Abstract
Osterix (Osx) is an osteoblast-specific transcription factor that is essential for bone formation. MicroRNAs (miRNAs) are ~22-nucleotide-long noncoding RNAs that play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression. They can also control osteoblast-mediated bone formation and osteoclast-related bone remodeling. The vital roles of Osx and miRNAs during bone formation have been well studied, but very few studies have discussed their co-functions and the relationships between them. In this review, we outline the significant functions of Osx and miRNAs on certain cell types during osteogenesis and illustrate their roles during tooth development. More importantly, we discuss the relationship between Osx and miRNAs, which we believe could lead to a new treatment for skeletal and periodontal diseases.
MeSH Keywords: Fractures, Bone; MicroRNAs; Tooth Calcification
Background
Osx, which was first discovered by Nakashima et al. in 2002 [1], is a zinc-finger transcription factor belonging to the specificity protein (Sp) family. To date, thousands of studies have investigated it and established the essential role of Osx in bone formation. Bone could not be formed if osteoblast-specific marker genes fail to express in Osx mutant embryos. After birth, if Osx is inactivated, mice will have multiple skeletal phenotypes, including nearly no bone formation, absence of resorption of mineralized cartilage, and defects in osteocyte maturation and function [1]. However, the functions of Osx on other cells differentiated from human mesenchymal stem cells have not been studied.
MiRNAs are a family of small, non-coding RNAs that can regulate expression of other genes by binding to or regulating the translation process of some specific mRNAs. The first miRNA, lin-4, was found in 1993 by Lee et al. to control the developmental time of C. elegans [2], and in 2000 the second miRNA, Let-7, was discovered by Reinhart’s group [3]. Since then, hundreds of miRNAs have been functionally linked to the development of specific tissues, such as eyes, blood vessels, muscle, nerve, fat, cartilage, and bone [4–13]. The co-function of miRNAs and Osx during skeletal development is also a popular research topic, but there has been little attention to the role in tooth development.
It is well acknowledged that bone formation consists of 2 developmental processes: intramembranous ossification and endochondral ossification. Osteoblast progenitors in mesenchymal condensations of endochondral and membranous skeletal elements first differentiate through 1 or several steps into preosteoblasts, which are still bipotential. Preosteoblasts then differentiate in 1 or more steps into mature osteoblasts and mature chondrocytes [1]. Osteoblasts form bones directly, while chondrocytes form a cartilage template that is later replaced by bone (Figure 1). In this review, we will focus on the roles of Osx and miRNAs on other cell types that are involved in bone formation, such as chondrocytes, osteoclasts, and adipocytes. We also address tooth development because it is similar to bone formation. Finally, we consider the relationships between Osx and miRNAs and prospects for further research.
Figure 1.
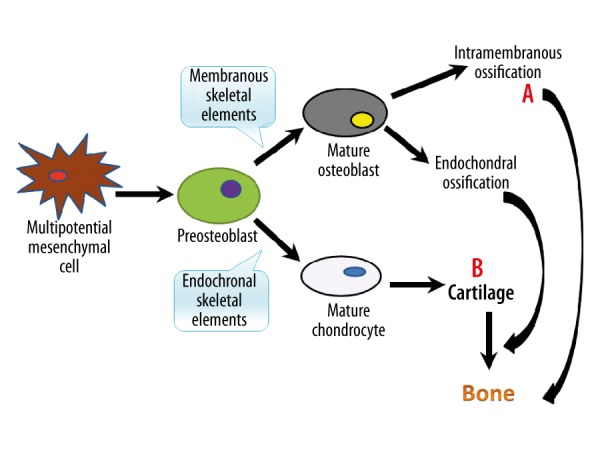
Model of bone formation. Bone can be formed by either intramembranous ossification or endochondral ossification. A – Osteoblasts form bones directly. B – Chondrocytes form a cartilage template that is later replaced by bone.
Role of Osx and miRNAs in Osteoblasts
Osx has been shown to have an essential role in osteoblast differentiation and bone formation [1]. Numerous studies, both in vitro and in vivo, have investigated its roles and mechanisms during these processes [14–19], but few studies have focussed on miRNAs. For example, miRNA-138 was verified to be able to modulate osteogenic differentiation of human mesenchymal stem cells (hMSCs). It significantly inhibits osteoblastic differentiation, and reduced levels of miRNA-138 boosted it. An in vivo experiment also showed that miR-138 negatively regulates osteoblast differentiation and bone formation [20]. Other miRNAs, such as miRNA-194, miRNA-210, miRNA-204, miRNA-24, miRNA-23, miRNA-145, miRNA-375, and miRNA-150, have also been confirmed to participate in this process [21–26]. Zhou et al. showed that miR-17-92 cluster critically regulates bone metabolism, mostly through its function in osteoblasts [27]. miRNAs also play important roles, both positive and negative, in regulating osteoblast differentiation and bone formation.
Role of Osx and miRNAs in Chondrocytes
Except for a few of the bones, such as craniofacial bones, which are formed by intramembranous ossification, the majority of the bones in our bodies, such as long bones, are formed by endochondral ossification, which needs a cartilage intermediate. This is why endochondral ossification is so crucial for skeletal development and growth. Osx transcripts were first found transiently in differentiating chondrocytes of E13.5 embryos [1] and in the primary cultures of chondrocytes [28]. Latter studies also showed that Osx appeared to function as a molecular switch between the osteoblast and chondrocyte fates [29], suggesting that Osx could play a role in differentiation of chondrocytes and terminal maturation of osteoblasts. Another 2 studies also demonstrated that Osx is a positive regulator of chondrocyte differentiation [30,31]. Kaback et al. verified that Osx inhibited chondrocyte maturation while promoting osteoblast differentiation in the MLB13MYC Clone 17 cell line [32]. The above evidence and other studies [33,34] show that Osx plays an essential role in late-stage endochondral ossification, but its exact function in chondrocytes needs further research.
Unlike Osx, which only has 1 or 2 clear functions in chondrocytes, hundreds of miRNAs are able to influence every process of chondrocyte differentiation and function. MiRNAs are generated from long primary transcripts (pri-miRNAs) after multi-step processing. First, primRNAs are processed into small hairpin pre-miRNAs by the microprocessor complex consisting of Drosha and DGCR8, and then are further processed by the RNase III, Dicer, into miRNAs [35]. Global reduction in miRNAs by deleting Dicer in growth plate chondrocytes reduced cellular proliferation and accelerated chondrocyte differentiation, causing a severe skeletal growth defect and early postnatal lethality [36]. Ablating the miRNA biogenesis pathway by deleting Drosha or DGCR8 in growth plate chondrocytes caused a lethal skeletal defect similar to that of Dicer deletion [37]. Both of these studies confirmed the essential role of total miRNAs in normal skeletogenesis. Some specific miRNAs, such as miRNA-23b, are able to induce mesenchymal stem cells (MSCs) to differentiate into chondrocytes by targeting protein kinase A signaling [38]. Others miRNAs (e.g., miRNA-1, miRNA-140, miRNA-145, and miRNA-365) have been identified to stimulate chondrocyte proliferation and differentiation [39,40] or regulate skeletal development [41–45]. Finally, silencing of miRNA-34a by miR-34a-specific LNA anti-sense can prevent cartilage degradation via inhibiting chondrocyte apoptosis, which gave us a new sight into the functions of miRNAs in cartilage homeostasis and structural integrity [46]. In summary, miRNAs are involved in every process of cartilage formation.
Role of Osx and miRNAs in Osteoclasts
Bone volume is determined by the balance between bone formation and osteolysis (Figure 2). Osteoblasts, which lead to bone formation, have been verified to be regulated by Osx and miRNAs, but the role of osteoclasts, which lead to osteolysis, is unclear. Multinucleated functional osteoclasts were found in the cartilage matrix in Osx null mutants, indicating that mutation of Osx has no effect on osteoclasts [1]. Similar results also indicated that Osx deficiency in osteoblasts did not affect osteoclast differentiation and function [47,48]. However, Cao et al. found that Osx inhibited interleukin (IL)-1a expression [49]. Chen et al. showed that the level of expression of the receptor activator of nuclear factor-nB ligand (RANKL) was clearly down-regulated in Osx-null mice [50] and Cao et al. found that OSX was unable to suppress RANKL expression [51]. IL-1a is a cytokine with potent stimulatory effects on osteoclastogenesis and RANKL is critical to osteoclast formation [52], suggesting that Osx has different effects on osteoclasts. Conflicting results from different researchers means that more studies about their mechanisms of action are neccesary.
Figure 2.
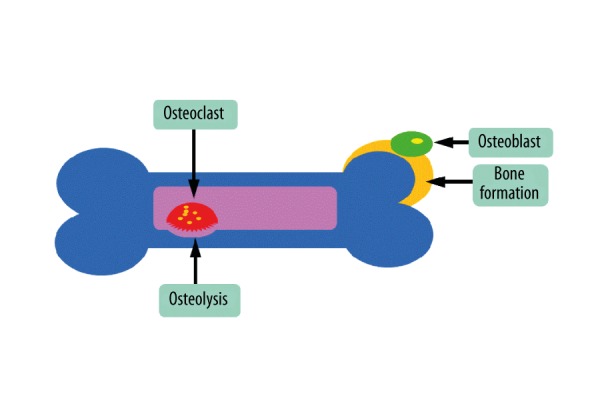
Bone volume is determined by the balance between bone formation and osteolysis.
It is easy to understand that miRNAs have various functions on osteoclasts. In a recently review about this, Tang et al. listed roles of several miRNAs relevant in osteoclasts and related bone diseases, which showed that some miRNAs enhanced osteoclast differentiation and others inhibited osteoclastogenesis [53]. Global reduction in miRNAs by using DGCR8, Dicer1, and Ago2 siRNA gene silencing in Dicer-null mice caused aberrant osteoclast differentiation and function, decreased osteoclastogenesis, and decreased bone resorption [54]. In summary, the function of miRNAs on osteoclasts differs from miRNA to miRNA, while overall, miRNAs promote osteoclastogenesis.
Role of Osx and miRNAs in Adipocytes
Another important type of cell that bone marrow mesenchymal stem cells (BMSCs) could differentiate into, besides chondrocytes and osteoblasts, is adipocytes [55,56]. The potential and capacity of BMSCs differentiating into osteoblasts decrease and adipocytes increase as people grow old [57–59]. Therefore, knowing the potential mechanism enables a better understanding of osteoblastogenesis. During the past decade some research has been conducted on the regulation of miRNAs in adipocyte differentiation [60–62]. Regarding the balance between adipogenesis and osteogenesis, Huang et al. found that miRNA-22 was able to regulate adipogenic and osteogenic differentiation of human adipose tissue-derived mesenchymal stem cells (hADMSCs) in opposite directions. After transfecting hADMSCs with miRNA-22 mimics, followed by culturing them in the adipogenic induction medium or osteogenic induction medium, they found that miRNA-22 negatively regulated adipogenic differentiation of hADMSCs while acting as a positive regulator in osteogenic differentiation [63]. In contrast, overexpression of miR-188 in mice reduced bone formation and increased bone marrow fat accumulation, and miR-705 and miR-3077-5p acted as inhibitors of MSCs osteoblast differentiation and promoters of adipocyte differentiation [64,65]. In conclusion, some miRNAs can regulate the age-related switch between osteoblast and adipocyte differentiation, which might indicate a new strategy for treating age-related bone loss and senile osteoporosis.
Although Osx is essential for osteoblast differentiation, little is known about its functions on adipocytes. Cheng et al. showed that osteoblast homeoprotein Msx2 up-regulated OSX expression in myofibroblasts and suppressed adipogenic differentiation of C3H10T1/2 cells [66]. Another study, Mikami et al. showed that dexamethasone (Dex) modulated osteogenesis and adipogenesis. They treated rat calvaria-derived cells with Dex and bone morphogenetic protein (BMP)-2 for various periods of time, and found that when cells were differentiating into adipocytes, OSX expression was inhibited. When cells were committed to osteogenic differentiation, OSX expression was not inhibited by Dex [67]. MiRNA-637 was also verified to promote the formation of adipocytes and, conversely, inhibit that of osteoblasts by direct suppression of OSX expression [68]. From these studies we hypothesized that the expression level of OSX was correlated with adipocyte differentiation because its reduction decreased the differentiation of osteoblasts so that the stem cells differentiated into adipocytes. However, the verification of this hypothesis and the detailed mechanism need further investigation.
Role of Osx and miRNAs in Tooth Development
Another mineralized organ, teeth, share many properties with bone, and there is great similarity between the process of tooth formation and intramembranous bone formation (Figures 1, 3). Cementoblasts, the cells that form cementum, are quite similar to osteoblasts, a cell type controlling bone formation [69,70]. In addition to its ability to promote mineral nodules in vitro [71], cementoblasts can also express genes such as bone sialoprotein (BSP) and osteocalcin (OCN). Runx2 and Osx, 2 key transcriptional factors in osteogenesis [1,72–74], are present in cementoblasts [75]. Therefore, Osx and miRNAs are also needed for odontoblast differentiation and tooth development.
Figure 3.

The model of dentin formation is quite similar to intramembranous bone formation.
Dentin sialophosphoprotein (DSPP) is a phosphorylated protein representing a major component of non-collagenous dentin extracellular matrix (DECM), which is highly expressed in odontoblasts and essential for dentinogenesis [76,77]. At later stages of the mouse embryo, DSPP transcription is weakly present in osteoblasts, but is strongly present in odontoblasts, where OSX is highly expressed. In mouse odontoblast-like cells, OSX overexpression increased DSPP transcription level, indicating that the Dspp gene could be a direct target of Osx activation and illustrating the function of Osx during dentinogenesis [78].
Because cementoblasts possess many characteristics similar to those of osteoblasts, our group, for the first time, used mouse models to elucidate the role of Osx in the formation of cellular cementum, and demonstrated that conditional deletion of Osx in mouse 2.3 Col1a1Cre in embryos, or with CMV-Cre ERT2 in adults, led to a sharp reduction in cellular cementum formation, while overexpression of OSX by transgenic mouse greatly accelerated the formation of cellular cementum [79]. We also found that Osx regulated the differentiation of cementoblasts by maintaining a low level of Wnt-β-catenin signalling via positive regulation of DKK1 (Figure 4) [80]. These results further support the vital function of Osx in tooth development. Therefore, we hypothesized that Osx plays a critical role in periodontal regeneration and Osx combined with gene-activated matrix (GAM) might be effective in the regeneration of cementum [81].
Figure 4.
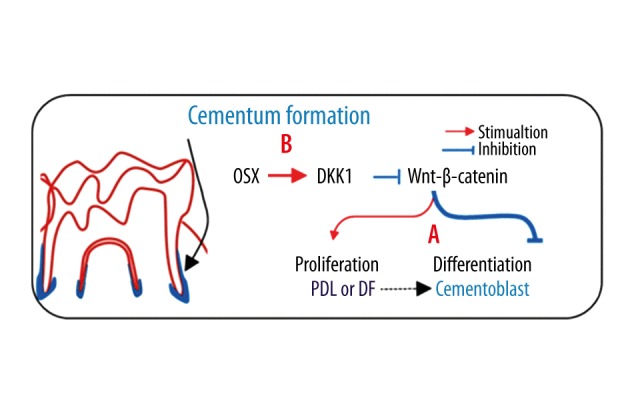
The negative regulation of Wnt-β-catenin by OSX is in part through activation of DKK1. A – Wnt-β-catenin is able to stimulate PDL or DF proliferation and inhibit cementoblast differentiation. B – OSX negatively regulates Wnt-β-catenin through activating DKK1 – a potent inhibitor of Wnt signaling. PDL – periodontal ligament. DF – dental follicles.
Recently, Sun et al., in a review about the fine-tuning role of miRNA in odontoblast differentiation and disease, summarized all the reported miRNAs that regulate odontoblast differentiation and listed them together [82]. However, there is still no research about miRNAs and cementogenesis. Expression patterns of miRNAs during periodontal disease [83,84] and certain miRNAs in different periodontal tissues [85,86] have been verified, but the exact roles these miRNAs play and the mechanisms behind these patterns need further study. These studies provided candidates for further analysis of miRNAs in periodontal infection and of miRNAs during tooth development. We know that miR-146a expression induces periodontal ligament cells (PDLCs) differentiation. IPDL-miR-146a cells, which express much higher levels of miR-146a than normal PDLCs, showed an up-regulation of ALP, OCN, and OPN mRNA expression and more mineralization nodules [87], but whether it induces PDLCs to differentiate into cementoblasts or osteoblasts remains unknown. Therefore, the relationship between miRNAs and tooth development needs further investigation. Combined with the information presented above, we made a schematic that shows functions of Osx and certain miRNAs during tooth development (Figure 5).
Figure 5.
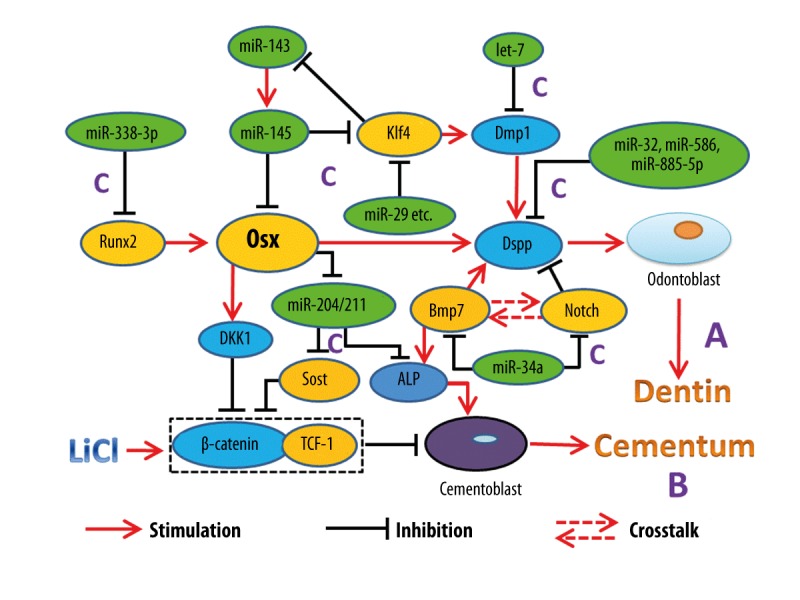
Role of Osx and certain miRNAs in tooth development. A – The role of Osx in dentinogenesis is shown by its regulation on Dspp. B – Osx regulates the differentiation of cementoblasts through Wnt-β-catenin pathway. C – Fine-tuning role of miRNA in tooth development.
Relationships between Osx and miRNAs
Since both Osx and miRNAs have such vital functions in skeletal development, it is easy to understand that Osx has correlations with some miRNAs. Researchers have established that miRNAs are able to target Osx. For instance, miRNA-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix [68]. Since osteoporosis and the increased risk of fragile fractures are one of the characteristic manifestations of aging, the results of this might contribute to this bone-associated metabolic disease. MiRNA-143 and miRNA-145 can suppress osteogenic differentiation by targeting Sp7/osterix [88,89] and form a feedback loop with KLF4 and OSX in odontoblasts. MiRNA-145 inhibits expression of Klf4 and Osx genes after they induce odontoblast differentiation, and then KLF4 represses miRNA-143 transcription, followed by miR-143 regulating odontoblast differentiation, in part through the miR-145 pathway [90]. Other miRNAs, such as miRNA-125b, miRNA-31, hsa-miR-135b, and miRNA-214, can also target Osx [91–94].
In addition, miRNAs could regulate Osx by targeting its upstream molecules or inhibitors. Runx2 is a potential target of miR-338-3p. Overexpression of miRNA-338-3p caused a decrease in the expression of Runx2 at mRNA and protein levels, while Runx2 expression increased after treatment with miR-338-3p inhibitors [95]. Several other miRNAs, such as miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-204, miR-205, miR-217, miR-218, and miR-335, were also found to target Runx2 during osteogenic differentiation [96–100]. As Runx2 acts upstream of Osx [1], the function of these miRNAs can be used indirectly to affect Osx. MiRNA-322 is involved in osteoblast differentiation by targeting Tob2. Overexpression of miR-322 decreases Tob2 mRNA and protein expression and leads to increased levels of the transcription factor Osx [101]. As Tob2 regulated Osx mRNA degradation, this was the first time that a miRNA was shown to increase the level of Osx, which could be a new molecular mechanism for controlling osteogenesis using the crucial role of Osx.
Since so many miRNAs can target Osx, we wondered whether is Osx involved in the regulation of miRNA expression. In addition to the feedback loop among miR-143, miR-145, KLF4, and Osx that we have presented previously [90], miRNA-93 is also able to form a feedback loop with Osx to regulate osteoblast mineralization [102]. Another result from Chen’s group suggested that Osx down-regulated expression of a group of miRNAs, including miR-133a and miR-204/211, but up-regulated expression of another group of miRNAs, such as miR-141/200a. Based on the knowledge that MiR-133a and miR-204/211 target Runx2, and because sclerostin and alkaline phosphatase (ALP) are 2 additional targets of miR-204/211 and miR-141/200a can target the transcription factor Dlx5, they postulated that Osx had the ability to modulate RUNX2, sclerostin, ALP, and DLX5 proteins through regulation of specific miRNAs [103]. Although some miRNAs have been demonstrated to have their own promoters and are transcribed independently [104], there appear to be few molecules that can regulate miRNAs. This result opens a new door to further study the interactions between Osx and miRNAs. The relationships between Osx and miRNAs that we presented above are listed in Figure 6.
Figure 6.
Relationships between Osx and miRNAs. A – Some miRNAs are able to target Osx. B – Some miRNAs regulate Osx by targeting its upstream molecules or inhibitors. C – Osx can regulate expression of some miRNAs.
Conclusions and Perspectives
Bone formation is a multi-step process that involves many cell types and various transcription factors, cytokines, and nucleotides. Osteoblasts are the most important cells during this process, while chondrocytes, osteoclasts, and adipocytes also play certain roles in skeletal formation and aging-related transformations. Hence, simply studying the functions of Osx and miRNAs with osteoblasts is far from sufficient. In this review, we mainly presented their functions with chondrocytes, osteoclasts, and adipocytes in order to introduce this subject and help generate new research ideas.
Two skeletal diseases, congenital dysplasia or aging-related degenerations, have been of concern to researchers for years. As one of the most important transcription factors during bone formation, Osx plays an important role in both conditions, as do miRNAs, which regulate gene expression involved in a wide variety of biological actions. Thus, it is easy to understand that any mutation or dysregulation that occurs on Osx or one of the miRNAs might be able to cause certain diseases. A better understanding of the mechanisms that regulate bone formation may yield new methods to treat these illnesses, and we hope that the present review will contribute to this.
Like many other organs in the human body, permanent teeth are non-reproductive. Although teeth are quite similar to bone, there has been little research on the role of Osx and miRNAs on the formation of cementum and dentine. Because of the high incidence of periodontitis, which leads to tooth loss by destroying alveolar bone and cementum, it is urgent to find ways to rescue periodontal tissues. However, little is known about the functions of Osx and miRNAs during tooth development. More importantly, we summarized the relationships between Osx and miRNAs and found that they can regulate each other. Since bone formation needs several pathways that comprise those factors, knowing their relationships and the mechanisms by which they regulate and control each other will make it easier to adjust these pathways to influence skeletal structures and treat skeletal diseases.
However, there are still many unanswered questions. For example, controversy still exists about the functions of Osx in chondrocytes and osteoclasts, and more research is needed to determine if Osx positively or negatively regulates osteoclast genesis and chondrocyte differentiation. The differentiations from BMSCs to osteoblasts and to adipocytes are in 2 opposite directions, and it is unknown whether this balance is to some degree regulated by Osx. Moreover, since the regeneration of cementum and dentin is the priority in protecting against periodontitis and consequent tooth loss, there remains much to learn about the functions of Osx and miRNAs during tooth development. This review summarizes current knowledge about the roles of Osx and miRNAs and their interactions, and shows directions for future research.
Footnotes
Source of support: The study was partly supported by grants from the National Natural Science Foundation of China to ZC (81170933, 81570946), the Chenguang Youth Project of Science and Technology of Wuhan City to ZC (2014072704011255), the Natural Science Foundation of Hubei Province to ZC (2015CFB259), and the Young Medical Talent Project of the Health Department of Wuhan (2014–2016)
Conflict of interests
The authors have no conflict of interests related to this study.
References
- 1.Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Zou H, Ding Y, Wang K, et al. MicroRNA-29A/PTEN pathway modulates neurite outgrowth in PC12 cells. Neuroscience. 2015;291:289–300. doi: 10.1016/j.neuroscience.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang Z, Qin X, Hu H, et al. Down-regulation of microRNA-155 attenuates retinal neovascularization via the PI3K/Akt pathway. Mol Vis. 2015;21:1173–84. [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Zhang L, Gao Y, et al. The multiple roles of microrna-223 in regulating bone metabolism. Molecules. 2015;20(10):19433–48. doi: 10.3390/molecules201019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umeda M, Terao F, Miyazaki K, et al. MicroRNA-200a regulates the development of mandibular condylar cartilage. J Dent Res. 2015;94(6):795–802. doi: 10.1177/0022034515577411. [DOI] [PubMed] [Google Scholar]
- 8.Izzotti A, Ceccaroli C, Longobardi MG, et al. Molecular damage in glaucoma: from anterior to posterior eye segment. The microRNA role. Microrna. 2015;4(1):3–17. doi: 10.2174/2211536604666150707124640. [DOI] [PubMed] [Google Scholar]
- 9.Hou C, Meng F, Zhang Z, et al. The role of microRNA-381 in chondrogenesis and interleukin-1-beta induced chondrocyte responses. Cell Physiol Biochem. 2015;36(5):1753–66. doi: 10.1159/000430148. [DOI] [PubMed] [Google Scholar]
- 10.Guller I, McNaughton S, Crowley T, et al. Comparative analysis of microRNA expression in mouse and human brown adipose tissue. BMC Genomics. 2015;16(1):820. doi: 10.1186/s12864-015-2045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda T, Ochi H, Sunamura S, et al. MicroRNA-145 regulates osteoblastic differentiation by targeting the transcription factor Cbfb. FEBS Lett. 2015;589(21):3302–8. doi: 10.1016/j.febslet.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 12.Duran BO, Fernandez GJ, Mareco EA, et al. Differential microRNA expression in fast- and slow-twitch skeletal muscle of piaractus mesopotamicus during growth. PLoS One. 2015;10(11):e0141967. doi: 10.1371/journal.pone.0141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–25. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha KM, Yasuda H, Zhou X, de Crombrugghe B. Osterix and NO66 histone demethylase control the chromatin of Osterix target genes during osteoblast differentiation. J Bone Miner Res. 2014;29(4):855–65. doi: 10.1002/jbmr.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu F, Friedman MS, Luo W, et al. The transcription factor osterix (SP7) regulates BMP6-induced human osteoblast differentiation. J Cell Physiol. 2012;227(6):2677–85. doi: 10.1002/jcp.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun-Feng W, Matsuo N, Sumiyoshi H, Yoshioka H. Sp7/Osterix up-regulates the mouse pro-alpha3(V) collagen gene (Col5a3) during the osteoblast differentiation. Biochem Biophys Res Commun. 2010;394(3):503–8. doi: 10.1016/j.bbrc.2010.02.171. [DOI] [PubMed] [Google Scholar]
- 17.Fu H, Doll B, McNelis T, Hollinger JO. Osteoblast differentiation in vitro and in vivo promoted by Osterix. J Biomed Mater Res A. 2007;83(3):770–78. doi: 10.1002/jbm.a.31356. [DOI] [PubMed] [Google Scholar]
- 18.Tu Q, Valverde P, Chen J. Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem Biophys Res Commun. 2006;341(4):1257–65. doi: 10.1016/j.bbrc.2006.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YJ, Kim HN, Park EK, et al. The bone-related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene. 2006;366(1):145–51. doi: 10.1016/j.gene.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Eskildsen T, Taipaleenmaki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA. 2011;108(15):6139–44. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Chen S, Deng C, et al. MicroRNA-204 targets Runx2 to attenuate BMP-2-induced osteoblast differentiation of human aortic valve interstitial cells. J Cardiovasc Pharmacol. 2015;66(1):63–71. doi: 10.1097/FJC.0000000000000244. [DOI] [PubMed] [Google Scholar]
- 22.Liu XD, Cai F, Liu L, et al. microRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. Biol Chem. 2015;396(4):339–47. doi: 10.1515/hsz-2014-0268. [DOI] [PubMed] [Google Scholar]
- 23.Li J, He X, Wei W, Zhou X. MicroRNA-194 promotes osteoblast differentiation via downregulating STAT1. Biochem Biophys Res Commun. 2015;460(2):482–88. doi: 10.1016/j.bbrc.2015.03.059. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Wu C, Dong Y, et al. MicroRNA-24 regulates osteogenic differentiation via targeting T-cell factor-1. Int J Mol Sci. 2015;16(5):11699–712. doi: 10.3390/ijms160511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du F, Wu H, Zhou Z, Liu YU. microRNA-375 inhibits osteogenic differentiation by targeting runt-related transcription factor 2. Exp Ther Med. 2015;10(1):207–12. doi: 10.3892/etm.2015.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong CL, Liu HZ, Zhang ZC, et al. The influence of microRNA-150 in osteoblast matrix mineralization. J Cell Biochem. 2015;116(12):2970–79. doi: 10.1002/jcb.25245. [DOI] [PubMed] [Google Scholar]
- 27.Zhou M, Ma J, Chen S, et al. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine. 2014;45(2):302–10. doi: 10.1007/s12020-013-9986-y. [DOI] [PubMed] [Google Scholar]
- 28.Yagi K, Tsuji K, Nifuji A, et al. Bone morphogenetic protein-2 enhances osterix gene expression in chondrocytes. J Cell Biochem. 2003;88(6):1077–83. doi: 10.1002/jcb.10467. [DOI] [PubMed] [Google Scholar]
- 29.Hilton MJ, Tu X, Cook J, et al. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development. 2005;132(19):4339–51. doi: 10.1242/dev.02025. [DOI] [PubMed] [Google Scholar]
- 30.Oh JH, Park SY, de Crombrugghe B, Kim JE. Chondrocyte-specific ablation of Osterix leads to impaired endochondral ossification. Biochem Biophys Res Commun. 2012;418(4):634–40. doi: 10.1016/j.bbrc.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omoteyama K, Takagi M. The effects of Sp7/Osterix gene silencing in the chondroprogenitor cell line, ATDC5. Biochem Biophys Res Commun. 2010;403(2):242–46. doi: 10.1016/j.bbrc.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Kaback LA, Soung do Y, Naik A, et al. Osterix/Sp7 regulates mesenchymal stem cell mediated endochondral ossification. J Cell Physiol. 2008;214(1):173–82. doi: 10.1002/jcp.21176. [DOI] [PubMed] [Google Scholar]
- 33.Xing W, Cheng S, Wergedal J, Mohan S. Epiphyseal chondrocyte secondary ossification centers require thyroid hormone activation of Indian hedgehog and osterix signaling. J Bone Miner Res. 2014;29(10):2262–75. doi: 10.1002/jbmr.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura R, Wakabayashi M, Hata K, et al. Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J Biol Chem. 2012;287(40):33179–90. doi: 10.1074/jbc.M111.337063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105(6):1949–54. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T, Papaioannou G, Mirzamohammadi F, et al. Early postnatal ablation of the microRNA-processing enzyme, Drosha, causes chondrocyte death and impairs the structural integrity of the articular cartilage. Osteoarthritis Cartilage. 2015;23(7):1214–20. doi: 10.1016/j.joca.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ham O, Song BW, Lee SY, et al. The role of microRNA-23b in the differentiation of MSC into chondrocyte by targeting protein kinase A signaling. Biomaterials. 2012;33(18):4500–7. doi: 10.1016/j.biomaterials.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Guan YJ, Yang X, Wei L, Chen Q. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011;25(12):4457–66. doi: 10.1096/fj.11-185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, Wei X, Guan Y, et al. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28:3930–41. doi: 10.1096/fj.13-249318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyaki S, Papaioannou G, Mirzamohammadi F, et al. MicroRNA-140 provides robustness to the regulation of hypertrophic chondrocyte differentiation by the PTHrP-HDAC4 pathway. J Bone Miner Res. 2014;30(6):1044–52. doi: 10.1002/jbmr.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papaioannou G, Inloes JB, Nakamura Y, et al. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc Natl Acad Sci USA. 2013;110(35):E3291–300. doi: 10.1073/pnas.1302797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287(2):916–24. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–28. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24(11):1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abouheif MM, Nakasa T, Shibuya H, et al. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology. 2010;49(11):2054–60. doi: 10.1093/rheumatology/keq247. [DOI] [PubMed] [Google Scholar]
- 47.Baek WY, Lee MA, Jung JW, et al. Positive regulation of adult bone formation by osteoblast-specific transcription factor osterix. J Bone Miner Res. 2009;24(6):1055–65. doi: 10.1359/jbmr.081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baek WY, de Crombrugghe B, Kim JE. Postnatally induced inactivation of Osterix in osteoblasts results in the reduction of bone formation and maintenance. Bone. 2010;46(4):920–28. doi: 10.1016/j.bone.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y, Jia SF, Chakravarty G, et al. The osterix transcription factor down-regulates interleukin-1 alpha expression in mouse osteosarcoma cells. Mol Cancer Res. 2008;6(1):119–26. doi: 10.1158/1541-7786.MCR-07-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S, Feng J, Zhang H, et al. Key role for the transcriptional factor, osterix, in spine development. Spine J. 2014;14(4):683–94. doi: 10.1016/j.spinee.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 51.Cao Y, Zhou Z. Osterix, a transcription factor for osteoblast differentiation, mediates antitumor activity in murine osteosarcoma. Cancer Res. 2005;65:1124–28. doi: 10.1158/0008-5472.CAN-04-2128. [DOI] [PubMed] [Google Scholar]
- 52.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 53.Tang P, Xiong Q, Ge W, Zhang L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. 2014;11(11):1355–63. doi: 10.1080/15476286.2014.996462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284(7):4667–78. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekiya I, Larson BL, Vuoristo JT, et al. Adipogenic differentiation of human adult stem cells from bone marrow stroma (MSCs) J Bone Miner Res. 2004;19(2):256–64. doi: 10.1359/JBMR.0301220. [DOI] [PubMed] [Google Scholar]
- 56.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–47. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 57.Shen W, Chen J, Gantz M, et al. MRI-measured pelvic bone marrow adipose tissue is inversely related to DXA-measured bone mineral in younger and older adults. Eur J Clin Nutr. 2012;66(9):983–88. doi: 10.1038/ejcn.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeung DK, Griffith JF, Antonio GE, et al. Osteoporosis is associated with increased marrow fat content and decreased marrow fat unsaturation: A proton MR spectroscopy study. J Magn Reson Imaging. 2005;22(2):279–85. doi: 10.1002/jmri.20367. [DOI] [PubMed] [Google Scholar]
- 59.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–89. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim SY, Kim AY, Lee HW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392(3):323–28. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Gerin I, Bommer GT, McCoin CS, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299(2):E198–206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Esau C, Kang X, Peralta W, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–65. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 63.Huang S, Wang S, Bian C, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21(13):2531–40. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li CJ, Cheng P, Liang MK, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–22. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao L, Yang X, Su X, et al. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng SL, Shao JS, Charlton-Kachigian N, et al. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278(46):45969–77. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 67.Mikami Y, Lee M, Irie S, Honda MJ. Dexamethasone modulates osteogenesis and adipogenesis with regulation of osterix expression in rat calvaria-derived cells. J Cell Physiol. 2011;226(3):739–48. doi: 10.1002/jcp.22392. [DOI] [PubMed] [Google Scholar]
- 68.Zhang JF, Fu WM, He ML, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011;22(21):3955–61. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Somerman MJ, Shroff B, Agraves WS, et al. Expression of attachment proteins during cementogenesis. J Biol Buccale. 1990;18(3):207–14. [PubMed] [Google Scholar]
- 70.Bosshardt DD. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res. 2005;84(5):390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 71.D’Errico JA, Berry JE, Ouyang H, et al. Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol. 2000;71(1):63–72. doi: 10.1902/jop.2000.71.1.63. [DOI] [PubMed] [Google Scholar]
- 72.Ducy P, Starbuck M, Priemel M, et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13(8):1025–36. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ducy P, Zhang R, Geoffroy V, et al. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 74.Zhou X, Zhang Z, Feng JQ, et al. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci USA. 2010;107(29):12919–24. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hirata A, Sugahara T, Nakamura H. Localization of runx2, osterix, and osteopontin in tooth root formation in rat molars. J Histochem Cytochem. 2009;57(4):397–403. doi: 10.1369/jhc.2008.952192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D’Souza RN, Cavender A, Sunavala G, et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12(12):2040–49. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- 77.Butler WT. Dentin matrix proteins. Eur J Oral Sci. 1998;106(Suppl 1):204–10. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 78.Chen S, Gluhak-Heinrich J, Wang YH, et al. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88(10):904–9. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao Z, Zhang H, Zhou X, et al. Genetic evidence for the vital function of Osterix in cementogenesis. J Bone Miner Res. 2012;27(5):1080–92. doi: 10.1002/jbmr.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao Z, Liu R, Zhang H, et al. Osterix controls cementoblast differentiation through downregulation of Wnt-signaling via enhancing DKK1 expression. Int J Biol Sci. 2015;11(3):335–44. doi: 10.7150/ijbs.10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu R, Cao Z. Osterix combined with gene-activated matrix: A potential integrated strategy for achieving cementum regeneration. Dent Hypotheses. 2015;6:10–13. [Google Scholar]
- 82.Sun Q, Liu H, Chen Z. The fine tuning role of microRNA-RNA interaction in odontoblast differentiation and disease. Oral Dis. 2015;21(2):142–48. doi: 10.1111/odi.12237. [DOI] [PubMed] [Google Scholar]
- 83.Ogata Y, Matsui S, Kato A, et al. MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients. J Oral Sci. 2014;56(4):253–60. doi: 10.2334/josnusd.56.253. [DOI] [PubMed] [Google Scholar]
- 84.Xie YF, Shu R, Jiang SY, et al. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci. 2011;3(3):125–34. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sipert CR, Morandini AC, Dionisio TJ, et al. MicroRNA-146a and microRNA-155 show tissue-dependent expression in dental pulp, gingival and periodontal ligament fibroblasts in vitro. J Oral Sci. 2014;56(2):157–64. doi: 10.2334/josnusd.56.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie YF, Shu R, Jiang SY, et al. MicroRNA-146 inhibits pro-inflammatory cytokine secretion through IL-1 receptor-associated kinase 1 in human gingival fibroblasts. J Inflamm. 2013;10(1):20. doi: 10.1186/1476-9255-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hung PS, Chen FC, Kuang SH, et al. Chang, miR-146a induces differentiation of periodontal ligament cells. J Dent Res. 2010;89(3):252–57. doi: 10.1177/0022034509357411. [DOI] [PubMed] [Google Scholar]
- 88.Li E, Zhang J, Yuan T, Ma B. MiR-143 suppresses osteogenic differentiation by targeting Osterix. Mol Cell Biochem. 2014;390(1–2):69–74. doi: 10.1007/s11010-013-1957-3. [DOI] [PubMed] [Google Scholar]
- 89.Jia J, Tian Q, Ling S, et al. miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett. 2013;587(18):3027–31. doi: 10.1016/j.febslet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 90.Liu H, Lin H, Zhang L, et al. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem. 2013;288(13):9261–71. doi: 10.1074/jbc.M112.433730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi K, Lu J, Zhao Y, et al. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. 2013;55(2):487–94. doi: 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 92.Goettsch C, Rauner M, Pacyna N, et al. miR-125b regulates calcification of vascular smooth muscle cells. Am J Pathol. 2011;179(4):1594–600. doi: 10.1016/j.ajpath.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, et al. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19(6):877–85. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- 94.Baglio SR, Devescovi V, Granchi D, Baldini N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013;527(1):321–31. doi: 10.1016/j.gene.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 95.Sun Q, Liu H, Lin H, et al. MicroRNA-338-3p promotes differentiation of mDPC6T into odontoblast-like cells by targeting Runx2. Mol Cell Biochem. 2013;377(1–2):143–49. doi: 10.1007/s11010-013-1580-3. [DOI] [PubMed] [Google Scholar]
- 96.Liu H, Sun Q, Wan C, et al. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229(10):1494–502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA. 2011;108(24):9863–68. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tome M, Lopez-Romero P, Albo C, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18(6):985–95. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105(37):13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gamez B, Rodriguez-Carballo E, Bartrons R, et al. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem. 2013;288(20):14264–75. doi: 10.1074/jbc.M112.432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang L, Cheng P, Chen C, et al. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. 2012;27(7):1598–606. doi: 10.1002/jbmr.1621. [DOI] [PubMed] [Google Scholar]
- 103.Chen Q, Liu W, Sinha MK, et al. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One. 2013;8:e58104. doi: 10.1371/journal.pone.0058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ozsolak F, Poling LL, Wang Z, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22(22):3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]



