Abstract
Hypoxia has been a research focus in cancer because of its important role in maintaining tumor microenvironments. Previous studies have demonstrated that the expression of several miRNAs was altered under hypoxic conditions, suggesting their crucial roles in the development of cancer. In the present study, the expression of 22 miRNAs reported to be significantly upregulated in cervical cancer tissues was examined. We found that four of these miRNAs were upregulated in response to hypoxia in HeLa cervical cancer cells. MiR-152 was upregulated to the greatest extent and was also found to be upregulated by hypoxia in C33A cells and tumor, but not in non-tumor cervical tissues. Moreover, we found that hypoxia-inducible factor-1α regulated the expression of miR-152 in HeLa cells through a hypoxia-responsive element. A bioinformatic tool predicted that WNT1 and ERBB3 were target genes of miR-152. This was confirmed by dual luciferase assays and Western blots. Overexpression of miR-152 repressed WNT1 and ERBB3 expression and decreased proliferation of HeLa cells. Collectively, these data indicate an important role for miR-152 in regulating the hypoxic response of tumor cells.
Keywords: Hypoxia, microRNA, cervical cancer, cell proliferation
Introduction
Cervical cancer is one of the most common cancers in females with about 530,000 new cases and more than 250,000 deaths each year.1,2 Infection with human papillomavirus appears to be a necessary factor in the development of almost all cases of cervical cancer. Over the past 100 years, the incidence of cervical cancer has been changing in developed countries. In the early 1950s, the incidence began to increase because of better diagnostic tools. However, in the early 1990s the incidence decreased because of an improved clinical approach and better use of prevention measures. However, cervical cancer is the leading cause of death from cancer in many low resource countries where widespread screening by cervical cytology is unavailable.
Hypoxia, defined as an insufficient oxygen supply to tissues, results from a reduction in oxygen availability, inadequate oxygen transport, or the inability of tissues to utilize oxygen. Hypoxia plays an important role in the tumor microenvironment by allowing the development and maintenance of cancer cells. However, the regulatory mechanisms by which tumor cells adapt to hypoxic conditions are not yet well understood. The hypoxia-inducible factors (HIFs) are a family of transcription factors that have been identified as important regulators of the cellular response to hypoxia.3 Under normoxic conditions, the alpha subunit of HIF-1 is hydroxylated at prolines 402 and 564 by prolyl-4-hydroxylases (PHDs), targeting HIF-1α for proteasome destruction mediated by the von Hippel–Lindau protein, an E3 ubiquitin ligase.4,5 Under hypoxic conditions, the activity of PHDs decreases, and HIF-1α is stabilized and transcriptionally regulates a large number of target genes involved in adaptation and protection against low oxygen conditions.6
MicroRNAs (miRNAs) are a type of short non-coding RNA that suppress the expression of protein coding genes by partial complementary binding, especially to the 3′ untranslated regions (UTRs) of mRNAs. Alterations in miRNA expression are involved in the initiation, progression, and metastasis of human cancers, and it is thought that miRNAs function as both tumor suppressors and oncogenes in cancer development.7,8 Previous studies have identified several miRNAs that are expressed differentially in response to hypoxia (hypoxia regulated miRNAs) and are involved in cancer initiation and development.
In the current study, we examined hypoxia-induced changes in the expression of 22 miRNAs that were reported to be significantly upregulated in cervical cancer tissues. We found that the expression of miR-152 was upregulated significantly and that its transcriptional induction was dependent on HIF-1α. Bioinformatics tools predicted potential target genes of miR-152, and dual luciferase assays and Western blots confirmed that both WNT1 and ERBB3 were identified as the target genes of miR-152. Functionally, we found that ectopic expression of miR-152 promoted tumor growth, linking HIF regulation to the regulation of the growth of tumor cells through miR-152.
Materials and methods
Cell lines and culture conditions
HeLa and C33A cervical cancer cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. Low oxygen treatments were performed in a hypoxia chamber (<0.02% O2; Sheldon Corp., Cornelius, OR) or in a Ruskinn Invivo2 Hypoxia Work Station (2% and 0.5% O2) (Biotrace International Plc., Bridgend, UK). All protein and RNA extracts were harvested in the hypoxia chamber.
Tissue samples
Forty-six patients with invasive cervical cancer (age 42–65 years; median, 52 years), all treated at the Department of Obstetrics and Gynecology, Beijing Friendship Hospital, between October 2010 and November 2013, were included in this study. Cervical cancer biopsy specimens were obtained and kept frozen at −70 ℃ without any additives. MiR-152 expression was assessed in all cancer specimens and 21 normal cervical tissue samples.
Western blotting
Protein extracts were boiled in sodium dodecyl sulfate (SDS)/β-mercaptoethanol sample buffer. Twenty micrograms of samples were loaded into each lane of 8% polyacrylamide gels. The proteins were separated by electrophoresis and blotted onto polyvinylidene fluoride membranes (Amersham Pharmacia Biotech, St. Albans, Herts, UK) by electrophoretic transfer. The membranes were incubated with a mouse anti-HIF-1α monoclonal antibody (Abcam, Cambridge, MA), rabbit anti-ERBB3 monoclonal antibody (Abcam), rabbit anti-WNT1 polyclonal antibody (Abcam), or mouse anti-β-actin monoclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA) for 1 h at 37 ℃. Protein–antibody complexes were detected using horseradish peroxidase conjugated rabbit anti-mouse or goat anti-rabbit immunoglobulin G (IgG). Detection was carried using an enhanced chemiluminescence kit (Pierce, Appleton, WI). The β-actin signal was used as a loading control.
RNA extraction and detection of miRNAs
Quantitative RT-PCR (qRT-PCR) analysis was used to determine the relative expression levels of 22 candidate miRNAs. Total RNA was extracted from cells using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. miRNA expression levels were detected using TaqMan miRNA RT-PCR. Single-stranded cDNA was synthesized using the TaqMan miRNA reverse transcription kit (Applied Biosystems, Foster City, CA) and then amplified using TaqMan Universal PCR Master Mix together with miRNA-specific TaqMan MGB probes (Applied Biosystems). U6 small nuclear RNA was used for normalization. Each sample in each group was measured in triplicate and the experiment was repeated at least three times.
Northern blot analysis
Fifty micrograms of total RNA per sample were subjected to electrophoresis on a 15% denaturing Tris-borate-EDTA (TBE)-urea polyacrylamide gel. RNA was electrophoretically transferred to nylon membranes (Hybond N+; Amersham Pharmacia Biotech). Ten picomoles of the oligo probes was labeled using T4 polynucleotide kinase (NEB, Ipswich, MA) and γ32P-ATP, and purified in a G-25 column (Amersham Biosciences, Piscataway, NJ). After being UV-cross-linked and baked at 80 ℃ for 30 min, the membrane was pre-hybridized at 42 ℃ for 4 h and then hybridized with 32P-labeled miR-152 or U6 probes at 40 ℃ overnight. Membranes were washed and exposed to phosphor imager screens (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). All experiments were repeated at least three times.
Reporter assay
To detect the HIF-1α binding region, amplified products were ligated into the Hind III and XhoI sites of pGL3-Basic Vector (Promega, Madison, WI). The full length 3′ UTRs of ERBB3 and WNT1 were cloned into a pmirGLO vector (Promega) by PCR amplification of genomic DNA. All transfections were carried out using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Luciferase activity was measured using the Dual Luciferase assay system (Promega) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay
HeLa cells were cultured under normoxia or 0.5% oxygen for 4 and 20 h, respectively. The ChIP assay was performed using either nonspecific mouse IgG or an anti-HIF-1α antibody (Abcam). 5′-AGGAGGCCAACTGTGTTGTGGTC-3′ (forward) and 5′-CTTATCTGCTACTCTATTTCTAG-3′ (reverse) sequences were used for PCR to amplify the miR-152 promoter fragment containing the functional hypoxia-responsive element (HRE) identified through serial deletion assays. To amplify the control fragment on the miR-152 promoter, the following sequence pair was used: 5′-AGATCACCTGAGGTCGGGAGTTC-3′ (forward) and 5′-GGGTTCAAGCAATTCTCCTGC-3′ (reverse). The PCR products were resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
Cell proliferation assay
HeLa cells (5 × 103) were seeded in 96-well plates in DMEM and allowed to attach overnight. The cells were then transfected with a miR-152 mimic or miR-152 inhibitor. A short nonsense double strand RNA was used as the miRNA mimic control and a single strand nonsense RNA was used as the inhibitor control. Twenty microliters of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (5 mg/mL) (Sigma-Aldrich, St. Louis, MO) were added into each well 48 h after transfection, and the cells were incubated for a further 4 h. The absorbance was recorded at 490 nm with a 96-well plate reader after the addition of dimethyl sulfoxide to dissolve the formazan. The results were confirmed using an EdU cell proliferation assay.
Statistical analysis
Data were analyzed by using SPSS Statistical Package version 16. Independent two group’s analyses are used t-test. Results of tissue miR-152 levels were analyzed by using Mann–Whitney U test. P < 0.05 was considered statistically significant.
Results
Hypoxia inducible miRNAs in HeLa cells
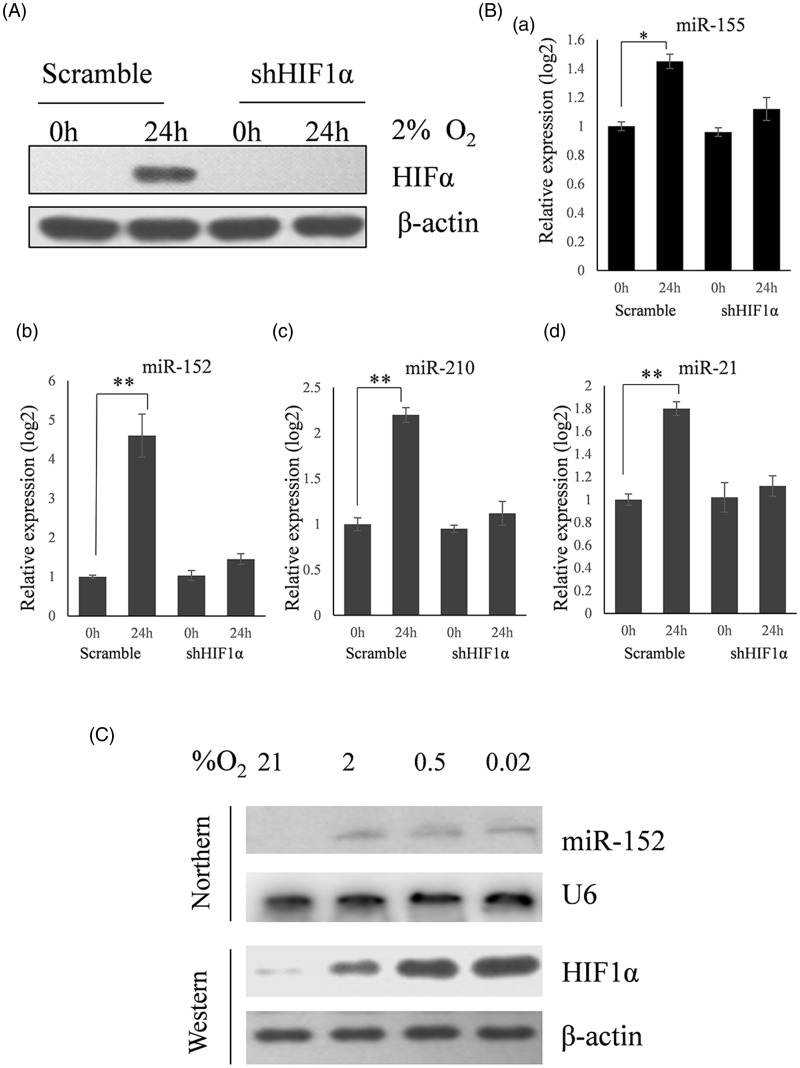
Twenty-two candidate miRNAs, reportedly upregulated by hypoxia in human cancer cells,9,10 were analyzed in HeLa cervical cancer cells. miRNA expression was detected in paired normoxia/hypoxia (2% oxygen) samples cultured for 24 h (Table 1). The expression of HIF-1α protein (Figure 1A) and four miRNAs (miR-155, miR-152, miR-21, and miR-210) (Figure 1Ba–d) were significantly induced by hypoxia. The expression of miR-152 increased more than 12-fold, which was the largest increase observed. This result was verified by a northern blot showing an oxygen concentration-dependent change in miR-152 miRNA (Figure 1C), and miR-152 was chosen for further study.
Table 1.
The relative expression of 22 miRNA candidates in Hela cells under normoxia and hypoxia conditions
| miRNA name | Relative expression (N/H) | P value |
|---|---|---|
| miR-10b | 1.35 | 0.087 |
| miR-103 | 0.89 | 0.37 |
| miR-107 | 0.94 | 0.24 |
| miR-125a | 1.45 | 0.072 |
| miR-152 | 12.13 | 0.0013 |
| miR-155 | 1.37 | 0.045 |
| miR-181b | 0.79 | 0.33 |
| miR-203 | 1.23 | 0.092 |
| miR-206 | 1.05 | 0.081 |
| miR-21 | 1.74 | 0.0087 |
| miR-210 | 2.30 | 0.0059 |
| miR-213 | 1.32 | 0.13 |
| miR-23 | 0.89 | 0.22 |
| miR-27a | 1.12 | 0.16 |
| miR-30a | 1.02 | 0.36 |
| miR-30c | 0.83 | 0.18 |
| miR-30d | 0.68 | 0.045 |
| miR-322 | 1.33 | 0.21 |
| miR-373 | 0.82 | 0.075 |
| miR-451 | 0.97 | 0.11 |
| miR-512 | 1.14 | 0.064 |
| miR-93 | 0.80 | 0.053 |
Figure 1.
MiR-152 is a predominant hypoxia-responsive miRNA. (A) Western blot showing the effects of hypoxia and HIF-1α siRNA on the expression of HIF-1α. (B) qRT-PCR analyses of the expression of candidate miRNAs. HeLa cells treated with scrambled or HIF-1α siRNA (shHIF1α) were cultured for 24 h. One plate of each remained in normoxia while the other was exposed to 2% O2 for an additional 24 h. RNA was then harvested and used for qRT-PCR analyses. Four miRNAs were upregulated by hypoxia with MiR-152 increasing to the greatest extent. The induction of all four miRNAs was dependent on HIF-1α (*P < 0.05, **P < 0.01) (n = 3). (C) Northern blot (miR-152) and Western blot (HIF1α) analyses showing induction under different levels of hypoxia. HeLa cells were split into four plates 24 h before treatment. One plate was then exposed to either 21%, 2%, 0.5%, or 0.02% oxygen for 24 h. Small nuclear RNA U6 was used as a loading control for the northern blot and β-actin for the Western blot
The in vivo and in vitro induction of MiR-152, dependence upon HIF-1α
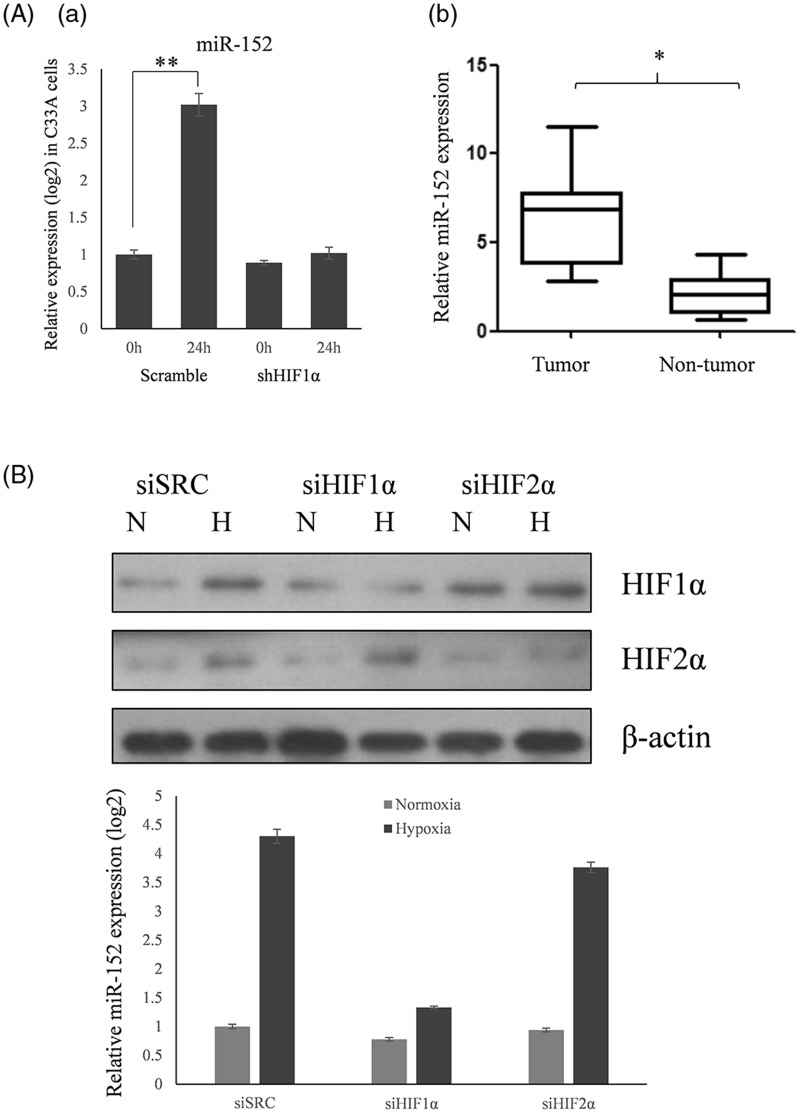
To investigate whether the induction of miR-152 by hypoxia was cell line-specific, qRT-PCR was used to assess the expression of miR-152 in C33A cells exposed to 2% oxygen for 24 h, as well as in normal and cervical cancer tissues. MiR-152 was induced in C33A cells (Figure 2Aa) and upregulated in cervical cancer, but not normal, human tissues (Figure 2Ab).
Figure 2.
MiR-152 expression in C33A cells, and tumor and non-tumor cervical tissues, and the effect of HIF-1α siRNA. (A) (a) The expression of miR-152 in C33A cells, and the effect of HIF-1α siRNA on this expression, was determined by qRT-PCR. Data are presented as means ± SD (**P < 0.01) (n = 3). (b) The expression of miR-152 tumor and non-tumor cervical tissues was determined by qRT-PCR (*P < 0.05). (B) miR-152 expression was assayed by qRT-PCR using RNAs harvested from HeLa cells treated with siRNAs against HIF-1α (siHIF-1α), HIF-2α (siHIF-2α), or a scrambled control siRNA (siSRC) under normoxia (N) or 2% O2 (H) for 24 h. RNA levels were normalized against small nuclear RNA U6. Representative blots of three replicates are shown. Data are presented as means ± SD
HIF-1α and HIF-2α are the two major HIFs expressed broadly in human tissues. Despite high conservation in their overall amino acid sequences, domain structures, and hypoxia-dependent mechanisms of activation, HIF-1α and HIF-2α have distinct physiological roles. In order to address the role of HIF-1α versus HIF-2α in miR-152 expression, they were downregulated in HeLa cells by transfections using specific siRNAs, or a scrambled control siRNA. The cells transfected with scrambled control siRNA, or siRNA against HIF-2α, showed significant induction of miR-152 expression under 2% oxygen. However, this induction was abolished in HeLa cells that were transfected with siRNA against HIF-1α (Figure 2B). These results indicate that miR-152 expression is HIF-1α-dependent.
Characterization of the miR-152 promoter
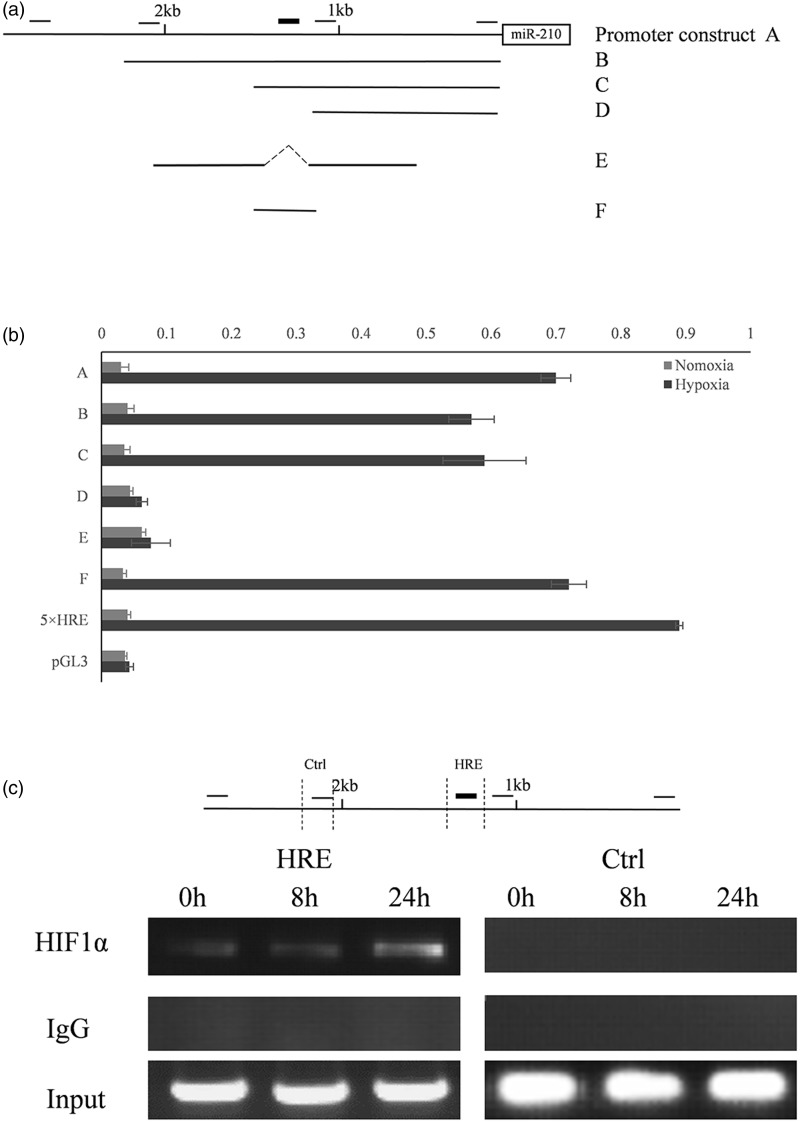
HIF-1α regulates target gene activity by binding to an HRE on the genomic DNA. Because our data indicated that miR-152 is a HIF-1α-regulated gene, we sought to determine how this regulation occurs. A 2.6 kb promoter sequence immediately upstream of the miR-152 stem-loop structure was cloned into a pGL3-basic vector (Figure 3A). By searching the 2.6 kb promoter sequence, five potential HRE sites were identified according to the consensus sequence (A/G) CGTG. In order to pinpoint the functional HRE, we made five serial deletion constructs and performed reporter assays to determine the effect of the deletions on reporter gene activity. We found that a 149-bp fragment harboring an HRE element is responsible for the induction of the miR-152 promoter under hypoxia. Mutation of this HRE site completely abolished the responsiveness of the promoter to HIF-1α (Figure 3B).
Figure 3.
Identification of the functional HRE in the miR-152 promoter region. (A) miR-152 promoter analysis. The location of the cloned promoter relative to the miR-152 coding region is shown. The open box indicates the coding region of miR-152 and the solid line indicates the 2.6 kb promoter that was cloned. The small black bars on the top represent the relative locations of the five potential HREs. Promoter fragments were cloned into the pGL3-Basic vector. This is a 5 × HRE construct that includes five copies of the human VEGF HRE that serves as a positive control. Serial deletion constructs (identified by A–F) are schematically represented by the lines. (B) Dual luciferase activity assay. Serial deletion constructs were co-transfected into HeLa cells with the pRL-TK vector. Their relative promoter activities in normoxia (gray bars) and 2% oxygen (black bars) are displayed. Data are expressed as means ± SD (n = 3). (C) ChIP analysis of the miR-152 promoter. Upper panel: the relative locations of the ChIP primers on the miR-152 promoter. Lower panel: ChIP results. HeLa cells were cultured under normoxia or 0.5% oxygen for 8 and 24 h, respectively. PCR was performed with primers specific to the functional miR-152 HRE. A region ∼800 bp upstream of the HRE site served as a control (Ctrl). IgG is the samples derived for the immunoprecipitated negative control. HIF-1α is the DNA immunoprecipitated with a HIF-1α antibody. Input is the DNA derived from samples prior to immunoprecipitation
To determine whether HIF-1α can bind to the miR-152 promoter directly, we designed two pairs of primers for ChIP. The first pair flanked the functional HRE element identified by serial deletion while the other was approximately 800 bp upstream of this HRE and was used as a control. We found that HIF-1α binds specifically to the functional HRE element, but not the control promoter sequence (Figure 3C).
Identification of miR-152 target genes
miRNAs can regulate their target genes by inhibiting protein translation and/or degrading mRNAs by binding to the 3′ untranslated region. To identify the biological functions of miR-152 upregulated in response to hypoxia, we first predicted the targets using miRecords (http://mirecords.biolead.org/), an online bioinformatics tool. Eleven algorithms were integrated and showed that two oncogenes, WNT1 and ERBB3, may be direct targets of miR-152.
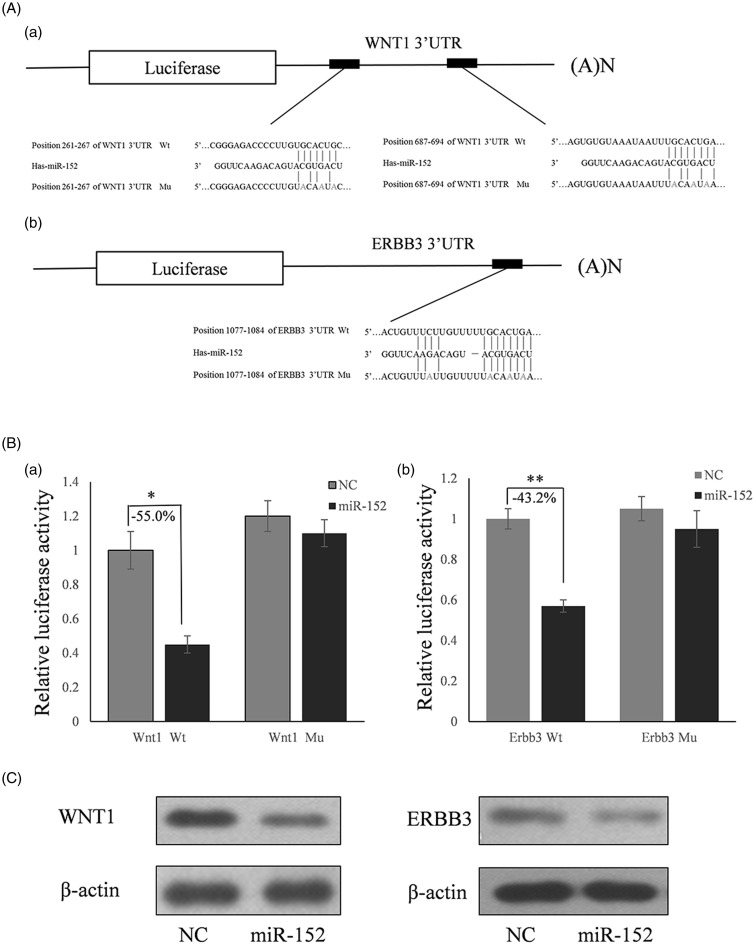
To determine whether WNT1 and ERBB3 were the target genes of miR-152, full length human WNT1 and ERBB3 3′ UTRs were cloned downstream of the firefly luciferase reporter gene in the pmirGLO vector (designated as pGLO-WNT1 and pGLO-ERBB3) for the dual luciferase assay (Figure 4Aa and Ab). A seed sequence mutation clone was used to further confirm the binding sites for miR-152. HeLa cells were co-transfected with pGLO-WNT1 or pGLO-ERBB2 and miR-152 mimics or scrambled negative control RNA. Compared with the miRNA control, luciferase activity was significantly suppressed by miR-152 for WNT1 (55%; P < 0.05) (Figure 4Ba) and ERBB3 (43.2%; P < 0.01) (Figure 4Bb). However, there were no significant differences among the miR-152 seed sequence mutation groups. These results indicate that miR-152 targets the 3′ UTR of WNT1 and ERBB3, leading to changes in the translation of firefly luciferase.
Figure 4.
miR-152 decreases WNT1 and ERBB3 expression in HeLa cells. (A) Schematic diagram for constructing the predicted miR-152 binding sites and inserting them into the pmirGLO vector. (B) Dual luciferase assay to confirm that miR-152 targets the 3′ UTR of WNT1 and ERBB3. HeLa cells were co-transfected with control or miR-152 mimic miRNAs, and wild type (Wt) or mutated (mu) WNT1/ERBB3 constructs. The results are shown as the ratio of firefly to Renilla luciferase activity. Data are expressed as means ± SD (*P < 0.05, **P < 0.01) (n = 3). (C) Western blots showing WNT1 and ERBB3 protein levels in HeLa cells transfected with a miR-152 mimic or miR control
MiR-152 regulates endogenous WNT1 and ERBB3 expression
Although WNT1 and ERBB3 were identified as target genes for miR-152, it was unknown whether miR-152 could regulate endogenous WNT1 and ERBB3 expression. HeLa cells were transfected with a miR-152 mimic to determine whether the overexpression of miR-152 could repress the expression of endogenous WNT1 and ERBB3. Compared with the corresponding control, the levels of WNT1 and ERBB3 proteins were significantly suppressed by a miR-152 mimic (Figure 4C).
MiR-152 overexpression suppresses the proliferation of cervical cancer cells
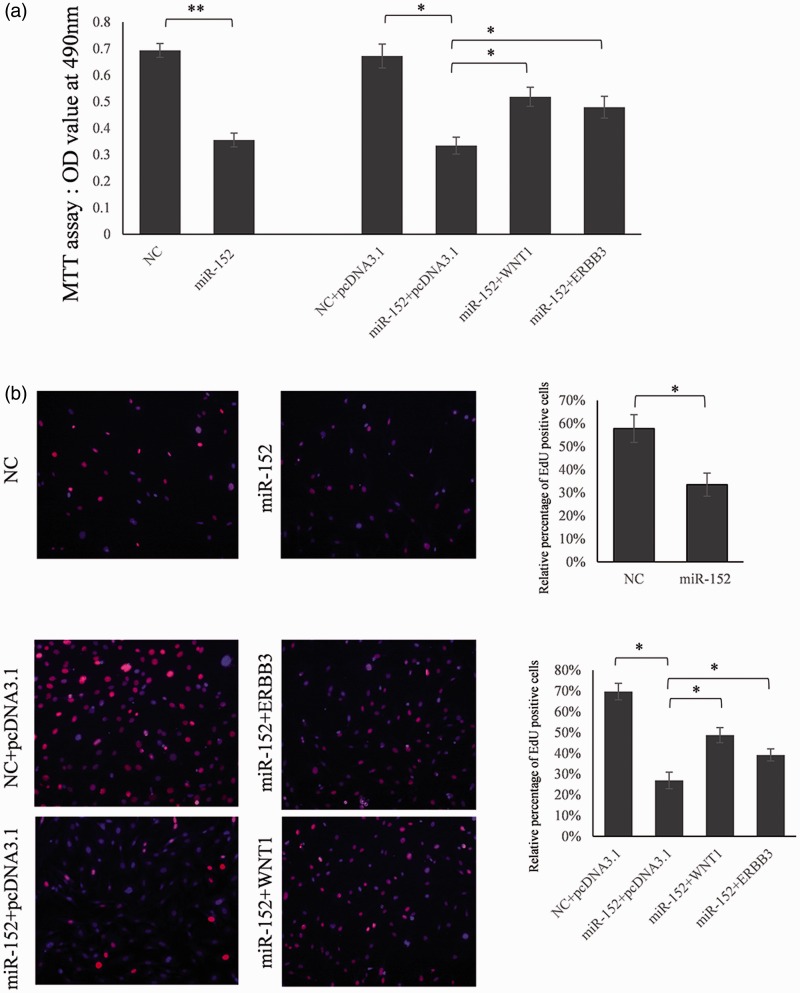
To determine whether miR-152 may have tumor-suppressive functions, we investigated the effect of WNT1 and ERBB3 on miR-152-mediated cell proliferation using the MTT (Figure 5A) and EdU (Figure 5B) cell proliferation assays. HeLa cells transfected with miR-152 showed less proliferation than cells transfected with control miRNA (Figure 5). This indicated that miR-152 inhibited cervical cancer cell proliferation. When co-transfected with the WNT1 or ERBB3 expression vector and miR-152 mimic, proliferation was partially restored compared with control cells (Figure 5). This implied that miR-152 mediated the suppression of cell growth partially through inhibiting the expression of WNT1 and ERBB3.
Figure 5.
Proliferation of cervical cancer cells is repressed by miR-152 partially through targeting WNT1 and ERBB3. (A) HeLa cells were transfected with a miR-152 mimic or miR-control, or a miR-152 mimic plus pcDNA3.1-WNT1/pcDNA3.1-ERBB3. The MTT method was used to assess cell proliferation. (B) EdU cell proliferation assay. The incorporation of EdU is expressed as the ratio of EdU-positive cells to the total number of Hoechst 33342-positive cells. Data are expressed as means ± SD (*P < 0.05, **P < 0.01) (n = 3). (A color version of this figure is available in the online journal.)
Discussion
In solid tumors, oxygen concentrations are significantly lower than in surrounding normal tissue. Thus, hypoxia has been a focus of research because of its important role in maintaining tumor microenvironments.11 Since the discovery of the first miRNA 20 years ago, the roles miRNAs play in gene regulation have been increasingly appreciated, especially in recent years.12 Previous studies discovered a number of miRNAs that are differentially expressed in response to hypoxia, such as miR-210, miR-155, miR-372/373, miR-21, and miR-10b, which were upregulated in response to hypoxia.10,13,14 In the current study, we scanned 22 candidate miRNAs that are upregulated in human cervical cancer. We found that miR-152, miR-21, miR-210, and miR-155 were upregulated significantly in response to hypoxia. Of these four miRNAs, the expression of miR-152 has the highest change (12.1 fold) in response to hypoxia; subsequent work, therefore, focused on the function of miR-152.
MiR-152 is a tumor suppressor in hepatocellular, ovarian, and endometrial cancer that works by repressing cancer cell proliferation. However, the structure of the miR-152 promoter has not been analyzed and the HRE responsible for the robust induction of miR-152 by hypoxia is not known.15–17 Here, we found that the expression of miR-152 was increased in clinical cervical cancer tissue samples as well as in cervical cancer cells cultured in several different low oxygen concentrations. By examining a series of miR-152 promoter deletion constructs, we identified a functional HRE and demonstrated that its induction by hypoxia was specific for HIF-1α.
The functions of miRNAs are reflected mainly in the suppression of target genes. In an effort to identify the target genes of miR-152, we first used an online bioinformatics tool that predicted WNT1 and ERBB3 may be the direct targets of miR-152 (WNT1 is one of molecules in Wnt pathway and ERBB3 is a member in ErbB pathway).18 this result was supported by six algorithms. Subsequently, we confirmed that the expression of WNT1 and ERBB3 was repressed by miR-152 in HeLa cells, suggesting that miR-152 may play a role in the development and/or progression of cervical cancer. This conclusion was further supported by the finding that overexpressing miR-152 inhibited the proliferation of HeLa cells, an effect that could be partially reversed by WNT1 or ERBB3. Overall, these results indicate that miR-152 plays a role as a tumor suppressor through repressing cell proliferation by targeting WNT1 and ERBB3.
In summary, the results of our study show that miR-152 is a robust hypoxia-inducible miRNA that represses cell proliferation during hypoxia. Our findings reveal a new mechanism of gene regulation by HIF-1α through a miRNA pathway, and illustrate the complexity of gene regulation by hypoxia. More importantly, ectopic expression of miR-152 decreases the proliferation of cervical cancer cells. Thus, miR-152 may account for at least part of the inhibitory effect of HIF-1α on cancer cell growth.
Author contributions
LL and LNS performed the experiments. LNS and XHT analyzed the data. XLT made the study design. XLT wrote this paper. All authors have approved the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Bosch FX, de Sanjose S. Chapter 1: Human papillomavirus and cervical cancer – burden and assessment of causality. J Natl Cancer Inst Monogr 2003; 31: 3–13. [DOI] [PubMed] [Google Scholar]
- 2.Pimenta JM, Galindo C, Jenkins D, Taylor SM. Estimate of the global burden of cervical adenocarcinoma and potential impact of prophylactic human papillomavirus vaccination. BMC Cancer 2013; 13: 553–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev 1998; 8: 588–94. [DOI] [PubMed] [Google Scholar]
- 4.Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1 alpha. Mol Cell Biol 2005; 25: 6415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WJ. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292: 464–8. [DOI] [PubMed] [Google Scholar]
- 6.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, Brown PO. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med 2006; 3: e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene 2010; 29: 5761–71. [DOI] [PubMed] [Google Scholar]
- 8.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs–the micro steering wheel of tumour metastases. Nat Rev Cancer 2009; 9: 293–302. [DOI] [PubMed] [Google Scholar]
- 9.Shen G, Li X, Jia YF, Piazza GA, Xi Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol Sin 2013; 34: 336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 2009; 35: 856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, Mami-Chouaib F, Chouaib S. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res 2012; 72: 4629–41. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–54. [DOI] [PubMed] [Google Scholar]
- 13.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol 2011; 31: 4087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 2009; 69: 1221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, Susumu N, Aoki D, Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res 2011; 71: 6450–62. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Zhao F, Wang ZN, Song YX, Chang H, Chiang Y, Xu HM. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep 2012; 27: 447–54. [DOI] [PubMed] [Google Scholar]
- 17.Huang S, Xie Y, Yang P, Chen P, Zhang L. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by regulating Wnt1 in HepG2 cells. PLoS One 2014; 9: e81730–e81730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, MacLeod CL, Shyamala G, Gillgrass AE, Cardiff RD. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol 2002; 161: 1087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]