Abstract
Adhesion and migration of T cells are controlled by chemokines and by adhesion molecules, especially integrins, and play critical roles in the normal physiological function of T lymphocytes. Using an RNA interference screen we have identified the WNK1 kinase as a regulator of both integrin-mediated adhesion and T cell migration. We demonstrate that WNK1 is a negative regulator of integrin-mediated adhesion, whereas it acts as a positive regulator of migration via OXSR1 and STK39 kinases and the SLC12A2 ion co-transporter. WNK1-deficient T cells home less efficiently to lymphoid organs, and migrate more slowly through them. Our results reveal that a pathway hitherto known only to regulate salt homeostasis in the kidney functions to balance T cell adhesion and migration.
T cells continuously recirculate between lymphoid organs via the blood and lymphatic systems, entering lymph nodes through specialized high endothelial venules (HEV)1. Initial selectin-mediated interactions result in T cells rolling on the endothelium. Subsequently, chemokine CCL21 signaling through CCR7 on T cells leads to activation of cell-surface integrin adhesion molecules such as LFA-1 (αLβ2), which binds to its ligands ICAM1 and ICAM2, leading to firm adhesion between the T cell and HEV. T cells then transmigrate across the endothelium and enter the parenchyma of the lymph node where they migrate rapidly under the influence of the CCR7 ligands CCL19 and CCL21, a process that is much less dependent on integrins2–4. Finally, T cells exit lymph nodes through lymphatic vessels and return into circulation via the venous system.
This recirculation is critical in allowing T cells to scan lymphoid tissue for antigen-presenting cells (APCs) bearing cognate antigen in the form of peptide:MHC complex that can bind to its T cell receptor (TCR). Binding of antigen to the TCR results in signaling that stops T cell migration, and activates LFA-1 causing firm adhesion between the T cell and the APC. The formation of such T cell:APC conjugates is necessary for T cell activation and thus initiation of T cell immune responses.
The activation of LFA-1 by chemokine receptors is triggered by “inside-out” signals from the receptor that lead to conformational changes in the integrin5–7. These convert the integrin from a conformation with low affinity for ligand, to an extended closed, and then extended open high affinity conformation. The inside-out signal also leads to binding of Talin and Kindlin-3 to the cytoplasmic domain of the β subunit of LFA-1, association of F-actin via Talin, and LFA-1 clustering which increases its avidity for ligand5. Importantly, for stable high affinity binding, both LFA-1 and its ligands need to be immobilized, such that binding of ligand to LFA-1 results in the exertion of force.
Inside-out signals from the TCR result in LFA-1 activation through similar mechanisms, however, in the absence of immobilized ligand, they do not change the integrin conformation8. Once again, for stable adhesion the ligand needs to be immobilized on the APC, and LFA-1 needs to be anchored to the actin cytoskeleton, such that binding of LFA-1 to ICAM1 results in a force which promotes high affinity binding. A critical signaling molecule that transduces inside-out signals from both chemokine receptors and the TCR is the RAP1 GTPase5. Active RAP1-GTP binds to RIAM and RAPL effector proteins, which in turn promote binding of Talin to the β subunit of LFA-1, and LFA-1 clustering respectively9–11.
In order to identify new proteins that may contribute to the activation of LFA-1, we used an RNA interference (RNAi) screen to identify kinases that regulate integrin-dependent binding of T cells to APCs. Here we report that the WNK1 kinase is a negative regulator of both chemokine receptor and TCR-induced LFA-1 activation and subsequent adhesion, and that it does so via RAP1. Conversely, we show that WNK1 is a positive regulator of T cell migration through the OXSR1 and STK39 kinases and the SLC12A2 ion co-transporter. In the absence of WNK1 T cells home less efficiently to lymphoid organs and migrate more slowly through them. Our results reveal that a pathway previously shown to regulate salt homeostasis in the kidney12,13, functions to balance T cell adhesion and migration.
Results
WNK1 is a negative regulator of integrin-mediated adhesion
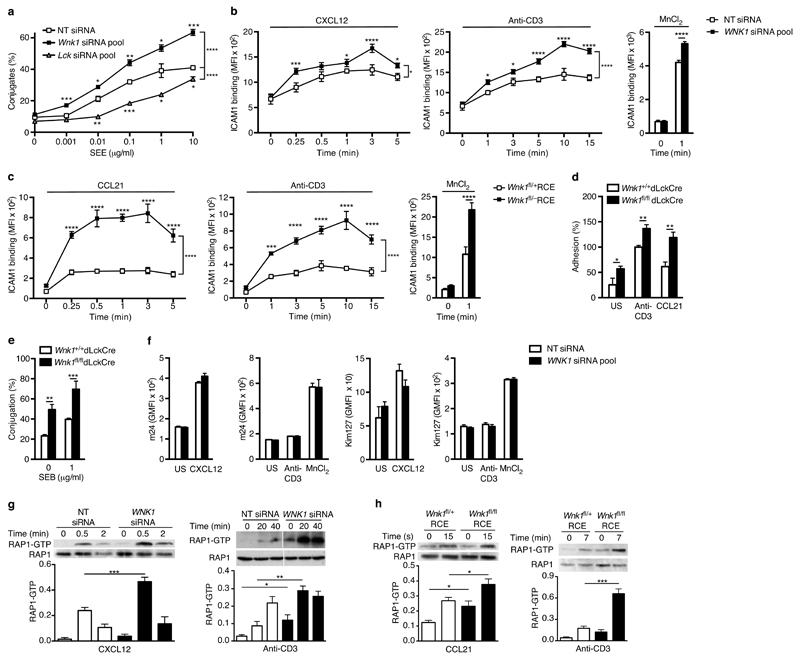
To identify novel signaling pathways regulating adhesion of T cells, we carried out an RNAi screen in which we knocked down expression of 719 kinase and kinase-related genes individually in the Jurkat T cell leukaemia cell line, and analyzed the ability of the cells to form antigen-specific conjugates with the Nalm6 B cell line in response to the superantigen Staphylococcal enterotoxin E (SEE) (data not shown). One of the validated hits from the screen was the WNK1 gene coding for the WNK1 serine/threonine kinase, knockdown of which resulted in increased T cell-B cell conjugation. Jurkat T cells with reduced amounts of WNK1 protein exhibited increased conjugation across a broad range of SEE concentrations (Fig. 1a, Supplementary Fig. 1a,b). In contrast, knockdown of the LCK tyrosine kinase, a critical positive regulator of TCR signaling, resulted in decreased conjugation.
Figure 1.
WNK1 is a negative regulator of T cell adhesion. (a) Conjugate formation between Jurkat T cells transfected with non-targeting (NT) siRNA, or siRNA pools against WNK1 or LCK and NALM6 B cells pulsed with different concentrations of SEE. (b) Binding of soluble ICAM1 complexes to Jurkat cells in response to treatment with CXCL12, anti-CD3 or MnCl2 for the indicated times. (c) Binding of ICAM1 to mouse CD4+ T cells from control or WNK1-deficient mice in response to CCL21, anti-CD3, or MnCl2 for the indicated times. (d) Adhesion of mouse CD4+ T cells to plate-bound ICAM1 in response to the indicated stimuli, normalized to the adhesion of control cells in response to anti-CD3. (e) Conjugation of mouse CD4+ T cells to CH27 cells pulsed with SEB. (f) Binding of antibodies specific for LFA-1 conformations with high or intermediate affinity for ligand (m24 or Kim127) on Jurkat T cells treated with the indicated stimuli; GMFI, geometric mean fluorescence intensity; US, unstimulated. (g) Immunoblots of RAP1 and RAP1-GTP pulled down from Jurkat T cells transfected with indicated siRNA and stimulated with CXCL12 or anti-CD3 for the indicated times; graphs show level of RAP1-GTP normalized to RAP1. (h) Immunoblots of RAP1 and RAP1-GTP pulled down from mouse CD4+ T cells stimulated with CCL21 or anti-CD3 for the indicated times; graphs show quantitated immunoblot signals for RAP1-GTP normalized to RAP1. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-way ANOVA (a,b,c), Mann-Whitney (d,e), unpaired t-test (f,g,h)). Sample sizes: a (3), b (4, control; 6, mutant), c (6), d (5), e (6, control; 5, mutant), f (6, except 3,in right panel), g (4), h (4). Data are from one experiment representative of 3 (a,b,c,e,f right panel) or 4 (g,h) experiments or pooled from 2 (d, f leftmost 3 panels) independent experiments. Graphs show mean±SEM.
Antigen-induced adhesion between T cells and APCs is mediated primarily by binding of integrins such as LFA-1 on T cells to their ligands such as ICAM1 on APCs14. To test whether the increased T:APC conjugation observed in WNK1-deficient Jurkat T cells was caused by increased TCR-induced integrin-mediated adhesion, we evaluated the ability of the cells to bind ICAM1 complexes, or to adhere to plate-bound ICAM1. In addition, we broadened the study by investigating the effect of WNK1 deficiency on chemokine-induced integrin-mediated adhesion. We found that knockdown of WNK1 resulted in increased adhesion of Jurkat T cells to both soluble and plate-bound ICAM1 following stimulation through either CXCR4 or the TCR, despite no alteration in surface CXCR4, TCR or LFA-1 (Fig. 1b, Supplementary Fig. 1c-f). In contrast, knockdown of LCK resulted in decreased adhesion. Notably, time course analysis showed that the binding of ICAM1 was already significantly increased in WNK1-deficient Jurkat T cells 15 s or 1 min after stimulation through CXCR4 or TCR respectively, suggesting that WNK1 plays a role in the early stages of adhesion.
To investigate whether WNK1 also regulates adhesion in primary T cells, we generated mice with WNK1-deficient T cells, using either the dLck-Cre transgene15 for conditional deletion of a loxP-flanked allele of Wnk1 (Wnk1fl)16 at late stages of thymocyte development or ROSA26-CreERT2 (RCE), a tamoxifen-inducible Cre17. We generated bone marrow chimeras by reconstituting irradiated RAG1-deficient mice with marrow cells from both strains. This served to increase numbers of mice from which WNK1-deficient T cells could be isolated, and for the Wnk1fl/-RCE strain, it limited complete loss of Wnk1 to lymphocytes. In both strains there was efficient loss of Wnk1 mRNA in CD4+ and CD8+ T cells (Supplementary Fig. 2a-c). We found that in response to stimulation through either CCR7 or TCR, WNK1-deficient CD4+ T cells showed increased binding of ICAM1 complexes and increased adhesion to ICAM1-coated plates (Fig. 1c,d). Furthermore, consistent with increased TCR-induced adhesion through LFA-1, the mutant T cells were more efficient at forming antigen-specific conjugates with APCs (Fig. 1e). Mutant T cells had slightly reduced surface TCR and CCR7, but slightly elevated LFA-1 (Supplementary Fig. 2d,e). Once again time course analysis showed increased ICAM1 binding at all timepoints following stimulation through CCR7 or TCR. Thus, as in Jurkat T cells, WNK1 is a negative regulator of both chemokine receptor and TCR-induced integrin-dependent adhesion in primary mouse T cells, acting at the earliest stages of adhesion.
WNK1 is a negative regulator of RAP1
Next we investigated the mechanism by which WNK1 regulates integrin-mediated adhesion. Using antibodies that recognize either an intermediate or high affinity conformation of human LFA-1, we were able to detect an increase in both of these conformations on Jurkat T cells in response to CXCL12 (Fig. 1f). In contrast, as expected8, stimulation of the cells through the TCR showed no detectable change in LFA-1 affinity. Notably, the CXCL12-induced increase in intermediate or high affinity LFA-1 conformations was not affected by loss of WNK1, thus WNK1 does not regulate the chemokine-induced affinity change in LFA-1 (Fig. 1f). In contrast, we noted increased adhesion to ICAM1 in both WNK1-deficient Jurkat cells and primary mouse T cells in response to Mn2+, a cation that directly induces the high affinity conformation of LFA-1 (Fig. 1b,c,f). This suggests that WNK1 may be regulating other processes that contribute to LFA-1-mediated adhesion such as anchoring of the integrin to the actin cytoskeleton or its clustering.
We extended the analysis to intracellular signaling pathways and found no change in chemokine or TCR-induced activation of ERK kinases, no change in TCR-induced calcium fluxes in WNK1-deficient Jurkat T cells, and no change in chemokine-induced actin polymerization in WNK1-deficient mouse CD4+ T cells (Supplementary Fig. 3a-c). However both Jurkat T cells and primary mouse CD4+ T cells deficient in WNK1 showed a large increase in both chemokine- and TCR-induced activation of RAP1, a critical regulator of integrin-mediated adhesion5,14 (Fig. 1g,h and Supplementary Fig. 1g). In view of previous studies showing that expression of constitutively active RAP1 in T cells results in increased integrin-mediated adhesion18, we conclude that WNK1 may be acting as a negative regulator of RAP1 activation, and thereby influencing integrin-mediated adhesion.
WNK1 is a positive regulator of T cell migration
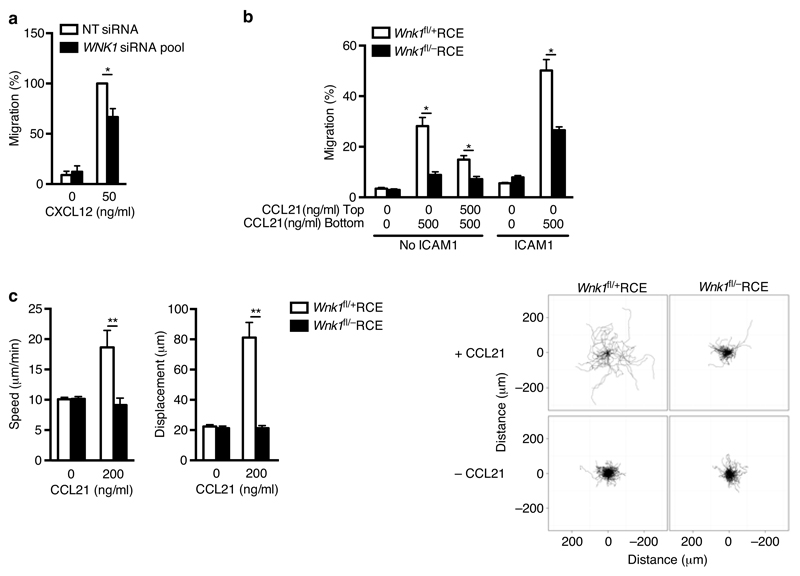
Since the processes of T cell adhesion and migration are closely related, with increased adhesion potentially leading to decreased migration19, we also examined the effect of WNK1 knockdown on cell migration using a Transwell assay. We found that WNK1-deficient Jurkat T cells migrated less efficiently in response to the chemokine CXCL12, despite expressing normal levels of CXCR4 (Fig. 2a, Supplementary Fig. 1f,h). Furthermore, WNK1-deficient primary mouse CD4+ T cells also showed less efficient chemotaxis in response to a gradient of CCL21 both in the absence or presence of ICAM1, and less chemokinesis in response to equal concentrations of CCL21 below and above the membrane (Fig. 2b). Time-lapse imaging showed that in response to CCL21, WNK1-deficient CD4+ T cells migrated much more slowly (Fig. 2c, Supplementary Video 1). Thus WNK1 is a positive regulator of CCL21-induced migration in primary mouse CD4+ T cells and this may be independent of LFA-1-mediated adhesion.
Figure 2.
WNK1 is a positive regulator of T cell migration. (a) Migration of Jurkat T cells transfected with NT siRNA or an siRNA pool against WNK1 in response to CXCL12, normalized to CXCL12-induced migration of control cells. (b) Migration of mouse CD4+ T cells from top to bottom chamber of a Transwell plate in response to indicated concentrations of CCL21 in presence or absence of ICAM1. (c) Speed and displacement of mouse CD4+ T cells measured by time-lapse microscopy in response to indicated concentration of CCL21. Flower plots show overlay of migration tracks of individual T cells. *P < 0.01, **P < 0.0001 (Mann-Whitney). Sample sizes: a (5), b (6), c (567, control unstimulated; 67, control with CCL21; 450, mutant, unstimulated; 293, mutant with CCL21). Data are from one experiment representative of 2 (b,c) experiments or pooled from 5 (a) independent experiments. Graphs show mean±SEM.
WNK1 is required for efficient homing into lymph nodes
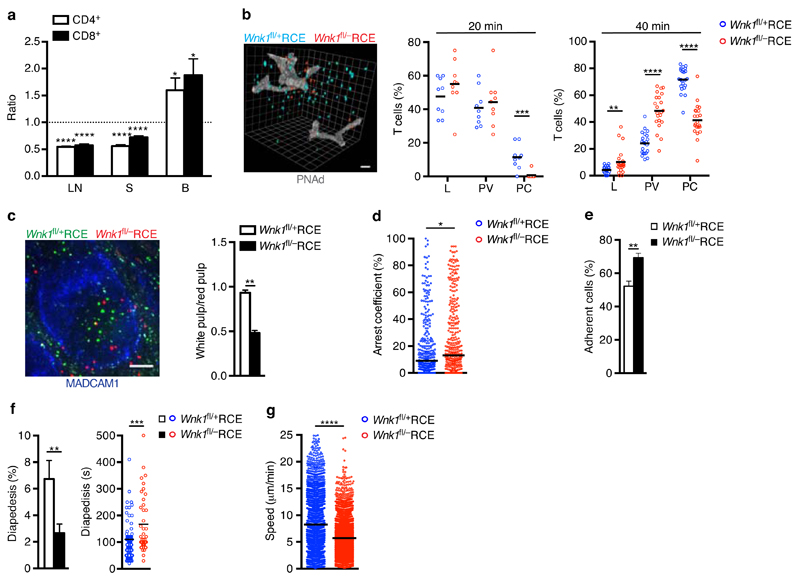
Next we evaluated whether WNK1 also regulates physiological T cell trafficking. We transferred a mixture of control and WNK1-deficient T cells intravenously into wild-type mice and analyzed their homing into lymphoid organs. We found that 1 h after transfer, WNK1-deficient CD4+ and CD8+ T cells homed less efficiently into both lymph nodes and the spleen, with more cells remaining in the blood (Fig. 3a). 3-D histological analysis showed that 20 or 40 min after transfer a larger fraction of the mutant T cells were located in the lumen of the blood vessels and in perivascular regions of the lymph nodes whereas fewer had entered the parenchyma, suggesting that WNK1-deficient T cells are less efficient at extravasating across the endothelium and at migrating away from the vasculature into the parenchyma (Fig. 3b, Supplementary Video 2). Similarly, histology of the spleen showed that 1 h after transfer, more WNK1-deficient T cells were located in the red pulp compared to the white pulp where most T cells reside, suggesting that WNK1-deficient T cells are defective at crossing the marginal sinus-lining cells that lymphocytes need to cross in order to gain entry to the white pulp (Fig. 3c). To dissect the integrin-dependent adhesion and transmigration steps required to enter lymph nodes, we imaged control and mutant T cells migrating on a monolayer of endothelial cells under flow and transmigrating through it20. We found that WNK1-deficient T cells arrested for a longer fraction of time on the endothelial cells and more of them resisted detachment from endothelial cells, in agreement with their hyperadhesive phenotype (Fig. 3d,e, Supplementary Video 3). Conversely, fewer mutant T cells were able to cross through the endothelial monolayer and took longer to do so (Fig. 3f). Lastly, the mutant T cells migrated more slowly on the endothelial monolayer (Fig. 3g).
Figure 3.
WNK1 is required for efficient homing into lymph nodes. (a) Ratio of WNK1-deficient to control mouse T cells in lymph node (LN), spleen (S) and blood (B) 1 h after i.v. injection into C57BL/6J recipients normalized to input ratio. (b) Percentage of T cells located in the lumen (L), perivascular area (PV) or parenchyma (PC) of LN as determined by 3D-histology 20 and 40 min after transfer; image shows location of T cells (blue and red) and HEVs revealed by expression of PNAd (white) 40 min after transfer; scale bar 20 μm. (c) Ratio of CD4+ T cells in the white to red pulp of the spleen 1 h after T cell transfer. Image shows T cells (red and green) in the spleen, MADCAM1 (blue) indicates boundary between red and white pulp; scale bar 100 μm. (d) Percentage of time control and WNK1-deficient T cells arrested (<2 µm/min) while migrating over an endothelial cell monolayer under flow conditions. (e) Percentage of cells that remained adherent on endothelial cells 10 min after arrest. (f) Percentage of cells that underwent diapedesis through endothelial cell monolayer and duration of diapedesis. (g) Speed of cell migration on the endothelial cell monolayer. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (1-sample t-test (a), Mann-Whitney (b,c,d,e,g, and f, time of diapedesis), Wilcoxon matched-pairs test (f, % diapedesis). Sample sizes: a (12, lymph node and spleen; 11, blood), b (9, 20 min; 22, 40 min), c (6), d (343, control; 364, mutant), e (9), f (8, % diapedesis; 73 control and 40 mutant, time of diapedesis), g (2508, control; 3846, mutant). Data are from one experiment representative of 2 experiments (d), or pooled from 2 independent experiments (a,b,c,e,f,g). Graphs show mean±SEM except d where horizontal lines indicate median.
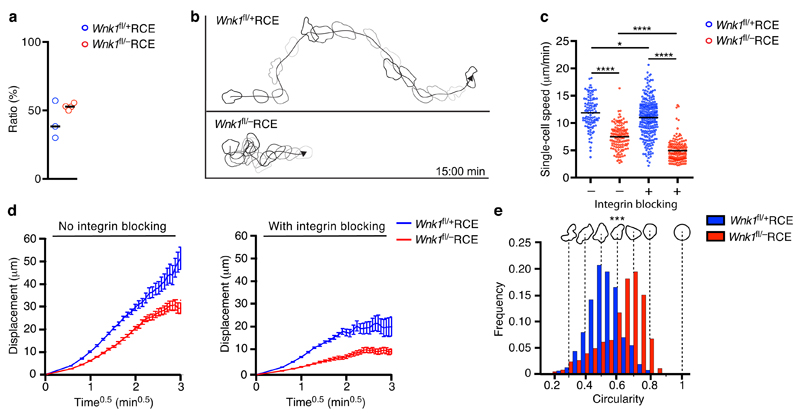
Finally, we transferred control or mutant T cells into wild-type mice and used intravital imaging to follow their movement in the lumen of the HEV and within the parenchyma of lymph nodes. Imaging of rapid T cell adhesion in lymph node HEVs uncovered a similar sticking fraction of firmly adherent cells in both WNK1-deficient and control T cells, suggesting that absence of WNK1 does not affect the ability of T cells to adhere firmly to endothelium (Fig. 4a). In contrast, analysis of cells in the parenchyma of the lymph nodes showed that WNK1-deficient T cells migrated more slowly than control T cells, had a lower motility coefficient (66.7 μm2/min and 34.5 μm2/min in control and mutant cells respectively) and were more rounded (Fig. 4b-e, and Supplementary Video 4). In order to evaluate if this reduced migration was caused by increased integrin-mediated adhesion, we transferred T cells into wild-type mice and then 15-18 h later blocked both LFA-1 and VLA-4 integrins. We found that WNK1-deficient T cells still migrated more slowly than control T cells, had a reduced motility coefficient (22.4 μm2/min and 3.4 μm2/min in control and mutant cells respectively) and displayed larger turning angles, a characteristic feature of slower migrating cells21 (Fig. 4c,d and Supplementary Fig. 4). Indeed the mutant cells migrated even more slowly in the presence of integrin blocking antibodies showing that their decreased migration speed was not due to hyperadhesiveness via integrins. Thus we conclude that WNK1 is a critical regulator of homing and migration of T cells in vivo. The defective overall homing of WNK1-deficient T cells may be caused by less efficient detachment from HEVs and lower diapedesis, despite comparable primary adhesion. In contrast, the reduced parenchymal migration of WNK1-deficient T cells is independent of increased integrin-mediated adhesion.
Figure 4.
WNK1 is required for migration within lymph nodes in vivo. (a) Ratio of firmly adherent to rolling or adherent T cells determined by intravital microscopy of LN HEVs after transfer of control and WNK1-deficient T cells into C57BL/6J recipient mice. (b-e) Migration of T cells in LN parenchyma analyzed by 2-photon intravital microscopy after transfer of control and WNK1-deficient T cells into C57BL/6J mice in the presence or absence of integrin blocking. (b) Tracks of a control and WNK1-deficient T cell indicating cell shapes over a 15 min period. (c) Migration speed. (d) Mean displacement of T cells as a function of time0.5. (e) Relative frequency of cell shape of T cells. *P < 0.05, **P < 0.001, ***P < 0.0001 (unpaired t-test (a), Mann-Whitney (c,e)). Sample sizes: a (3, control; 4, mutant), c (103, control with no blocking; 134, mutant with no blocking; 257, control with integrin blocking; 149, mutant with integrin blocking), e (103, control; 134, mutant). Data are pooled from 2 independent experiments. Graphs show mean±SEM.
Chemokine receptors and TCR activate the WNK1 pathway
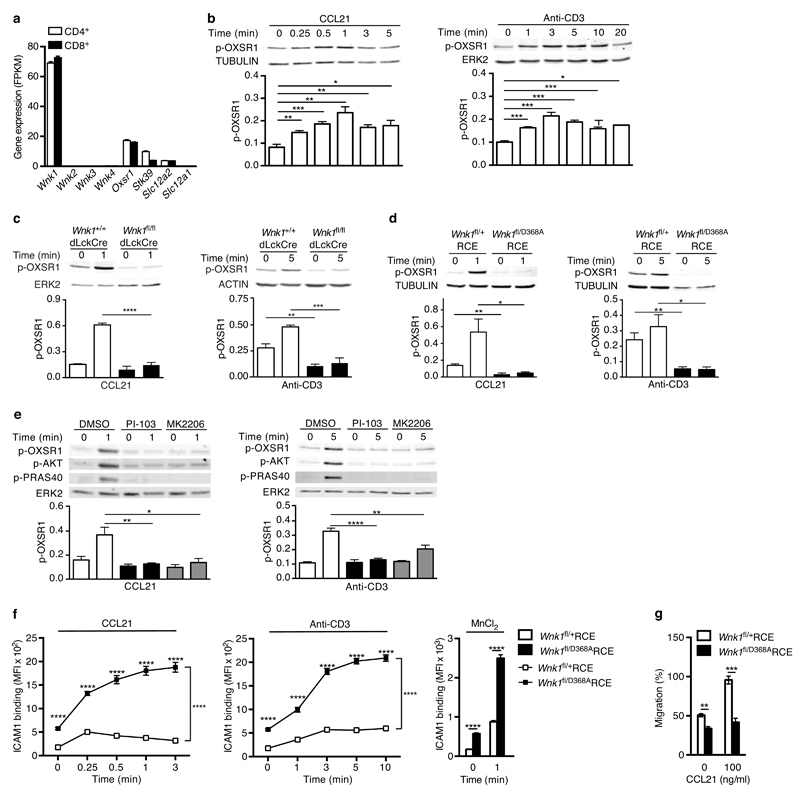
While WNK1 has not been previously studied in lymphocytes, its function has been extensively investigated in kidney tubular epithelial cells where it controls the uptake of sodium, potassium and chloride ions12. In particular, WNK1 phosphorylates and activates two related kinases OXSR1 and STK39, which in turn phosphorylate and activate the SLC12A2 (NKCC1) and SLC12A1 (NKCC2) Na+/K+/Cl- co-transporters, thereby stimulating uptake of these ions22. In view of the unexpected role for WNK1 in T cell adhesion and migration we wondered if the WNK1-OXSR1/STK39-SLC12A2/SLC12A1 pathway might also function in T cells, and, if so, whether it might contribute to the adhesion and migration of the cells. Analysis of RNAseq data showed that both CD4+ and CD8+ primary mouse T cells expressed Wnk1 but no other members of the Wnk-family (Fig. 5a). Both subsets of T cells also expressed Oxsr1, Stk39 and Slc12a2, but not Slc12a1. To investigate if the WNK1-OXSR1/STK39-SLC12A2 pathway functions in T cells, we stimulated primary mouse CD4+ T cells through CCR7 or the TCR and immunoblotted cell extracts with antibodies specific for WNK1 phosphorylation sites on OXSR1 and STK39 (Ser325 and Ser383 respectively)23. We found that both stimuli resulted in rapid phosphorylation of OXSR1, which was abrogated in WNK1-deficient T cells (Fig. 5b, c). Thus both CCR7 and the TCR transduce signals through WNK1 leading to the phosphorylation and, presumably, activation of OXSR1.
Figure 5.
Chemokine receptors and TCR activate WNK1. (a) Expression of the indicated genes in mouse T cells determined by RNAseq and expressed as fragments per kilobase per million reads (FPKM). (b-e) Immunoblots of mouse CD4+ T cells analyzed for phosphorylation of OXSR1 (p-OXSR1), Tubulin, Actin or ERK2 following stimulation with CCL21 or anti-CD3 for the indicated times. Levels of p-OXSR1 quantitated by normalizing to Tubulin, ERK2 or Actin; (b) p-OXSR1 in wild-type T cells; (c) p-OXSR1 in WNK1-deficient and control T cells; (d) p-OXSR1 in T cells expressing kinase-inactive WNK1-D368A or control cells; (e) p-OXSR1 in wild-type T cells treated with PI3-kinase inhibitor (PI-103), AKT inhibitor (MK2206) or vehicle (DMSO) and additionally analyzed for phosphorylation of AKT (p-AKT) or PRAS40 (p-PRAS40). (f) Binding of ICAM1 to mouse CD4+ T cells expressing kinase-inactive WNK1-D368A or control cells in response to CCL21, anti-CD3, or MnCl2 for the indicated times. (g) CCL21-induced migration of T cells of the indicated genotypes through a Transwell system normalized to the CCL21-induced migration of control cells. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-way ANOVA (f), Mann-Whitney (g), unpaired t-test (b,c,d,e)). Sample sizes: a (3), b (4), c (4, CCL21; 3, anti-CD3), d (4), e (4, CCL21; 7, anti-CD3), f (6), g (9). Data are pooled from 2 (f,g), 3 (a, c anti-CD3), 4 (b, c CCL21, d, and e CCL21) or 7 (e anti-CD3) independent experiments. Graphs show mean±SEM.
To evaluate whether WNK1 kinase activity was required for its regulation of T cell adhesion and migration we generated a novel strain of mice with a mutation of Asp368 to alanine (D368A) in the kinase domain of the protein, a change that had been previously shown to eliminate kinase activity23 (Supplementary Fig. 5a). Mice homozygous for this Wnk1D368A allele did not survive to birth, similar to that previously reported for mice with a total loss of WNK124 (Supplementary Fig. 5b). To analyze T cells expressing only kinase-inactive WNK1, we generated compound heterozygous mice with both a conditional and kinase-inactive allele of Wnk1, and a tamoxifen-inducible Cre (Wnk1fl/D368ARCE); treatment of such mice with tamoxifen results in T cells that express only kinase-inactive WNK1. As controls we used Wnk1fl/+RCE mice in which tamoxifen injection leads to deletion of the conditional allele of Wnk1, leaving an intact wild-type allele. T cells from both strains express similar amounts of Wnk1 (Supplementary Fig. 5c).
T cells expressing kinase-inactive WNK1 had very strongly reduced phosphorylation of OXSR1, and showed no inducible OXSR1 phosphorylation in response to stimulation through either CCR7 or TCR (Fig. 5d). These results show that the kinase activity of WNK1 is required to transduce CCR7 and TCR signals leading to OXSR1 phosphorylation on Ser325, and since this site is a known WNK1 substrate, the results strongly suggest that both CCR7 and TCR signaling lead to activation of WNK1 kinase activity.
CCR7 and TCR signaling activates WNK1 via PI3-kinase and AKT
Next we investigated how CCR7 and the TCR might activate WNK1, despite engaging largely different signaling pathways. We noted that previous studies had suggested that AKT may activate WNK125, and since both chemokine receptors and the TCR are known to activate AKT via phosphoinositide-3-kinase (PI3-kinase), we tested the potential involvement of these kinases in the activation of WNK1. We found that treatment of T cells with PI-103 and MK2206, inhibitors of PI3-kinase and AKT respectively, blocked both CCR7- and TCR-induced phosphorylation of AKT and PRAS40, a known substrate of AKT, demonstrating that both inhibitors were acting as expected (Fig. 5e). Importantly, both inhibitors also blocked CCR7 and TCR-induced phosphorylation of OXSR1 (Fig. 5e). Taken together these results suggest that both receptors transduce signals via PI3-kinase and AKT leading to the activation of WNK1.
WNK1 kinase activity regulates adhesion and migration
Finally, we investigated if WNK1 kinase activity was required for the regulation of integrin-mediated adhesion or migration in T cells. We found that CD4+ T cells expressing kinase-inactive WNK1 showed increased binding of ICAM1 following stimulation through CCR7 or TCR, and in response to treatment with Mn2+ to activate integrins from the outside (Fig. 5f), and showed decreased CCL21-induced migration (Fig. 5g). These results are very similar to those seen in WNK1-deficient T cells, and thus show that the kinase activity of WNK1 is required for its negative regulation of integrin-mediated adhesion and positive regulation of migration.
OXSR1, STK39 and SLC12A2 do not regulate adhesion via LFA-1
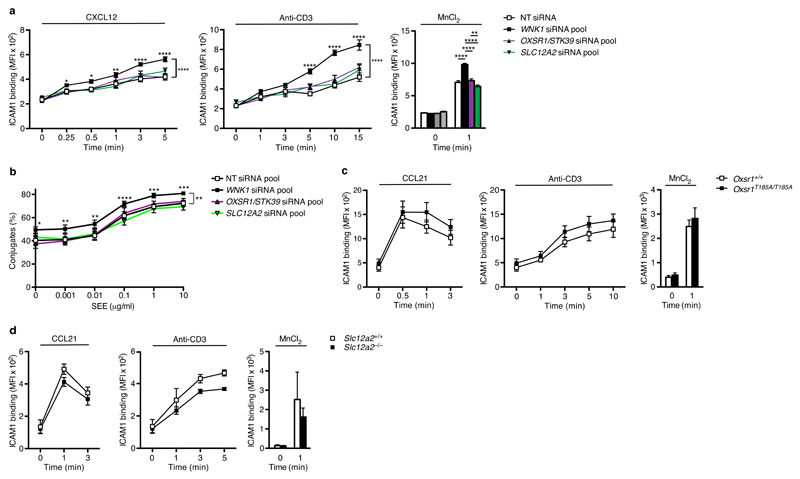
We next investigated if the OXSR1/STK39-SLC12A2 pathway, the best characterized pathway downstream of WNK1, is also involved in these processes. Knockdown of OXSR1, STK39, both OXSR1 and STK39 together, or SLC12A2 in Jurkat T cells had no effect on CCR7- or TCR-induced binding of ICAM1 complexes or adhesion to plate-bound ICAM1, or on conjugation of Jurkat T cells to SEE-pulsed APCs (Fig. 6a,b, and Supplementary Fig. 1a,e). Extending these studies to primary mouse T cells, we investigated CD4+ T cells taken from mice bearing a point mutation in the Oxsr1 gene, replacing Thr185 with alanine (T185A)13. This residue is phosphorylated by WNK1, leading to activation of OXSR122, and thus OXSR1-T185A can no longer be activated by WNK1. We also examined SLC12A2-deficient CD4+ T cells26. We found that T cells expressing OXSR1-T185A or deficient in SLC12A2 showed no alteration in CCR7- or TCR- or Mn2+-induced binding of ICAM1 complexes (Fig. 6c, d). Thus while the WNK1 kinase is a negative regulator of integrin-mediated adhesion, it does not do this through OXSR1, STK39 or SLC12A2.
Figure 6.
OXSR1, STK39 and SLC12A2 do not regulate integrin-mediated adhesion. (a) ICAM1 binding to Jurkat T cells transfected with NT siRNA, or siRNA pools against indicated genes following stimulation with CXCL12, anti-CD3 or MnCl2 for the indicated times. (b) Conjugation of Jurkat T cells to NALM6 B cells pulsed with indicated dose of SEE. (c) Binding of ICAM1 to mouse CD4+ T cells of the indicated genotypes following stimulation with CCL21, anti-CD3 or MnCl2 for the indicated times. (d) Binding of ICAM1 to mouse CD4+ T cells of the indicated genotypes following stimulation with CCL21, anti-CD3 or MnCl2 for the indicated times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-way ANOVA). Sample sizes: a (6), b (4), c (8), d (5, control cells; 6, mutant cells). Data are pooled from 2 (c), 3 (a), or 4 (b) independent experiments. Graphs show mean±SEM.
OXSR1, STK39 and SLC12A2 regulate migration
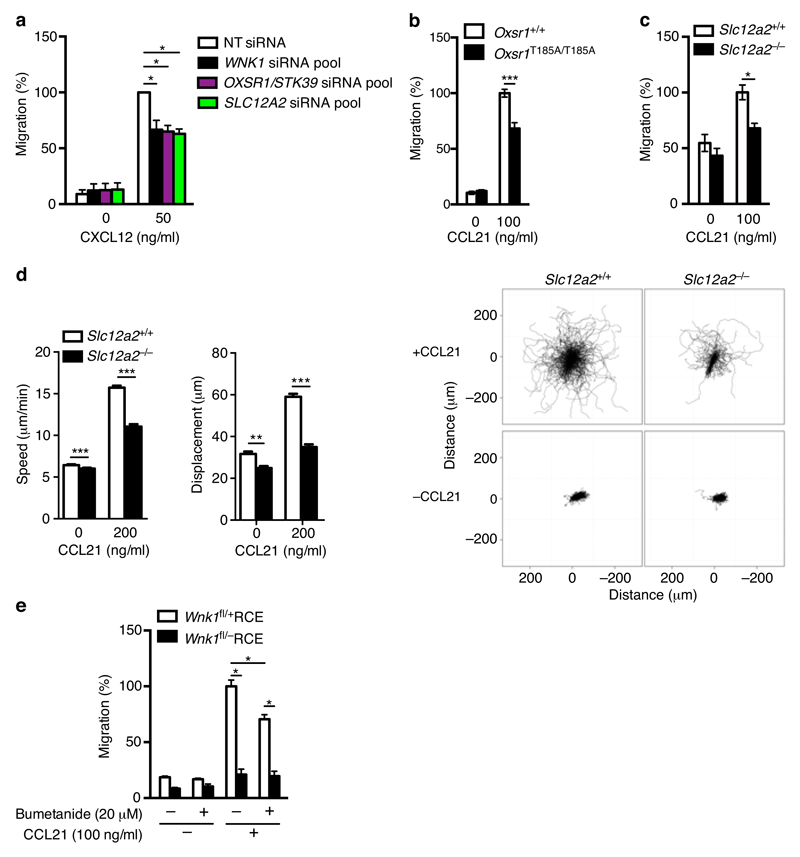
In contrast to the lack of effect on adhesion, knockdown of OXSR1, STK39 or both together, or of SLC12A2 resulted in reduced CCL21-induced chemotaxis of Jurkat T cells (Fig. 7a and Supplementary Fig. 1h). Furthermore, CCL21-induced chemotaxis was decreased in mouse CD4+ T cells expressing OXSR1-T185A or deficient in SLC12A2 (Fig. 7b,c), and time-lapse imaging showed that SLC12A2-deficient T cells migrated more slowly in response to CCL21 (Fig. 7d and Supplementary Video 5), despite unaltered surface levels of CCR7 (Supplementary Fig. 2f,g). Finally, we asked if the movement of ions through the SLC12A2 co-transporter was required for its function in T cell migration, by treating cells with bumetanide, an inhibitor of SLC12A2 ion transport. We found that bumetanide decreased CCL21-induced chemotaxis of wild-type CD4+ T cells, though the decrease was not as large as in WNK1-deficient T cells (Fig. 7e). Taken together these results suggest that WNK1 regulates T cell migration through the OXSR1/STK39-SLC12A2 pathway, and that it does so in part by controlling ion movement through SLC12A2.
Figure 7.
OXSR1, STK39 and SLC12A2 positively regulate chemokine-induced migration. (a) CXCL12-induced migration of Jurkat T cells transfected with the indicated siRNAs normalized to migration of cells transfected with NT siRNAs. Data for NT and WNK1 siRNA is the same as that shown in Fig. 2a and is repeated here for clarity. (b) CCL21-induced migration of mouse CD4+ T cells of the indicated genotypes. (c) CCL21-induced migration of mouse CD4+ T cells of the indicated genotypes. (d) Speed and displacement of mouse CD4+ T cells measured by time-lapse microscopy in response to CCL21. Flower plots show overlay of migration tracks of individual T cells. (e) CCL21-induced migration of mouse CD4+ T cells of the indicated genotypes in the presence or absence of bumetanide. *P < 0.01, **P < 0.001, ***P < 0.0001 (Mann-Whitney). Sample sizes: a (5), b (8), c (10, control cells; 11, mutant cells), d (630 control cells no CCL21; 1773 control cells with CCL21; 643 mutant cells no CCL21; 1105, mutant cells with CCL21), e (6). Data are representative of 2 (d) independent experiments or were pooled from 2 (b,e), 4 (c), or 5 (a) independent experiments. Graphs show mean±SEM.
Discussion
Using an RNAi screen we have identified a novel pathway regulating T cell adhesion and migration in vivo. WNK1 had previously been shown to control salt reabsorption in the kidney in part through activation of OXSR1 and STK39 and the SLC12A2 and SLC12A1 co-transporters22. Thus it was a surprise to find that WNK1 regulates T cell adhesion, that the WNK1-OXSR1/STK39-SLC12A2 pathway controls cell migration, and that this pathway can be activated by stimulation of the cells through the TCR or CCR7. Our results suggest that both receptors transduce signals via PI3-kinase and AKT that lead to the rapid activation of WNK1. AKT has been reported to phosphorylate WNK1 on Thr6027,28, however this phosphorylation does not lead to activation of WNK1 kinase activity, so it remains unclear how AKT activates WNK1.
We show here that WNK1 is a negative regulator of LFA-1 mediated adhesion and that WNK1 kinase activity is required for this. We showed that WNK1 does not regulate chemokine-induced conformational changes of LFA-1. Instead we found that WNK1-deficient cells had a large increase in activated RAP1, which we propose may be responsible for the increased adhesion. In agreement with this, T cells expressing a constitutively active RAP1 are hyperadhesive18. Interestingly, in this study hyperadhesion was seen in T cells treated with Mg2+ and EGTA, which induces a high affinity conformation of LFA-1 similar to Mn2+ treatment, and thus the increased RAP1-GTP in WNK1-deficient T cells may account for the increased adhesion in response to Mn2+. Active RAP1-GTP binds to RIAM and RAPL effector proteins, which in turn induce binding of Talin to the β subunit of LFA-1, and LFA-1 clustering respectively, thereby promoting adhesion9–11. It is unclear how WNK1 may regulate RAP1 activation, but it is possible that it may directly or otherwise regulate RAP1-specific GEFs or GAPs.
Our results also show that WNK1 is a positive regulator of T cell migration in response to chemokines. This function requires WNK1 kinase activity but is distinct from its role as a negative regulator of adhesion for two reasons. Firstly, genetic studies show that the OXSR1/STK39-SLC12A2 pathway downstream of WNK1 is required for the regulation of migration, but not adhesion. Secondly, if the reduced migration of WNK1-deficient T cells was due to increased adhesion, blocking adhesion should allow mutant T cells to migrate more rapidly. However intravital imaging of T cells in lymph nodes showed that blocking integrin binding did not cause WNK1-deficient T cells to speed up – on the contrary, they migrated even more slowly, suggesting that their hyperadhesive phenotype partially rescues the migration defect. Our results show that WNK1 regulates migration through the OXSR1/STK39-SLC12A2 pathway, and that at least part of this may be mediated by the movement of ions across the membrane through SLC12A2. An interesting possibility is that ion transport contributes to cell migration by causing osmotic uptake of water and hence changes in cell volume that are required for cell movement29. In this regard we note the recently proposed ‘osmotic engine model’ in which polarized uptake of ions and water at the leading edge of a cell, and efflux of ions and water at the trailing edge results in cell movement30. Supporting a role for SLC12A2 in cell migration, the co-transporter localizes to the leading edge of migrating glioma cells, and inhibition of its ion transport function results in reduced migration31. It will be interesting to investigate localization of the proteins in the WNK1-OXSR1-STK39-SLC12A2 pathway in migrating T cells. Finally, we note that WNK1 is enriched in the ‘adhesome’ of fibroblasts and is upregulated in migrating fibroblasts, suggesting that it may play a role in these processes in other cell types32,33.
In summary, we have identified WNK1 as a novel regulator of T cell adhesion and migration, which, to the best of our knowledge is the first regulator of its kind that inversely controls these two processes. In contrast, for example, T cells deficient in RAC1 and RAC2 GTPases show a decrease in both integrin-mediated adhesion and migration34. T cells with a hyperactive LFA-1 show increased adhesiveness, but reduced migration35,36, however this reduction in migration is a consequence of hyperadhesiveness and seen only in the presence of integrin ligands, whereas interstitial migration in lymph nodes is unaffected. By contrast, loss of WNK1 results in increased adhesion and decreased migration, but the latter phenotype is independent of the adhesion phenotype. Thus WNK1 inversely and independently regulates adhesion and migration and may act to balance these two related processes.
Online Methods
Mice
Mice with a conditional allele of Wnk1 (Wnk1tm1Clhu, Wnk1fl), expressing the dLck-Cre transgene (Tg(Lck-cre)#Nik), with a tamoxifen-inducible Cre in the ROSA26 locus (Gt(ROSA)26Sortm1(cre/ESR1)Thl, ROSA26-CreERT2, RCE), mice expressing OXSR1-T185A mice (Oxsr1T185A, Oxsr1tm1.1Arte), or RAG1-deficient (Rag1tm1Mom) were maintained on a C57BL/6JNimr background and have been described before13,15–17,37. Mice with a deleted allele of Wnk1 (Wnk1tm1.1Clhu, Wnk1-) were generated by crossing Wnk1fl/+ mice with C57BL/6J.129S4-Tg(Prm-cre)70Og/Nimr mice that delete in the male germline38. SLC12A2-deficient mice were maintained on an FVB/N background (Slc12a2tm1Ges)26. Both male and female mice were used. In all cases control and experimental mice were age- and gender-matched. No randomization or blinding was done. All mice were bred and maintained at the MRC National Institute for Medical Research (now The Francis Crick Institute). All experiments were carried out under the authority of a Project Licence granted by the UK Home Office.
Generation of Wnk1D368A mice
ES cells (C57BL/6J background) were targeted by standard methods with a targeting vector consisting of two regions of homology to the Wnk1 gene containing exon 3 and exon 4 respectively (genomic coordinates in GRCm38 Mmu6:119991956-120000272 and Mmu6:119983864-119991955), separated by an Frt-flanked neomycin resistance gene (Neo). Exon 3 in the targeting vector was mutated at two base pairs (Mmu6:119992389-119992390) to change aspartate 368 to alanine (wild-type sequence 5’-GGAGACCTT-3’, D368A mutant 5’-GGAGCTCTT-3’, mutated bases shown in bold). ES cells bearing the correctly targeted allele (Wnk1D368A-neo, Wnk1tm1Tyb) with both the Neo gene and the D368A mutation were used to generate mice by standard procedures. Wnk1D368Aneo mice were crossed to mice expressing the Flp recombinase in the germline (B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ) to delete the Neo gene and generate mice expressing WNK1-D368A (Wnk1D368A, Wnk1tm1.1Tyb), which were maintained on a C57BL/6JNimr background. Correct targeting was verified by Southern blotting, PCR and DNA sequencing. Construction of the targeting vector, targeting of ES cells and generation of mice was carried out by Biocytogen (Worcester, MA).
Radiation chimeras
To generate radiation chimeras, either bone marrow cells were harvested from Wnk1+/+dLck-Cre, Wnk1fl/fldLck-Cre, Wnk1fl/+RCE, Wnk1fl/-RCE, Wnk1fl/flRCE, Wnk1fl/D368ARCE, or from Slc12a2+/+ or Slc12a2-/- mice, or fetal livers were harvested from E14.5 embryos generated by intercrossing Oxsr1T185A/+ mice. RAG1-deficient animals (5-8 weeks of age) were irradiated with 5Gy using a 137Cs-source, and then reconstituted intravenously with at least 1x106 bone marrow cells/recipient or 0.5x106 fetal liver cells/recipient. All chimeric animals received Baytril in their drinking water (0.02%, Bayer Healthcare) for at least 4 wk post-transplantation. If required, 8-20 wk after reconstitution chimeric mice were injected intraperitoneally for 3 d with 2 mg/day of tamoxifen (Sigma) resuspended at 20 mg/ml in corn oil (Sigma) and analyzed 7 d after start of tamoxifen treatment.
RNA interference
E6.1 Jurkat T cells (gift from R. Zamoyska, negative for Mycoplasma) were transfected using a 96-well Shuttle Nucleofector device (Lonza) and the SE Cell Line 96-well Nucleofector Kit (Lonza). For each nucleofection 2x106 Jurkats were spun down, resuspended in 20 µl of Nucleofector solution containing 300 ng of total short interfering RNAs (siRNAs, Life Technologies), transfected with program 96-CL-120 and then placed into culture for 72 h.
Quantitative PCR
Jurkat cells were processed using the Cells to Ct Kit (Life Technologies). For mouse T cells, up to 1x105 CD44low splenic T cells from spleens of control or Wnk1 deficient animals were sorted directly into RLT lysis buffer, total RNA was extracted with an RNAEasy Plus Micro Kit (Qiagen) and cDNA was synthesized with a Superscript III kit (Life Technologies). Samples were analyzed on an ABI 7900 using TaqMan gene expression assays (Life Technologies). Data was normalized to HPRT1 and analyzed using the comparative threshold cycle method.
RNA sequencing (RNAseq)
CD4+ and CD8+ splenic T cells from female C57BL/6J mice were sorted into Trizol (Life Technologies) using a Beckman Coulter MoFlo XDP cell sorter. RNA was isolated using Trizol and cleaned up using RNEasy mini kit (Qiagen). Stranded polyA-enriched libraries were made using the Stranded TruSeq RNA Sample Preparation Kit (Illumina) and sequenced in the HiSeq 2500 (Illumina), collecting 22-60 million paired-end reads of 100 bases per sample. Paired-end reads were aligned against the mouse genome (build mm9) by Tophat v2.0.9 (default settings, except stranded protocol was used) followed by assessment of expression values in FPKM (fragments per kb per million reads). Data has been deposited in the Sequence Read Archive (SRA, accession number SRP059425).
Flow Cytometry, antibodies, cytokines and other materials
Flow cytometry was carried out using standard techniques with pre-titred antibodies. Antibodies for stimulations and flow cytometry against the following proteins were obtained from BioLegend or eBioscience: B220 (RA3-6B2), CCR7 (4B12), CD3ε (2C11 or OKT3), CD4 (RM4-5 or GKL-5), CD8 (53-6.7), CD11b (M1/70), CD11c (N418), CD19 (MB19-1), CD25 (3C7 or PC-61.5), CD44 (IM7), CD62L (MEL-14), LFA-1 (M17/4), TCRβ (H57-597 or IP26) and Vβ8 (F23.1). Further reagents: anti-CXCR4 (ID9, BD Pharmingen); anti-human LFA-1 (38, Abd Serotec); anti-Armenian hamster antibody (619-101-002, Rockland); murine TNF-α (PromoKine); murine CCL21 (PeproTech and R&D Systems); human and mouse ICAM1-Fc, human CXCR4 (R&D systems); cell dyes CMFDA, CFSE, CMTMR, CMAC and CTV (Life Technologies). Unless otherwise stated antibody dilutions were 1:200 for flow cytometry and T cell enrichment, and 1:1000 for immunoblotting experiments. Dilutions or concentrations for all other antibodies can be found in the appropriate methods section. All antibodies listed here and in subsequent sections have been validated by their suppliers and references can be found on their website or on the online validation databases Antibodypedia and 1DegreeBio. To measure intermediate and high affinity conformations of LFA-1, Jurkat T cells were transfected with siRNAs as described earlier and 72 h later were stimulated for 20 s at 37 °C in CaCl2 and MgCl2 containing HBSS, 0.5% BSA with either 10 µg/ml anti-CD3 or 0.2 µM CXCL12 in the presence of 10 µg/ml Kim127 or m24 antibodies (kind gifts of Nancy Hogg). The reaction was stopped with ice-cold CaCl2 and Mg Cl2 containing HBSS, 0.5% BSA and Kim127 and m24 binding was revealed by staining with anti-mouse IgG1 APC and analyzed by flow cytometry. To measure F-actin, T cells were rested for 2 h at 37°C in RPMI/10% FCS. T cells at 5 x 106 cells/ml were stimulated with CCL21 (10 μM) at 37°C for the indicated times, fixed in cold 4% PFA and stained intracellularly with Phalloidin-FITC (Millipore) before flow cytometric analysis.
Purification of T cells
Naïve CD44lowCD4+ T cells from peripheral and mesenteric lymph nodes were purified by negative selection. Single-cell suspensions were incubated with biotin-conjugated antibodies against CD8, CD11b, CD11c, CD25, CD44 and B220, and were then washed and incubated with Streptavidin-conjugated magnetic beads (Dynabeads, Life Technologies). Purity of CD44lowCD4+ T cells (typically 90%) was confirmed by flow cytometry. Alternatively T cells were isolated using an EasySep Mouse T cell Negative Selection Kit (StemCell). CD44+ lymphocytes were depleted using anti-CD44 magnetic beads in a 1:10 cells:beads ratio (Life Technologies). CD44low T cells purity (typically >95%) was verified by flow cytometry.
Conjugation
Jurkat T cells and Nalm6 B cells (DSMZ, negative for Mycoplasma) were labeled with 0.25 µM CMFDA and 1 µM CMTMR, respectively, for 20 min in PBS at 37°C. Jurkats (0.4x105 cells) were mixed with 3x105 Nalm6 cells in the absence or presence of staphylococcal enterotoxin E (Toxin Technology Inc.) in a total volume of 20 µl RPMI in 96 well v-bottom plates. After 20 min conjugation was stopped by adding 80 µl of 4% PFA. Samples were analyzed by flow cytometry. Percentage of conjugates was calculated by dividing the number double positive conjugates by the total number of Jurkat cells.
Mouse T cells: 2x105 CD44lowCD4+ T cells purified from Wnk1+/+dLck-Cre or Wnk1fl/fldLck-Cre mice were mixed with 2x105 CH27 B cells in the absence or presence of 1 µg/ml staphylococcal enterotoxin B (Sigma) in a total volume of 20 µl RPMI in 96 well V-bottom plates. After 20 min conjugation was stopped by adding 80 µl of 4% PFA and cells were stained with antibodies against CD4, CD44, Vβ8, CD19 and B220 and analyzed by flow cytometry. Percentage of conjugates was calculated by dividing the number of conjugates (Vβ8+CD19+B220+) by the total number of Vβ8+ cells.
Adhesion assays
Jurkat T cells: Black 96-well Maxisorp plates (Thermo Fisher) were coated overnight with 5 µg/ml recombinant human ICAM1-Fc in PBS at 4°C and blocked with 2% BSA in PBS for 2 h at room temperature. Jurkat T cells were labeled with 0.25 µM CMFDA in PBS for 20 min at 37°C and placed in the wells with appropriate stimulus in 100 µl HBSS at 106 cells/ml and incubated for 20 min at 37°C. The plates were analyzed on a 96-well fluorescence reader (Tecan Safire2) to determine the total fluorescence signal per well. After at least 3 washes with HBSS the fluorescence was measured again and percentage adhesion was calculated by dividing the fluorescence after washing with that before washing. Adhesion was normalized to the response of cells transfected with NT siRNAs.
Primary mouse T cells: V-bottom plates were either left uncoated or were coated overnight with 100 ng/ml recombinant mouse ICAM1-Fc in PBS at 4°C and blocked with 2% BSA in PBS for 2 h. Primary mouse CD44lowCD4+ T cells were labeled with 1 µM CMFDA for 20 min in PBS at 37°C, rested for 3 h in IMDM, 5% FCS at 37°C, and 2x104 cells were added to each well in 100 µl HBSS, 0.5% FCS containing the appropriate stimulus and incubated for 20 min at 37°C. The cells were stimulated with CCL21 (100 ng/ml) or anti-CD3 (2C11, 10 μg/ml). The plate was then centrifuged at room temperature for 10 min at 200 g and the fluorescence intensity at the bottom of the well was determined using a Tecan Safire2 plate reader. Data was normalized to the fluorescence signal in wells of control cells treated with anti-CD3ε (100% adhesion) and to unstimulated control cells in wells with no ICAM1-Fc, but blocked with BSA (0% adhesion).
Binding of ICAM1 complexes to Jurkat cells or primary mouse T cells was analyzed as described39. Soluble ICAM1-Fc-F(ab’)2 complexes were generated by diluting APC-labeled goat anti-human IgG F(ab’)2 fragments (109-135-098, Jackson Immunoresearch) 1:6.25 with ICAM1-Fc (200µg/ml final) in HBSS and incubated for 30 min in HBSS at 4°C. Splenocytes were rested for 3 h in IMDM, 5% FCS at 37°C, centrifuged and resuspended in HBSS, 0.5% BSA. Each adhesion reaction (50 µl) contained 20x106 cells/ml, 25 µg/ml ICAM1 complex and the appropriate stimulus and was incubated at 37°C for the indicated times. Jurkat cells were stimulated with CXCL12 (0.2 μM), anti-CD3 (10 μg/ml) or MnCl2 (2.5mM). Mouse T cells were stimulated with CCL21 (100 ng/ml), anti-CD3 (OKT3, 10 μg/ml) or MnCl2 (2.5mM). Cells were fixed in PFA for 20 min and binding of ICAM1 complexes to T cells analyzed by flow cytometry.
Migration
Migration assays were carried out in 96-well Transwell plates, containing polycarbonate filters (5 µm pore size, Corning). Transwell filters were coated overnight with 100 ng/ml human or 500 ng/ml mouse ICAM1-Fc in PBS (for Jurkat cells or mouse T cells respectively) and blocked with PBS, 2% BSA for 2 h. The receiver plate was filled with IMDM, 0.5% BSA, containing the appropriate stimulus, and 4x104 cells in IMDM, 0.5% BSA were added to each well of the filter plate. For SLC12A2 inhibitor experiments cells were pre-incubated with either DMSO (1:1000) or with 20 µM Bumetanide (Santa Cruz) in DMSO for 20 min and all subsequent steps were performed in the presence of DMSO or the inhibitor. After 3 h (Jurkat cells) or 1-2 h (mouse T cells) the filter plate was removed, EDTA was added to each well (40 mM final concentration) and the cells were transferred to 96 well V-bottom plates, spun, resuspended in PBS, 0.5% BSA, and analyzed on a flow cytometer. Percentage migration was calculated by dividing the number of cells that migrated through the filter by the total number of cells that added to each well. For mouse T cells, purified T cells were added to the top well and after migration, staining with antibodies to CD4, CD8 and CD44 was used to analyze migration of CD4+CD44low naïve T cells. For Jurkat cells, migration was normalized to the response of cells transfected with NT siRNAs. For mouse T cells, where data was combined from several experiments, migration was normalized to the response of WT cells to CCL21.
Chemokinesis
8-well Chamber Slides (Lab-Tek) were coated overnight with 3 µg/ml ICAM1-Fc and blocked for 2 h with PBS, 2% BSA. Mouse CD4+CD44low T cells were labeled with 1 µM CMFDA in PBS at 37°C, and rested in phenol-red free IMDM, 0.5% BSA, 20 mM HEPES at 37°C for at least 3 h. 1x105 cells were placed on the slides in the absence or presence of 200 ng/ml CCL21 and allowed to settle for 1 h. Cell migration was imaged with temperature control every 10 s over a 20 min period. Migration was analyzed using the Trackmate plugin in Fiji40. Flower plots were generated using a custom-made script in R software (http://www.r-project.org/) available upon request.
In vivo homing
C57BL/6J mice were injected intravenously with a 1:1 mixture of splenocytes from Wnk1fl/+RCE and Wnk1fl/-RCE radiation bone marrow radiation chimeras, labeled with 1 µM CMFDA or 1 µM CTV. Dyes were swapped in repeat experiments. After 1 h, blood, spleen and lymph nodes were harvested, stained with antibodies against CD4, CD8 and CD44 and analyzed by flow cytometry to determine the ratio between CMFDA and CTV labeled CD44lowCD4+ or CD8+ T cells.
3D immunohistology
CD44low T cells from Wnk1fl/+RCE and Wnk1fl/-RCE chimeric mice were labeled with CMAC or CMTMR for 15-20 min at 37°C, with dyes swapped between experiments. 3-4 x 106 cells were washed and injected intravenously at a 1:1 ratio into 5-10 week old C57BL/6J recipient mice. After 20 min, further adhesion of T cells to HEV was prevented by intravenous injection of anti-CD62L (Mel-14, 100 μg/mouse; Nanotools) in combination with Alexa Fluor 633-conjugated anti-PNAd (MECA-79, BD Biosciences, 15 µg/mouse) for visualization of HEVs by staining of PNAd. After a further 20 min (40 min after transfer) mice were sacrificed and perfused using 10 ml cold PBS and 10 ml cold 4% PFA. PLNs were processed as described3. Images were visualized with Volocity software and T cells were counted manually and differentiated between cells in the lumen of HEV (“luminal”), attached to the outer MECA-79 signal (“perivascular”) and cells in the parenchyma of the peripheral lymph node (“parenchymal”). Alternatively mice were analyzed 20 min after transfer of T cells without injection of Mel-14.
Spleen histology
C57BL/6J mice were injected intravenously with a 1:1 mixture of CD44lowCD4+ T cells purified by negative depletion from Wnk1fl/+RCE and Wnk1fl/-RCE radiation bone marrow chimeras and labeled with 1 µM CMFDA and 5 µM CMTMR, respectively. Dyes were swapped in repeat experiments. After 1 h spleens were harvested and embedded in optimum cutting temperature compound (BDH) and frozen. 8 µm sections were fixed with 4% PFA and blocked with 2% goat serum in PBS. To identify the white pulp sections were stained with anti-MADCAM1 (1:50, MECA-367, eBioscience) and an Alexa Fluor 647-goat anti-rat IgG secondary antibody (1:400, A-21247, Life Technologies). Images were acquired using an Olympus VS120 slide scanner and analyzed by drawing regions of interest around the spleen and the white pulp areas and automated cell counting with the spot detector plugin in Icy41. Numbers of cells were first normalized to the red and white pulp area, respectively and a ratio of cell density in the white pulp to red pulp was then calculated.
T cell interaction with endothelial cells under physiological shear stress
Primary mouse brain microvascular endothelial cells (pMBMECs) were isolated from C57BL/6J mice as previously described42, cultured for 16–18 h in the presence of TNFα (10 ng/ml) and covered with 1 µM CCL21 prior to the experiment. All experiments were performed in migration assay medium (DMEM, 5% calf serum, 25 mM HEPES) at 37°C. CD44low T cells isolated from Wnk1fl/+RCE and Wnk1fl/-RCE mice were labeled with CFSE or CMTMR for 15-20 min at 37°C and mixed at a 1:1 ratio for perfusion into a custom made flow chamber mounted on the pMBMEC monolayer. T cells accumulated on the monolayer at a flow rate of 0.1 dyn/cm2 for 4 min, followed by physiological shear at 1.5 dyn/cm2 for 12-15 min. In vitro time-lapse imaging was performed with an inverted microscope (AxioObserver, Zeiss) as described43 and analyzed using Imaris 8.0 software. Arrest coefficient was defined as percentage of time that cells migrated ≤2 μm/min. Diapedesis events were evaluated manually, starting with protrusion formation of arrested T cells below the pMBMEC monolayer until conclusion of diapedesis assessed by phase contrast changes of transmigrated T cells (“phase bright” on top of and “phase dark” below the pMBMEC monolayer).
Two-photon intravital microscopy
CD44low T cells were fluorescently labeled with CellTracker® Orange or blue or with e670 or CFSE as described44, with dyes swapped between experiments. 2-3 x 106 cells were adoptively transferred 18-24 h prior to imaging into recipient mice. 2PM imaging of right popliteal lymph nodes was as described44 using the TrimScope system (LaVision Biotec, Germany) and a 20X objective (NA 0.95, Olympus) and a MaiTai Ti:Sa laser tuned to 780-840 nm. For four-dimensional analysis of cell migration, 11-16 z-stacks of 250–300 × 250–300 μm x–y sections with 4 μm z spacing were acquired every 20 s for 20-30 min. Volocity software (Perkin-Elmer) was used for cell migration analysis. The average track velocity and turning angles (defined as the angle between the two velocity vectors before and after a measurement time point) were calculated from the x, y, and z coordinates of cell centroids as described3. Motility coefficient was calculated from slope of displacement against √(time) in Fig. 4d. Skeletal length and circularity were measured only from cells whose migrating cell body was entirely visualized in the imaged z-volume, 3D movies were converted into 2D projection and analyzed with Volocity software. In experiments where integrins were blocked, mice were injected with 100 μg/mouse of anti-LFA-1 (FD448.1) and anti-α4 integrin (PS/2; both from Nanotools, Germany) antibodies 15-18 h after transfer of T cells. Two-photon intravital imaging of popliteal lymph nodes was performed 12-15 h after integrin blockade, when antibodies had saturated integrins on transferred T cells (not shown).
Measurement of sticking fraction
Inguinal lymph nodes of C57BL/6 recipient mice were surgically exposed for wide-field fluorescent microscopy and 1-1.5 x 107 cells CFSE-labeled CD44low T cells from Wnk1fl/+RCE or Wnk1fl/-RCE mice were separately transferred i.v. From off-line video rate image analysis, the ratio (sticking fraction) of firmly adherent over combined rolling and firmly adherent cells was determined as described3.
Calcium flux
Jurkat cells were labeled with 2 mM Indo-1-AM at room temperature for 45 min and stimulated by the addition of 5 µg/ml anti-CD3ε antibody. Ca2+ flux was measured on an LSRII flow cytometer.
Immunoblotting
Jurkat cells: 2x106 Jurkat T cells were stimulated in RPMI at 37°C either CXCL12 (0.2 μM) or anti-CD3ε antibody (1 µg/ml). Reactions were stopped by adding ice-cold PBS and cells were spun down and lysed in RIPA buffer containing protease inhibitors. Samples were analyzed by standard immunoblotting techniques using anti-phospho ERK1/2 (44-680G, Life Technologies) and anti-ERK1/2 (44-654G, Life Technologies) as a loading control.
Primary mouse T cells
CD44lowCD4+ T cells from peripheral and mesenteric lymph nodes of indicated mouse lines were purified by negative selection as described earlier. Stimulation of the cells through the TCR by crosslinking with antibody against CD3ε (10 μg/ml) and subsequent immunoblotting analysis were performed as described previously45. For stimulation with CCL21 (500 ng/ml for all studies except RAP1 activation where 100 ng/ml was used) cells were first rested for 3 h in IMDM, 5% FCS at 37 °C. For the inhibitor studies cells were first pre-incubated for 1 h in the presence of either DMSO (1:1000), 1 µM PI-103 (BioVision) or 2 µM MK2206 (Cambridge Bioscience Ltd.) both in DMSO and then stimulated as described above in the presence of the same inhibitors. The following antibodies were used for detection of proteins by Western blotting: anti-phospho ERK1/2 (44-680G, Life Technologies) and anti-ERK1/2 (44-654G, Life Technologies), anti ERK2 (C-14, Santa Cruz), anti-pSer325-OXSR1/pSer383-STK39 (D. Alessi, University of Dundee, anti-ACTIN (C-11, Santa Cruz). To measure activation of RAP1, RAP1-GTP was pulled down using the EZ-Detect Rap1 activation kit (Thermo Scientific) according to the manufacturer’s instructions, and this pull down as well as total cell lysates were analyzed by immunoblotting SDS-PAGE gels with anti-RAP1. Signals for phosphorylated proteins or RAP1-GTP were normalized to loading controls and then to the sum of the signals across multiple experiments, as described46.
Supplementary Material
Acknowledgements
We thank Dario Alessi (University of Dundee) for Oxsr1T185A mice, antibodies and discussions, Rose Zamoyska (University of Edinburgh) for cells, Donald Bell (Francis Crick Institute) for help with image analysis, Michael Abadier (Theodor Kocher Institute) for isolation and cultivation of pMBMECs, Nancy Hogg (Francis Crick Institute) for antibodies, and Lisa Satlin and Cindy Else (Mount Sinai Medical Center) for access to Slc12a2 mutant mice (supported by the NIH NIDDK Grant P30 DK079307, PITTSBURGH CENTER FOR KIDNEY RESEARCH, Core B). VT was funded by the MRC programme number U117527252 and by the Francis Crick Institute which receives its core funding from the MRC, Cancer Research UK and the Wellcome Trust. RK and VT were funded by Wellcome Trust grant 089185 and BBSRC grant BB/L00805X/1, and C-LH was funded by NIH grant DK59530. RL was funded by the Swiss Multiple Sclerosis Society.
Footnotes
Accession code
RNAseq data has been deposited in the Sequence Read Archive (SRA, accession number SRP059425).
Author Contributions
RK, FT, LV, TB, KF, RL designed and performed experiments and analyzed data; JX and C-LH provided a mouse strain; JVS and VLJT designed experiments and analyzed data; RK, FT, RL, JVS, VLJT wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Girard JP, Moussion C, Forster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 2.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 3.Boscacci RT, et al. Comprehensive analysis of lymph node stroma-expressed Ig superfamily members reveals redundant and nonredundant roles for ICAM-1, ICAM-2, and VCAM-1 in lymphocyte homing. Blood. 2010;116:915–925. doi: 10.1182/blood-2009-11-254334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolf E, et al. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat Immunol. 2007;8:1076–1085. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- 5.Alon R, Feigelson SW. Chemokine-triggered leukocyte arrest: force-regulated bidirectional integrin activation in quantal adhesive contacts. Curr Opin Cell Biol. 2012;24:670–676. doi: 10.1016/j.ceb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Shulman Z, et al. Lymphocyte crawling and transendothelial migration require chemokine triggering of high-affinity LFA-1 integrin. Immunity. 2009;30:384–396. doi: 10.1016/j.immuni.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24:107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feigelson SW, et al. Occupancy of lymphocyte LFA-1 by surface-immobilized ICAM-1 is critical for TCR- but not for chemokine-triggered LFA-1 conversion to an open headpiece high-affinity state. J Immunol. 2010;185:7394–7404. doi: 10.4049/jimmunol.1002246. [DOI] [PubMed] [Google Scholar]
- 9.Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat Immunol. 2006;7:919–928. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- 10.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 11.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM Activates Integrins by Linking Talin to Ras GTPase Membrane-targeting Sequences. J Biol Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormick JA, Ellison DH. The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev. 2011;91:177–219. doi: 10.1152/physrev.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafiqi FH, et al. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol Med. 2010;2:63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burbach BJ, Medeiros RB, Mueller KL, Shimizu Y. T-cell receptor signaling to integrins. Immunol Rev. 2007;218:65–81. doi: 10.1111/j.1600-065X.2007.00527.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang DJ, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol. 2005;174:6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]
- 16.Xie J, et al. Endothelial-specific expression of WNK1 kinase is essential for angiogenesis and heart development in mice. Am J Path. 2009;175:1315–1327. doi: 10.2353/ajpath.2009.090094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Luca C, et al. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest. 2005;115:3484–3493. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebzda E, Bracke M, Tugal T, Hogg N, Cantrell DA. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat Immunol. 2002;3:251–258. doi: 10.1038/ni765. [DOI] [PubMed] [Google Scholar]
- 19.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. 2008;9:960–969. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 20.Steiner O, Coisne C, Engelhardt B, Lyck R. Comparison of immortalized bEnd5 and primary mouse brain microvascular endothelial cells as in vitro blood-brain barrier models for the study of T cell extravasation. J Cereb Blood Flow Metab. 2011;31:315–327. doi: 10.1038/jcbfm.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiuri P, et al. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 22.Alessi DR, et al. The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci Signal. 2014;7:re3. doi: 10.1126/scisignal.2005365. [DOI] [PubMed] [Google Scholar]
- 23.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zambrowicz BP, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng CJ, Huang CL. Activation of PI3-kinase stimulates endocytosis of ROMK via Akt1/SGK1-dependent phosphorylation of WNK1. J Am Soc Nephrol. 2011;22:460–471. doi: 10.1681/ASN.2010060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flagella M, et al. Mice lacking the basolateral Na-K-2Cl cotransporter have impaired epithelial chloride secretion and are profoundly deaf. J Biol Chem. 1999;274:26946–26955. doi: 10.1074/jbc.274.38.26946. [DOI] [PubMed] [Google Scholar]
- 27.Jiang ZY, et al. Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J Biol Chem. 2005;280:21622–21628. doi: 10.1074/jbc.M414464200. [DOI] [PubMed] [Google Scholar]
- 28.Vitari AC, et al. WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem J. 2004;378:257–268. doi: 10.1042/BJ20031692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuddapah VA, Sontheimer H. Ion channels and transporters in cancer. 2. Ion channels and the control of cancer cell migration. Am J Physiol Cell Physiol. 2011;301:C541–549. doi: 10.1152/ajpcell.00102.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stroka KM, et al. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas BR, Sontheimer H. Inhibition of the Sodium-Potassium-Chloride Cotransporter Isoform-1 reduces glioma invasion. Cancer Res. 2010;70:5597–5606. doi: 10.1158/0008-5472.CAN-09-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiller HB, Friedel CC, Boulegue C, Fassler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faroudi M, et al. Critical roles for Rac GTPases in T cell migration to and within lymph nodes. Blood. 2010;116:5536–5547. doi: 10.1182/blood-2010-08-299438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park EJ, et al. Distinct roles for LFA-1 affinity regulation during T-cell adhesion, diapedesis, and interstitial migration in lymph nodes. Blood. 2010;115:1572–1581. doi: 10.1182/blood-2009-08-237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semmrich M, et al. Importance of integrin LFA-1 deactivation for the generation of immune responses. J Exp Med. 2005;201:1987–1998. doi: 10.1084/jem.20041850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 38.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstandin MH, et al. A novel flow-cytometry-based assay for quantification of affinity and avidity changes of integrins. J Immunol Methods. 2006;310:67–77. doi: 10.1016/j.jim.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Chaumont F, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 42.Lyck R, et al. Culture-induced changes in blood-brain barrier transcriptome: implications for amino-acid transporters in vivo. J Cereb Blood Flow Metab. 2009;29:1491–1502. doi: 10.1038/jcbfm.2009.72. [DOI] [PubMed] [Google Scholar]
- 43.Steiner O, et al. Differential roles for endothelial ICAM-1, ICAM-2, and VCAM-1 in shear-resistant T cell arrest, polarization, and directed crawling on blood-brain barrier endothelium. J Immunol. 2010;185:4846–4855. doi: 10.4049/jimmunol.0903732. [DOI] [PubMed] [Google Scholar]
- 44.Soriano SF, et al. In vivo analysis of uropod function during physiological T cell trafficking. J Immunol. 2011;187:2356–2364. doi: 10.4049/jimmunol.1100935. [DOI] [PubMed] [Google Scholar]
- 45.Reynolds LF, et al. Vav1 transduces T cell receptor signals to the activation of phospholipase C-γ1 via phosphoinositide 3-kinase-dependent and -independent pathways. J Exp Med. 2002;195:1103–1114. doi: 10.1084/jem.20011663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Degasperi A, et al. Evaluating strategies to normalise biological replicates of Western blot data. PLoS One. 2014;9:e87293. doi: 10.1371/journal.pone.0087293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.