Abstract
We developed a high-throughput bead-based suspension array for simultaneous detection of 20 respiratory tract pathogens in clinical specimens. Pathogen-specific genes were amplified and hybridized to probes coupled to carboxyl-encoded microspheres. Fluorescence intensities generated via the binding of phycoerythrin-conjugated streptavidin with biotin-labeled targets were measured by the Luminex 100 bead-based suspension array system. The bead-based suspension array detected bacteria in a significantly higher number of samples compared to the conventional culture. There was no significant difference in the detection rate of atypical pathogensatypical pathogens or viruses between the bead-based suspension array and real-time PCR. This technology can play a significant role in screening patients with pneumonia.
Keywords: Acute respiratory tract infections, unexplained pneumonia, bead-based suspension array, pathogen detection, high-throughput screening
Introduction
Severe acute respiratory tract infections have historically had a very detrimental impact on human health and social stability,1,2 and continue to cause devastating outbreaks.3–5 Although the importance of early and rapid diagnosis is recognized, the complexity of pathogens associated with these infections is a major challenge. Conventional diagnostic methods such as bacterial cultures, immunological tests and real-time PCR assays have the disadvantages of low sensitivity, single detection at a time, and delayed result availability.
Recent molecular techniques used a sensitive and accurate multiplex reverse transcription PCR (RT-PCR) assay for pneumonia and sepsis,6 and the Respiratory Multicode-Plx Assay system which detected a large number of respiratory viruses with high sensitivity and accuracy.7 Gene chip technology includes solid phase arrays or loop-mediated isothermal amplification systems and liquid bead-based suspension arrays. A loop-mediated isothermal amplification system developed cooperatively by the Peking University People’s Hospital and CapitalBio Corporation in China is currently being tested in clinical trials.8 The bead-based suspension array (liquid chip) platform developed by the Luminex Corporation in the United States is a rapid, high-throughput system for multi-analyte detection.4,5,8–11 The system is based on polystyrene microspheres internally labeled with a unique dye combination. The microspheres are coated with thousands of copies of a probe for a specific target which is amplified from clinical samples using 5′-biotin-labeled primers. The fluorescent emission from the microspheres is evaluated using a 635 nm/10 inW red polar laser while a 532 nm/13 inW yttrium aluminum garnet (YAC) laser is used to measure the target analyte by exciting the streptavidin-phycoerythrin (SP-PE) fluorescent reporter bound to the surface of microspheres.10 Target quantification is done by determining the mean fluorescence intensity (MFI) for each encoded microsphere by high speed computing.12
A suspension array-based respiratory virus detection kit (xTAG RVP) developed by the Luminex corporation received FDA approval in 2008.13,14 This kit does not detect various common respiratory tract bacteria or atypical pathogens. Moreover, the kit has not yet been used in China, where significant variation in viruses from different geographical regions contributes to inconsistent detection sensitivity. In the present study, we aimed to utilize the bead-based suspension array technology to develop a high-throughput screening system which can detect common viruses, bacteria as well as atypical pathogens from respiratory tract infections in Chinese patients.
Materials and methods
Patient specimens
This study enrolled 333 subjects who were hospitalized for severe acute respiratory infections at the Department of Respiratory Medicine, Fujian Provincial Hospital between March 2012 and May 2015. Sputum samples were collected from 293 patients and alveolar lavage fluid specimens from 40 patients. This study was reviewed and approved by the Institutional Review Board of Fujian Provincial Hospital (IRB number: K2012-003-01), and informed consent was obtained from the patients.
Inclusion and exclusion criteria
Patients who were able to provide an accurate and complete medical history were included in this study. The inclusion criteria were:
Occurrence of fever accompanied by sore throat, stuffy nose, running nose, headache, fatigue, cough, expectoration, difficulty in breathing.
Presence of congested and swollen throat, conjunctival congestion, sores and ulcers in the throat, oral cavity or gingiva or fine crackles are heard on auscultation.
Pulmonary infection evidenced by presence of spotted, patchy high-density shadows in lungs or disordered and thickened bronchi on chest imaging by chest radiography or CT.
Symptoms or physical signs of respiratory tract infections and routine blood tests indicating normal, elevated or lowered white blood cell counts.
Presence of tumors, COPD, bronchial dilation, and other organ-based diseases, such as heart, liver, kidney diseases (including organ failure) were not excluding factors.
Patients with definite or suspected pulmonary tuberculosis, pulmonary fungal infections or non-infectious diseases not accompanied by lower respiratory tract infection were excluded.
Nucleic acid extraction from sputum specimens and reverse transcription
Bacterial concentrations in sputum specimens were calculated as follows: bacterial load in sputum samples (copies/ml) = template concentration (copies/µl) × 100/0.6.14 Sputum DNA was prepared and quantified as previously described.15,16 Viral nucleic acids were extracted using the MiniBEST Viral RNA/DNA Extraction Kit (Ver.5.0; TakaRa, Shiga, Japan) and were then reverse transcribed into cDNA with the TUREscipt 1st Strand cDNA Synthesis Kit (Aidlab Biotechnologies Co., Beijing, China). Supplementary Table 1 shows the limits of detection for each pathogen based on serial dilution.
Design of primers and probes
Six kinds of primers and probes corresponding to sequences from the SARS coronavirus, the Influenza A virus, the highly pathogenic avian influenza A H5N1, the human metapneumovirus, and the human bocavirus were designed based on previous studies6,15,17–21 and optimized for our research purposes. Fourteen pathogen sequences, including human cytomegalovirus transmembrane protein gene (X04650), human adenovirus type 6 hexon protein gene (DQ149613), human parainfluenza virus 1 HN gene (U70942), human parainfluenza virus 2 HN gene (AB367954), human parainfluenza virus 3 HN gene (AB623457), human respiratory syncytial virus nucleocapsid gene (X00001), Mycoplasma pneumoniae P1 gene (KF154759), Chlamydia pneumoniae rpoB gene (KC305894), Staphylococcus aureus tuf gene (HM352930), Streptococcus pneumoniae ply gene (GU968401), Klebsiella pneumoniae phoE gene (AF009172), Acinetobacter baumannii Oxa gene (JQ342838), Pseudomonas aeruginosa gryB gene (FJ652722), and Stenotrophomonas maltophilia 23S rRNA gene (HE798556), were retrieved from GenBank and aligned with pathogen sequences from patient samples using the Cluster Omega software in order to confirm pairwise identities.
Some primer sequences were obtained from literature,6,13–15,17,22,23 and optimized to meet our research needs. Primers and probes were designed using Primer Premier 5.0 software and synthesized by Invitrogen Trading (Shanghai) Co., Ltd, Shanghai, China. Primers were labeled with biotin at the 5′ end and all probes were 5′-labeled with NH2C6. The sequences of primers and probes used in our study are shown in Table 1.
Table 1.
Primer and probe sequences for liquid chip arrays of acute contagious respiratory disease pathogens
| Pathogen | Name | Sequences | Length (bp) | Reference |
|---|---|---|---|---|
| SARS | SARS-F1B | Biotin-CTAACATGCTTAGGATAATGG | 368 | This study |
| SARS-R1 | biotin-CAGGTAAGCGTAAAACTCATC | |||
| SARS-P | NH2-TTTTTTTTTTATGCTACAACTGCTTATGCTA | |||
| Influenza A virus | FluA-M-F65 | biontin-CCGAGATCGCACAGAGACTTGAAGAT | 336 | This study |
| FluA-M-R400 | biontin-GGCAAGTGCACCAGCAGAATAACT | |||
| FluA-F65/R400-1 | NH2-TTTTTTTTTTAAGGGGATTTTAGGATTTGTGTTCA | |||
| Influenza A virus subtype H5N1 | H5HA-R1138 | biotin- CTCCCCTGCTCATTGCTATG | 219 | This study |
| H5HA-F920 | biotin- GCCATTCCACAACATACACCC | |||
| H5HA | NH2-TTTTTTTTTTATGCCCCAAATATGTGAAATCAAAC | |||
| N1-F2 | biotin-CAAGTGCTTGCCATGATG | 367 | ||
| N1-R2 | biotin-TCAGGATAACAGGAGCACTC | |||
| Human Bocavirus | HBOV-F | biotin-TAACACTTGGCACGCACAGC | 265 | This study |
| HBOV-R | biotin-TCCCTCGTCTTCATCACTTGGT | |||
| HBOV-P | NH2-TTTTTTTTTTTCATCAGGAACA CCCAATCAGC | |||
| Human Metapneumovirus | HMPV –F | biotin-GAAGAGCTAACCGTGTACTAAGTGATG | 165 | This study |
| HMPV –R | biotin-CTTTGCTGCCTGTAGAGGATGA | |||
| HMPV –P | biotin-CTTTGCTGCCTGTAGAGGATGA | |||
| Human cytomegalovirus | HCMV-F | Biotin-AAGTTTGTGCCCCAACGGTA | 149 | This study |
| HCMV-R | Biotin-GCGTGCTTTTTAGCCTCTGC | |||
| HCMV-P | 5′NH2C12-AAACAGCGTGACGATGACCTGC | |||
| Adenovirus | AD-F | Biotin-CGCAGTGGTCTTACATGCACA | 295 | This study |
| AD-R | Biotin-ACGCCGCGGATGTCAAAGT | |||
| AD-P | 5′NH2C6-TTTTTTTTTTGCCTGAATAACAAGTTTAGAA | |||
| Human parainfluenza virus 1 | PIV1-F | Biotin-CCTTGGAGCGGAGTTGTTAAG | 317 | This study |
| PIV1-R | Biotin-CCGGTAATTTCTCATACCTATG | |||
| PIV1-P | 5′NH2C6-TTTTTTTTTTGGAAAGACCAAATCTCATCG | |||
| Human parainfluenza virus 2 | PIV2-F | Biotin-ATGGAATCAATCGCAAAAGC | 234 | This study |
| PIV2-R | Biotin-GATGATAGATCCCGCTTCCA | |||
| PIV2-P | 5′NH2C6-TTTTTTTTTTGCTGAACTGAGACTTGC | |||
| Human parainfluenza virus 3 | PIV3-F | Biotin-CTCGAGGTTGTCAGGATATAG | 189 | This study |
| PIV3-R | Biotin-CTTTGGGAGTTGAACACAGTT | |||
| PIV3-P | 5′NH2C6-TTTTTTTTTTGATCTCTCATACTTTTAACAT | |||
| Respiratory syncytial virus, | RSV-F | Biotin-CAAGTTGTTGAGGTTTATGAATATGC | 273 | This study |
| RSV-R | Biotin-TTCTGCTGTCAAGTCTAGTACACTGTAGT | |||
| RSV-P | 5′NH2C6-TTTTTTTTTTTCAATTTCCTCACTTCTCCA | |||
| M. pneumoniae | MP-F | Biotin-TGCCATCTACCCGCGCTTA | 300 | Kumar et al.6 |
| MP-R | Biotin-GTGATCTGCCCGGTTTGGTC | |||
| MP-P | 5′NH2C6-TTTTTTTTTTTAACAAACCACGTATGAAC | |||
| Chlamydia pneumonia | CP-F | Biotin-AGTTGAGCATATTCGTGAGG | 127 | Maass et al.17 |
| CP-R | Biotin-TTTATTTCCGTGTCGTCCAG | |||
| CP-P | 5′NH2C6-TTTTTTTTTTAGACTTTAACTTGGCGAA | |||
| Staphylococcus aureus | SA-F | Biotin–ATGGAAGTTCGTGACTTATTAAGC | 313 | Kumar et al.6 |
| SA-R | Biotin–AACAGTTGTTTTAGATGTGTCATGT | |||
| SA-P | 5′NH2C6-TTTTTTTTTTTGATTCTGACAAACCATT | |||
| Streptococcus pneumonia | SP-F | Biotin–GTGATATTTCTGTAACAGCTACC | 354 | Jeong et al.15 |
| SP-R | Biotin–GAGAATTCCCTGTCTTTTCAAA | |||
| SP-P | 5′NH2C6-TTTTTTTTTTAAGTGGAAGACCCCAGCAAT | |||
| Klebsiella pneumonia | KP-F | Biotin-CTGGATCTGACCCTGCAGTA | 68 | Wang et al.18 |
| KP-R | Biotin-CCGTCGCCGTTCTGTTTC | |||
| KP-p | 5′NH2C6-TTTTTTTTTTAAAAACGAAGGCCGTGA | |||
| Acinetobacter baumannii | AB-F | Biotin-TCGTGCTTCGACCGAGTAT | 248 | Nomanpour et al.19 |
| AB-R | Biotin-AACCAACACGCTTCACTTCC | |||
| AB-P | 5′NH2C6-TTTTTTTTTTACCATCCCACTTAAATAC | |||
| Pseudomonas aeruginosa | PA-F | Biotin-GGCGTGGGTGTGGAAGTC | 185 | Lee et al.20 |
| PA-R | Biotin-GTGGCGATCTTGAACTTCTT | |||
| PA-P | 5′NH2C6-TTTTTTTTTTGCTTCACCAACAACAT | |||
| Stenotrophomonas maltophilia | SM-F | Biotin-CAGCCTGCGAAAAGTA | 532 | Whitby et al.21 |
| SM-R | Biotin-TTAAGCTTGCCACGAACAG | |||
| SM-P | 5′NH2C6-TTTTTTTTTTGAGGGGAGTGAAATAGAA |
Establishment of bead-based suspension array detection system
Development of PCR system
DNA from various samples was amplified with the DBI Bestar Taq DNA Polymerase PCR kit (Shanghai Xinghan Sci&Tech Co., Shanghai, China) using 20 pairs of forward and reverse primers. PCR amplification was performed in a 20 µl reaction volume which included dNTPs (2 µl, 2 mM), Bestar Taq Buffer (4.5 µl 10×), Bestar Taq DNA Polymerase (0.8 µl, 2.5 U/µl), forward and reverse primer pairs (1 µM, 2 µl), and 2 µl template strand DNA. Cycling conditions consisted of one cycle of denaturation at 95℃ for 5 min, followed by 38 cycles of denaturation at 94℃ for 30 s, annealing at 58℃ for 30 s, and extension at 72℃ for 30 s. The final extension was done at 72℃ for 10 min. Six primer pairs for viruses (HCMV, AD, PIV1, PIV2, PIV3, RSV) were mixed to develop the first set of multiplex PCR amplification reactions and six primer pairs (FluA, H5N1, SARS, HBOV, HMPV) to develop the second set. Three primer pairs targeting non-viral pathogens (SP, SM, MP) were mixed to develop the third set, and five primer pairs targeting non-viral pathogens (SA, AB, KP, CP, PA) were mixed to develop the fourth set.
Coupling of microspheres to probes
For the coupling reaction, microspheres (50 µl equivalent to 0.623 × 106 beads) were washed thoroughly using sterile water, centrifuged and resuspended in 50 µl MES. The corresponding probes (2 µl of 10 µM) were added to the microsphere suspension, followed by 2.5 µl of freshly prepared 1-Ethyl-3-(3-Dimethyllaminopropyl)- carbodiimide Hydrochloride (EDC). The reaction mix was vortexed and incubated in the dark for 40 minutes at 37℃. Subsequently, 1 ml of 0.02% Tween-20 was added to the coupled microspheres, the mix was centrifuged and the pellet of coupled microspheres was resuspended in 50 µl 1 × Tris-EDTA, and stored in the dark at 4℃.
Cut-off determination of fluorescence for bead-based suspension array and diagnostic criteria for pathogens
Fluorescence intensity values of the resulting hybrids from the amplified DNA products of respiratory tract pathogens were read with a Luminex 100 suspension array analyzer instrument. MFI was defined as the arithmetic mean of the fluorescence signal detected from the encoded microspheres for their respective pathogens. A fluorescence value of ≥200 detected from the positive control DNA (104 copies/µl) was defined as positive, and a value of <150 was defined as negative. Samples with fluorescence values in the grey zone (values < 200 but > 150) were defined as probable positive, and were required to be retested or verified by other methods.24 The sample was defined as positive if the fluorescence value from a repeat detection was > 150, and negative if the value was < 150. The exact fluorescence levels for each pathogen are given in Supplementary Table 2. In addition, multiplex PCR amplification products from samples in the grey zone were analyzed using agarose gel electrophoresis, and samples were classified as positive based on the presence of pathogen-specific bands.
Detection of respiratory tract pathogens
Products amplified from the same nucleic acid sample using 4 different sets of multiplex PCR were mixed. Twenty kinds of encoded microspheres coupled to their respective probes were vortexed and used to prepare 25 µl of hybridization reaction mixture containing 0.1 µl of each probe-coupled microspheres and 4 µl of mixture comprising four sets of multiplex PCR amplification products. The hybridization mixture was denatured at 95℃ for 5 min, and subsequently incubated at the hybridization temperature of 46℃ for 60 min. Fluorescent reporter (75 µl of 4 µg/ml solution) was added to the samples in a 96-well plate, the plate was sealed and further incubated at 46℃ for 15 min. Fluorescence was measured using a suspension array analyzer.
Choice of the gold standards
Bacterial detection from sputum cultures using the VITEK® 2 microbial ID/AST testing system (bioMérieux, Marcy l'Etoile, France) served as the gold standard for bacteria. For viruses and atypical pathogens (including M pneumoniae and C. pneumoniae in this study), the gold standard of detection was real-time, fluorescence-based quantitative real-time PCR using the DBI-2040 Bestar Taq DNA Polymerase PCR kit purchased from Shanghai ZJ Bio-Tech Co., Ltd, Shanghai, China.
Statistical analysis
Gender distribution, incidence of severe pneumonia, and results from bead-based suspension arrays, sputum cultures or quantitative reverse transcriptase-polymerase chain reactions (real-time-PCR) were expressed as counts. The accuracy of the bead-based suspension array was tested by calculating the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), with 95% confidence intervals (CIs) using the findings from sputum culture and real-time-PCR as the gold standards. The indices were calculated as follows:
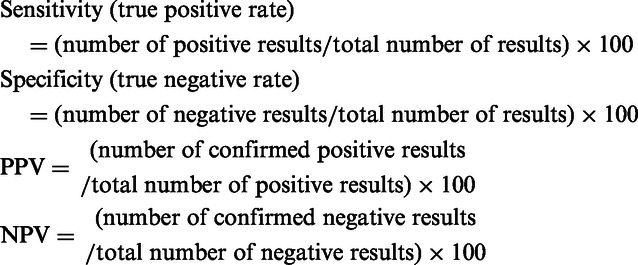
A higher value for sensitivity, specificity, PPV, or NPV, reflected good accuracy of the bead-based suspension array in identifying bacteria, virus and Chlamydia. The difference in accuracy between the suspension array and the gold standards was examined using the McNemar’s test. Conditional logistic regression was carried to test the effect of different technologies on positive detections of bacteria, viruses, and non-classic pathogens. The significance level was defined as 0.05. All statistical analyses were two-sided, and performed using the SAS version 9.2 software (SAS Inc., Cary, NC, USA).
Results
Patient characteristics
In this study, we integrated the bead-based suspension array system for rapid detection of 20 pathogens associated with acute respiratory tract infections. The study population had 223 males and 110 females (Table 2). The age range of patients was 13–96 years and the mean age was 63.2 ± 16.3 years. Although 17% of the patients reported severe pneumonia, only 10.8% of the sputum smears had positive cultures. A total of 115 patients (34.5%) were positive for viral infections, and 10 patients (3%) were positive for M. pneumoniae (In this study, atypical pathogens include M. pneumoniae and C. pneumoniae. As a result, M. pneumonia was detected in 10 cases, while C. pneumoniae was not detected among all examined cases by standard methods. So in this study, only the data of M. pneumoniae were shown and analyzed.) (Table 2). Of the 39 strains of bacteria detected, 29 (74.4%) were Gram-negative bacilli and 10 (25.6%) were Gram-positive cocci.
Table 2.
Baseline characteristics of 333 patients
| Variables | N/mean ± standard deviation | Percentage/ range |
|---|---|---|
| Age, years | 63.2 ± 16.3 | (13, 96) |
| Male | 223 | (67.0) |
| Severe pneumonia | 56 | (16.8) |
| Laboratory test * | ||
| Bacteria (+) | 36 | (10.8) |
| Virus (+) | 104 | (31.2) |
| M. pneumoniae (+) | 10 | (3.0) |
| White blood cell, 109† | 8.8 ± 4.3 | (1.3, 32.7) |
| Neurophil cells in total white blood cells, %† | 69.3 ± 15.2 | (6.5, 98.4) |
| Lymphocytes in total white blood cells, %† | 20.1 ± 13.1 | (0.7, 114.5) |
| C-reactive protein, mg/L† | 59.0 ± 74.9 | (0.0, 404.0) |
Laboratory tests were done by sputum smear for bacteria, and quantitative reverse transcriptase-polymerase chain reaction for virus as well as M. pneumoniae.
†Seven patients had missing data for white blood cell counts; 9 patients for N cell counts; 63 patients for lymph node, and 43 patients for C-reactive protein.
Analysis of coinfections
Analysis of bacterial infections indicated 126 positive patients (37.8%) by liquid phase chip detection; 93 of these patients (73.8%) had single infections and 33 (26.2%) had multiple infections. Among the 33 patients with multiple infections, 25 (19.8%) had infections by 2 bacteria (PA + KP: n = 6; PA + SP: n = 4; KP + SP: n = 3; SA + AB: n = 3; SA + SM: n = 2; PA + AB: n = 1; PA + SM: n = 1; PA + SA: n = 1; SP + AB: n = 1; SP + SM: n = 1; KP + SA: n = 1; KP + SM: n = 1), and 8 (6.3%) had infections by 3 bacteria (KP + PA + AB: n = 2; KP + PA + SP: n = 1; KP + AB + SP: n = 1; KP + AB + SM: n = 1; KP + SM + SP: n = 1; PA + SB + SA: n = 1; PA + SP + SA: n = 1).
Analysis of viral infections indicated 115 positive patients (34.5%); 95 of these patients (82.6%) had single infections and 20 (17.4%) had multiple infections (Flu-A +HCMV: n = 10 [8.7%]; Flu-A + AD: n = 2 [1.7%]; Flu-A + RSV: n = 2 [1.7%]; Flu-A + PIV1: n = 2 [1.7%]; Flu-A + N1: n = 1 [0.9%]; Flu-A + PIV2: n = 1 [0.9%]; HCMV +PIV3: n = 1 [0.9%]; HCMV + H5: n = 1 [0.9%]).
Among all 333 patients, we identified diverse infections in 35 samples (10.5%) in which there was evidence of bacteria or atypical pathogenic mircobes with a virus.
Analysis of sensitivity and specificity
The bead-based suspension array detected bacteria in a significantly higher number of samples (126/333; 37.8%) compared to conventional culture (36/333: 10.8%, P < 0.001). There was no significant difference in the detection rate of atypical pathogens (means only M. pneumoniae here) between the bead-based suspension array and real-time PCR (9/333 (2.7%) vs. 10/333 (3.0%), respectively (P = 0.657). There was also no significant difference in the viral detection rate between the bead-based suspension array and real-time PCR (115/333 (34.5%) vs. 104/333 (31.2%), respectively (P = 0.181). The bead-based array had a sensitivity of 70.0% and a specificity 99.4% for M. pneumoniae compared to the gold standards, respectively. The bead-based array had a sensitivity of 73.1% and a specificity of 83.0% for viruses compared to the gold standards, respectively. The NPV was 87.2% for viruses and 99.1% for M. pneumoniae. Although the PPV was not high as the NPV, it was still higher than 65% regardless of the type of microorganism (Table 3).
Table 3.
Sensitivity and specificity of the bead-based suspension array as compared with the sputum culture for bacteria, and quantitative reverse transcriptase-polymerase chain reaction (real-time-PCR) assay for virus and M. pneumoniae
| Sputum culture for bacteria* | RT-PCR for virus |
RT-PCR for M. pneumoniae |
|||||
|---|---|---|---|---|---|---|---|
| Tested result | Positive | Positive | Negative | P † | Positive | Negative | P † |
| Positive | 28 | 76 | 39 | 0.222 | 7 | 2 | 0.999 |
| Negative | 8 | 28 | 190 | 3 | 321 | ||
| Total | 36 | 104 | 229 | 10 | 323 | ||
| Sensitivity (95% CI) | 77.8% | 73.1% (63.5%–81.3%) | 70% (34.8%–93.3%) | ||||
| Specificity (95% CI) | NA | 83.0 (77.5%–87.6%) | 99.4% (97.8%–99.9%) | ||||
| PPV (95% CI) | NA | 66.1% (56.7%–74.7%) | 77.8% (40.0%–97.2%) | ||||
| NPV (95% CI) | NA | 87.2% (82.0%–91.3%) | 99.1% (97.3%–99.8%) | ||||
CI: confidence interval; PPV: positive predictive value; NPV: negative predictive value; NA: not available.
Only patients with positive outcome were included in the analysis due to difficulty in obtaining valid sputum culture.
†McNemar test was performed.
We compared the sensitivity and specificity of the bead-based suspension array with that of real-time-PCR for eight different viruses (Table 4). There was no significant difference between the two methods in accuracy of detection of PIV2, PIV3, AD, Flu-A, or N1. However, the bead-based suspension array test had a significantly poorer accuracy for HCMV (P < 0.001) and RSV (P = 0.016) compared to real-time-PCR.
Table 4.
Sensitivity and specificity of the bead-based suspension array as compared with quantitative reverse transcriptase-polymerase chain reaction (real-time-PCR) assay* for HCMV, PIV1, PIV2, PIV3, RSV, AD, Flu-A, N1†
| Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|---|---|---|---|---|
| HCMV | 13.0% (4.9%–26.3%) | 98.6% (96.5%–99.6%) | 60.0% (26.2%–87.8%) | 87.6% (83.5%–91.0%) |
| PIV1 | NA | 99.4% (98.6%–100.0%) | NA | 100.0% (98.9%–100.0%) |
| PIV2 | 100.0% (15.8%–100.0%) | 99.4% (97.8%–99.9%) | 50.0% (6.8%–93.2%) | 100.0% (98.9%–100.0%) |
| PIV3 | 50.0% (6.8%–93.2%) | 99.7% (98.3%–99.9%) | 66.7% (9.4%–99.2%) | 99.4% (97.8%–99.9%) |
| RSV | 12.5% (3.2%–52.7%) | 100.0% (98.9%–100.0%) | 100.0% (2.5%–100.0%) | 97.9% (95.7%99.2%) |
| AD | 25.0% (6.3%–80.6%) | 99.4% (97.8%–99.9%) | 33.3% (8.4%–90.6%) | 99.1% (97.4%–99.8%) |
| Flu-A | 82.4% (56.6%–96.2%) | 97.5% (95.1%–98.9%) | 63.6% (40.7%–82.8%) | 99.0% (97.2%–99.8%) |
| N1 | 50.0% (1.3%–98.7%) | 100.0% (98.9%–100.0%) | 100.0% (2.5%–100.0%) | 99.7% (98.3%–99.9%) |
AD: adenovirus; CI: confidence interval; Flu-A: influenza A virus; HCMV: human cytomegalovirus; N1: influenza A virus subtype N1; NPV: negative predictive value; NA: not available (due to no positive event); PPV: positive predictive value; PIV1: human parainfluenza virus 1; PIV2: human parainfluenza virus 2; PIV3: human parainfluenza virus 3; RSV: respiratory syncytial virus.
Bold values indicate significantly different from gold standard, P < 0.05.
McNemar test was performed.
Discussion
In this study, our high-throughput bead-based suspension array had a significantly higher efficiency of detection of bacteria compared to conventional culture. However, there was no significant difference in the positivity rates for atypical pathogens or viruses between the bead-based suspension arrays and real-time PCR assays.
The demanding culture conditions, complexity of culture techniques, and the long detection time for viruses and atypical pathogens have resulted in the emergence of new diagnostic tools such as bead-based suspension arrays which have shown promise in the detection of Vibrio species, human genital papillomaviruses, biothreat agents, and bacterial pathogens implicated in food-borne illnesses.25–28 The advantages of our present bead-based suspension array include: (1) the ability to simultaneously detect viruses, atypical pathogens as well as bacteria, in contrast with most current detection methods which are used to detect either only viruses or bacteria,7,28 (2) rapid, high-throughput detection where detection of 96 samples can be completed in 7–8 h, (3) semi-quantitative detection, (4) small amounts of sample required, and (5) economy and ease of use compared to solid-phase arrays.29
Our final annealing temperature of 58℃ for target DNA amplification was higher than the annealing temperature recommended by the manufacturer. Higher annealing temperatures may cause decreased amplification efficiency, but a higher specificity. Based on the guidelines suggested by Luminex, the best capture probe lengths are 18–20 bp, and can be extended to 22–24 bp under special occasions in order to obtain stronger fluorescence signal.30 In the present study, we used probes of around 14 to 18 bp, which would generate lower fluorescence signals for some pathogens, but would reduce non-specific signals. The shorter probe sequences used in our study resulted in a hybridization temperature lower than previously described.31 We used a relatively longer hybridization period (60 min) to provide sufficient reaction time for interaction of probes with their target fragments, as well as for amplified target gene fragments to compete with non-specific sequences for binding with the probes.
Our detection technology was semi-quantitative. Fluorescence values of the same pathogen could be compared between different samples, although fluorescence values could not be compared between groups of different pathogens. The lowest concentrations that could be detected by our technology ranged from 100/µl to 104/µl, and detection efficiency was affected by various conditions, including amplification efficiency of primers, binding of the probe and its target sequence, hybridization temperature, the concentration of SA-PE, hybridization time, Tm of the probe, percentage of GC content in the probe, and the primer binding site on target fragments.
The higher bacterial detection rates of bead-based suspension arrays compared to conventional culture in our study could be because the positivity rates of bacterial cultures are influenced by factors such as prior antibiotic use, shapes of sputum samples, and whether the samples are delivered to the lab in a timely manner. Previous results reported a high incidence of pathogenic Gram-negative bacilli in patients with acute respiratory tract infections.32 These data were consistent with our study where the majority of bacterial strains detected (68.2%) were Gram-negative bacilli. This could be because most of the hospitalized patients in our study were older, had complications and had reported self-medication with antibiotics. The rate of virus infections detected in our study was higher than previously reported incidence rates of community-acquired pneumonia (5% to 15%). Of the viral strains found in our study, there are 24 strains of Flu-A (46.7%). This could be because our cases mostly comprised hospitalized patients during the winter-spring season who may experience seasonal influenza epidemics more frequently.
The infection rate of HCMV in the normal population was previously shown to be 30%–100%,33,34 which was consistent with our present data. M. pneumoniae have been shown to cause approximately 10% to 15% of all acute upper respiratory tract infections in children and teenagers.35–38 C. pneumoniae causes infections in over 50% of adults (age ≥ 20) while half of the patients were asymptomatic.31,39 In contrast, a total of five cases were positive for M. pneumoniae, four cases were positive for C. pneumoniae in our study and this low incidence could be because most of our cases were elderly patients with various complications. However, our low detection rate of atypical pathogens could be due to the insufficient sample size being tested.
We found that PA and KP were the most common pathogens in patients with multiple bacterial infections. In contrast, de Roux et al.40 reported that multiple infections were usually caused by SP and other bacteria. This difference might be ascribed to our use of SP-sensitive antibiotics before the sample collection. Our results also showed that 17.4% of patients had infections with two viruses, higher than previously reported by Drews et al.,41 possibly due to the higher sensitivity of our assays. In the present study, Flu-A and HCMV were the most common viruses in the multiple-strain infections. It is possible that an outbreak of influenza A infection weakened immunity, and led to activation of colonized cytomegaloviruses and the replication of viral DNA. Most studies of infections caused by bacteria and viruses regard the viral infection as secondary to the bacterial infection. This indicates that concomitant viral infection should be considered if there is no response after long-term antibiotic therapy.42
In summary, we showed that bead-based suspension array technology had a significantly higher sensitivity for bacterial detection and was not inferior to real-time PCR for the detection of virus and atypical pathogens in patients with acute respiratory tract pathogens. The use of bead-based suspension array technology has some limitations. Non-specific binding can cause interference. Moreover, since the Luminex technology platform is an open system, it is possible that PCR products may cause contamination in labs. Our present study also had some limitations. The positive rates for Mycoplasma and Chlamydia detection were not high enough for further evaluation. Furthermore, we did not analyze our data for false-positives. It is important to validate our findings in larger sample sizes in order to facilitate the transition of this technology to a clinical setting.
Supplementary Material
Supplementary Material
Acknowledgements
This study was supported by Leading Project of Fujian Science and Technology Department Funds (2016Y0015); Subject of Fujian Provincial Development and Reform Commission (no.: 2060404); Key projects of Fujian Provincial Hospital (no. 2014080).
Authors’ contributions
Y-SC: guarantor of integrity of the entire study, definition of intellectual content, manuscript review; H-RL: study concepts, study design, literature research, data analysis, manuscript editing; WZ: experimental studies, statistical analysis; Z-DH: experimental studies, data analysis, manuscript preparation; X-HLin: experimental studies, data acquisition; M-QL: data acquisition; W-SH: data acquisition; L-PH: clinical studies, data acquisition; X-LY: clinical studies, data acquisition; N-LXu: clinical studies; ML: clinical studies; B-SX: clinical studies; X-NS: experimental studies; J-FX: experimental studies; YW: experimental studies; MH: experimental studies; Y-AW: experimental studies; X-LHu: clinical studies.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 2006; 12: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cockburn WC, Delon PJ, Ferreira W. Origin and progress of the 1968-69 Hong Kong influenza epidemic. Bull World Health Organ 1969; 41: 345–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Groneberg DA, Zhang L, Welte T, Zabel P, Chung KF. Severe acute respiratory syndrome: global initiatives for disease diagnosis. QJM 2003; 96: 845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan SJ, Jacobson RM, Dowdle WR, Poland GA. 2009 H1N1 influenza. Mayo Clin Proc 2010; 85: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memish ZA. MERS-CoV. An emerging viral zoonotic disease: three years after and counting. Recent Pat Antiinfect Drug Discov 2014; 9: 159–60. [PubMed] [Google Scholar]
- 6.Kumar S, Wang L, Fan J, Kraft A, Bose ME, Tiwari S, Van Dyke M, Haigis R, Luo T, Ghosh M, Tang H, Haghnia M, Mather EL, Weisburg WG, Henrickson KJ. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J Clin Microbiol 2008; 46: 3063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL, Shult PA, Prudent JR, Gern JE. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol 2007; 45: 2626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y, Deng R, Wang C, Deng T, Peng P, Cheng X, Wang G, Qian M, Gao H, Han B, Chen Y, Hu Y, Geng R, Hu C, Zhang W, Yang J, Wan H, Yu Q, Wei L, Li J, Tian G, Wang Q, Hu K, Wang S, Wang R, Du J, He B, Ma J, Zhong X, Mu L, Cai S, Zhu X, Xing W, Yu J, Deng M, Gao Z. Etiologic diagnosis of lower respiratory tract bacterial infections using sputum samples and quantitative loop-mediated isothermal amplification. PLoS One 2012; 7: e38743–e38743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA 2002; 99: 15687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battaglia A, Schweighardt AJ, Wallace MM. Pathogen detection using a liquid array technology. J Forensic Sci 2011; 56: 760–5. [DOI] [PubMed] [Google Scholar]
- 11.Schweighardt AJ, Battaglia A, Wallace MM. Detection of anthrax and other pathogens using a unique liquid array technology. J Forensic Sci 2014; 59: 15–33. [DOI] [PubMed] [Google Scholar]
- 12.Hulse RE, Kunkler PE, Fedynyshyn JP, Kraig RP. Optimization of multiplexed bead-based cytokine immunoassays for rat serum and brain tissue. J Neurosci Methods 2004; 136: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol 2007; 45: 2965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. xTAG RVP assay: analytical and clinical performance. J Clin Virol 2007; 40(Suppl 1): S39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong ES, Lee KS, Heo SH, Seo JH, Choi YK. Rapid identification of Klebsiella pneumoniae, Corynebacterium kutscheri, and Streptococcus pneumoniae using triplex polymerase chain reaction in rodents. Exp Anim 2013; 62: 35–40. [DOI] [PubMed] [Google Scholar]
- 16.Sun K, Wang Q, Huang XH, Zhen MC, Li W, Zhang LJ. Establishment of multiplexed, microsphere-based flow cytometric assay for multiple human tumor markers. Acta Pharmacol Sin 2007; 28: 2011–8. [DOI] [PubMed] [Google Scholar]
- 17.Maass M, Krause E, Engel PM, Kruger S. Endovascular presence of Chlamydia pneumoniae in patients with hemodynamically effective carotid artery stenosis. Angiology 1997; 48: 699–706. [DOI] [PubMed] [Google Scholar]
- 18.Wang IK, Lai HC, Yu CJ, Liang CC, Chang CT, Kuo HL, Yang YF, Lin CC, Lin HH, Liu YL, Chang YC, Wu YY, Chen CH, Li CY, Chuang FR, Huang CC, Lin CH, Lin HC. Real-time PCR analysis of the intestinal microbiotas in peritoneal dialysis patients. Appl Environ Microbiol 2012; 78: 1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomanpour B, Ghodousi A, Babaei A, Abtahi H, Tabrizi M, Feizabadi M. Rapid, cost-effective, sensitive and quantitative detection of Acinetobacter baumannii from pneumonia patients. Iran J Microbiol 2011; 3: 162–9. [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CS, Wetzel K, Buckley T, Wozniak D, Lee J. Rapid and sensitive detection of Pseudomonas aeruginosa in chlorinated water and aerosols targeting gyrB gene using real-time PCR. J Appl Microbiol 2011; 111: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitby PW, Carter KB, Burns JL, Royall JA, LiPuma JJ, Stull TL. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol 2000; 38: 4305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta 2006; 363: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earley MC, Vogt RF, Jr., Shapiro HM, Mandy FF, Kellar KL, Bellisario R, Pass KA, Marti GE, Stewart CC, Hannon WH. Report from a workshop on multianalyte microsphere assays. Cytometry 2002; 50: 239–42. [DOI] [PubMed] [Google Scholar]
- 24.Deregt D, Gilbert SA, Dudas S, Pasick J, Baxi S, Burton KM, Baxi MK. A multiplex DNA suspension microarray for simultaneous detection and differentiation of classical swine fever virus and other pestiviruses. J Virol Methods 2006; 136: 17–23. [DOI] [PubMed] [Google Scholar]
- 25.Tracz DM, Backhouse PG, Olson AB, McCrea JK, Walsh JA, Ng LK, Gilmour MW. Rapid detection of Vibrio species using liquid microsphere arrays and real-time PCR targeting the ftsZ locus. J Med Microbiol 2007; 56(Pt 1): 56–65. [DOI] [PubMed] [Google Scholar]
- 26.Wallace J, Woda BA, Pihan G. Facile, comprehensive, high-throughput genotyping of human genital papillomaviruses using spectrally addressable liquid bead microarrays. J Mol Diagn 2005; 7: 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson WJ, Erler AM, Nasarabadi SL, Skowronski EW, Imbro PM. A multiplexed PCR-coupled liquid bead array for the simultaneous detection of four biothreat agents. Mol Cell Probes 2005; 19: 137–44. [DOI] [PubMed] [Google Scholar]
- 28.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J Microbiol Methods 2003; 53: 245–52. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–8. [DOI] [PubMed] [Google Scholar]
- 30.Spiro A, Lowe M, Brown D. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl Environ Microbiol 2000; 66: 4258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S. The microimmunofluorescence test for Chlamydia pneumoniae infection: technique and interpretation. J Infect Dis 2000; 181(Suppl 3): S421–5. [DOI] [PubMed] [Google Scholar]
- 32.Chen YS, Wang DX, Li HR, Chen YW. The applications of loop-mediated isothermal amplification in the detection of common pathogens causing lower respiratory tract infections. Chin J Tubercul Respir Dis 2011; 17: 1–1. [Google Scholar]
- 33.Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis 2004; 4: 725–38. [DOI] [PubMed] [Google Scholar]
- 34.Retalis P, Strange C, Harley R. The spectrum of adult adenovirus pneumonia. Chest 1996; 109: 1656–7. [DOI] [PubMed] [Google Scholar]
- 35.Krunkosky TM, Jordan JL, Chambers E, Krause DC. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb Pathog 2007; 42: 98–103. [DOI] [PubMed] [Google Scholar]
- 36.Halbedel S, Stulke J. Tools for the genetic analysis of Mycoplasma. Int J Med Microbiol 2007; 297: 37–44. [DOI] [PubMed] [Google Scholar]
- 37.Samransamruajkit R, Jitchaiwat S, Wachirapaes W, Deerojanawong J, Sritippayawan S, Prapphal N. Prevalence of Mycoplasma and Chlamydia pneumonia in severe community-acquired pneumonia among hospitalized children in Thailand. Jpn J Infect Dis 2008; 61: 36–9. [PubMed] [Google Scholar]
- 38.Eun BW, Kim NH, Choi EH, Lee HJ. Mycoplasma pneumoniae in Korean children: the epidemiology of pneumonia over an 18-year period. J Infect 2008; 56: 326–31. [DOI] [PubMed] [Google Scholar]
- 39.Reischl U, Lehn N, Simnacher U, Marre R, Essig A. Rapid and standardized detection of Chlamydia pneumoniae using LightCycler real-time fluorescence PCR. Eur J Clin Microbiol Infect Dis 2003;22:54–7. [DOI] [PubMed]
- 40.de Roux A, Ewig S, García E, Marcos MA, Mensa J, Lode H, Torres A. Mixed community-acquired pneumonia in hospitalised patients. Eur Respir J 2006; 27: 795–800. [DOI] [PubMed] [Google Scholar]
- 41.Drews AL, Atmar RL, Glezen WP, Baxter BD, Piedra PA, Greenberg SB. Dual respiratory virus infections. Clin Infect Dis 1997; 25: 1421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet 2011; 377: 1264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


