Abstract
The ability to write a stable record of identified molecular events into a specific genomic locus would enable the examination of long cellular histories and have many applications, ranging from developmental biology to synthetic devices. We show that the type I-E CRISPR-Cas system of E. coli can mediate acquisition of defined pieces of synthetic DNA. We harnessed this feature to generate records of specific DNA sequences into a population of bacterial genomes. We then applied directed evolution to alter the recognition of a protospacer adjacent motif by the Cas1-Cas2 complex, which enabled recording in two modes simultaneously. We used this system to reveal aspects of spacer acquisition, fundamental to the CRISPR-Cas adaptation process. These results lay the foundations of a multimodal intracellular recording device.
Main Text
DNA has the potential to encode, preserve, and propagate information (1). The precipitous drop in DNA sequencing cost has now made it practical to read out this information with high throughput (2). However, the ability to write arbitrary information into DNA, in particular within the genomes of living cells, has been restrained by a lack of biologically compatible recording systems that can exploit anything close to the full encoding capacity of nucleic acid space.
A number of approaches aimed at recording information within cells have been explored (3). These systems can be broadly divided into those that alter transcription through feedback loops and toggles (4–14), and those that encode information permanently into the genome, most often using recombinases to store information via the orientation of DNA segments (15–19). Although the majority of these systems are effectively binary, efforts have also been made toward analogue recording systems (20) and digital counters (21). Despite these efforts, the recording and genetic storage of little more than a single byte of information (18) has remained out of reach.
Immunological memory is essential to an organism’s adaptive immune response, and hence must be an efficient and robust form of recording molecular events in living cells. The CRISPR-Cas system is a recently understood form of adaptive immunity used by bacteria and archaea (22). This system records past infections by storing short sequences of viral DNA within a genomic array. These acquired sequences are referred to as protospacers in their native viral context, and spacers once they are inserted into the CRISPR array. New spacers are integrated into the CRISPR array ahead of older spacers (23). Over time, a long record of spacer sequences can be stored in the genomic array, arranged in the order in which they were acquired. Thus, the CRISPR array functions as a high capacity temporal memory bank of invading nucleic acids.
We harnessed the CRISPR-Cas system to record specific and arbitrary DNA sequences into a bacterial genome. We could generate a record of defined sequences, recorded over many days, and in multiple modalities. In exploring this system, we also elucidated fundamental aspects of native CRISPR-Cas spacer acquisition and leveraged this knowledge to enhance the recording system.
A type I-E CRISPR-Cas system accepts synthetic spacers in vivo
Overexpression of the E. coli type I-E CRISPR-Cas proteins Cas1 and Cas2 is sufficient to drive acquisition of new spacers in a strain containing two genomic CRISPR arrays but lacking endogenous Cas proteins (BL21-AI) (23). We replicated this result (Fig. 1A), and similarly found that new spacers were consistently integrated into the first position of array I directly adjacent to the leader with a consistent size of 33 bases (fig. S1A–B). These spacers were drawn in roughly equal number from the cell’s own genome and from the plasmid used to overexpress Cas1 and Cas2 (Fig. 1B). Considering the overall DNA content of the cell, this ratio of genome-to-plasmid-derived spacers represents a substantial bias toward the plasmid as a protospacer source (24). Despite this bias, new spacers were drawn from a diverse range of sites around the genome and plasmid (Fig. 1C) and, besides the overrepresentation of a 5′ AAG protospacer adjacent motif (PAM), there was no way to predict a priori the full sequence of a new spacer without sequencing the expanded array.
Fig. 1.
Acquisition of synthetic spacers. (A) Schematic of the minimal elements of the type I-E CRISPR acquisition system, used including Cas1, Cas2, and array with leader (L), repeat (R), and spacer (S) along with PCR detection of an expanded array following the overnight induction of Cas1-Cas2. (B) Origin of new spacers (plasmid or genome) mean ±SEM. (C) Genome- and plasmid-derived spacers following overnight induction are mapped back to the approximate location of their protospacer (marked in red). (D) Array expansion (top) and specific acquisition of synthetic oligo protospacer (bottom) following electroporation. Top schematic shows the experimental outline. Schematics under each gel show specific PCR strategy. (E) Sequence-specific acquisition in either the forward (top) or reverse (bottom) orientation following electroporation with various single- and double-stranded oligos. 5′PT indicates phosphorothioate modifications to the oligos at the 5′ ends. (F) Time course of expansion following electroporation, mean ±SEM. (G) Percent of arrays expanded by spacer source as a function of electroporated oligo concentration, mean ±SEM. (H) Position of new spacers relative to the leader, mean ±SEM. (I) Size of new spacers in base-pairs, mean ±SEM. All gels are representative of ≥ 3 biological replicates, * indicates p<0.05, additional statistical details in Table S1.
To extend the function of the CRISPR acquisition system into a synthetic device for recording molecular events, it is necessary to direct the system to capture spacers of specific, defined sequence. In vitro, Cas1 and Cas2 can mediate integration of synthetic 33-bp DNA oligos into plasmid-based arrays (25). We reasoned that similarly supplying an exogenous source of protospacers to the system within a cell might direct sequence-specific spacer acquisition in-vivo. We therefore passaged an overnight culture of E. coli BL21-AI containing arabinose- and isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible Cas1 and Cas2 genes with or without arabinose and IPTG for two hours. We then electroporated the cells with a complementary pair of 33 base oligos (protospacer ps33), which matched the sequence of the most abundant M13-derived spacer found after phage infection of a native type I-E system (26). After incubating the cells for another two hours after transformation, we checked the genomic array for expansion and specific integration of the synthetic protospacer into the array by PCR (Fig. 1D). By using the reverse sequence of the supplied oligo as the reverse primer, we also observed amplification of specifically-sized PCR products that confirmed acquisition of the oligo-supplied sequence when Cas1 and Cas2 were induced or (more weakly) uninduced, but never for the case in which the oligos were not supplied. We confirmed the specific ps33 nucleotide sequence was present within a fraction of the expanded arrays by Sanger sequencing. These results demonstrate that the CRISPR-Cas system acquired a sequence-specific spacer.
To better understand both the properties of this synthetic system, as well as the fundamental properties of Cas1-Cas2-mediated spacer acquisition, we we altered the oligos that we provided via electroporation. The system required both complementary strands for acquisition, and the double-stranded protospacer could insert in either direction (Fig. 1E). We modified the 5′ ends of the oligos with phosphorothioate bonds to help resist degradation by cellular nucleases, but found no differences in acquisition efficiency (Fig. 1E). We tested whether RNA could serve as a protospacer by supplying either one or both of the oligo strands as RNA, but detected no sequence-specific integration of RNA oligos (fig. S1D).
To investigate these results more quantitatively, we performed a PCR across the array (as in Fig. 1D) and subjected the resulting amplicon to high-throughput sequencing on an Illumina MiSeq platform. We quantified the percentage of all arrays that were expanded at the completion of an experiment, as well as the spacer source. Coupled with qPCR, we generated a time course of spacer acquisition (Fig. 1F). Sequence-specific acquisitions occurred as early as 20 minutes after electroporation, reaching ~4% of all arrays by two hours. The oligo concentration required to achieve spacer acquisition was determined by testing a two-fold dilution series (Fig. 1G, fig. S1E). Whether oligos were delivered or acquired as spacers had no effect on the genome- or plasmid-derived spacers. Thus, protospacer availability in the cell may be a limiting factor in spacer acquisition. On the other hand, the addition of an additional CRISPR array on the expression plasmid had little to no effect on the acquisition frequency of new spacers into the endogenous genomic array (Fig. 1G). Like genome- and plasmid-derived spacers, the synthetic spacers were inserted into the first (or occasionally first and second) positions of the array, and the great majority were of 33 bases (Fig. 1H, I). Loss of previously acquired spacers has been reported both in the presence (27, 28) and absence (29, 30) of selective pressure. While our analysis was restricted to the leader-proximal spacers, we did find rare instances in which the previous first spacer was deleted (0.096% of arrays sequenced ±0.012 SEM).
PAMs modify the efficiency and directionality of spacer acquisition
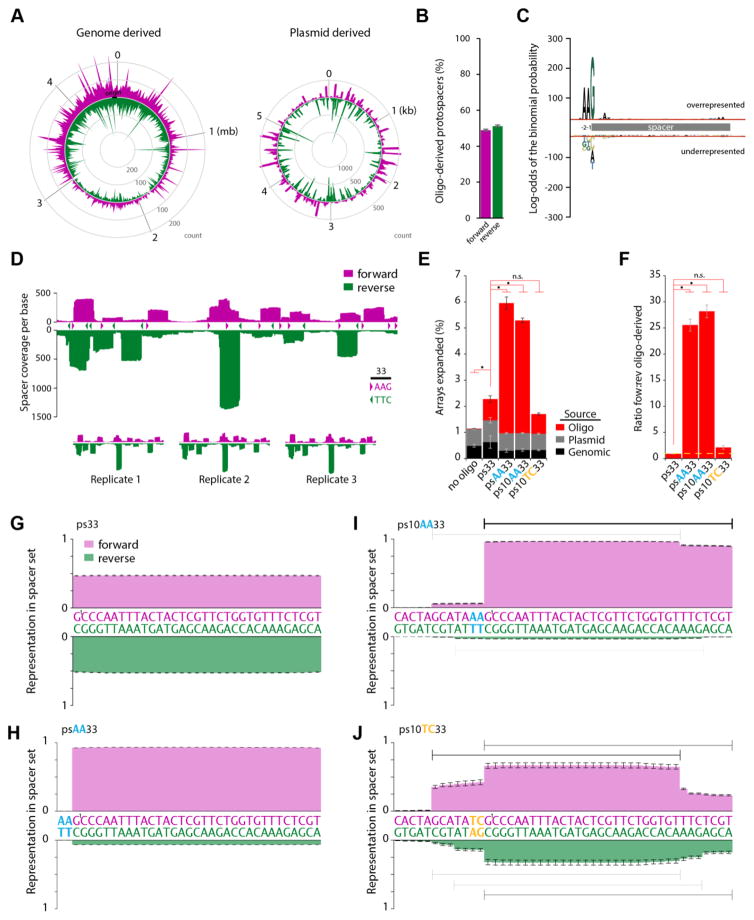
Data from sequencing millions of expanded arrays showed that genome- and plasmid-derived protospacers were drawn in equivalent numbers from the forward and reverse strands overall, with the only apparent bias being toward the genomic origin of replication (Fig. 2A). Similarly, oligo-derived protospacers were found in equal proportions in the forward and reverse orientation in the array (Fig. 2B). When we further examined the context of the genomic- and plasmid-derived protospacers, we found strong evidence for a PAM on the 5′ end of the protospacer consisting of two adenines at positions -2 and -1 from the spacer and a strong bias for a guanine as the first spacer base (Fig. 2C). This is largely consistent with previous characterizations of the E. coli type I-E system (31, 32). An interior sequence motif at the 3′ end of the spacer termed the acquisition affecting motif or “AAM” has also been reported for this system (31). We find spacer sequences that are consistent with the presence of this interior motif, but the frequency of its occurrence is minor compared with the 5′ PAM.
Fig. 2.
PAMs modify the efficiency and orientation of spacer acquisition. (A) Genome- (count/10 kb) and plasmid- (coverage/base) derived spacers mapped to their protospacer location on the forward (purple) or reverse (green) strands. (B) Direction of oligo-derived spacers in the forward (purple) or reverse (green) orientation, mean ±SEM. (C) Representative sequence pLOGO (44) generated based on 896 unique genome- and plasmid-derived protospacers. Five bases of the protospacer are included at each end of the spacer. (D) Plot of the summed spacer coverage mapped to the plasmid among three replicates at each nucleotide for a 553 nucleotide stretch. Carrots demarcate canonical PAMs on the forward (purple) or reverse (green) strand. Scale bar is 33 bases. Individual replicates are shown below. (E) Percent of arrays expanded by spacer source for different oligo protospacers, mean ±SEM. (F) Ratio of oligo-derived spacers acquired in the forward vs reverse orientation for different oligo protospacers, mean ±SEM. (G–J) Normalized representation of oligo-derived spacers by base acquired in the forward and reverse direction for each oligo. Bars in I and J are 33 bases long to show dominant and minority spacers drawn from the oligo protospacers. For all panels, * indicates p<0.05, additional statistical details in Table S1.
Although there is no bias in forward- or reverse-strand-derived protospacers from the genome or plasmid on the whole, a sharper picture emerged at the level of individual nucleotides. For example, examining one small stretch of the plasmid (~550 bases), asymmetric peaks of spacer coverage – that is, the cumulative count of each time a given nucleotide was observed within an acquired spacer – emerged (Fig. 2D). Plotting the forward and reverse PAMs along the same stretch of plasmid revealed that, in addition to biasing toward specific sequences for acquisition, the PAM also specified the orientation of integration into the array. Although nearly every protospacer that contained a PAM was acquired as a spacer, not all were acquired at the same frequency (Fig. 2D).
The presence of Chi sites – an eight base motif where double-strand break repair is more likely to occur – within a genome or plasmid bias the frequency of protospacer acquisitions (24). However, we wondered whether the sequence of the protospacer itself might also bias acquisition frequency. We ranked every PAM (AAG)-containing potential protospacer in the plasmid according to the frequency at which it was acquired into the genomic array (fig. S2A). We searched for characteristics among protospacers including GC percentage and free energy that might explain the difference in acquisition frequency, but failed to identify a correlation (fig. S2B, C). For a direct test, we selected and synthesized three protospacer sequences (including their 15-bp flanking regions): one each from the high (psH), middle (psM), and low (psL) end of the frequency spectrum (fig. S2A). We then electroporated each of these oligo protospacers into cells expressing Cas1-Cas2 from an alternate plasmid that did not include these particular sequences. psL was acquired much less frequently than psH or psM (fig. S2F). To determine whether this was caused by the sequence of the spacer itself or a flanking region, we swapped the 15-bp flanking regions of psH with those of psL, and vice versa (psH/L and psL/H, respectively). Again, the psL/H spacer was acquired at a lower frequency than was psH/L, independent of the flanking regions. These results indicate the sequence of the protospacer itself influences the efficiency of acquisition. We do not know, however, the mechanism of this effect, whether by a direct effect on the acquisition process itself or by indirect effects such as sequence dependent interactions with endogenous nucleotides, competing proteins, or degradation.
Given that spacers are selected from the genome and plasmid according to an adjacent sequence, we wondered whether the inclusion of a PAM in our synthetic protospacer ps33 would alter acquisition frequency. We designed three additional oligo protospacers: psAA33, in which two adenines were included at the 5′ end of ps33 to create the entire canonical AAG PAM; ps10AA33, which includes an additional ten 5′ nucleotides; and ps10TC33, in which the AA of the PAM was mutated to TC to create a non-canonical PAM (PAMNC). Using these oligos, we found that the inclusion of a PAM greatly increased the efficiency of sequence-specific acquisition (Fig. 2E). Whether preceded by ten extra nucleotides or not, oligos with the AAG PAM (psAA33 and ps10AA33) were acquired at greater than 5 times the frequency of those that did not include a PAM (ps33). Conversely, including the TCG PAMNC did not change acquisition frequency relative to ps33 (Fig. 2E).
In line with what has been previously observed for the PAM motif in CRISPR adaptation – that it is consistently localized to the leading rather than trailing end of the integrated spacer (24, 31, 33–36) – the inclusion of a PAM also altered the orientation frequency of oligo-derived spacer acquisition. Whereas ps33 and ps10TC33 were acquired equally in both orientations, psAA33 and ps10AA33 were acquired almost exclusively in the forward orientation (Fig. 2F–J, fig. S3A). Consistent with the type I-E preference for an AAG PAM, psAA33 and ps10AA33 were consistently inserted with nucleotide G1 as the first base of the spacer (Fig. 2H, I). In contrast, ps10TC33 lacked a single dominant spacer product, and was inserted at several different PAMsNC (Fig. 2J). We verified that both Cas1 and Cas2 were necessary for synthetic spacer integration, whereas Cas2 nuclease activity was not required (25) (fig. S3B, C). Therefore, the inclusion of a PAM in synthetic protospacers dictates both the efficiency and orientation of the spacer that is acquired by the Cas1-Cas2 complex.
A molecular recording over time
We tested whether we could harness the acquisition of specific spacer sequences to record a series of synthetic spacers into a population of cells over time. As an initial test, we recorded three unique elements (1 × 3) into a single culture of E. coli by sequentially electroporating a series of three different oligo protospacer sequences into the culture, over a period of three days (one protospacer each day) (fig. S4A ). After sequencing a population of the arrays on day three, we could reconstruct the order in which the spacers were delivered (fig. S4B and C, and discussed in detail below). To further probe the limits of this system, we recorded fifteen distinct elements (3 × 5): three sets of five protospacers, electroporated three-at-a-time over five days (Fig. 3A). The analysis of both the 1 × 3 and 3 × 5 recordings are conceptually similar so we will discuss the latter in detail (fig. S4B and Fig. 3B, respectively).
Fig. 3.
A molecular recording over time. (A) Experimental outline of the 3 × 5 recording. Over five days, three sets of five oligo protospacers (fifteen elements) were electroporated (one protospacer from each of the three sets each day) into cells expressing Cas1-Cas2. Time points at which cells were sampled for sequencing are numbered 1–6. (B) Schematic illustrating all possible pairwise ordering of new spacers. G/P denotes a spacer derived from the genome or plasmid. Ordering rules are shown below. In the case of y=z, * indicates a tolerance within ± 20% of the mean of both values. (C) At each of the six sample points (marked in A), percent of all arrays expanded with synthetic spacers from each of the indicated rounds, mean ±SEM. (D) Single, double, and triple expansions for each round, mean ±SEM. (E) Percent of all expansions at sample point six, broken down by electroporation round and set. Open circles are individual replicates, filled bars are mean ±SEM. (F) Results of ordering rule analysis for one replicate across each set. For all 120 permutations, results of the tested rule are shown (green indicates pass, red indicates fail). For all sets, only one permutation passed all rules and in every case that permutation matched the actual order in which the oligos were electroporated (as indicated by check mark). Additional statistical details in Table S1.
For the 3 × 5 recording, all oligo protospacers consisted of 35 nucleotides, beginning with a 5′ AAG PAM followed by a 5-base-barcode (unique to each of the 3 sets) and 27 more bases (unique to each of the 15 protospacers). At the end of the 3 ×5 recording, nearly a quarter of all arrays in the cell population contained at least one oligo-derived spacer, with spacers from each round of electroporation represented in roughly equivalent proportions (Fig. 3C, D). Individual variations among the spacer acquisition frequency were more heavily driven by spacer nucleotide sequence than by the round in which they were acquired (Fig. 3E), while loss of recorded spacers after acquisition was rare (0.076% ±0.182 S.E.M.).
Because of the low probability of acquiring spacers from every round in any single array (Fig. 3D), successful readout of the recording required analysis of a population of arrays. Therefore, we sequenced the first three spacers of each array (moving in from the leader), and considered only the order of pairs of newly acquired spacers (Fig. 3B). For any given synthetic spacer pair within the same set, the order should follow a predictable rule: among all arrays that contain any two new spacers, a spacer electroporated in an earlier round will always be found further from the leader than a spacer introduced at a later round. We also gained information by considering the arrangement of oligo-derived spacers in relation to newly acquired genome- and plasmid-derived spacers. Because the endogenous spacers will accumulate over time, synthetic spacers from an earlier round will be paired more often with a new genome/plasmid spacer in one direction (toward the leader) than in the other (relative to the synthetic spacer), and vice versa for oligo-derived spacers from a later round. With five possible spacers (in each set), we considered all possible pairwise comparisons and generated 15 ordering rules from which we can reconstruct the order of the entire set (Fig. 3B). We took the sequences of arrays after the completion of the 3 × 5 recording and passed them through an algorithm that, with the only sequence-based input being the sequence of the CRISPR repeat, would predict all oligo-derived spacer sequences, assign them to a set based on the barcodes, and then test all possible permutations of the sequence against the 15 ordering rules. For each set, only one permutation satisfied all 15 ordering rules, and in every case that permutation matched the actual order of electroporated oligos (Fig. 3F). Although we analyzed ~2 million reads for each replicate, we found that order could be correctly reconstructed in most cases with 20,000 reads or fewer. Thus, we could reliably record and read out the fifteen element recording.
Cas1-Cas2 PAM recognition can be modified
The ability to control not only the sequence of new spacers, but also the orientation of new spacer integration would enable recording of information in multiple modalities simultaneously. Because the addition of a 5′ AAG PAM on our synthetic spacers controlled the orientation of new acquisitions (Fig. 2F), we sought to modify integration orientation by altering PAM recognition of Cas1-Cas2. To do this, we performed the directed evolution approach shown in Figure 4A. First, we generated a large library of random Cas1-Cas2 mutants by error-prone PCR (fig. S5A, B), and inserted this library into a plasmid upstream of a minimal CRISPR array. After cloning the plasmid library into BL21-AI, we induced and transformed mutants with a protospacer bearing the canonical 5′ AAG PAM on the forward strand, and a non-canonical 5′ TCG PAMNC on the reverse strand. After outgrowth, we selected mutants using a forward primer ahead of the Cas1-Cas2 mutant genes, and a reverse primer matching the PAMNC spacer sequence to yield specific amplification of only those mutants that had acquired the spacer in the (reverse) PAMNC orientation. A subset of these selected mutants were then tested for PAM specificity, and a separate subset were subjected to another round of selection for refinement before testing. For testing, individually selected mutant clones were induced overnight, and their expanded arrays were analyzed by sequencing. Specifically, we analyzed the PAMs of the all genome- and plasmid-derived spacers to determine what, if any, PAM specificity remained. Wild-type Cas1-Cas2 acquires spacers from AAG PAM protospacers at nearly the same frequency as from all other (non-AAG) PAM protospacers combined (Fig. 4B). In contrast, the majority of mutants we selected acquired non-AAG protospacers at a greater frequency than AAG protospacers (Fig. 4B). There was no gain in non-AAG acquisition frequency from the extra step of refinement (fig. S5C), so mutants from both subsets are shown together (Fig. 4B and fig. S5D).
Fig. 4.
Directed evolution of PAM recognition. (A) Schematic of the directed evolution. (B) Testing of selected mutants, plotting 5′ AAG versus non-AAG PAM protospacers normalized to count per 100,000 sequences. Scatter plot shows 65 induced mutants (open black circles), three induced wild-type replicates (open green circles), an uninduced wild-type (open red circle), the average of the induced mutants (filled black circle), and the average of the induced wild-types (filled green circle) ±SEM. Scatter plot to the right is an inset of the larger plot. (C) Heatmap of protospacer PAM frequency over the entire sequence space for wild type Cas1-Cas2 (wt), mutants that increase or maintain AAG PAM specificity (m-27 and m-24), and mutants that lose AAG PAM specificity (m-74, m-80, m-89). Numbers in the upper right correlate to numbers in B. (D) A subset of selected mutants re-assayed in triplicate as well as a subset of single point mutants chosen from the original selection. All points are the average of three replicates ±SEM. (E) Crystal structure of Cas1-Cas2 complex bound to a protospacers (38). Inset highlights, in magenta, residues in the Cas1 active site that (when mutated) decrease PAM specificity. The protospacer PAM complementary sequence (T30 T29 C28, numbering as in PDB ID 5DQZ) is also noted. Additional statistical details in Table S1.
To visualize shifts in PAM specificity, we plotted a heat map showing the normalized frequency of observed PAMs among all potential PAMs for wild type Cas1-Cas2 and several selected mutants (Fig. 4C). Wild type Cas1-Cas2 had strong selectivity for the canonical AAG PAM. A minority of mutants also retained (m-24) or even increased (m-27) this preference. However, many more mutants showed reduced or, in the case of the three mutants shown (m-74, m-80, m-89), nearly no specificity for the canonical PAM. From the sequence of these selected mutants, we chose a subset of single-point mutations for follow-up analysis based on repeated observations in the data set or location in the crystal structure of the Cas1-Cas2 complex (37–39) (Table S3; Fig. 4E). Most of the single-point mutants tested in isolation also reduced the PAM specificity compared to that of wild-type (Fig. 4D and fig. S5D). These results demonstrate that PAM recognition by the Cas1-Cas2 complex can be modified by many different mutations without drastically reducing spacer acquisition efficiency.
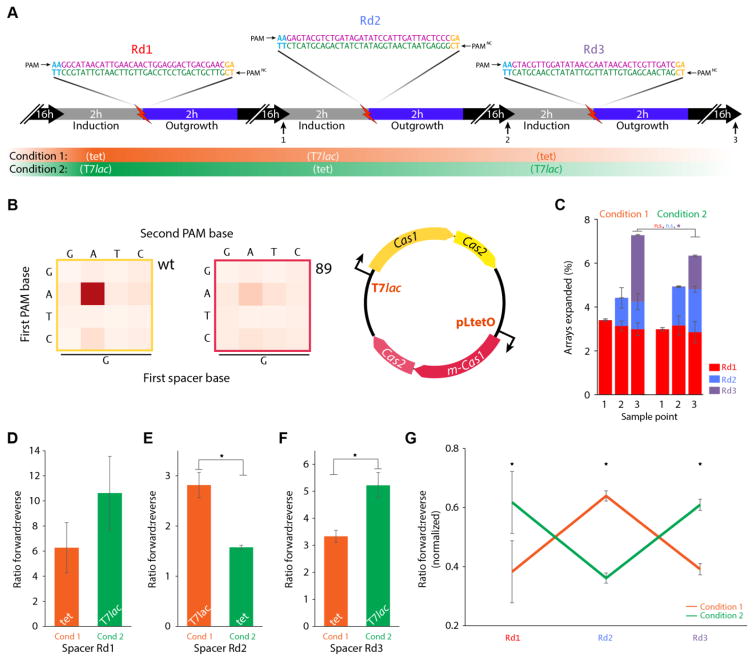
Recording in a second modality
As a proof-of-concept, we selected a PAMNC Cas1-Cas2 mutant (m-89, Fig. 4C and fig. S5D) to add an extra modality to the 1 × 3 recording (fig. S4). We subjected bacteria to three sequential rounds of electroporation, with each oligo protospacer containing a 5′ AAG PAM on the forward strand, and a 5′ TCG PAMNC on the reverse (Fig. 5A). We controlled expression of wild type Cas1-Cas2 and m-89 using different inducible promoters (pLTetO and pT7lac, respectively) on the same plasmid (Fig. 5B). We split the bacteria between two conditions, each alternating between T7lac and tet induction from round-to-round. We found that cells of both conditions acquired spacers from each round at similar frequencies, indicating that transcription and integration activity of the wild type and m-89 Cas1-Cas2 were both adequate (Fig. 5C). At the completion of the recording, we compared the orientation of each spacer between the two conditions. The ratio of forward to reverse oriented spacers shifted toward PAMNC (reverse) during tet induction (Fig. 5D, F). After normalization for the total spacer orientation ratio for each spacer, we could clearly discriminate which cultures had been exposed to each inducer at each time point based only on the direction of integration (Fig. 5G). Thus, this system can simultaneously record in two modalities.
Fig. 5.
Recording in an additional mode. (A) Outline of the recording process. Three different synthetic protospacers (each containing a 5′ AAG PAM on the forward strand, and a 5′ TCG PAM on the reverse) were electroporated over three days (one protospacer each day) into two bacterial cultures under different induction conditions (shown below timeline). Sampling time points are numbered 1–3. (B) Schematic of the plasmid construct used, showing wild-type and PAMNC mutant (m-89) Cas1-Cas2 driven by independently inducible promoters (T7lac and pLtetO, respectively). The heatmap shows 5′ PAM specificity for wild-type (boxed in yellow) and mutant m-89 (boxed in red). (C) At each of the three sample points (marked in B), percent of expanded arrays with spacers from each of the indicated rounds for the two conditions, mean ±SEM. (D–F) Ratio of synthetic spacers acquired in the forward versus reverse orientation for each round under each condition, mean ±SEM. (G) Ratio of forward to reverse integrations normalized to the sum of both possible orientations for each of the two conditions, mean ±SEM. For all panels, * indicates p<0.05, additional statistical details in Table S1.
Discussion
We developed a CRISPR-Cas-based system to record molecular events into a genome in the form of essentially arbitrary synthetic DNA sequences. Although the information is only partially encoded within any given cell, the complete record remains distributed across a population of cells. To read out the recordings, we used high-throughput sequencing, and only considered the pairwise order of any two new spacer sequences within single CRISPR arrays. From these many binary comparisons, a complete record of events could then be assembled, faithfully decoding the distributed memory fully preserved within the cell population. An important consideration of this system is that, despite the necessary destruction of cells for read out at the end of the recording, the encoding process is not destructive. Thus, as opposed to sequential sampling of a population to generate a record of events, the current approach does not require that cells be destroyed while the experiment is ongoing. Moreover, since the recording is distributed across a population, only a fraction of the population needs to be sampled to retrieve the recording.
We uncovered details of the native CRISPR-Cas adaptation system. Integration of synthetic oligo sequences in vivo by the Cas1-Cas2 protein complex enabled us to directly assess detailed aspects of protospacer acquisition. Because the frequency of spacers acquired from the genome and plasmid is largely unaltered in the presence of oligo-derived acquisition (Figs. 1G, 2E), we conclude that the availability of adequate protospacers is likely one limiting aspect of the adaptation system. The presence of a 5′ AAG PAM modulated both the frequency and orientation of spacer acquisition, and the interior sequence of the protospacer influenced acquisition efficiency.
Directed evolution allowed us to experimentally modify PAM recognition of the Cas1-Cas2 complex, which enabled us to generate a record in multiple modalities simultaneously. This directed evolution method required no structural information and should be generally applicable to evolving other activities of CRISPR-Cas proteins by coupling them to the spacer acquisition process (e.g. modifying target site specificity).
There are challenges to directly comparing between different cellular recording approaches. For instance, some are rewriteable (4–7, 9–14, 17, 20, 21) while others, similar to our system, create permanent records (15, 17–21). To date, the highest permanent storage capacity of a synthetic in vivo recording device was achieved using 11 orthogonal recombinases, capable of 211 (2,048) unique states, capturing 1.375 bytes of information within a single cell (18). In our 3 × 5 recording, we encoded 15 individual elements within a population of cells. However, because this system can record arbitrary defined sequences, the number of possible states is expanded dramatically. With an invariable G at the beginning of the spacer and a 5 base set identifier, 27 bases remain that could encode information, yielding 427 possible unique sequences per spacer. It was possible to encode the order within each set to at least five elements, resulting in a unique state capacity for each set based on the permutation P(427,5) = 1.9 × 1081, or 5.7 × 1081 combining the three sets and assuming set independence. If we include interdependence between each set, total unique states would rise to (427)15 or ~7 × 10243. As a point of comparison, the number of atoms in the observable universe is estimated at 1 × 1080.
Moving from theoretical to practical considerations, the information capacity of a given recording in our system depends on the degree to which the sequence of the protospacer is constrained. If there are no sequence constraints on the protospacer and thus any arbitrary sequence is available, then the 15 recorded spacers (in the 3 × 5 recording paradigm) each contain 27 bases of recording potential at four bases per byte yielding 101.25 bytes per recording. Throughout our experiments, we were able to vary the nucleotide identity at every one of these 27 positions in our oligo protospacers. However, we have not explicitly tested, nor is it practical to test, all possible protospacers for viability. Moreover, we have shown that the sequence of the protospacer can influence acquisition frequency so it is reasonable to assume that not all possible sequences will be suitable protospacers.
We can set an absolute lower limit on the information capacity of the 3 × 5 recording presented here by assuming that the particular sequences that we used in the recording are the only possible sequences that could be used. In that case, we can encode information only in the order of the sequences recorded in three sets of five possible spacers, disallowing repetition. In this case the bits per set is given by log2(P(5,5)) = ~6.9 bits or ~2.59 bytes summing all three sets.
However, to assume that no other sequences are allowable is conservative. For instance, considering just the new spacers that were observed in this work, there were 48,773 unique genome-derived, 186 unique plasmid-derived, and 23 unique oligo-derived spacers of 33 bases that included an AAG PAM in their protospacer context. Using this pool of validated sequences in our recording paradigm would yield log2(P(48982,5)) = ~77.9 bits per set or ~29.21 bytes of potential encoding capacity for all three sets. Again, this estimation is certainly over-constrained as these sequences are drawn from an incredibly small subset of all possible sequences. Nonetheless, in the interest of being cautious, we can say that the recording capacity of the 3 × 5 paradigm is not less than 2.59 bytes nor more than 101.25 bytes and likely falls somewhere between 29.21 and 101.25 bytes. By also considering the ability to control spacer orientation (an extra modality), we could potentially encode an additional 5 bits per set. Of course, this only reflects the information of our current recordings, which we arbitrarily limited to 15 spacers. Native species have been found with as many as 458 spacers in a single cell (S. tokodaii) (40). This illustrates the potential space to encode complex biological phenomena, such as the transcriptional time course of many genes in a cell by reverse transcription of mRNA protospacers (41). We anticipate such a recording system will be valuable in applications that require tracing long histories of in vivo cellular activity, including development, lineage, and activity in the brain (42, 43).
Supplementary Material
Acknowledgments
S.L.S is a Shurl and Kay Curci Foundation Fellow of the Life Sciences Research Foundation and received additional support from the National Institute on Aging (5T32AG000222). The project was supported by grants from the National Institute of Mental Health (5R01MH103910) to G.M.C., the National Institute of Neurological Disorders and Stroke (5R01NS045523) to J.D.M., and an Allen Distinguished Investigator Award from the Paul G. Allen Family Foundation to J.D.M.
Sequence data will be deposited into the National Center for Biotechnology Information Sequence Read Archive database as appropriate and all plasmids will be deposited into Addgene upon publication.
Footnotes
Supplementary Materials file includes:
Materials and Methods
Additional Author Notes
Additional references in supplementary materials (44–46).
References and Notes
- 1.Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. 2012;337:1628. doi: 10.1126/science.1226355. published online EpubSep 28 . [DOI] [PubMed] [Google Scholar]
- 2.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. published online EpubOct . [DOI] [PubMed] [Google Scholar]
- 3.Burrill DR, Silver PA. Making cellular memories. Cell. 2010;140:13–18. doi: 10.1016/j.cell.2009.12.034. published online EpubJan 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, Murray AW. Positive-feedback loops as a flexible biological module. Current biology: CB. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. published online EpubApr 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA. Rational design of memory in eukaryotic cells. Genes & development. 2007;21:2271–2276. doi: 10.1101/gad.1586107. published online EpubSep 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrill DR, Inniss MC, Boyle PM, Silver PA. Synthetic memory circuits for tracking human cell fate. Genes & development. 2012;26:1486–1497. doi: 10.1101/gad.189035.112. published online EpubJul 1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. published online EpubJan 20 . [DOI] [PubMed] [Google Scholar]
- 8.Greber D, El-Baba MD, Fussenegger M. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic acids research. 2008;36:e101. doi: 10.1093/nar/gkn443. published online EpubSep . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. published online Epub May 30. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. published online EpubJun 1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilaboa N, Fenna M, Munson J, Roberts SM, Voellmy R. Novel gene switches for targeted and timed expression of proteins of interest. Molecular therapy: the journal of the American Society of Gene Therapy. 2005;12:290–298. doi: 10.1016/j.ymthe.2005.03.029. published online EpubAug . [DOI] [PubMed] [Google Scholar]
- 12.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci U S A. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. published online EpubJul 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrill DR, Silver PA. Synthetic circuit identifies subpopulations with sustained memory of DNA damage. Genes & development. 2011;25:434–439. doi: 10.1101/gad.1994911. published online EpubMar 1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu M, Su RQ, Li X, Ellis T, Lai YC, Wang X. Engineering of regulated stochastic cell fate determination. Proc Natl Acad Sci U S A. 2013;110:10610–10615. doi: 10.1073/pnas.1305423110. published online EpubJun 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One. 2008;3:e2815. doi: 10.1371/journal.pone.0002815). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon TS, Clarke EJ, Groban ES, Tamsir A, Clark RM, Eames M, Kortemme T, Voigt CA. Construction of a genetic multiplexer to toggle between chemosensory pathways in Escherichia coli. Journal of molecular biology. 2011;406:215–227. doi: 10.1016/j.jmb.2010.12.019. published online EpubFeb 18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. published online EpubJun 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Nielsen AA, Fernandez-Rodriguez J, McClune CJ, Laub MT, Lu TK, Voigt CA. Permanent genetic memory with >1-byte capacity. Nat Methods. 2014;11:1261–1266. doi: 10.1038/nmeth.3147. published online EpubDec . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448–452. doi: 10.1038/nbt.2510. published online EpubMay . [DOI] [PubMed] [Google Scholar]
- 20.Farzadfard F, Lu TK. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346:1256272. doi: 10.1126/science.1256272. published online EpubNov 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. published online EpubMay 29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. published online EpubMar 23 . [DOI] [PubMed] [Google Scholar]
- 23.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research. 2012;40:5569–5576. doi: 10.1093/nar/gks216. published online EpubJul . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. published online EpubApr 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. published online EpubMar 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature communications. 2012;3:945. doi: 10.1038/ncomms1937). [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Molecular microbiology. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. published online EpubSep . [DOI] [PubMed] [Google Scholar]
- 28.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogenMycoplasma gallisepticum. PLoS genetics. 2012;8:e1002511. doi: 10.1371/journal.pgen.1002511. published online EpubFeb . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. Journal of bacteriology. 2008;190:1401–1412. doi: 10.1128/jb.01415-07. published online EpubFeb . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Molecular microbiology. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. published online EpubJan . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yosef I, Shitrit D, Goren MG, Burstein D, Pupko T, Qimron U. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci U S A. 2013;110:14396–14401. doi: 10.1073/pnas.1300108110. published online EpubAug 27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA biology. 2013;10:716–725. doi: 10.4161/rna.24325. published online EpubMay . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading, England) 2009;155:733–740. doi: 10.1099/mic.0.023960-0. published online EpubMar . [DOI] [PubMed] [Google Scholar]
- 34.Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife. 2015;4 doi: 10.7554/eLife.08716). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shmakov S, Savitskaya E, Semenova E, Logacheva MD, Datsenko KA, Severinov K. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic acids research. 2014;42:5907–5916. doi: 10.1093/nar/gku226. published online EpubMay . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nature reviews. Microbiology. 2014;12:479–492. doi: 10.1038/nrmicro3279. published online EpubJul . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nature structural & molecular biology. 2014;21:528–534. doi: 10.1038/nsmb.2820. published online EpubJun . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y. Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. published online EpubNov 5 . [DOI] [PubMed] [Google Scholar]
- 39.Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature. 2015;527:535–538. doi: 10.1038/nature15760. published online EpubNov 26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPI: a CRISPR interactive database. Bioinformatics (Oxford, England) 2009;25:3317–3318. doi: 10.1093/bioinformatics/btp586. published online EpubDec 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silas S, Mohr G, Sidote DJ, Markham LM, Sanchez-Amat A, Bhaya D, Lambowitz AM, Fire AZ. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science. 2016;351:aad4234. doi: 10.1126/science.aad4234. published online EpubFeb 26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marblestone AH, Zamft BM, Maguire YG, Shapiro MG, Cybulski TR, Glaser JI, Amodei D, Stranges PB, Kalhor R, Dalrymple DA, Seo D, Alon E, Maharbiz MM, Carmena JM, Rabaey JM, Boyden ES, Church GM, Kording KP. Physical principles for scalable neural recording. Frontiers in computational neuroscience. 2013;7:137. doi: 10.3389/fncom.2013.00137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alivisatos AP, Andrews AM, Boyden ES, Chun M, Church GM, Deisseroth K, Donoghue JP, Fraser SE, Lippincott-Schwartz J, Looger LL, Masmanidis S, McEuen PL, Nurmikko AV, Park H, Peterka DS, Reid C, Roukes ML, Scherer A, Schnitzer M, Sejnowski TJ, Shepard KL, Tsao D, Turrigiano G, Weiss PS, Xu C, Yuste R, Zhuang X. Nanotools for neuroscience and brain activity mapping. ACS nano. 2013;7:1850–1866. doi: 10.1021/nn4012847. published online EpubMar 26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods. 2013;10:1211–1212. doi: 10.1038/nmeth.2646. published online EpubDec . [DOI] [PubMed] [Google Scholar]
- 45.Colwell RK, Coddington JA. Estimating terrestrial biodiversity through extrapolation. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 1994;345:101–118. doi: 10.1098/rstb.1994.0091. published online EpubJul 29 . [DOI] [PubMed] [Google Scholar]
- 46.Rogers JK, Guzman CD, Taylor ND, Raman S, Anderson K, Church GM. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic acids research. 2015;43:7648–7660. doi: 10.1093/nar/gkv616. published online EpubSep 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.