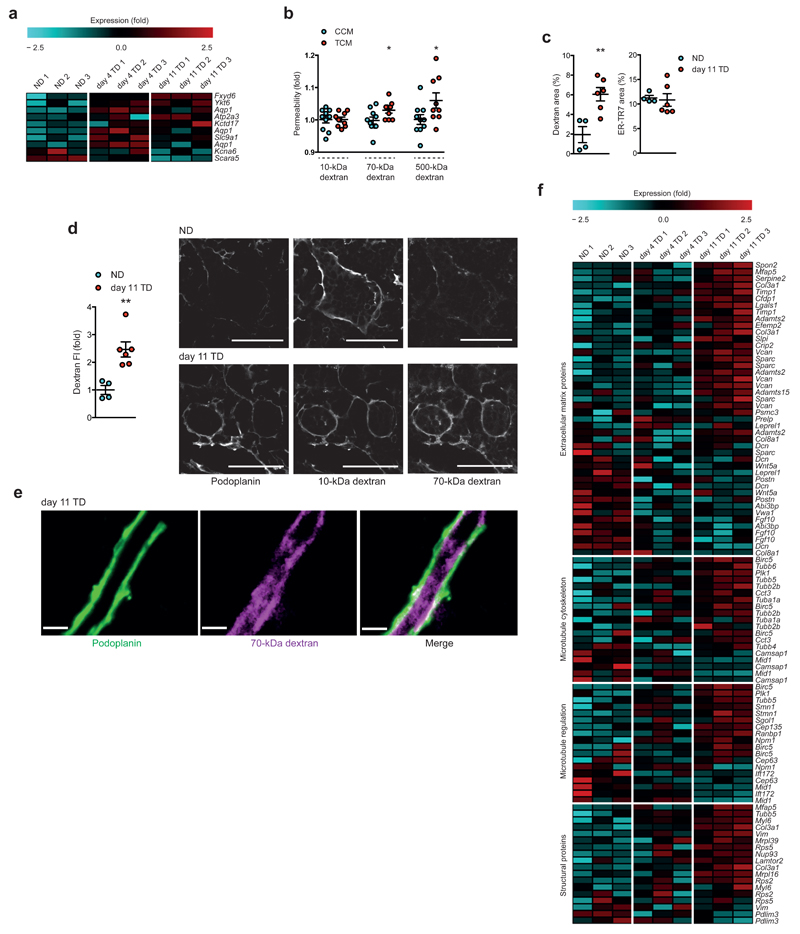
Profiling of TDLN FRCs also hinted at previously undocumented behavior, in particular, a number of channels and ion transporters are deregulated in day 4 or day 11 TDLNs (Fig. 6a). For example, Aqp1 is highly upregulated in FRCs of day 4 TDLNs, before downregulation at day 11 (verified by qRT-PCR in Supplementary Fig. 5f). To investigate if changes to channels or ion transporters can impact fluid transport through the conduit system, we first utilized an FRC culture model to measure dextran passage in vitro. While no changes in the transport of 10-kDa dextran were measured, significantly greater transit of 70-kDa and 500-kDa dextran were observed following exposure to TCM indicating a less selective barrier (Fig. 6b). In vivo, dextran transport into size-restricted conduits was analyzed and measured in control or tumor-bearing mice. In both NDLNs and TDLNs, 10-kDa dextran freely entered into conduits. In contrast, quantification of 70-kDa dextran revealed transport further into paracortical areas of day 11 TDLNs (Fig. 6c, as a comparison, ER-TR7 coverage did not change; and Fig. 6d), where it was restricted to the FRC-lined conduits (Fig. 6e). 500-kDa was observed beyond the subcapsular sinus in TDLNs, but was undetectable in the paracortical area (data not shown).
Figure 6.
Modified transporter repertoires within TDLN FRCs translate to altered solute transport throughout the conduit system. (a) Heatmap of genes related to ion/solute conduction or membrane permeability. (b) In vitro measurement of relative permeability of 10, 70 and 500-kDa dextran transport through CCM- or TCM-treated FRC monolayers. (c) Quantification of 70-kDa dextran (left) and ER-TR7 (right) area fraction/cortical area in NDLNs and day 11 TDLNs. (d) Quantification of 70-kDa dextran as relative fluorescence intensity/area (FI; left) and confocal image of 10-kDa, 70-kDa and podoplanin in NDLNs and day 11 TDLNs. Each channel is gray scaled. (e) High-power confocal image of dextran-filled conduit from a day 11 TDLN. (f) Network analysis of top deregulated genes for day 11 TDLNs. Top networks are shown as heat maps of the genes for each time point. Scale bars (d) 50 μm (e) 3.4 μm. Each symbol represents an individual replicate (b) or LN (c,d). Small horizontal lines indicate mean ± s.e.m. *P <0.05, **P <0.01 (two-tailed unpaired t-test (b-d). Data are representative of two independent experiments performed in quintuplicate (b); or one experiment, n=4 NDLNs and n=6 TDLNs in C57BL/6 female mice (c,d).
As a result of the altered TDLN environment, significant changes in cell assembly machinery would be expected to underlie the restructuring and enlargement of the FRC networks, along with the need for enhanced interaction between FRCs and associated matrix proteins essential to the conduit. Thickened collagen cores but reduced branches that were seen suggests the increased numbers of FRCs in TDLNs supports the increased diameter of the conduit. In doing so, FRCs will form contacts with a larger number of, and encounter a greater area of neighboring cells. This was confirmed by performing network analysis. Such analyses cluster gene sets into functionally and spatially linked networks, highlighting families of genes that are both significantly deregulated and involved in the same biological pathways, expressed together, or that physically interact. In the case of TDLNs, four significantly relevant gene groups heavily involved in cell structure, shape and extracellular matrix were interlinked. (Fig. 6f, with interaction networks shown in Supplementary Fig. 6). Taken together with earlier data showing larger diameter conduits at day 11 (Fig. 1g), altered transporter repertoires point to a perturbation of conduit capacity, whereby conduits of TDLNs are more permissive for fluid entry and transit, potentially enabling greater penetration of soluble tumor-derived factors to deeper areas of the LN.
Discussion
Lymph nodes function as a major immunological hub, essential for immune homeostasis and generation of appropriate immune responses, yet LNs are also the first site of metastasis for many cancers that manage to avoid immune-mediated clearance. It is increasingly accepted that LNs receive and respond to tumor-derived signals generating a pro-tumor niche, but it remains unclear as to how these responses manifest and what in the LN drives them. The stromal populations of the LN not only provide structural support but are essential to its maintenance and physiological function1–3, 6, 8–12, 16, 23–28. While studies show tumor-derived factors contribute to LN lymphangiogenesis29, 30, 31, the LN fibroblasts, FRCs, and the conduit network they form have not been thoroughly investigated in the context of the tumor. Here we describe that proliferation, remodeling and transcriptional reprogramming of FRCs occurs in TDLNs. This in turn impacts FRC-driven chemokine signaling, trafficking events, immune localization and transport, all of which have the potential to contribute to impaired function within TDLNs.
Consistent with previous studies, TDLNs enlarged29, 31, 32, and was supported by expansion and structural reorganization of stromal compartments. Beyond gross network remodeling, FRCs specifically within TDLNs displayed strikingly altered transcriptional profiles. Key to LN function, FRC-derived CCL21 and IL-7 were downregulated. Perturbation of these essential guidance cues can contribute to abnormal immune homing, localization and survival. It has been previously reported that TDLNs exhibit reduced CCL2133, 34. Consistent with these findings we observed smaller T cell zones, accumulation of B cells around HEVs, a loss of the B-T cell zone boundary, and frequently observed T cells within B cell follicles. These features phenocopy plt/plt mice, where spontaneous loss of LN-specific CCL19 and CCL21 isoforms translate to fewer T cells and impaired immune responses2, 12, 26, 35. Our results also draw parallels with other pathological states such as infection, where reduced LN CCL21, and disruption to FRC networks underpin aberrant homing and mislocalization of leukocytes to support immune evasion by Salmonella or virus particles16, 27, 36. Thus we speculate that similar functional impairments to immune responses may exist in TDLNs where FRC networks and guidance cues are disrupted. This, however, remains to be determined. Moreover, our B cell observations are consistent with recent reports that have demonstrated accumulation of B cells in TDLNs. Although the activation status or subtypes of the B cells occupying TDLNs remains to be determined, it is possible that they may function as regulatory B cells adding a further dimension to the local immune suppressive environment. It should also be noted that pathogen-related inflammation didn’t underlie our observations. Firstly, stromal modifications were consistent in two independent melanoma models, one of which is genetically driven rather than allografted. Secondly, a comparison of our array with data from Malhotra et al., in which responses of FRCs to LPS-mediated inflammation were characterized20, shows that the response of the FRCs in TDLNs is tumor-dependent. In particular, key factors such as CCL21 and IL7 were inversely regulated between the two pathological settings (data not shown).
Consistent with previous studies on LN stroma, FRCs of TDLNs acquired a more activated status37, 38. This gene signature in particular is reminiscent of fibroblasts found within the tumor microenvironment. These hyper-activated fibroblasts modulate ECM to support a tumor, and are now emerging as key immune modulatory intermediates. CAFs produce a host of factors that recruit immune populations including myeloid-derived cells and Tregs to the tumor, but also promote their polarization to a more suppressive phenotype39–43. Having observed accumulation of similar populations in TDLNs it cannot be excluded that following sustained exposure to tumor-derived factors, FRCs of TDLNs adopt a more CAF-like state to generate a supportive, immune suppressive niche.
Within TDLNs, wider conduits and enhanced collagen deposition point to increased stiffness of the node32, but the remodeling of the collagen core may also contribute to the size exclusion properties of the conduits5, 6, that in TDLNs is disrupted. This, combined with deregulated junction properties and protein pores of the FRCs lining these channels, suggests an altered integrity of the conduit network. These changes have the potential to lead to rapid, but poorly controlled delivery of tumor-derived factors, debris and antigen to the deeper areas of the LN upsetting the functional status quo. Moreover, the process of lymphangiogenesis both at primary tumors and connected LNs enhances the drainage capacity, and consequently these stromal populations experience raised fluid flux and shear stresses. Mechanical cues such as these rather than chemical, tumor-derived signals (data not shown) may also act as a stimulus for FRC proliferation44–47 or synergize to drive the transcriptional reprogramming. We have not excluded this in the present study, but this avenue warrants more in depth investigation using in vitro studies in which the effects of biophysical stimuli i.e. flow can be isolated.
In summary, using functional assays and comparative transcriptome analysis of FRCs in NDLNs and TDLNs from multiple tumor models, we demonstrate that FRCs immediately downstream of tumors acquire unique transcriptional programs. Together with structural remodeling, these deregulated pathways and adapted FRC traits contribute to modified immune composition and aberrant localization that may ultimately translate to a more suppressive, pro-tumor environment.
Online Methods
Animal Experiments
All experiments involving mice were performed in accordance with UK Home Office regulations under Project License PPL 80/2574. For mouse models, G*Power was used to estimate samples sizes required to achieve 80% power with 5% threshold. For syngeneic tumors, 2.5 × 105 B16.F10 melanoma cells were inoculated subcutaneously into the shoulder region of immune competent 8-9 week old female C57BL/6 mice assigned to control or tumor groups randomly. Animals were only excluded if tumors failed to form or if health cencerns were reported. Control mice were injected with 50 μl PBS. Tumor size was monitored with calipers and the volume was calculated based on the ellipsoid formula π/6 × (length × width2). Animals were culled after 4, 7, 11 or 14 days and tumor/PBS draining (brachial) and non-draining (axillary) lymph nodes were isolated for subsequent analysis. For EdU in vivo proliferation assays, mice received 200 μl of 500 μg/ml 5-ethynyl-2’-deoxyuridine (EdU, Thermo Scientific) in PBS via intra peritoneal injection 24 h before tumor cell injection and every 48 h thereafter. LNs were isolated at day 4, 7, and 11 after tumor cell injection and analyzed by flow cytometry. For dextran transport assays, day 11 tumor-bearing mice received lysine fixable dextrans 10,000 MW (Texas Red) 70,000 MW (biotin) and 500,000 MW (FITC; all Life Technologies; at a final concentration of 5 mg/ml in PBS) via subcutaneous injection into the forelimb. 10 min later brachial lymph nodes were harvested and fixed in 4% PFA overnight before transferring into a 30% sucrose solution and preserving in OCT (TissueTek). For homing assays, splenocytes were isolated from C57BL/6-Tg(CAG-EGFP)131Osb/LeySopJ mice (Jackson Laboratory) and 5×106 cells were injected into the tail vein of wt C57BL/6 mice. After 18h LNs were isolated and immune cell contents were analyzed based on GFP+, CD8α, CD4, CD3ε, CD45 and CD45R staining (Table 1). For spontaneous murine melanoma, tumors were induced in 5-10 weeks old TyrCreERBrafCAPtenlox (B6.Cg-Braftm1Mmcm Ptentm1Hwu Tg(Tyr-cre/ERT2)13Bos/BosJ, Jackson Laboratory) mice48 following topical application of 4-hydroxy tamoxifen in DMSO (25 mg/ml) for 4 consecutive days to either shoulder or flank. Non-carrier littermates were used as controls. As for injected tumors, draining lymph nodes (inguinal and brachial LNs) were harvested for subsequent analysis.
Table 1.
Antibodies for Flow cytometry
Flow cytometry
Lymph nodes were mechanically disrupted and digested in a 500 μl mixture of 1 mg/ml collagenase A (Roche) and 0.4 mg/ml DNase I (Roche) in PBS at 37 °C for 30 min with 600 rpm rotation. Following centrifugation at 1000 rpm for 5 min the supernatant was discarded and replaced with 500 μl of PBS containing 1 mg/ml Collagenase D (Roche) and 0.4 mg/ml DNase I. The mixture returned to 37 °C for 20 min with 600 rpm rotation prior to addition of EDTA (final concentration 10 mM). Suspensions were passed through a 70 μm mesh prior to immunostaining with fixable viability dye live/dead violet (Molecular Probes) and combinations of fluorescently conjugated antibodies (Table 1). For intracellular staining FoxP3/Transcription Factor Fixation/Permeabilization Kit (eBioscience) guidelines were followed. For EdU proliferation assays, the Click-iT Flow Cytometry Assay Kit Alexa Fluor 488 (Thermo Scientific) was followed as per manufacturers guidelines. Flow cytometry was performed on CyAn ADP (Beckman Coulter) and LSR Fortessa (BD Biosciences) analyzers. Offline analysis was performed with FlowJo software (Treestar).
Cell processing post sorting for RNA analysis
For RNA processing, LNs cell suspensions were sorted on a High speed Influx Cell Sorter (100 μm nozzle, BD Biosciences) into RNA protect Cell Reagent (QIAGEN). RNA was isolated with the RNeasy plus micro Kit (QIAGEN) and RNA quality and quantity was analyzed with a Bioanalyzer (Agilent Technologies). Only RNA samples with a RIN value above 8 and total concentration of 100 pg/μl were further processed for whole transcriptome amplification via the Ovation PicoSL WTA V2 Kit (NuGEN). qRT-PCR was performed using TaqMan assays (Pdpn Mm01348912_g1, Il7 Mm01295803_m1, Ccl21a Mm03646971_gH, S100a4 (FSP1) Mm00803372_g1, Thy1 Mm00493682_g1, Aqp1 Mm00431834_m1, Tyrp1 Mm00453201_m1, Dct Mm01225584_m1) and a StepOne Real Time PCR System instrument (both Life Technologies).
Microarray hybridization and data normalization
RNA was assessed for concentration and quality using a SpectroStar (BMG Labtech) and a Bioanalyzer (Agilent Technologies). Microarray experiments were performed at Cambridge Genomic Services, University of Cambridge, using the MouseWG-6v2 Expression BeadChip (Illumina) according to the manufacturer’s instructions. Briefly, Total RNA was amplified using the Ovation Pico WTA V2 kit (NuGEN) and subsequently labeled using the Biotin Il kit (NuGEN). The concentration, purity and integrity of the resulting cRNA were measured using the Nanodrop ND-1000 (Thermo Scientific) and by Bioanalyzer. cRNA was then hybridized to the MouseWG-6 v2 Beadchip overnight followed by washing, staining and scanning using the Bead Array Reader (Illumina). Raw microarray data was preprocessed using the “lumi” bioconductor49 in R. After filtering for significance, data was transformed using the Variance Stabilization Transformation50 from lumi, and normalized using quantile normalization. The data discussed in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; GEO Series accession: GSE73728)
Assessment of metastasis
RNA was isolated from whole TDLNs as described above and qRT-PCR was performed using TaqMan assays (TYRP1 Mm00453201_m1 and DCT Mm01225584_m1) and a StepOne Real Time PCR System instrument (both Life Technologies). A standard curve was performed in parallel in which 1×106 lymph node cells in suspension were spiked with exact numbers of B16.F10 tumor cells (titrated by index FACS sorting from 100,000 down to a single tumor cell).
Principal component analysis
Principal Component Analysis (PCA) was performed using the R “stats” package. Log2 transformed data was used as input for the analysis. PCA was performed on all probes with a p-value > 0.05, and a fold change in either day 4 or day 11 > 1.5. Many-dimensional data from the gene array was reduced into linearly uncorrelated variables (Principal Components). The three largest principal components were then plotted against one another to assess the relationships between different biological repeats of each sample. PCA was visualized with the python library “matplotlib” (“matplotlib: v1.4.3”, and can be found on zenodo.org).
Correlational analysis
Correlational Analysis was performed using the R “stats” package on all probes with a p-value > 0.05, and a fold change in either day 4 or day 11 > 1.5.
Hierarchical clustering
Hierarchical clustering was performed for the probes with the greatest differences in expression (all of those with a p-value > 0.05, and a fold change in either day 4, or day 11 of greater than 1.5). Clustering was performed with the R “stats” package and utilizes the Euclidean distance between each sample, which are then clustered using the Ward algorithm.
Heat map analysis
For displaying differential probe expression values in heatmaps the TIGR MultiExperiment Viewer (TIGR MeV)51 tool for microarray data analysis was used. The data was row centered and normalized.
Analysis of functional enrichment in gene signatures
Microarray data was analyzed and interpreted with the help of Ingenuity Pathway Analysis (IPA, QIAGEN) and GSEA52, 53 (Broad Institute), focusing on canonical pathways and disease and biofunction (IPA) and KEGG canonical pathways (GSEA). Restricting the analysis for probes at a fold change of ± 1.5 and p value 0.05 obtained after raw data processing. Results were collated and displayed in heatmaps.
Cell Culture
B16.F10 (CRL 6475, ATCC) cells were maintained in DMEM with 10 % fetal bovine serum (Sigma Aldrich) and 100 U/ml penicillin-streptomycin (both Life Technologies). FRCs were maintained in RPMI (R875, Sigma Aldrich) with 10 % fetal bovine serum (Sigma Aldrich), 10 mM HEPES, 100 U/ml penicillin-streptomycin (both Life Technologies), 15 μM beta-mercaptoethanol (Sigma-Aldrich). For tumor cell conditioned medium (TCM) or control conditioned medium (CCM) production; B16.F10 cells or FRCs were grown until 50 % confluent in full growth medium, subsequently the medium was exchanged for RPMI-1640 containing 2 % FBS and medium was harvested after 24 h. After sterile filtration, medium was stored at -80 °C. For treatments TCM or CCM were mixed with 50 % full growth medium and performed over 7 days. RNA extraction was performed using RNeasy Mini Kit (QIAGEN). 1 μg of RNA was used for reverse transcription using First Strand cDNA synthesis Kit (Thermo Scientific) and qRT-PCR was performed as mentioned above.
Isolation and culture of FRCs
FRCs were isolated from murine lymph nodes of C57BL/6 as previously described54, 55. Cells were characterized based on their expression levels of PDPN and VCAM-1 and their lack of expression of CD45 and CD31.
In vitro contraction assay
After 7 days of preconditioning in either TCM or CCM, 1.5 × 105 FRCs were seeded into collagen gels at a final concentration of 2 mg/ml (Rat tail collagen, BD Biosciences) into 24 well plates. Following 20 min of polymerization at 37 °C, 500 μl of full medium was added and the gel was detached from culture dish using a pipette tip. Gels were allowed to contract for 24 h and the difference in size was quantified using ImageJ.
In vitro permeability assay
After 7 days of preconditioning in TCM or CCM, 6.5 × 104 FRCs were seeded onto PCF 0.4 μm pore transwell inserts (Millipore) and cultured overnight in full growth media. To ensure the presence of confluent monolayers, inserts were stained with CellMaskTM plasma membrane stain (Molecular Probes). Inserts were transferred to a fresh 24 well plate and 500 μl basal media was added to the lower chamber. 200 μl media containing 2 % FBS and dextrans at 10,000 MW (Cascade Blue) 70,000 MW (TRITC) and 500,000 MW (FITC; all Life technologies) at a final concentration of 1 mg/ml were added to the upper chamber and fluorescence was measured of the lower chambers’ media after 22h.
Immunofluorescence
Lymph nodes were embedded in OCT medium (TissueTek). 10 μm frozen sections were fixed in ice-cold acetone for 2 min, blocked with 5 % chicken serum, 2 % BSA in PBS, and incubated with the primary anti-mouse antibodies at 4 °C overnight (Table 2). For all stainings, Alexa Fluor secondary antibodies (Thermo Scientific) were used at a dilution of 1:300. Sections were mounted in ProLong Gold (Thermo Scientific) and confocal images were taken using either a Leica SP5 or Zeiss LSM 880 confocal microscope and processed with Volocity (Perkin Elmer) or FIJI (ImageJ). Skeleton and gap analyses are based on collagen I staining and confined to the T cell paracortical area of the LN. The analysis was performed using a macro for FIJI (ImageJ). Conduit thickness was measured by 0.1 μm z-stack Airyscans of the collagen I core rotated to display ellipse area of cross section for NDLNs and TDLNs and measured in Volocity (Perkin Elmer). For dextran studies, fluorescence intensity quantifications were performed in FIJI (ImageJ). 70,000 MW dextran (biotin, SA-647 Alexa Fluor (Thermo Scientific)) was quantified over total T cell paracortical area and back normalized on area size. Following FCs were calculated to the average of all NDLNs (Ctrl). For calculating area fraction in FIJI, the Otsu threshold was used and adjusted according to ER-TR7 area coverage.
Table 2.
Antibodies used for immunofluorescence
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad). For comparisons of three or more groups, data were subjected to one-way or two-way ANOVA analysis, followed by post hoc test (Dunnett’s when comparing every mean to a control mean, or Tukey’s multiple comparisons test when comparing every mean to every other mean). When two groups were compared, a two-tailed unpaired Student’s t-test was applied. Data are represented as mean ± SEM, and p ≤ 0.05 was considered significant.
Network Analysis
In order to study the relatedness of the top expressed genes – a network analysis was performed using the Multiple Association Network Integration Algorithm (MANIA)56. A large dataset of roughly 300 gene association networks are used to correlate genes, and create gene networks from a list of highly deregulated probes. The 599 most deregulated 11d TDLN probes were used as an input for the algorithm, and connections between probes visualized using Cytoscape57. The networks generated using the MANIA algorithm include 3 edge types between nodes: Predicted Edges, whereby functional relationships are implied due to orthology with other organisms, Co-localization Edges, whereby linked gene products are expressed in the same cellular location, and Co-expression edges, where the expression levels of gene products are similar across conditions in a previously published gene expression study. Genes are linked through calculated edges, and networks of linked genes generated.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Shields Group for constructive comments and continued scientific discussion, Ares Facility QU, E23 and T29 staff members for animal husbandry and technical assistance, R. Butler for support with image analysis and algorithm development, and Cambridge Genomic Services for microarray services and post-analysis advice. We wish to thank the CIMR flow cytometry core facility for advice and support in flow cytometry and cell sorting applications. Work was supported by Medical Research Council core funding (J.S) and Royal Society grant UF130039 (B.A.H).
Footnotes
Database Accession Numbers
The data discussed in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; GEO Series accession: GSE73728)
Author Contributions
A.R. planned and performed majority of experiments and associated analysis; L.H. performed in vitro experiments; D.S. performed in silico analysis; B.A.H. contributed to in silico analysis and data interpretation; J.S. conceived project, planned and performed experiments and contributed to data interpretation. A.R. J.S. and D.S. co-wrote the paper. All authors contributed to editing of manuscript and critical review.
Competing Financial Interest
The authors declare no competing financial interest.
References
- 1.Bajenoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nature immunology. 2007;8:1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
- 3.Schumann K, et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 2010;32:703–713. doi: 10.1016/j.immuni.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Roozendaal R, Mebius RE, Kraal G. The conduit system of the lymph node. International immunology. 2008;20:1483–1487. doi: 10.1093/intimm/dxn110. [DOI] [PubMed] [Google Scholar]
- 5.Gretz JE, Norbury CC, Anderson AO, Proudfoot AE, Shaw S. Lymph-borne chemokines and other low molecular weight molecules reach high endothelial venules via specialized conduits while a functional barrier limits access to the lymphocyte microenvironments in lymph node cortex. J Exp Med. 2000;192:1425–1440. doi: 10.1084/jem.192.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roozendaal R, et al. Conduits mediate transport of low-molecular-weight antigen to lymph node follicles. Immunity. 2009;30:264–276. doi: 10.1016/j.immuni.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sixt M, et al. The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity. 2005;22:19–29. doi: 10.1016/j.immuni.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Luther SA, Tang HL, Hyman PL, Farr AG, Cyster JG. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12694–12699. doi: 10.1073/pnas.97.23.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 10.Katakai T, et al. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. International immunology. 2004;16:1133–1142. doi: 10.1093/intimm/dxh113. [DOI] [PubMed] [Google Scholar]
- 11.Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J Exp Med. 2004;200:783–795. doi: 10.1084/jem.20040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn MD, et al. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher AL, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J Exp Med. 2010;207:689–697. doi: 10.1084/jem.20092642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nature immunology. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 15.Nichols LA, et al. Deletional self-tolerance to a melanocyte/melanoma antigen derived from tyrosinase is mediated by a radio-resistant cell in peripheral and mesenteric lymph nodes. Journal of immunology. 2007;179:993–1003. doi: 10.4049/jimmunol.179.2.993. [DOI] [PubMed] [Google Scholar]
- 16.Mueller SN, et al. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15430–15435. doi: 10.1073/pnas.0702579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Boer M, van Dijck JA, Bult P, Borm GF, Tjan-Heijnen VC. Breast cancer prognosis and occult lymph node metastases, isolated tumor cells, and micrometastases. J Natl Cancer Inst. 2010;102:410–425. doi: 10.1093/jnci/djq008. [DOI] [PubMed] [Google Scholar]
- 18.Morton DL, et al. Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med. 2006;355:1307–1317. doi: 10.1056/NEJMoa060992. [DOI] [PubMed] [Google Scholar]
- 19.Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124:922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malhotra D, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nature immunology. 2012;13:499–510. doi: 10.1038/ni.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox TR, Erler JT. Molecular pathways: connecting fibrosis and solid tumor metastasis. Clin Cancer Res. 2014;20:3637–3643. doi: 10.1158/1078-0432.CCR-13-1059. [DOI] [PubMed] [Google Scholar]
- 22.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 23.Cremasco V, et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nature immunology. 2014;15:973–981. doi: 10.1038/ni.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher AL, Acton SE, Knoblich K. Lymph node fibroblastic reticular cells in health and disease. Nat Rev Immunol. 2015;15:350–361. doi: 10.1038/nri3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forster R, Braun A, Worbs T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends in immunology. 2012;33:271–280. doi: 10.1016/j.it.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Gunn MD, et al. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J Exp Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller SN, et al. Regulation of homeostatic chemokine expression and cell trafficking during immune responses. Science. 2007;317:670–674. doi: 10.1126/science.1144830. [DOI] [PubMed] [Google Scholar]
- 28.Yang CY, et al. Trapping of naive lymphocytes triggers rapid growth and remodeling of the fibroblast network in reactive murine lymph nodes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E109–118. doi: 10.1073/pnas.1312585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrell MI, Iritani BM, Ruddell A. Tumor-induced sentinel lymph node lymphangiogenesis and increased lymph flow precede melanoma metastasis. The American journal of pathology. 2007;170:774–786. doi: 10.2353/ajpath.2007.060761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirakawa S, et al. VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood. 2007;109:1010–1017. doi: 10.1182/blood-2006-05-021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirakawa S, et al. VEGF-A induces tumor and sentinel lymph node lymphangiogenesis and promotes lymphatic metastasis. J Exp Med. 2005;201:1089–1099. doi: 10.1084/jem.20041896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohner NA, et al. Lymph node biophysical remodeling is associated with melanoma lymphatic drainage. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015 doi: 10.1096/fj.15-274761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carriere V, et al. Cancer cells regulate lymphocyte recruitment and leukocyte-endothelium interactions in the tumor-draining lymph node. Cancer research. 2005;65:11639–11648. doi: 10.1158/0008-5472.CAN-05-1190. [DOI] [PubMed] [Google Scholar]
- 34.Soudja SM, et al. Disrupted lymph node and splenic stroma in mice with induced inflammatory melanomas is associated with impaired recruitment of T and dendritic cells. PloS one. 2011;6:e22639. doi: 10.1371/journal.pone.0022639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MartIn-Fontecha A, et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J Exp Med. 2003;198:615–621. doi: 10.1084/jem.20030448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St John AL, Abraham SN. Salmonella disrupts lymph node architecture by TLR4-mediated suppression of homeostatic chemokines. Nature medicine. 2009;15:1259–1265. doi: 10.1038/nm.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acton SE, et al. Podoplanin-rich stromal networks induce dendritic cell motility via activation of the C-type lectin receptor CLEC-2. Immunity. 2012;37:276–289. doi: 10.1016/j.immuni.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Astarita JL, et al. The CLEC-2-podoplanin axis controls the contractility of fibroblastic reticular cells and lymph node microarchitecture. Nature immunology. 2015;16:75–84. doi: 10.1038/ni.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Augsten M, et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer research. 2014;74:2999–3010. doi: 10.1158/0008-5472.CAN-13-2740. [DOI] [PubMed] [Google Scholar]
- 40.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 41.Harper J, Sainson RC. Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Seminars in cancer biology. 2014;25:69–77. doi: 10.1016/j.semcancer.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Kraman M, et al. Suppression of Antitumor Immunity by Stromal Cells Expressing Fibroblast Activation Protein-alpha. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 43.Shields JD, Kourtis IC, Tomei AA, Roberts JM, Swartz MA. Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science. 2010;328:749–752. doi: 10.1126/science.1185837. [DOI] [PubMed] [Google Scholar]
- 44.Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. I. Role of interstitial pressure and convection. Microvasc Res. 1989;37:77–104. doi: 10.1016/0026-2862(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 45.Dafni H, Landsman L, Schechter B, Kohen F, Neeman M. MRI and fluorescence microscopy of the acute vascular response to VEGF165: vasodilation, hyper-permeability and lymphatic uptake, followed by rapid inactivation of the growth factor. NMR Biomed. 2002;15:120–131. doi: 10.1002/nbm.724. [DOI] [PubMed] [Google Scholar]
- 46.Goldman J, et al. Cooperative and redundant roles of VEGFR-2 and VEGFR-3 signaling in adult lymphangiogenesis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:1003–1012. doi: 10.1096/fj.06-6656com. [DOI] [PubMed] [Google Scholar]
- 47.Hompland T, Ellingsen C, Ovrebo KM, Rofstad EK. Interstitial Fluid Pressure and Associated Lymph Node Metastasis Revealed in Tumors by Dynamic Contrast-Enhanced MRI. Cancer Res. 2012;72:4899–4908. doi: 10.1158/0008-5472.CAN-12-0903. [DOI] [PubMed] [Google Scholar]
- 48.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 51.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 52.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fletcher AL, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J Exp Med. 2010;207:689–697. doi: 10.1084/jem.20092642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirosue S, et al. Steady-state antigen scavenging, cross-presentation, and CD8+ T cell priming: a new role for lymphatic endothelial cells. Journal of immunology. 2014;192:5002–5011. doi: 10.4049/jimmunol.1302492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.