Abstract
DNA methylation is an epigenetic DNA modification catalyzed by DNA methyltransferase 1 (DNMT1). The purpose of this study was to investigate DNMT1 gene and protein expression and the effects of methylation status on tumor suppressor genes in human non-small cell lung cancer (NSCLC) cell lines grown in vitro and in vivo. Human lung adenocarcinoma cell lines, A549 and H838, were grown in vitro and inoculated subcutaneously into nude mice to form tumors and were then treated with the DNA methylation inhibitor, 5-aza-2′-deoxycytidine, with and without treatment with the benzamide histone deacetylase inhibitor, entinostat (MS-275). DNMT1 protein expression was quantified by Western blot. Promoter methylation status of tumor suppressor genes (RASSF1A, ASC, APC, MGMT, CDH13, DAPK, ECAD, P16, and GATA4) was evaluated by methylation-specific polymerase chain reaction. Methylation status of the tumor suppressor genes was regulated by the DNMT1 gene, with the decrease of DNMT1 expression following DNA methylation treatment. Demethylation of tumor suppressor genes (APC, ASC, and RASSF1A) restored tumor growth in nude mice. The results of this study support a role for methylation of DNA as a potential epigenetic clinical biomarker of prognosis or response to therapy and for DNMT1 as a potential therapeutic target in NSCLC.
Keywords: DNA methylation, DNA methyltransferase 1, epigenetic, histone deacetylase, non-small cell lung cancer, tumor suppressor gene
Introduction
Lung cancer is an important cause of morbidity and mortality worldwide, and the incidence is increasing, with more than 1,000,000 new cases of lung cancer diagnosed worldwide each year.1,2 Non-small cell lung cancer (NSCLC) is the most common type of lung cancer.3 The five-year survival rate for NSCLC remains poor, and the pathogenesis of this malignancy is a complex biological process involving multiple steps and factors that are still being studied. In recent years, with the development of molecular biology techniques, the mechanisms involved in the initiation and progression of NSCLC, including the role of epigenetic factors, have been given more attention.
DNA methylation, nucleosome positioning, modifications of histones, and non-coding microRNA expression are all forms of epigenetic modification with potential application to therapy, particularly in oncology.4 Furthermore, the importance of epigenetic changes in lung cancer development has also been recognized.5 These epigenetic changes result in stable inherited phenotypes with DNA methylation, histone modifications, and miRNA6 playing a key role in transcription and post-transcription gene expression in lung cancer and other types of malignancy.7,8 DNA methylation typically occurs at the 5′-position of the cytosine ring within CpG dinucleotides. The most common location for CpG within the human genome is the CpG island. It is now believed that more than half of mammalian genes contain CpG islands, located in the gene promoter, the non-encoding region, and the first exon. In normal cells, DNA methylation does not occur. In lung cancer, DNA methylation may occur throughout the genome with both low levels of methylation and high levels of methylation of the gene promoter region.9,10 The DNA methyltransferase 1 (DNMT1) gene is considered to be primarily responsible for the maintenance of this epigenetic methylation modification that results in copies of the methylation patterns being copied during DNA replication. In carcinogenesis, DNA methylation affects the local protective mechanism of the CpG island, resulting in proto-oncogene activation and chromosome instability.11 The study of these epigenetic alterations in the lung cancer genome, such as DNA methylation, may lead to the identification of novel cancer-related genes. The aim would then be to target these genes for future treatment of lung cancer. Several previous studies have demonstrated an association between DNA methylation and histone deacetylation in silencing genes, including tumor suppressor genes, which supports the importance of continuing to study DNA methyltransferase (DNMT) inhibitors and histone deacetylase (HDAC) inhibitors,12,13 Juergens et al.14 have focused on the therapeutic potential of DNMT gene inhibitors in the treatment of solid tumors including NSCLC. However, as yet there have been no studies to determine whether blocking DNA methylation and/or histone deacetylation of the DNMT1 gene has an effect on NSCLC cells. There is also a potential role for studies on CpG island hypermethylation as a potential prognostic molecular biomarker in cancer, including NSCLC.
DNMT1 is now known to be the primary enzyme responsible for copying methylation patterns following DNA replication.15 The epigenetic process of DNA methylation plays a key role in the genesis of lung cancer.16 DNA methylation and histone deacetylation are associated with transcriptional suppression of gene expression.17,18 The inhibitor of DNA methylation, 5-AZA-CdRm prevents the completion of the DNA methylation reaction and has been reported to be demethylation treatment in several different cancer types.19–21 5-Aza-2′-deoxycytidine (5-AZA-CdR) has recently gained approval from the US Food and Drug Administration for the treatment of myelodysplastic syndrome and is one of the first epigenetic drugs approved for treatment.22 However, HDAC inhibitors can induce differentiation, growth arrest, and apoptosis of tumor cells.23 Entinostat (MS-275) is the most well developed HDAC inhibitor and has shown promising results in recent clinical trials in solid tumors.24 A recent study investigated the combination of decitabine with an HDAC inhibitor; the results increased tumor cell apoptosis in lung cancer cell lines when treated with combined decitabine and an HDAC inhibitor when compared with an HDAC inhibitor alone.25 However, the effects of using the combination of 5-AZA-CdR and MS-275 in human lung cancer cells remain unclear.
Because of the remaining questions, in this study, we chose to investigate the expression and function of the DNMT1 gene in NSCLC cells using an in vitro cell culture model and an in vivo model of tumor inoculation in nude mice. Tumor cell treatment with 5-AZA-CdR and MS-275 was chosen in order to examine the relationship between tumor suppressor gene expression and tumor growth.
Materials and methods
Cell culture and epigenetic treatments
The human NSCLC cell lines, A549 and H838, were purchased and established from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines were grown in 90% RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco) in a humidified incubator with 5% CO2 and 95% air at 37℃. Cells were passaged once the total confluence was reached about 80%. Lung cancer cell lines (A549 and H838) were split to low density (less than 30% confluence) 12 h before treatment. Growth medium, conditioned with 5-Aza (Sigma–Aldrich) at 500 nM, was exchanged every 24 h. After 72 h treatment, MS275 (500 nM, Sigma–Aldrich) was added for 24 h.
RNA extraction and relative quantitation by real-time PCR
RNA was isolated by Trizol reagent Kit (Life Technologies, Gaithersburg, MD, USA) and stored at −80℃ before use. RNA quantity and quality were evaluated by spectrophotometric analysis and gel electrophoresis; 5 µg of total RNA was used to synthesize the first-strand cDNA using the reverse transcriptase kit (Invitrogen, USA). cDNA was used to examine the expression of DNMT1, APC, and RASSF1A and was synthesized by using PrimeScript™ RT reagent kit (TaKaRa) according to manufacturers’ protocols. Expression of DNMT1, RASSF1A, and APC was examined using SYBR® Premix Ex Taq™ II (TaKaRa), and GAPDH was served as internal reference. Results were represented as fold induction using the 2−ΔΔCt method. The PCR data were presented as fold-changes in normalized mRNA levels in control vs. experimental samples and the average of at least triplicate experiments, with standard error presented as error bars. Primers used to examine the expression of DNMT1, RASSF1A, and APC are listed in Table 1.
Table 1.
Primers for quantitative real-time RT-PCR
| ID | Sequence (5′–3′) | Amplified products (bp) |
|---|---|---|
| APC F1 | TCCTGTCCCTGTATCAGAGACT | |
| APC R1 | ACTGTGTTTGCTTGAGCTGCT | 82 bp |
| DNMT1 F1 | CCTCTATGGAAGGCTCGAGT | |
| DNMT1 R1 | TCACCACACGGTGCTGCTCT | 143 bp |
| GAPDH F | ACACCCACTCCTCCACCTTT | |
| GAPDH R | TTACTCCTTGGAGGCCATGT | 181 bp |
| RASSF1A F | ACAGCAACCTCTTCATGAGCT | |
| RASSF1A R | CAAGGAGGGTGGCTTCTTGCT | 116 bp |
Protein preparation and Western blotting
Cells were harvested 96 h after treatment and homogenized in RIPA buffer (PBS buffer with 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS of final concentration and protease inhibitor cocktail; Roche, Applied Science). The concentration of protein was quantified by the BCA protein assay from Pierce Bioscience (Rockford, IL, USA). The protein lysates (40 µg) were separated by 4–12% NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA, USA) and transferred to PVDF membranes (Millipore, Billerica, MA, USA). Following blocking with 5% non-fat milk powder and 0.1% Tweeen-20 in TBS, the membranes were incubated with mouse anti-DNMT1 antibody (Sigma–Aldrich), mouse anti-RASSF1A monoclonal antibody (Abgent), mouse anti-APC monoclonal antibody (Abgent), mouse anti-ASC monoclonal antibody (Abgent), mouse anti-β-actin (Sigma–Aldrich), rabbit and mouse IgG ECL antibody, and HRP-conjugated secondary antibodies (GE Healthcare). β-Actin antibody was used as a loading control. The blots were visualized using enhanced chemiluminescence (Sangon, Shanghai, China).
DNA extraction and methylation-specific PCR
Genomic DNA was extracted by a standard phenol/chloroform procedure. Briefly, DNA from A549 and H838 was modified by sodium bisulfite treatment using EZ DNA methylation kit (Zymo Research, CA, USA). Methylation-specific PCR (MSP) for the RASSF1A, ASC, APC, MGMT, CDH13, DAPK, ECAD, P16, GATA4 promoter, and CpG islands was carried out as per a previously published protocol. Primers were designed by Biosystems. The methylation-specific primers and the unmethylation-specific primers used in MSP are listed in Table 2.
Table 2.
Primers for MSP analysis
| ID | Sequence (5′–3′) |
|---|---|
| APC-U-F | GTGTTTTATTGTGGAGTGTGGGTT |
| APC-U-R | CCAATCAACAAACTCCCAACAA |
| APC-M-F | TATTGCGGAGTGCGGGTC |
| APC-M-R | TCGACGAACTCCCGACGA |
| RASSF1A-U-F | GGGGTTTGTTTTGTGGTTTTGTTT |
| RASSF1A-U-R | AACATAACCCAATTAAACCCATACTTCA |
| RASSF1A-M-F | GGGTTCGTTTTGTGGTTTCGTTC |
| RASSF1A-M-R | TAACCCGATTAAACCCGTACTTCG |
| ASC-U-F | TTG GTG GTT GTA GTG GGG TGA GT |
| ASC-U-R | CCA ACA CAT CA AAA TAA CAT CAC A |
| ASC-M-F | GCG GTT GTA GCG GGG TGA GC |
| ASC-M-R | CGC ATC CAA AAT AAC GTC GCG |
| CDH13-U-F | GGTTTTTATGGAAAATATGTTTAGTGTAGTTGT |
| CDH13-U-R | CAAAAAAACACACAAAACAAACA AAA TTCTCA |
| CDH13-M-F | CGGAAAATAT GTTTAGTGTAGTCGC |
| CDH13-M-R | GC ACA AAA CGA ACG AAA TTC TCG |
| DAPK-U-F | GGAGGATAGTTGGATTGAGTTAATGTT |
| DAPK-U-R | CAAATCCCTCCCAAACACCAA |
| DAPK-M-F | GGATAGTCGGATCGAGTTAACGTC |
| DAPK-M-R | CCCTCCCAAACGCCGA |
| ECAD-U-F | TGGTTGTAGTTATGTATTTATTTTTAGTGGTGT T |
| ECAD-U-R | ACA CCA AAT ACA ATC AAA TCA AAC CAA A |
| ECAD-M-F | TGT AGT TAC GTA TTT ATT TTT AGT GGC GTC |
| ECAD-M-R | CGA ATA CGA TCG AAT CGA ACC |
| GATA4-U-F | AGTTTTTTGTATAGTTTTGTAGTTTGTGTTTAGTG |
| GATA4-U-R | ACTAACTAACCTATAAAAATCACATACAAAAACA |
| GATA4-M-F | AGTTTTTTGTATAGTTTTGTAGTTTGTGTTTAGTG |
| GATA4-M-R | ACTAACTAACCTATAAAAATCACATACAAAAACA |
| MGMT-U-F | TTT GTG TTT TGA TGT TTG TAG GTT TTT GT |
| MGMT-U-R | AAC TCC ACA CTC TTC CAA AAA CAA AAC A |
| MGMT-M-F | TTT CGA CGT TCG TAG GTT TTC GC |
| MGMT-M-R | GCA CTC TTC CGA AAA CGA AAC G |
| P16-U-F | GTTATTAGAGGGTGGGGTGGATTGT |
| P16-U-R | ACTCCATACTACTCCCCACCACCAA |
| P16-M-F | GGGTGGGGCGGATCGC |
| P16-M-R | TACTACTCCCCGCCGCCGA |
Each MSP reaction incorporated approximately 100 ng of bisulfite-treated DNA, 25 pmoles of each primer, 100 pmoles dNTPs, 2.5 µL of 10 × PCR buffer, and 1 unit of JumpStart Red Taq Polymerase (Sigma) in a final reaction volume of 25 µL. For PCR, methylated (M) and unmethylated (U) primer pairs were initially denatured at 94℃ for 3 min followed by 35 cycles with a 1-min denaturation step, 30 s of annealing at 60℃ (unmethylation at 58℃), and 3 min of extension at 72℃. Final extension after 35 cycles was at 72℃ for 10 min, and the product was stored at 4℃. PCR products were separated on a 2% agarose gel and were visually scored as methylated or unmethylated according to the presence or absence of a PCR product.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc.) was used to evaluate cell growth of NSCLC cells according to the manufacturer’s protocol. Cells were re-plated in 96-well plates at each harvesting time point in three wells for each group. The CCK-8 solution was added to each well at a 1:10 dilution. Cells were incubated for 4 h, and the absorbance at 450 nm was measured using a multi-well plate reader. A CCK-8 kit was used to measure cell growth and make growth curve.
Tumor formation assays in an athymic BALB/c nude mouse model
Male athymic BALB/c nude mice aged four weeks were maintained under specific pathogen-free conditions and manipulated according to protocols approved by the Shanghai Medical Experimental Animal Care Commission. Four groups of mice (n = 5/group) were tested. Group 1 (Control) was injected with NSCLC cells following treatment with DMSO; Group 2 (5-AZA-CdR) was injected with NSCLC cells following treatment with 5-AZA-CdR 500 nM; Group 3 (MS-275) was injected with NSCLC cells following treatment with MS-275 500 nM; Group 4 (Combo) was injected with NSCLC cells following treatment with 5-AZA-CdR and with MS-275 500 nM. A549 and H838 cells were treated under the above four conditions, then harvested from six-well cell culture plates, washed with PBS, and resuspended at a concentration of 5 × 106 cells/mL. A volume of 0.2 mL of suspended cells was subcutaneously injected into a single side of the posterior flank of each mouse. Tumor growth was examined every seven days, and tumor volumes were calculated using the equation V = 0.5 × D × d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter).26 At four weeks post-injection, mice were euthanized, and the subcutaneous growth of each tumor was examined. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved according to the institutional ethical guidelines for animal experiments. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering in mice.27
Statistical analysis
Continuous data were presented as mean ± standard deviation; Student’s t-test and one-way or two-way ANOVA were applied in this study. Result were expressed as the mean ± SEM (standard error of the mean), n represents the number of independent experiments. A P value < 0.05 was considered statistically significant. All statistical analyses were performed using the SPSS version 17.0 (SPSS, USA). All experiments were performed in triplicate.
Results
Treatment of NSCLC cells with inhibitors of DNMT and HDAC reduced DNMT1 transcription expression
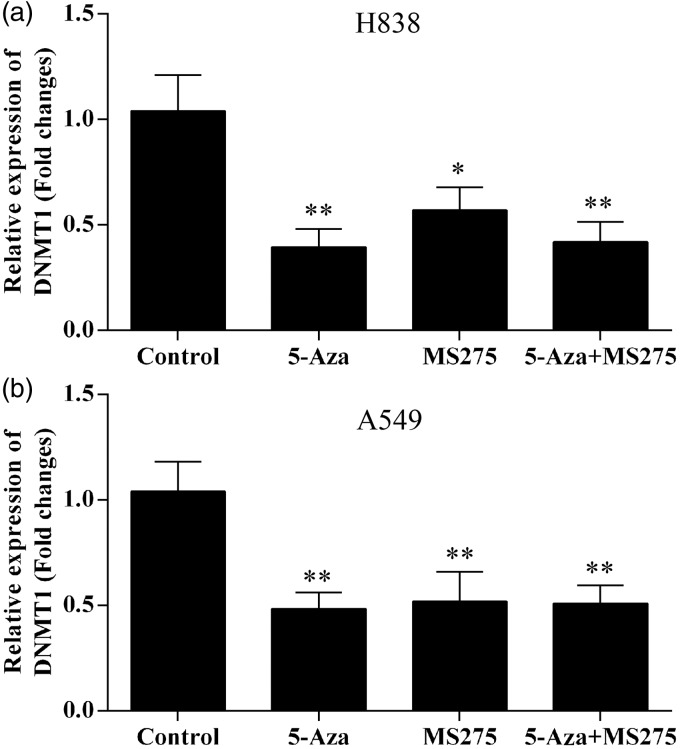
The level of expression of the DNMT1 gene in NSCLC cell lines was determined using quantitative reverse transcription-polymerase chain reaction (RT-PCR) using β-actin as a reference. NSCLC cells were treated with different doses of 5-AZA-CdR and/or the benzamide HDAC inhibitor, entinostat (MS-275), and were incubated for five days. Total RNA was isolated from untreated control cells, 5-AZA-CdR, MS-275, and the combination of 5-AZA-CdR + MS-275 treated cells. In both A549 and H838 cell lines, we demonstrated a significant decrease in DNMT1 mRNA expression levels by 5-AZA-CdR + MS-275 treatment, which was compared with 5-AZA-CdR alone or MS-275 alone (Figure 1). The combination of 5-AZA-CdR and MS-275 resulted in a decrease of the DNMT1 mRNA expression by one-half when compared with the effects of the individual treatments, indicating a potential cooperative effect.
Figure 1.
DNMT1 mRNA expression was significantly reduced following 5-AZA-CdR and/or MS-275 treatment in NSCLC cell lines. (a) The mRNA expression of DNMT1 in H838 cells. (b) The mRNA expression of DNMT1 in A549 cells. The mRNA expression of DNMT1 was normalized to GAPDH. Data represented as means ± SD.*p < 0.05; **p < 0.01
Treatment with 5-AZA-CdR and MS-275 inhibited DNMT1 expression in NSCLC cells
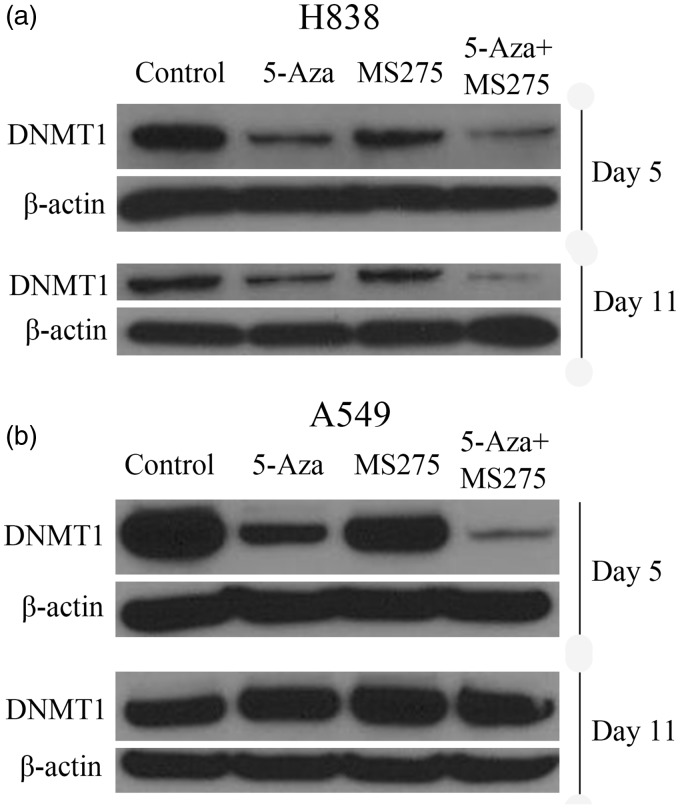
To show whether the mRNA expression of DNMT1 was consistent with DNMT1 protein expression, we performed Western blot assays. Total proteins were extracted on day 5 and day 11 following DNA methylation, and levels were compared with the expression of DNMT1 protein at these two time points. The results showed a significant decrease in DNMT1 protein expression in H838 and A549 cell lines following treatment with 5-AZA-CdR at 500 nM and the combination of 5-AZA-CdR + MS-275 treatment, when compared with the untreated control group on day 5 (Figure 2). However, treatment of NSCLC cells with 5-AZA-CdR alone reduced the expression of DNMT1 protein to a greater degree than with MS-275 alone (Figure 2). Specifically, in H838 cells, the expression of DNMT1 protein was gradually restored by day 11 (Figure 2(a)). DNMT1 protein expression was significantly reduced in all treatment groups in A549 cells (Figure 2(b)). Consistent with the RT-PCR results, treatment of A549 and H838 cells with 5-AZA-CdR and/or MS-275 reduced DNMT1 protein expression, but different cell lines had different sensitivity to the DNA methylation treatment.
Figure 2.
Treatment of NSCLC cells with 5-AZA-CdR and/or MS-275 decreased DNMT1 protein expression. (a) The expression of DNMT1 protein was reduced significantly in 5-AZA-CdR and combined groups in H838 cells on day 5 and day 11 following treatment. (b) DNMT1 protein expression was significantly reduced in all treatment groups in A549 cells. β-actin was used as loading control
Treatment with 5-AZA-CdR and MS-275 resulted in tumor suppressor gene demethylation in NSCLC cells
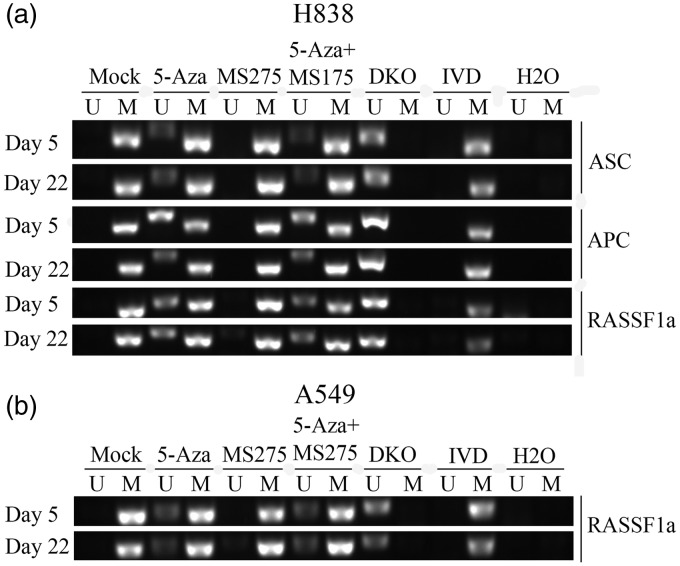
To determine whether the tumor suppressor genes (RASSF1A, ASC, APC, MGMT, CDH13, DAPK, ECAD, P16, and GATA4) were methylated, and if the methylation was linked with DNA hypermethylation in NSCLC cells following treatment, MSP assays were performed. The results showed that the methylation status of tumor suppressor genes changed. In H838 cells, the RASSF1A, ASC, and APC promoter CpG islands were demethylated from full methylation to partial methylation following 5-AZA-CdR alone and the combination of 5-AZA-CdR + MS-275 treatment (Figure 3(a)), but treatment with MS275 alone had no effect on tumor suppressor genes. Furthermore, in A549 cells, only RASSF1A gene was demethylated from full methylation to partial methylation (Figure 3(b)). In previous studies, RT-PCR and Western blotting were used to analyze the expression of DNMT1 gene and protein levels in A549 and H838 cell lines. There was reduced expression of both DNMT1 gene and protein levels following 5-AZA-CdR and/or MS-275 treatment when compared with the untreated cells.
Figure 3.
The impact of treatment with 5-AZA-CdR and MS-275 and tumor suppressor gene methylation status in NSCLC cell lines. (a) Methylation status of RASSF1A, C, APC promoter CpG islands before and after adding to 5-AZA-CdR or MS-275 as measured by MSP. De-methylation changes of tumor suppressor genes in H838 cells. H838 cell line was used as an unmethylated control, in vitro-methylated DNA (IVD), DKO (DNMT1 and DNMT3b knock-out), and H2O. Lane M: methylated amplicon. Lane U: unmethylated amplicon. (b) Methylation status of RASSF1A detected using MSP before and after adding to 5-AZA-CdR or MS-275. De-methylation changes of tumor suppressor genes in A549 cells. Lane M: methylated amplicon. Lane U: unmethylated amplicon
Treatment with 5-AZA-CdR and MS-275 induced tumor suppressor gene expression in NSCLC cells
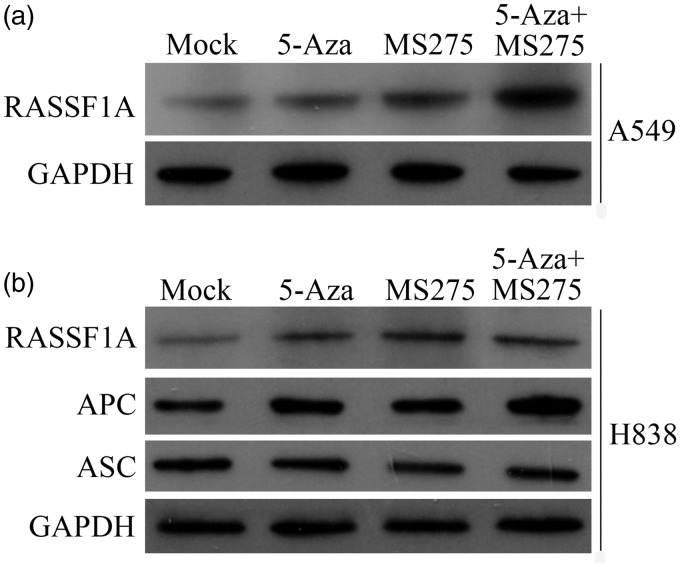
The expression levels of the tumor suppressor gene proteins (RASSF1A, ASC, and APC) in NSCLC cells following epigenetic treatment were measured using Western blots. The expression level of RASSF1A protein was significantly increased following treatment of the A549 cells (Figure 4(a)), and the expression levels of RASSF1A and APC protein were also increased following treatment of the H838 cells (Figure 4(b)). However, the expression level of ASC protein was not changed following treatment of the H838 cells (Figure 4(b)).
Figure 4.
The protein expression level of RASSF1A, ASC, and APC following 5-AZA-CdR and/or MS-275 treatment in NSCLC cell lines. (a) The protein expression level of RASSF1A in A549 cells. (b) The protein expression levels of RASSF1A, APC, and ASC in A549 cells
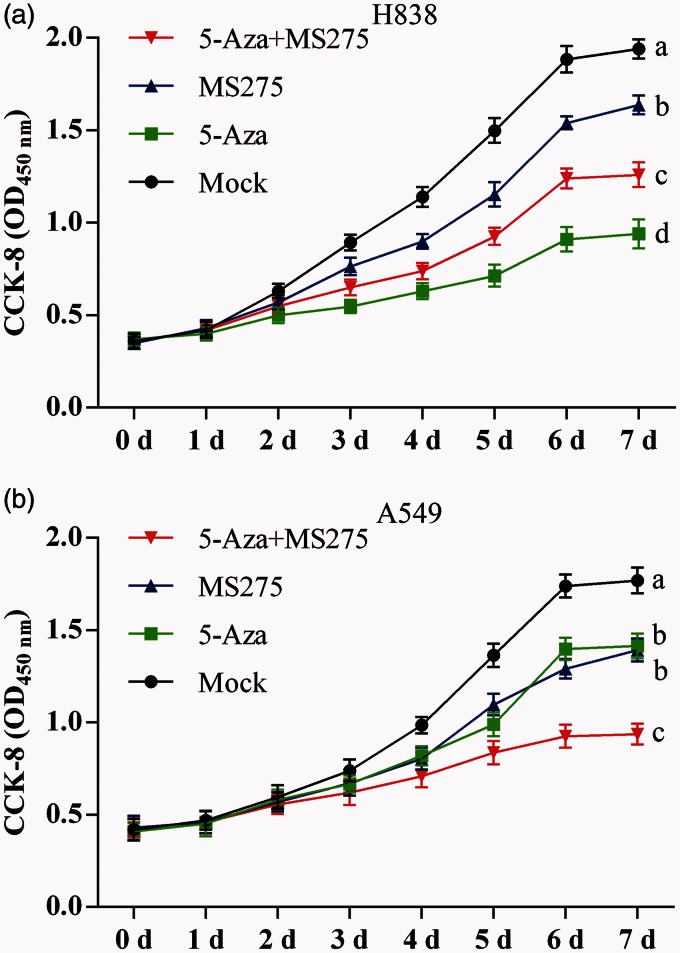
Treatment with 5-AZA-CdR and MS-275 inhibited proliferation in NSCLC cells
Optical density (OD) measurements of H838 and A549 cells were done using the CCK-8 to generate cell growth curves. In H838 cell lines, the cell proliferation declined following 5-AZA-CdR and/or MS-275 treatment from day 3 to day 7. The OD was lower in 5-AZA-CdR treated cells alone than with the combination of 5-AZA-CdR and MS-275 and lower in the combined treatment group compared with the group treated with MS-275 alone (Figure 5(a)). In A549 cell lines, the OD was significantly decreased in the combined 5-AZA-CdR and MS-275 treated group; however, there was no difference between the group treated with 5-AZA-CdR alone and the group treated with MS-275 alone, as shown in Figure 5(b).
Figure 5.
The proliferation curve for NSCLC cell lines following 5-AZA-CdR and/or MS-275 treatment by the CCK-8 assay. (a) Cell growth curve of H838 cells on day 5 to seed cells. (b) Cell growth curve of A549 cells on day 5 to seed cells. Data are represented as mean ± SD from three independent experiments. In each of two groups, the different letters means significant differences between the two groups.
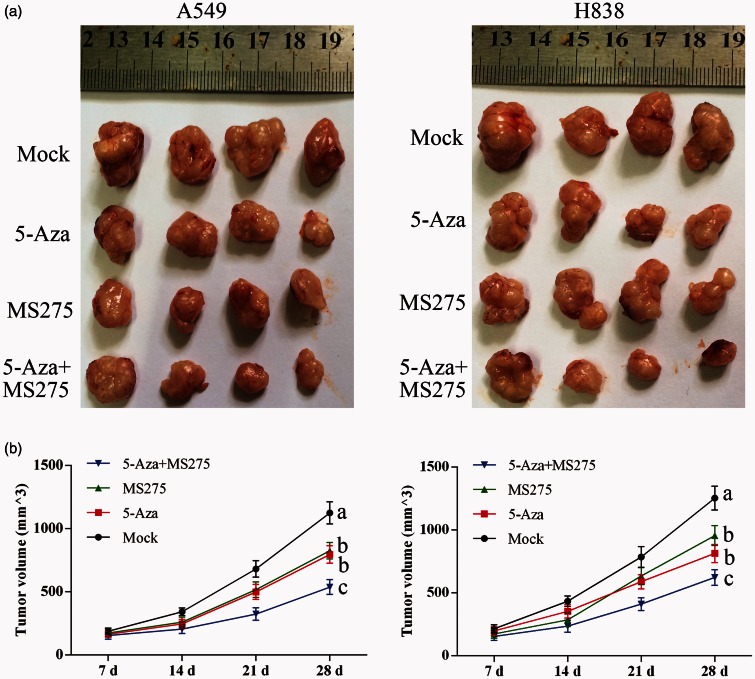
Treatment with 5-AZA-CdR and MS-275 inhibited tumor growth in vivo
H838 and A549 cell lines were inoculated into male nude mice. The tumor sizes were recorded at days 7, 14, 21, and 28. One month following tumor cell inoculation, the tumors formed in the 5-AZA-CdR and/or MS-275 treated group were reduced in size when compared with the untreated group (Figure 6).
Figure 6.
DNA methylation treatment with 5-AZA-CdR with MS-275 inhibited tumor growth in nude mice. (a) Tumors isolated from nude mice injected with NSCLC cells. (b) Growth curve of inoculated tumors. Data represented as means ± SD.
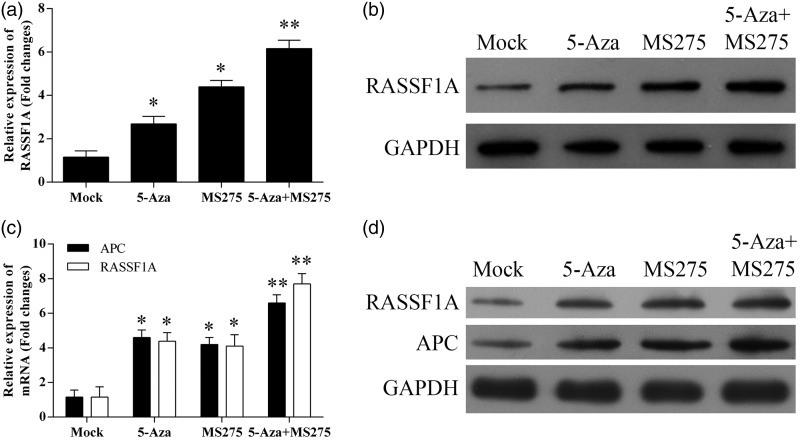
Treatment with 5-AZA-CdR and MS-275 restored expression of RASSF1A and APC in vivo
RT-PCR and Western blotting analysis of RASSF1A and APC expression were performed in selected tumor tissues (Figure 7). The results showed that the levels of RASSF1A and APC expression in H838 tumor cells treated with 5-AZA-CdR and/or MS275 treated group were greater than those of tumors formed in the non-treated control group. In treated A549 cells, only RASSF1A expression remained high.
Figure 7.
DNA methylation treatment restored expression of RASSF1A and APC in tumors isolated from nude mice injected with NSCLC cells. (a) The mRNA expression of RASSF1A in tumors isolated from nude mice injected with A549 cells. (b) The protein expression level of RASSF1A in tumors isolated from nude mice injected with A549 cells. (c) The mRNA expression levels of RASSF1A and APC in tumors isolated from nude mice injected with H838 cells. (d) The protein expression level of RASSF1A and APC in tumors isolated from nude mice injected with H838 cells. *p < 0.05; **p < 0.01
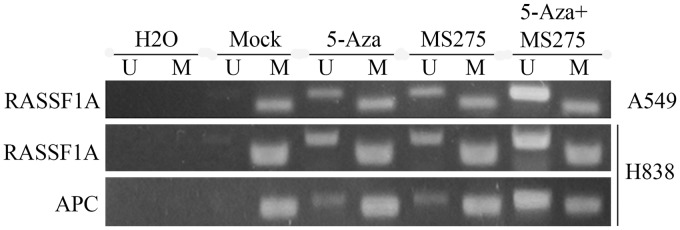
Treatment with 5-AZA-CdR and MS-275 resulted in the demethylation of RASSF1A and APC in vivo
MSP assays were used to analyze RASSF1A and APC methylation status in resected tumor tissues. Data showed that the methylation of RASSF1A and APC in promoter regions was restored in tumors isolated from nude mice injected with the H838 cell line when treated with 5-AZA-CdR and with MS275, compared with controls (Figure 8). However, in A549 cells, only RASSF1A was demethylated.
Figure 8.
DNA methylation treatment with 5-AZA-CdR and MS-275 resulted in the de-methylation of RASSF1A and APC in tumors isolated from nude mice injected with NSCLC cells. Lane M: methylated amplicon. Lane U: unmethylated amplicon
Discussion
In this study, we investigated the suppression of DNMT1 gene expression in NSCLC cell lines and inoculated lung cancer cells grown subcutaneously in nude mice. Studies were done following tumor cell treatment with 5-AZA-CdR, with and without treatment with the benzamide HDAC inhibitor, entinostat (MS-275). The findings have shown that DNMT1 protein expression was reduced following treatment with 5-AZA-CdR and/or MS-275 in NSCLC cells as measured by Western blotting; quantitative RT-PCR assays showed that the expression of the DNMT1 gene was significantly decreased following treatment with a combination of 5-AZA-CdR and MS-275. Downregulation of DNMT1 protein expression remained until day 11 in A549 cell lines but not in H838 cell lines, confirming that different tumor cell lines, and probably different lung cancers in different patients, are likely to show heterogeneity in their response to DNA methylation.
To assess the role of 5-AZA-CdR alone or in combination with MS-275 treatment, we analyzed the proliferation rates in A549 and H838 lung cancer cells in vitro. The combination of 5-AZA-CdR with MS-275 treatment reduced the tumor cell proliferation rates when compared with treatment with MS-275 alone, in both A549 and H838 cell lines. These data further support a role for DNA methylation in the inhibition of the proliferation of lung cancer cells. Several previous studies have demonstrated that SET7 protein affects DNMT protein enzymic activity via promoting degradation of the DNMT1 gene.28 A recent study has confirmed that the histone methyltransferase SET7 can methylate the histone and the lysine142 residues of DNMT1, which can reduce the expression of the DNMT1 gene by the proteasome-mediated pathway.28 It can, therefore, be assumed that methylation inhibitors can also inhibit the function of DNMT1 and also activate the histone methyltransferase, SET7. The SET7-mediated lysine methylation of DNMT1 is known to be degraded through the protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1).28
SET7 was present in the same molecular complexes as DNMT1 is supported by previous studies. One previous study has shown that ICBP90/UHRF1, also known as E3 ubiquitin-protein ligase, interacts with the DNMT1 gene before the cell enters the S-phase.29
A study by Vendetti and Rudin30 has confirmed increased methylation of the promoter region of the CDKM2A, CDH13, and RASSF1A genes in primary lung cancer tissue when compared with normal bronchial epithelium. A previously published study has shown that a combination of treatment with a DNMT inhibitor with an HDAC inhibitor restored expression of silenced tumor suppressor genes in cancer cells.31 Another previously published study has shown that the expression of the RASSF1A gene was restored by downregulating DNMT1 and DNMT3B in prostate cancer cells.32 In this study, we detected the methylation status of tumor suppressor genes by MSP and demonstrated that the promoter methylation status of tumor suppressor genes was associated with decreased cell growth following treatment with the DNA methylation inhibitor 5-AZA-CdR with or without MS-275 treatment. The in vivo studies on tumor inoculation into nude mice showed reduction in tumor size in the tumors treated with DNA methylation inhibitors. Western blotting and RT-PCR analysis of tumor suppressor gene expression, RASSF1A and APC showed that the tumors treated with the combination of 5-AZA-CdR and MS-275 showed significantly more tumor shrinkage. These data also support a role for tumor suppressor genes in the control of cell proliferation of NSCLC cells.
In conclusion, treatment of NSCLC cells in vitro and in vivo with the DNA methylation inhibitor, 5-AZA-CdR, with and without treatment with the benzamide HDAC inhibitor, MS-275, was studied, with the effects on the methylation status of several tumor suppressor genes (including APC and RASSF1A) and DNMT1 gene expression. The findings have shown a correlation between DNMT1 gene expression and DNA methylation; tumor suppressor genes were methylated in NSCLC, and this methylation was associated with DNMT1 gene expression. These results suggest that decreased DNMT1 at the transcript and protein level in response to epigenetic therapies is associated with treatment response in mice and may be used to support further development of preclinical rationale for therapeutic intervention.
Author contributions
The first three authors contributed equally to this paper. All authors participated in the design, interpretation of the studies, analysis of the data and review of the manuscript; BL conducted the experiments and wrote the manuscript.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81160296, 81160273) and Natural Science Foundation of Guangxi (2013GXNSFCB019005, 2013JJAA40484).
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 2.Ma J, Ward EM, Smith R, Jemal A. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 2013; 119: 1381–5. [DOI] [PubMed] [Google Scholar]
- 3.Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One 2012; 7: e35065–e35065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Canc 2011; 11: 726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piperi C, Vlastos F, Farmaki E, Martinet N, Papavassiliou AG. Epigenetic effects of lung cancer predisposing factors impact on clinical diagnosis and prognosis. J Cell Mol Med 2008; 12: 1495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol 2011; 409: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997; 389: 349–52. [DOI] [PubMed] [Google Scholar]
- 8.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature 1994; 371: 435–8. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986; 321: 209–13. [DOI] [PubMed] [Google Scholar]
- 10.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 1989; 83: 181–8. [DOI] [PubMed] [Google Scholar]
- 11.Day TK, Bianco-Miotto T. Common gene pathways and families altered by DNA methylation in breast and prostate cancers. Endocr Relat Canc 2013; 20: R215–32. [DOI] [PubMed] [Google Scholar]
- 12.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 1999; 23: 62–6. [DOI] [PubMed] [Google Scholar]
- 13.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 2000; 25: 269–77. [DOI] [PubMed] [Google Scholar]
- 14.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, Lee B, Tsai S, Delgado IE, Rudek MA, Belinsky SA, Herman JG, Baylin SB, Brock MV, Rudin CM. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Canc Discov 2011; 1: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Canc Res 2006; 66: 729–35. [DOI] [PubMed] [Google Scholar]
- 16.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, Franklin WA, Herman JG, Baylin SB, Bunn PA, Byers T. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Canc Res 2006; 66: 3338–44. [DOI] [PubMed] [Google Scholar]
- 17.Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6: 597–610. [DOI] [PubMed] [Google Scholar]
- 18.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 2005; 15: 490–5. [DOI] [PubMed] [Google Scholar]
- 19.Cacan E, Ali MW, Boyd NH, Hooks SB, Greer SF. Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance. PLoS One 2014; 9: e87455–e87455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu A, Xia J, Zuo J, Jin S, Zhou Y, Yao L, Huang H, Han Z. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol 2012; 29: 2701–9. [DOI] [PubMed] [Google Scholar]
- 21.Patties I, Jahns J, Hildebrandt G, Kortmann RD, Glasow A. Additive effects of 5-aza-2′-deoxycytidine and irradiation on clonogenic survival of human medulloblastoma cell lines. Strahlenther Onkol 2009; 185: 331–8. [DOI] [PubMed] [Google Scholar]
- 22.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, Ellerton J, Larson RA, Schiffer CA, Holland JF. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol 2002; 20: 2429–40. [DOI] [PubMed] [Google Scholar]
- 23.Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci U S A 1999; 96: 4592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yardley DA, Ismail-Khan RR, Melichar B, Lichinitser M, Munster PN, Klein PM, Cruickshank S, Miller KD, Lee MJ, Trepel JB. Randomized phase II, double-blind, placebo-controlled study of exemestane with or without entinostat in postmenopausal women with locally recurrent or metastatic estrogen receptor-positive breast cancer progressing on treatment with a nonsteroidal aromatase inhibitor. J Clin Oncol 2013; 31: 2128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu WG, Lakshmanan RR, Beal MD, Otterson GA. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Canc Res 2001; 61: 1327–33. [PubMed] [Google Scholar]
- 26.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013; 32: 1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. Animal research: reporting in vivo experiments: the ARRIVE guidelines. J Gene Med 2010; 12: 561–3. [DOI] [PubMed] [Google Scholar]
- 28.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 2011; 18: 42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, Schini-Kerth VB, Bronner C. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 2008; 27: 2187–97. [DOI] [PubMed] [Google Scholar]
- 30.Vendetti FP, Rudin CM. Epigenetic therapy in non-small-cell lung cancer: targeting DNA methyltransferases and histone deacetylases. Expert Opin Biol Ther 2013; 13: 1273–85. [DOI] [PubMed] [Google Scholar]
- 31.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999; 21: 103–7. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal S, Amin KS, Jagadeesh S, Baishay G, Rao PG, Barua NC, Bhattacharya S, Banerjee PP. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol Canc 2013; 12: 99–99. [DOI] [PMC free article] [PubMed] [Google Scholar]