Abstract
The ubiquitin system plays an important role in essentially every cellular process, regulating numerous pathways ranging from development, transcription, DNA damage response, cell cycle, and signal transduction. Its best studied role involves removal of faulty proteins or those that are not necessary anymore. Aberrations in the ubiquitin system have been implicated in various pathologies including cancer, where specific mutations in E3 ligases such as Mdm2, pVHL, and BRCA1 have been linked to disease progression, prognosis, and resistance to drugs. Yet, there are hundreds of E3 ligases in the human genome and our knowledge of their target proteins and their dynamic regulation in the cellular environment is largely limited. In addition, fundamental questions related to recognition and specificity in ubiquitin conjugation remain unanswered. It is thus of major importance to characterize the ubiquitin landscape under various cellular conditions, and study how the regulatory network is altered in health and disease. To do so, analytical tools that allow identification of ubiquitin substrates, the conjugation and removal of ubiquitin, and the nature of specific ubiquitin linkages that are formed are needed. In this mini-review, we discuss common proteomic methodologies applied to studying the ubiquitome, and specifically focus on our recently developed post-translational modification (PTM) profiling approach. PTM profiling is a functional assay, amenable to biochemical manipulation, which allows the detection of protein modifications in a high-throughput manner. We discuss in detail the advantages and limitations of this system, focusing primarily on examples for analyzing the ubiquitin system in cancer. Uncovering the intricate signaling dynamics governed by and regulating ubiquitin modifications should clearly evolve into a new paradigm in understanding the molecular basis of malignant transformation and the development of novel therapeutic modalities.
Keywords: Ubiquitin, ubiquitin-like molecules, E3 ligases, cancer, proteomics, protein microarrays
Introduction
Ubiquitin and ubiquitin-like post-translational modifications (PTMs)
Among more than 200 types of PTMs in humans, the ubiquitin and ubiquitin-like (Ubl) protein families comprise a class of evolutionarily conserved proteins that are reversibly covalently conjugated to other proteins to regulate a variety of fundamental cellular processes, such as maintenance of the human genome and proteome, cell division and differentiation, signal transduction, and targeting of proteins to their proper subcellular destinations.1–4 Aberrations in ubiquitin and Ubl pathways have been implicated in the pathogenesis of numerous human diseases such as cancer and immune system pathologies.5,6 While vast amount of information describing various ubiquitination events has accumulated over the years, we are still in search of a comprehensive understanding of the ubiquitin regulatory code and its dynamic changes in the rapidly changing cellular environment. Importantly, fundamental questions related to the biology of the ubiquitin system and its involvement in cancer pathology remain unanswered: What are the regulatory principles governing substrate specificity and recognition? Can a ubiquitin profile of protein substrates be indicative of a pathological state? Can such a profile teach us about the molecular pathways that are dysregulated in disease? Most importantly, perhaps, will be the ability to engage these problems to identify novel means for reshaping the cellular environment by controlling the enzymatic machinery of the ubiquitin system. To address these questions, significant achievements have been made in the past decade in the ability of mass spectrometry (MS) to analyze whole proteomes and map modification sites under various conditions. Yet, challenges and difficulties are warranting additional solutions, independent from MS analysis.
Of particular difficulty is the ability to identify the ubiquitin and Ubl regulatory network. While the Ubls share only modest primary sequence identity with ubiquitin, they are closely related to it in structure and similar to ubiquitin, their conjugation to the target substrate requires a multistep enzymatic cascade. The biochemical reaction of ubiquitin conjugation has been reviewed extensively. In brief, one of a few ubiquitin-activating enzymes, E1s, forms a high-energy thiol ester intermediate with the C-terminal Gly residue of ubiquitin that is then transferred to one of several ubiquitin conjugating E2 enzymes. In the final step of the cascade, one of several hundreds ubiquitin E3 ligases, that are the specific recognition elements of the system, forms an isopeptide bond between the C-terminal Gly of ubiquitin and – in most cases – an ɛ-NH2 group of an internal Lys of the substrate.7 This cascade may then be repeated, allowing additional ubiquitin molecules to attach sequentially to one another via an isopeptide bond involving one of the seven internal Lys residues in the ubiquitin moiety, thus generating a polyubiquitin chain. Polyubiquitinated substrates with specific internally bound ubiquitin moieties (via Lys48) are targeted for proteasomal degradation, while mono-ubiquitination, and conjugation via other Lys residues as well as by Ubls, have different cellular functions. For some Ubls, however, the enzymatic machinery that is involved in their conjugation is vaguely known. Hundreds of E3 ligases in the proteome confer substrate specificity. Hence, careful depiction of this highly complex regulatory system, while being of great significance, is still impeded by analytical challenges.
In this mini-review, we discuss the current available methodologies used in ubiquitination research. Specifically, we focus on a novel approach, designated PTM profiling, and discuss the advantages it presents in addressing the ubiquitin network landscape in health and disease.
Methodologies and approaches used in studying the ubiquitin network
Common approaches for studying ubiquitination include biochemical methods as well as advanced MS technologies. Yet, global analysis of alterations in ubiquitin substrates is still difficult. What are then the hurdles in systematically following such changes in ubiquitination? One difficulty is due to the traditional practice of biochemical assays, which primarily focus on analyzing single proteins and are not geared toward revealing global patterns of regulation. For example, screening ubiquitination or degradation activities using SDS-PAGE or Western blotting analyses is cumbersome and cannot be done on large scales. Furthermore, most cell-based assays, which provided informative molecular and mechanistic insights and revealed numerous functions of ubiquitin, are primarily focused on identifying degradation of targets rather than changes in ubiquitin signaling.8,9 Another obstacle is the highly reversible nature of these modifications, which can be easily lost during sample preparation. Yet, state-of-the-art MS analyses have greatly advanced the ubiquitin research field. What still holds MS analysis short of providing complete solutions for delineating the ubiquitome network? In brief, the identification of ubiquitinations by MS, in most cases, relies on tryptic digest of proteins prior to analysis.10 In ubiquitinated proteins, trypsin cleaves the attached ubiquitin after residue R74, resulting in peptides with an attached Gly–Gly (GG) remnant fragment of 140 Da. Thus, analysis for peptides containing such fragments can identify ubiquitinated proteins as well as their specific site of modification.11 Indeed, extensive descriptions of ubiquitination events have been acquired over the years using this approach.12–18 Nevertheless, a crucial disadvantage of this system is its ineffectiveness in detecting changes in polyubiquitination and specific linkage topologies of polyubiquitinated species, as these are also cleaved by trypsin prior to the MS analysis. In addition, rapidly degraded substrates, low abundant proteins, and highly reversible modifications are often undetectable using MS analysis. Recently, enrichment protocols using anti-GG antibodies have been developed;19 however, substrates subject to this analysis are still cleaved in vitro to peptides prior to analysis, restricting the ability to analyze them in the relevant cellular context, and limiting the detection of multiple modifications on a single protein.20 With the goal of achieving broad depiction of protein modifications, initially focused on phosphorylation, researchers have employed high-throughput platforms. Many such systems utilized different forms of protein microarrays, which allow studying thousands of proteins in parallel.21
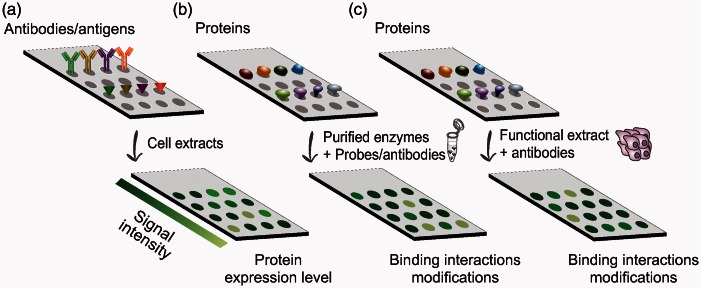
Protein arrays rely on immobilizing or capturing molecules (antibodies/antigens), or samples (in the case of reverse-phase protein arrays [RPPA]),22,23 on solid surfaces, exposing them to the studied reaction mixture, followed by probing with specific antibodies of interest (Figure 1). Designed to afford detection of dynamic changes across many samples, RPPA is a powerful tool in proteomic analysis and is used commonly in MS-independent assays for molecular profiling of specific proteins and biomarkers in clinical samples.24 However, utilizing RPPA for detecting ubiquitination of proteins in biological samples is still highly limited by the relatively small set of antibodies available for ubiquitinated substrates. Furthermore, protein modifications and functionality may be affected during sample preparation and immobilization, potentially limiting the interpretation of attained data. Efforts to develop activity-based assays, to test for the activity of enzymes rather than abundance, resulted in the production of functional protein arrays. In these platforms, native proteins are arrayed and incubated with cellular extracts/purified enzymes and probes or antibodies to identify binding properties as well as PTMs in various systems. In two pioneering studies, MacBeath and Schreiber25 and Zhu et al.26 demonstrated the use of attaching substrates to microarray surfaces to study kinase specificities. Their results show that a single substrate could be detected specifically among thousands of other proteins spotted on the array. The latter study further revealed novel protein–protein interactions presenting the great potential functional protein arrays have for large-scale studies of biochemical pathways. Another approach that has been developed is that of activity-based protein profiling (ABPP) utilizing chemical probes directed against the active site of specific enzymes which can indicate their functional state.27 ABPP systems have been designed to target multiple enzyme classes, including, among many, kinases and phosphatases, proteases and hydrolases, and histone deacetylases.27 This makes ABPP a useful applicable tool that can be used with numerous biological systems. It should be noted, however, that the specificity of such probes is often not high, and the methodology may disrupt other pathways or even be toxic to the cell. Interestingly, some studies have already used of in vitro ubiquitination assays on protein microarrays to identify substrates of a specific E3 ligase.28,29 Major drawbacks still exist, however, in the proficiency of protein arrays to analyze PTMs. First, common approaches rely on strictly biochemical in vitro assays which may be prone to artifacts due to the artificial environment of mixing purified components or cell extracts in a test tube. Second, commercial arrays rely on proteins that were expressed and purified by the manufacturer, rendering the analysis biased toward these proteins, which may not be expressed in every cell type or conditions, or inversely, are expressed in cells but are lacking in the array.
Figure 1.
Common types of protein microarrays. In abundance-based arrays (a), captured antibodies/antigens are probed with cell extracts to reveal reactive protein expression levels. Functional protein arrays are based on the immobilization of proteins on the array and probing them with specific antibodies (b). The PTM profiling microarray incorporates incubation of the spotted proteins with functional cell extracts before probing with specific antibodies of interest (c). Different signal intensities reflect the reactivity of modified proteins.
In the next sections, we shall discuss in detail the recently developed PTM profiling approach30–33 in the context of specific biological questions related to analysis of the enzymatic machinery and substrates of the ubiquitin system. We shall further illustrate its potential for analyzing the ubiquitin system in the context of cancer research.
PTM profiling as a tool for mapping the cellular PTM landscape
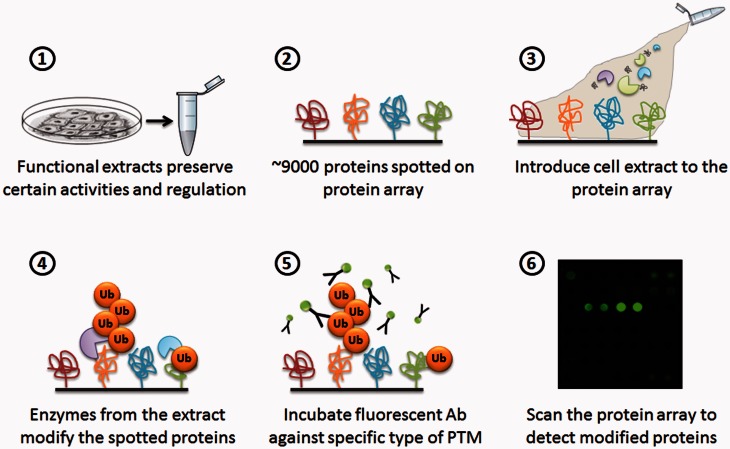
PTM profiling makes use of functional cellular extracts that may be prepared from cells that were “induced” by a specific signal such as DNA damage, cell cycle arrest, or specific activation of signaling pathways. In sharp contrast to most practiced methods, the extract preparation in this protocol is detergent free, rendering the samples biochemically active, and preserving many aspects of the biological context.34 The extracts are incubated on protein microarrays containing thousands of recombinant proteins that act as substrates for modifications executed by active enzymatic cascades during the incubation. Importantly, unlike conventional in vitro assays, the active extracts comprise a complex environment to facilitate the ubiquitination of target proteins. It is not necessary to supplement the system with exogenous enzyme to affect the reaction, and all conjugation events may be the consequence of intrinsic activities of the functional extract. However, in some cases, in order to promote/inhibit specific enzymatic activities, an enzyme or a mutant form of an enzyme may be added to the extract in order to analyze its effect on downstream signaling and modifications. The modified proteins are then collectively identified using an antibody against the specific modification of interest and scanned. Using quantification software, signal intensity values and signal-to-noise ratios are calculated per all spots in the array, as well as local background, and the reactivity level of each protein is then calculated (Figure 2).
Figure 2.
Schematic representation of the PTM profiling approach. Steps 1–6 represent the sequential process of the PTM profiling protocol.
What are the advantages and limitations of the PTM profiling approach in analyzing the ubiquitin system?
In addition to allowing for large-scale studies, this system is unique in that the biochemically active extracts preserve many regulatory events under conditions that are relatively close to those of the complex cellular environment. For example, certain E3 ligases require specific phosphorylation and/or dephosphorylation for their activation, which may be missed in reactions carried out by purified/recombinant components. Furthermore, the extracts themselves provide a plethora of potential substrates and competitors to the substrates immobilized to the surface, enhancing the specificity and the significant changes of PTMs one detects. Importantly, as proteins of low cellular abundance are represented equally to highly abundant substrates on the array, PTM profiling detects modifications irrespective of the endogenous cellular expression. We have found that the PTM profiling system is highly sensitive and that even low-reactivity spots may represent significant biological information. Another technical feature that may be advantageous in this system is the ability to detect several modifications simultaneously, for example, by probing the array with antibodies against more than one Ubl. In such a manner, the pattern of modifications by different Ubls can be systematically compared among different conditions, and dynamic regulations involving different Ubl modifications, or different chain linkages in ubiquitin (using specific antibodies against different linkages), can be identified. Therefore, PTM profiling may both uncover previously undetected conjugation events, as well as reveal fine alterations in the status of a given modification, which could reflect changes in cellular processes. Analyses should still be complemented however by corresponding MS assays to identify the specific residues that undergo modifications,35 a feature which PTM profiling is still largely missing. The main advantages and limitations of the PTM profiling system are summarized in Box 1.
Box 1.
Advantages and limitations of the PTM profiling system
| Advantages of PTM profiling: |
|---|
| ✓ High-throughput technology provides a platform for large-scale quantitative studies. |
| ✓ Conservation of biochemical function in the extract preserves a faithful biological context in the reaction. |
| ✓ Lack of correlation to protein abundance and rate of turnover facilitates identification of PTMs for both high- and low-abundance proteins equally. |
| ✓ Detection of more than a single Ubl per reaction allows for complex network assessments. |
| ✓ High assay sensitivity permits evaluation of small cell number samples. |
| ✓ Amenability to extract manipulations presents great potential for drug design studies targeting PTM alterations in various pathologies. |
| Limitations of PTM profiling: |
|---|
| × Sometimes difficulties to distinguish between substrate conjugation and conjugation to an interacting partner may arise. |
| × As the substrates are represented randomly on the microarray, validation of substrate expression in the extract of study is required. This can be easily achieved via RNA-seq. Alternatively, custom arrays representing known substrates can be prepared. |
| × Availability of only two-color labeling in the microarray scanning technologies (available nowadays) limits the analysis to two PTMs at a time and reduces the multiplexing potential of the system. However, advanced scanning technologies will most likely be developed in the near future which may be implemented to this protein microarray system. |
PTM profiling as a tool for deciphering regulatory principles of E3 ligases
With several hundreds of E3 ligases and tens of E2 enzymes in the human genome, the regulatory pathways involving ubiquitin and Ubl modifications could be of gigantic complexity. Identifying the enzymes and substrates that are involved in these modifications, including the assignment of myriad substrates to specific ligases, is an essential first step for deciphering the molecular pathways in which they operate. How can such an ambitious goal be approached? In past years, numerous ubiquitination enzymes were identified using genetic screens in model organisms such as S. cerevisiae or D. melanogaster. The introduction of the siRNA technology and screens substantially strengthened the ability to identify and characterize ubiquitination enzymes in human cells. Still, in spite of providing grounds for large-scale identification of regulators, success of such screens was often incomplete due to redundancy in the modification/regulation of substrates by more than a single enzyme. A different approach, widely used to delineate enzymatic Ubl cascades, relied on their recapitulation in vitro, using purified enzymatic components. While such reactions may provide mechanistic clues, they are often incomplete as they may lack components present in the cellular context. Scaffolds, adaptors, inhibitors, and de-conjugating enzymes are all crucial for regulating the specificity of conjugation events. With hundreds of modifying enzymes in the genome, we have limited ability to predict the constituents of each reaction.
How can PTM profiling be utilized to reveal the ubiquitin regulatory network?
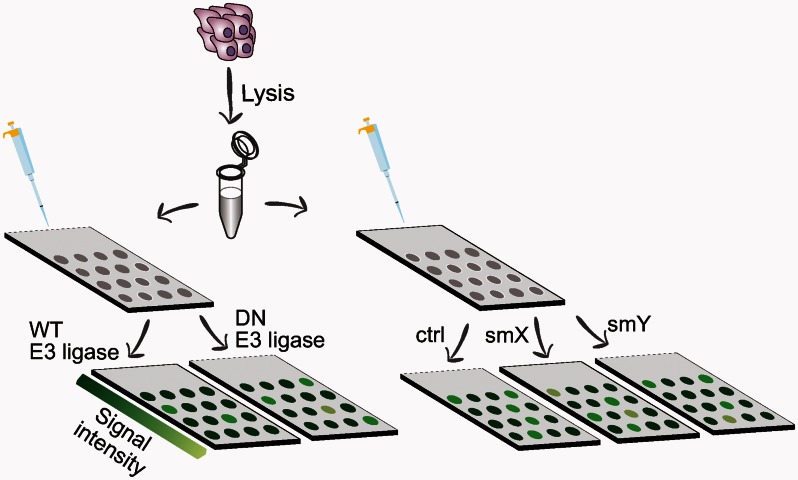
An imperative feature of the PTM profiling platform is its amenability to biochemical manipulations. The functional extract may be supplemented with substances such as chemicals, antibodies, mutant proteins/enzymes and substrates, inhibitors, and more, and the impartial effect of the perturbation on downstream modifications can be measured. For example, introduction of a purified dominant-negative ubiquitin ligase (e.g. catalytically dead HECT E3 mutant that can still bind the substrate) into extracts during incubation allows to systematically map its targets by analyzing the change in the ubiquitination signal compared to its wild-type counterpart. This competitive inhibition assay allows for the identification of substrates that are specific to that ligase, whereas promiscuous or non-specific substrates will presumably remain unaltered (Figure 3). In the same fashion, PTM profiling can be a useful tool for studying other ubiquitination regulators such as deubiquitinating enzymes (DUbs) by either comparing wild-type and mutant forms of such enzymes, or by probing the array first with ubiquitin and measuring the reactivity of the targets, and then looking at the disappearance of a signal by introducing the specific DUb. The effect of small molecule inhibitors can be evaluated in the same manner (Figure 3). Combining the information on ubiquitin cascades should yield a comprehensive view of the dynamic regulation of the immense ligase-substrate network operating in various physiological conditions.
Figure 3.
Possible manipulations of the PTM profiling platform. Cell extracts supplemented with exogenous wild-type (WT) or dominant-negative (DN) forms of E3 ligase (left), or with different small molecules (smX/Y) (right), will reveal specific protein substrates of different E3s. Represented is labeling with an antipolyubiquitin antibody and a fluorescently labeled secondary antibody for detection. Different signal intensities reflect the reactivity of modified proteins.
PTM profiling as a tool for identifying E3 ligases as targets for anticancer treatment
In the past two decades, tremendous achievements have been made in the ability to decode the genome and identify genes that are associated with disease. Yet, both DNA and RNA analyses data are often indirect and do not correlate with the actual activity of proteins in cells and tissues that is largely dependent on PTMs. Seminal studies published recently have addressed the problem of inference from genomic abnormalities to cancer phenotypes,36 showing that proteomic classification of tumor subtypes generated signatures that were clinically more relevant than those inferred from transcriptomic analyses. Since the control of signaling cascades is fundamentally dependent on PTMs, it is plausible that mapping the ubiquitin and Ubl landscape in cancer would yield significant insight(s) on the underlying mechanisms of cellular transformation.
Ubiquitin E3 ligases can be generally classified into two subfamilies: RING and HECT-domains, based on their mechanism of action and sequence homology of their E2-binding domains.37 There are ∼600 RING finger and ∼30 HECT E3 ligases in humans, and we are far from knowing the complete range of their biological functions and the principles of their recognition due to the limited knowledge of their targets. E3 ligases play key roles in regulating the cellular environment and aberrations in their expression or function are associated with various pathologies, including cancer. Indeed, several E3 ligases were demonstrated to be involved in cancer.37 Prominent examples are those of double minute 2 (MDM2) and S-phase kinase-associated protein 2 (SKP2), two E3 proto-oncogenes which promote the degradation of the p53 and p27 tumor suppressors, respectively, and are often overexpressed in malignancies.38–42 Moreover, certain oncogenic human papillomavirus E6 proteins bind to the HECT E3 ligase E6-AP and induce the degradation of p53.43 Increased complexity arises from the promiscuous nature of this system, where a single substrate can be targeted by more than one ligase. A striking example is the targeting of the NF-κB p105 precursor by two different E3 ligases, which result in completely different fates. While ubiquitination catalyzed by βTrCP induces complete degradation of p105, the KPC1 E3 ligase leads to processing of the precursor to the p50 active subunit of the transcription factor.44 Therefore, activation of one E3 cascade over the other may be crucial for cell-fate decisions, promoting or inhibiting the p50 tumor-suppressive effect. Still, the conditions that affect the induction of these two cascades are unknown. Further complexity arises from the fact that E3 ligases can target other E3 ligases, thus affecting several ubiquitination cascades.45 These examples demonstrate the significance of revealing and interpreting the ubiquitin regulatory network in the context of health and disease, to identify potential biomarkers and specific targets for intervention.
While inhibitors of the proteasome (Bortezomib®/Velcade®/PS-341 and Carfizomib®) are already in clinical practice as anticancer drugs for the treatment of multiple myeloma, they are not specific and may provoke side effects. In contrast, the substrate specificity of E3 ligases makes them attractive targets for anticancer treatment, which would preferentially target subsets of proteins. In such a manner, therapeutic interventions targeting oncogenic E3 ligases may restore the normal expression of tumor suppressors. The specificity of such treatment is expected to boost effectiveness and diminish side effects. Interestingly, the current treatment regimen of multiple myeloma also includes the drug thalidomide that affects the ubiquitin-proteasome system by functioning as a “molecular glue” targeting proteins for degradation by the SCF E3 ubiquitin ligase complex cereblon.46 Other E3-targeting anticancer pharmacological agents that are in preclinical/clinical trials include small molecules targeting the p53-MDM2 interaction, inhibitors of apoptosis proteins, SKP2, and the anaphase-promoting complex/cyclosome.47–53 Thus, E3 ligase-directed therapy is indeed a promising effective approach. In this respect, the HECT domain E3 ligases are attractive potential targets for anticancer intervention as they possess a catalytic site and directly catalyze substrate ubiquitination.37 However, a prerequisite for the design of anti-E3 therapeutics would be the comprehensive mapping of the E3s and their specific targets under different cellular conditions, and specifically in the context of different malignancies.
How can PTM profiling aid in identifying potential therapeutic targets?
A first step in classifying E3 ligases as potential targets for intervention can arise from high-throughput screening of cancer cells using PTM profiling. Functional extracts derived from diverse cancer cell lines or patient-derived cells may be manipulated by introducing mutants or inhibitors, as discussed in the section above, to target ubiquitin regulators. This approach could potentially reveal fundamental data regarding the specific E3 ligases and their substrates, which play a role in specific biological circumstances such as different cancer types, stage of malignancy, and the response to treatment. Such analysis now offers novel potential to systematically uncover multiregulated events, such as the one described for p105, and to assign the different regulations to specific biological conditions. Importantly, PTM profiling can also be used to systematically screen for small molecules or peptide inhibitors of E3 ligases, in cancer cell lines or patient-derived clinical samples. Taken together, PTM profiling can become a powerful tool used to extend our understanding of the oncogenic potential of HECT-E3s and other E3 ligases, potentially leading to the discovery of novel biomarkers and targets for cancer treatment.
Concluding remarks
Over the four decades since its initial discovery, tremendous knowledge was gained describing the importance of the ubiquitin system in regulation of diverse cellular processes in health and disease. Yet, our ability to analyze the ubiquitin system and understand the principles of its dynamic regulation in cells and tissues is still largely limited. Technological advancements in MS analyses provide means to examine the intricate changes of the specific ubiquitination substrates in various cells and tissues. The PTM profiling approach provides another dimension of analysis via probing the enzymatic activity of the ubiquitin system in cells and tissues, and thereby presents great scientific and therapeutic potential. Integrating the information gained from such technologies along with the available proteomic and genomic capabilities should take us closer to generating a comprehensive view of the ubiquitin regulatory network in health and disease.
Acknowledgements
We thank Anat Zimmer for graphical design. YM is supported by The Rising Tide Foundation; Israeli Centers of Research Excellence (I-CORE), Israel Science Foundation; European Research Council; Israel Cancer Research Fund; Gruber Foundation; Azrieli Institute for Systmes Biology; The Abramson Family Center for Young Scientists; Estate of David Levinson; Alan S. and Liz Jaffe, New York, NY; Dr Celia and Dr Lutz Zwillenberg-Fridman. YM the incumbent of the Leonard and Carol Berall Career Development Chair. Research in the laboratory of AC is supported by grants from the Dr Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF), the Israel Science Foundation (ISF), the I-CORE Program of the Planning and Budgeting Committee and the ISF (Grant1775/12), and the Deutsch-Israelische Projektkooperation (DIP). AC is an Israel Cancer Research Fund (ICRF) USA Professor.
Authors’ contributions
AEL, AC, and YM wrote the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Schulman BA. Twists and turns in ubiquitin-like protein conjugation cascades. Protein Sci 2011; 20: 1941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature 2009; 458: 422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol 2007; 19: 199–205. [DOI] [PubMed] [Google Scholar]
- 4.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem 2012; 81: 323–57. [DOI] [PubMed] [Google Scholar]
- 5.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov 2011; 10: 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim Biophys Acta 2004; 1695: 3–17. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67: 425–79. [DOI] [PubMed] [Google Scholar]
- 8.Williamson A, Werner A, Rape M. The colossus of ubiquitylation: decrypting a cellular code. Mol Cell 2013; 49: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science 2008; 322: 918–23. [DOI] [PubMed] [Google Scholar]
- 10.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol 2005; 7: 750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 2003; 21: 921–6. [DOI] [PubMed] [Google Scholar]
- 12.Beltrao P, Albanèse V, Kenner LR, Swaney DL, Burlingame A, Villén J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell 2012; 150: 413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emanuele MJ, Elia AEH, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen Y-N, Rush J, Hsu PW-C, Yen H-CS, Elledge SJ. Global identification of modular cullin-RING ligase substrates. Cell 2011; 147: 459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirzaei H, Rogers RS, Grimes B, Eng J, Aderem A, Aebersold R. Characterizing the connectivity of poly-ubiquitin chains by selected reaction monitoring mass spectrometry. Mol Biosyst 2010; 6: 2004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 2011; 10: M111.013284–M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011; 44: 325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendriks IA, D’Souza RCJ, Yang B, Verlaan-de Vries M, Mann M, Vertegaal ACO. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 2014; 21: 927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009; 138: 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 2010; 28: 868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics 2014; 14: 513–24. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science 2001; 293: 2101–5. [DOI] [PubMed] [Google Scholar]
- 22.Hultschig C, Kreutzberger J, Seitz H, Konthur Z, Büssow K, Lehrach H. Recent advances of protein microarrays. Curr Opin Chem Biol 2006; 10: 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc 2008; 3: 1796–808. [DOI] [PubMed] [Google Scholar]
- 24.Masuda M, Yamada T. Signaling pathway profiling by reverse-phase protein array for personalized cancer medicine. Biochim Biophys Acta 2015; 1854: 651–7. [DOI] [PubMed] [Google Scholar]
- 25.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science 2000; 289: 1760–3. [DOI] [PubMed] [Google Scholar]
- 26.Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. Analysis of yeast protein kinases using protein chips. Nat Genet 2000; 26: 283–9. [DOI] [PubMed] [Google Scholar]
- 27.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer 2010; 10: 630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol 2007; 3: 116–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews PS, Schneider S, Yang E, Michaels M, Chen H, Tang J, Emkey R. Identification of substrates of SMURF1 ubiquitin ligase activity utilizing protein microarrays. Assay Drug Dev Technol 2010; 8: 471–87. [DOI] [PubMed] [Google Scholar]
- 30.Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci USA 2009; 106: 2543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merbl Y, Kirschner MW. Post-translational modification profiling – a high-content assay for identifying protein modifications in mammalian cellular systems. Curr Protoc Protein Sci 2014; 77: 27.8.1–27.8.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merbl Y, Kirschner MW. Protein microarrays for genome-wide posttranslational modification analysis. Wiley Interdiscip Rev Syst Biol Med 2011; 3: 347–56. [DOI] [PubMed] [Google Scholar]
- 33.Merbl Y, Refour P, Patel H, Springer M, Kirschner MW. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell 2013; 152: 1160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson A, Jin L, Rape M. Preparation of synchronized human cell extracts to study ubiquitination and degradation. Methods Mol Biol 2009; 545: 301–12. [DOI] [PubMed] [Google Scholar]
- 35.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics 2013; 12: 3444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJC, Carr SA, Tabb DL, Coffey RJ, Slebos RJC, Liebler DC. Proteogenomic characterization of human colon and rectal cancer. Nature 2014; 513: 382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 2008; 14: 10–21. [DOI] [PubMed] [Google Scholar]
- 38.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–9. [DOI] [PubMed] [Google Scholar]
- 39.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitorp27. Nat Cell Biol 1999; 1: 193–9. [DOI] [PubMed] [Google Scholar]
- 40.Hershko D, Bornstein G, Ben-Izhak O, Carrano A, Pagano M, Krausz MM, Hershko A. Inverse relation between levels of p27(Kip1) and of its ubiquitin ligase subunit Skp2 in colorectal carcinomas. Cancer 2001; 91: 1745–51. [DOI] [PubMed] [Google Scholar]
- 41.Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev 2004; 18: 2602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203–6. [DOI] [PubMed] [Google Scholar]
- 43.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993; 75: 495–505. [DOI] [PubMed] [Google Scholar]
- 44.Kravtsova-Ivantsiv Y, Shomer I, Cohen-Kaplan V, Snijder B, Superti-Furga G, Gonen H, Sommer T, Ziv T, Admon A, Naroditsky I, Jbara M, Brik A, Pikarsky E, Kwon YT, Doweck I, Ciechanover A. KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 p105 to p50 restricts tumor growth. Cell 2015; 161: 333–47. [DOI] [PubMed] [Google Scholar]
- 45.de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 2011; 18: 1393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science 2010; 327: 1345–50. [DOI] [PubMed] [Google Scholar]
- 47.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–8. [DOI] [PubMed] [Google Scholar]
- 48.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LGGC, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 2004; 10: 1321–8. [DOI] [PubMed] [Google Scholar]
- 49.Fristedt Duvefelt C, Lub S, Agarwal P, Arngården L, Hammarberg A, Maes K, Van Valckenborgh E, Vanderkerken K, Jernberg Wiklund H. Increased resistance to proteasome inhibitors in multiple myeloma mediated by cIAP2 – implications for a combinatorial treatment. Oncotarget 2015; 6: 20621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramakrishnan V, Painuly U, Kimlinger T, Haug J, Rajkumar SV, Kumar S. Inhibitor of apoptosis proteins as therapeutic targets in multiple myeloma. Leukemia 2014; 28: 1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chauhan D, Neri P, Velankar M, Podar K, Hideshima T, Fulciniti M, Tassone P, Raje N, Mitsiades C, Mitsiades N, Richardson P, Zawel L, Tran M, Munshi N, Anderson KC. Targeting mitochondrial factor Smac/DIABLO as therapy for multiple myeloma (MM). Blood 2007; 109: 1220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood 2008; 111: 4690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lub S, Maes K, Menu E, Bruyne E, De, Vanderkerken K, Van Valckenborgh E. Novel strategies to target the ubiquitin proteasome system in multiple myeloma. Oncotarget 2015; 7: 6521–37. [DOI] [PMC free article] [PubMed] [Google Scholar]