Abstract
Humans are host to complex microbial communities that were previously termed “normal flora” and largely overlooked. However, it is increasingly understood that resident microbes contribute to both health and disease, including the risk for malignancy. Investigators are just beginning to define microbes, or groups of microbes, that contribute to the development of gastrointestinal malignancies and the mechanisms by which this occurs. Resident microbes can induce inflammation, leading to cell proliferation and altered stem cell dynamics, which can lead to alterations in DNA integrity and immune regulation and thus promote carcinogenesis. Studies in human patients and rodent models of cancer have identified alterations in the microbiota of the stomach, esophagus, and colon that increase the risk for malignancy. Manipulation of the composition and/or function of the gastrointestinal microbiota could allow for the prevention or treatment of certain gastrointestinal malignancies in the future.
Keywords: Helicobacter pylori, Gastric adenocarcinoma, Microbiota, Pyrosequencing, Colon cancer, Inflammation
Introduction
In the last 2 decades, there has been a remarkable shift in identifying and understanding the multitude of microbes that colonize the human body. Previously, the “normal flora” was felt to be largely a silent passenger, only declaring itself when it traveled outside of its usual niche. However, it is now recognized that the microbiome, which is comprised of bacteria, archaea, eukaryotes, and viruses, plays a key role in health and disease. Bacteria are the most abundant and well studied. The gastrointestinal (GI) microbiome is molded from birth by a multitude of interactions that can be distinct, such as the host genetic background, or variable, including diet, antibiotics, and other environmental exposures.1, 2
Cancer is the second leading cause of death in the United States and GI cancers represent a leading cause of morbidity and mortality.3 While genetic factors leading to an increased risk of cancer have been identified, such as adenomatous polyposis coli (APC) mutations that lead to familial adenomatous polyposis and E-cadherin (CDH1) mutations that lead to hereditary diffuse-type gastric cancer (GC), these mutations do not account for the majority of cases. In addition, the association of microbial infections with the risk for cancer development is well documented, including Helicobacter pylori with GC and hepatitis viruses with liver cancer.4 Even non-pathogenic GI tract microbes, once considered inert, have been found to play a role in chronic inflammation, altering cell proliferation and stem cell dynamics, and altering immune surveillance mechanisms.2, 5 The focus of this review is the role of the GI microbiome in the development of gastric and colonic malignancies with a brief discussion of esophageal malignancy.
Gastric Cancer
Gastric adenocarcinoma is the third leading cause of cancer-related death in the world.6 In developed countries, the incidence of gastric adenocarcinoma has significantly decreased over the past century;7, 8 however, the incidence rates of both proximal gastric and gastroesophageal junction adenocarcinomas have increased in both the United States and Europe.9, 10 Chronic infection with Helicobacter pylori is the strongest known risk factor for developing gastric adenocarcinoma.11
Helicobacter pylori
H. pylori is a Gram-negative bacteria that selectively colonizes the gastric epithelium. Infection is usually acquired in childhood and, in the absence of combination antibiotic therapy, can persist for the lifetime of the host.12 H. pylori has colonized humans for almost 100,000 years,13 and approximately half of the world’s population is infected with H. pylori, promoting speculation that H. pylori is an endogenous member of the gastric microbiota. Between 1–3% of H. pylori-colonized persons develop gastric adenocarcinoma,14 and factors that play a role in the pathologic outcome of H. pylori infection are varied, including strain-specific bacterial constituents, host genetic factors, environmental influences including diet, and alterations in the host microbiota.15
Bacterial and host factors affecting the propensity toward gastric cancer
One H. pylori virulence factor that influences GC risk is the cag pathogenicity island (cagPAI), which contains genes encoding proteins that form a type IV bacterial secretion system (T4SS).14 Another H. pylori virulence factor linked to the development of GC is the secreted toxin VacA.16, 17 All H. pylori strains contain vacA, but there are considerable differences in vacA sequences among strains. Strains containing type s1, i1, or m1 alleles within the 5′ region of the gene are highly associated with GC.18–20 Host polymorphisms in IL-1β and TNF-α as well as environmental factors such as a high salt diet and low iron levels in the context of H. pylori infection also influence gastric carcinogenesis.15
Although H. pylori infection is the strongest identified risk factor for developing GC, clinical trials suggest that other gastric microbiota constituents may influence disease progression. Antibiotic therapy directed against H. pylori was reported to significantly decrease the incidence of GC in a 15-year follow-up study of 3365 subjects. Of note, more than 50% of the antibiotic-treated individuals remained colonized by H. pylori at the 15-year follow-up.21 These findings suggest that antibiotic treatment may attenuate the development of GC by inducing alterations in the non-H. pylori microbiota.
The stomach microbiota in gastric pathogenesis
The stomach harbors a large and diverse bacterial community ranging from 101 to 103 colony forming units/g,22 which may influence gastric homeostasis and disease in conjunction with H. pylori infection.23
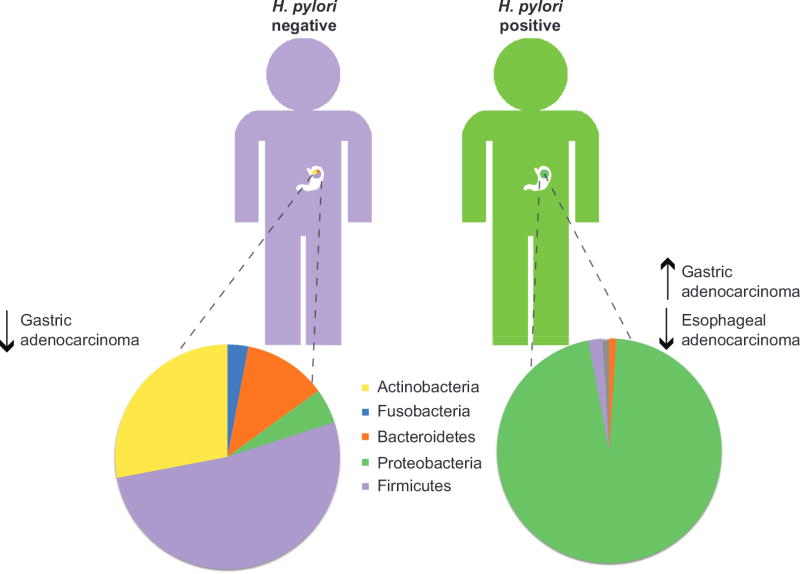
The composition of the gastric microbiome in H. pylori-negative individuals is highly diverse (Figure 1). Sequencing of DNA isolated from human gastric biopsies identified 128 phylotypes within 8 bacterial phyla of which Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria were the most abundant.2, 24 Utilizing a newer technology, tagged 454 pyrosequencing, analysis of H. pylori-negative biopsy samples identified 262 phylotypes representing 13 phyla.25 These findings lend further support to the gastric microbiota being highly diverse, despite significant variability in the microbial composition between individuals.24, 25 In contrast, the microbiota among H. pylori-infected individuals is much more uniform and H. pylori represents the most abundant phylotype present in the stomach of H. pylori-positive persons.24, 25 H. pylori DNA accounted for 93–97% of all sequence reads in H. pylori-positive persons and a total of 33 phylotypes were detected, over 200 less than in H. pylori-negative persons.25 Taken together, these data suggest that H. pylori colonization dramatically alters gastric microbiota diversity (Figure 1). Characterization of the human gastric microbiota using DNA microarrays detected 44 phyla with 4 dominant phyla: Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. Using this method, infection with H. pylori was shown to increase the relative abundance of non-H. pylori Proteobacteria, Spirochaetes, and Acidobacteria and decrease the relative abundance of Actinobacteria, Bacteroidetes and Firmicutes compared to uninfected stomachs.26 H. pylori infection accounted for 28% of the variance in the microbiota; however, the bacterial communities in both H. pylori-negative and -positive individuals remained highly complex.26
Figure 1.
Schematic representation of the gastric microbiome in H. pylori-negative versus H. pylori-positive individuals.
Studies examining differences in microbial composition and outcomes of GC are more limited. Development of atrophic gastritis, which induces hypochlorhydria due to parietal cell loss, is a key step in the histologic progression to intestinal-type GC, and can lead to overgrowth of non-Helicobacter microbiota which may promote the progression towards GC.27 Two recent studies have independently identified that proton pump inhibitor use may detrimentally alter the gut microbiota.28, 29
When comparing the microbiota of 10 GC patients to 5 dysplastic controls, the microbiota of GC patients was found to be equally as diverse as dysplastic patients. Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were identified. The microbiota was predominately composed of species of Streptococcus, Lactobacillus, Veillonella, and Prevotella. H. pylori were present in relatively low abundance.30 More recently, pyrosequencing has been used to compare the gastric microbiota in persons with chronic gastritis, intestinal metaplasia, and GC. Pyrosequencing identified 10 bacterial phyla and significant differences were observed in both the composition and diversity of the gastric microbiota in the histological progression towards GC. Bacilli and members of the Streptococcaceae family were significantly enriched in GC samples compared with chronic gastritis and intestinal metaplasia samples, while Epsilonproteobacteria and Helicobacteraceae family members were decreased.31
An interesting new study compared the gastric microbiota of subjects from two Colombian populations; one at high-risk, Tuquerres, and one at low-risk, Tumaco, of developing GC. Despite high variability in the microbial composition between individuals, significant correlations were found with the town of origin.32 Two operational taxanomic units, Leptotrichia wadei, which is associated with necrotizing enterocolitis and bacteremia, and a Veillonella sp., were significantly more abundant in Tuquerres. In the low-risk region of Tumaco, 16 operational taxanomic units, including a Staphylococcus sp, which is considered a constituent of the normal human microbiota, were significantly more abundant.32 These results provide a tantalizing opportunity to manipulate the microbiota of animal models to more closely represent the microbiota of either the high-risk or low-risk populations of Colombia and determine key players in cancer development.
Animal models to study the microbiome and gastric cancer
Inbred mice with defined genotypes are commonly used to model carcinogenesis; however, this can be limited by uncontrolled microbial diversity within mice despite identical genetic backgrounds.33, 34 To counter this, gnotobiotic mice allow for controlling the microbiome and adding back individual or collections of microorganisms.
Similar to humans, the most abundant phylotypes in the mouse stomach are Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria33 and infection of mice with H. pylori can alter the gastric microbiota.35 H. pylori infection induces gastritis in mice, and following H. pylori infection for 2 months the gastric microbiota in specific pathogen free (SPF) mice harbored reduced numbers of Lactobacillus species and increased bacterial diversity.35 An independent study, however, found that both acute and chronic infection of SPF C57BL/6 mice with H. pylori failed to cause significant shifts in the gastric microbial composition.36 It is possible that the inherent gastric microbial diversity of SPF mice may play a role in the outcome of H. pylori infection.
INS-GAS mice are genetically predisposed to GC, and chronic interaction between H. pylori and the gastric microbiota was found to influence disease progression in this model.37 In SPF INS-GAS mice, GC spontaneously developed.38, 39 However, in germfree (GF) INS-GAS mice, cancer was slower to develop.37 Moreover, H. pylori-infected GF INS-GAS mice developed less severe lesions and were slower to progress to gastrointestinal intraepithelial neoplasia than H. pylori-infected SPF INS-GAS mice.37 A detailed analysis using 454 sequencing of partial 16S ribosomal DNA amplicons revealed specific differences in phyla between H. pylori-infected and uninfected SPF INS-GAS mice. H. pylori colonization led to an expansion in the proportion of Firmicutes and decreased numbers of Bacteroidetes, while causing an overall increase in species diversity.37 In fact, only three species of commensal bacteria (ASF356 Clostridium species, ASF361 Lactobacillus murinus and ASF519 Bacteroides species) were required to promote gastric neoplasia in H. pylori-infected GF INS-GAS mice to the same extent as that reported in H. pylori-infected SPF INS-GAS mice.40
Esophageal adenocarcinoma and the microbiome
The incidence of esophageal adenocarcinoma has been rising rapidly in developed countries over the past 40 years, and this coincides with a decreasing incidence of H. pylori infection and GC, suggesting that gastric colonization with H. pylori may be protective against esophageal adenocarcinoma.41 This could reflect inhibition of acid secretion via enhanced production of IL-1β and TNF-α in response to H. pylori, or through loss of parietal cells in atrophic gastritis.23 Alternatively, changes in the gastric microbiota resulting from the loss of H. pylori may increase the risk for an individual to develop esophageal cancer (Figure 1).23
The esophageal microbiome is altered during inflammation and metaplasia. Using a 16S rRNA gene survey, 2 types of microbiota, termed type I and type II, were identified in the esophagus.42 The type I microbiome was dominated by Gram-positive bacteria and the genus Streptococcus, whereas the type II microbiome was composed of a higher percentage of Gram-negative bacteria, with the phyla Bacteroidetes, Proteobacteria, Fusobacteria, and Spirochaetes being the most abundant. The type II microbiome correlated with the histologic presence of esophagitis and Barrett’s esophagus, whereas the type I microbiome was associated with a histologically normal esophagus.42
In a recent study, 30 esophageal adenocarcinoma cases were compared to 39 control subjects using cultured biofilms. In control subjects, 56 species belonging to 19 genera were detected, while in esophageal adenocarcinoma, 73 species from 23 genera were identified. Despite finding more species in esophageal adenocarcinoma than controls, no statistical differences were reported.43 These findings provide an important framework for more detailed future studies delineating the structure and function of the esophageal microbiome and disease.
Colorectal Cancer
Colorectal cancer (CRC) is the third leading cause of cancer mortality in the U.S. and the risk of CRC increases with age.3 The vast majority of cases are sporadic; however, up to 25% of patients have a family history of CRC, but no evidence of an identified inherited syndrome.44 This underscores the complex interaction of multiple genetic and epigenetic events contributing to CRC pathogenesis. The initiation of CRC can be due to mutations in tumor-suppressor genes such as APC, catenin (cadherin-associated protein) beta 1 (CTNNB1), tumor protein p53 (TP53) and the oncogene Kirsten rat sarcoma viral oncogene homolog (KRAS), leading to a growth advantage in colonic epithelial cells progressing to adenomas and cancer.23, 44, 45 While these genetic mutations have clearly been linked to CRC development, the steps leading to the accumulation of these mutations and other epigenetic changes are not fully known. In addition to genetic alterations, microbial and environmental factors, including diet and lifestyle, have been shown in recent studies to play a role in promoting CRC.46, 47
The microbiome in patients with colonic polyps or CRC
Similar to the stomach and esophagus, the colon plays host to a complex and diverse population of microorganisms. These microorganisms outnumber human somatic and germ cells by at least an order of magnitude, and the collective microbial genome contains ≥100 times more genes than the human genome.48 The majority of the colonic microbiota is composed of Bacteroidetes and Firmicutes, though other components include Proteobacteria, Actinobacteria, and Fusobacteria.23, 49 An individual’s colonic microbiota is determined by factors such as environmental exposures, diet, and host genetics, although the identification of specific genetic factors leading to alterations in the microbiota is in a nascent stage.50
While the microbiome contributes to both immune system development and the release of key nutrients and energy from dietary intake, alterations in the microbiome related to chronic inflammation appear to play a role in promoting the increased risk of carcinogenesis seen in inflammatory bowel disease patients.51 There is also mounting evidence that the microbiome plays a role in sporadic CRC.52–59 (Table 1) Studies of the fecal microbiota in patients with either CRC or colonic polyps have shown decreased temporal stability, with increased diversity of the Clostridium leptum and C. coccoides subgroups versus control subjects, although not between patients with CRC versus colonic polyps.60 Studies assessing mucosa-associated bacteria showed that the predominant phyla in control patients were Firmicutes, Bacteroidetes, and Proteobacteria.61 While adenoma patients had a lower relative abundance of Bacteroidetes and a higher abundance of Proteobacteria, there was a trend toward increased diversity in adenoma patients versus those without adenomas.61 When similar studies assessed mucosa-associated microbiota in paired samples from patients with CRC (i.e., tumor tissue and adjacent non-tumor tissue), Coriobacteridae, Roseburia, Fusobacterium and Faecalibacterium, which are generally regarded as gut commensals, were overrepresented in tumor tissue samples.62 Multiple studies have assessed both the luminal and mucosa-associated microbiota in control, adenoma, and/or CRC patients.63, 64 However, no single microbial species has been identified as a causative agent leading to a working model that overall disturbances in the composition, diversity, or functional properties of the colonic microbiota dysregulate the balance between the epithelium and the immune system towards inflammation, dysplasia, and ultimately cancer.
Table 1.
Alterations in Human GI microbiota in patients with colonic adenoma and CRC
| Patients | Sample Site | Changes in Microbiota | Study (author, year, ref no.) |
|---|---|---|---|
| 20 controls, 20 polypectomy patients, and 20 CRC patients (all s/p resection) | Fecal samples (3 samples over a 3 month period for 20 polypectomy, 20 CRC patients, as well as 6 of the control patients) | There was reduced temporal stability and increased diversity for the microbiota of CRC and polyp patients. In addition, there was increased diversity of the Clostridium leptum and C. coccoides subgroups versus control subjects. |
Scanlan et. al., 2008, 60 |
| 21 adenoma patients and 23 controls | Rectal mucosal biopsies | Adenoma patients had a trend toward increased diversity and richness versus controls. There was lower relative abundance of Bacteroidetes and higher abundance of Proteobacteria. | Shen et. al., 2010, 61 |
| 6 CRC patients | Paired biopsies from tumor and adjacent non-tumor tissue in the surgical resection specimen | The tumor areas had overrepresentation of Coriobacteridae, Roseburia, Fusobacterium and Faecalibacterium, and underrepresentation of Firmicutes and Enterobacteriaceae. | Marchesi et al., 2011, 62 |
| 46 CRC patients and 56 controls | Fecal samples, rectal swab samples, and from the CRC patients 27 with paired tumor and adjacent non-tumor tissue | Overall the microbiota was similar between the paired tumor and non-tumor tissues though tumor tissues had lower bacterial diversity. Lactobacillales was enriched where Faecalibacterium was reduced in tumor tissue. In the mucosa-adherent microbiota, Bifidobacterium, Faecalibacterium, and Blautia were reduced in CRC patients, whereas Fusobacterium, Porphyromonas, Peptostreptococcus, and Mogibacterium were enriched. |
Chen et al., 2012, 56 |
| 29 adenoma patients, 31 CRC patients, 34 with symptoms but normal colonoscopy, and 31 asymptomatic controls | Colonic biopsies including paired biopsies from tumor and adjacent non-tumor tissue in CRC patients | When assessing for E. coli and E. coli-like bacteria, there was increased presence of intracellular E. coli in patients with adenoma and CRC. | Swidsinski et. al., 1998, 53 |
| 60 CRC patients and 119 controls | Fecal samples and colon/rectal biopsies from a subset of 22 CRC patients and 22 controls | Pyrosequencing on 6 CRC and 6 control samples indicated microbiota differences in CRC patients vs controls. Higher levels of Bacteroides/Prevetolla were detected in patients with CRC determined by quantitative polymerase chain reaction. |
Sobhani et. al., 2011, 54 |
| 104 CRC patients | Paired biopsies from tumor and adjacent non-tumor tissue in the surgical resection specimen | Whole genome sequences from 9 tumor/normal pairs revealed that Fusobacterium sequences were enriched CRC. Quantitative PCR and 16S rDNA sequence analysis of the remaining 95 CRC/normal tissue pairs confirmed the increased Fusobacterium, while Bacteroidetes and Firmicutes were depleted in tumors. |
Kostic et. al., 2012, 55 |
| 99 CRC patients | Paired biopsies from tumor and adjacent non-tumor tissue in the surgical resection specimen | There was increased Fusobacterium in tumor tissues. | Castellarin et. al., 2012, 57 |
| 10 CRC patients and 11 controls | Fecal samples | No significant differences in microbial community structure or diversity between CRC patients and controls. However, Bacteroides and Prevotella were relatively underrepresented whereas there were higher percentages of Akkermansia muciniphila in CRC patients. | Weir et. al., 2013, 58 |
| 344 patients with advanced adenomas (size > 10mm or villous, tubulovillus, or high grade dysplasia on pathology) and 344 controls | Fecal samples | Increased abundance of Enterococcus and Streptococcus species and decreased prevalence of Roseburia and Clostridium in the adenoma patients. | Chen et. al., 2013, 64 |
| 33 adenoma patients and 38 controls | Rectal biopsies | Increased microbial richness with increased abundance of Firmicutes, Bacteroidetes, and Proteobacteria in adenoma patients. | Sanapareddy et. al., 2012, 63 |
| 30 adenoma patients, 30 CRC patients, and 30 controls | Fecal samples | Microbial dysbiosis, enrichment of pathogenic bacteria was seen in adenoma and CRC patients compared to controls. In adenoma patients, there were higher relative abundances of Blautia, Ruminococcus, Clostridium, and Lachnospiraceae compared to CRC. CRC patients had higher relative abundances of Fusobacterium, Bacteroides, Phascolarctobacterium, and Porphyromonas compared to adenoma patients. |
Zackular et. al., 2014, 59 |
Diet and microbial metabolites
Epidemiologic studies have consistently indicated that diets with increased red meat and fat content (i.e., Western diet) increase CRC risk, while increased fiber intake is associated with decreased CRC risk.65, 66 This alteration in risk by dietary intake may be facilitated by the colonic microbiota, which, for example, promote health via metabolism of fiber to produce short-chain fatty acids (SCFA) such as acetate, propionate, and butyrate.65 Butyrate is the preferred energy source of colonic enterocytes and, along with propionate, has been shown to downregulate pro-inflammatory cytokines, such as IL-6 and IL-12, in colonic macrophages.67 In addition, butyrate and propionate can induce FOXP3+ regulatory T cells to control intestinal inflammation, thereby maintaining intestinal homeostasis.67, 68 The anti-inflammatory effects of SCFAs not only influence host cells, but may also contribute to homeostasis of the gut microbiota.67, 68
Recent studies have attempted to define the link between dietary intake, the gut microbiota, and CRC.52, 64 One study found lower levels of butyrate-producing bacteria and decreased fecal SCFAs in meat-eating African Americans, who are at higher risk for CRC, compared to native Africans.52 In addition, patients with advanced colorectal adenomas were found to have lower dietary fiber intake patterns and consistently lower SCFA levels versus controls.64 When categorized by dietary fiber intake, low dietary fiber intake remained associated with a deficiency in butyrate-producing bacteria, which could increase the risk for advanced adenomas.64
Animal models
Animal models of CRC allow for investigation of the potential links between GI microbes and risk for colonic neoplasia; specifically, the ability to study these models both in conventional or GF conditions. One of the most common models utilized is the adenomatous polyposis coli (APC) multiple intestinal neoplasia (Min) murine model of colon carcinogenesis. APCMin/+ mice possess a point mutation in the murine homolog of the human APC tumor-suppressor gene resulting in spontaneous adenomas, primarily in the small intestine.69 GF APCMin/+ mice have a reduced overall tumor burden including fewer colon tumors versus conventionally housed controls.70 Exposure to commensal strains such as enterotoxigenic Bacteroides fragilis and Citrobacter rodentium promotes colon tumor formation in APCMin/+ mice, while a non-toxigenic strain of B. fragilis does not.71 Furthermore, mono-association with Bacteroides vulgatus reduced colorectal tumorigenesis in IL10−/− mice versus conventionally housed controls.72 Thus, when the host genetic background is identical, exposure to different microbial species can lead to altered risk for colon carcinogenesis. This risk can also be modified by antibiotic exposure in models such as Nod2−/− mice. Nod2−/− mice have an altered GI microbiota when compared with wild-type (WT) mice, and develop more tumors following azoxymethane/dextran sulfate sodium treatment.73 Antibiotic exposure or fecal transplants from WT mice can abrogate this phenotype.73 Additionally, WT mice co-housed with Nod2−/− mice, exhibit increased tumorigenesis versus separately housed controls.73 Collectively, these findings suggest that microbial manipulation could be used to abrogate CRC risk if the specific exposures driving the risk are identified.
Summary
Many factors contribute to the development of GI cancers. Multiple inherited and acquired mutations in epithelial cells have been identified and alterations in risk have also been attributed to environmental exposures such as diet and now the microbiota. However, identifying whether specific microbes within the complex microbiota are driving the progression to cancer is challenging due to the multidimensional nature of the community, which can be altered by diet, antibiotics, and influenced by the genotype of the individual.
Ongoing analyses of the GI microbiota have found patterns that associate with both malignant and pre-malignant lesions, such as atrophic gastritis and colonic polyps. However, whether these alterations play a direct role in disease development or progression, or whether they are markers of underlying epithelial or immune cell dysregulation have yet to be determined. The GI microbiome harbors significant metabolic activity, which can alter dietary nutrients with both beneficial and potentially harmful sequelae. It is tempting to speculate that in the future, we may identify groups of bacterial taxa that can predict GI disease risk or outcome. These biomarkers could potentially be used to stratify patients towards particular therapeutic regimens, providing further options for utilizing the GI microbiome in the treatment and potentially prevention of GI tract malignancies.
Key Points.
The risk of developing gastric cancer is multifactorial and the microbiota has been identified as an important contributing factor.
Colon cancer risk is modified by the GI tract microbiota and environmental exposures including diet, in addition to known genetic factors.
With no single microbial causative agent identified, it is likely that an overall disturbance in the composition/metabolism of the colonic microbiota can promote cancer development.
Acknowledgments
Sources of Funding: National Institutes of Health R01 DK58587, R01 CA77955, P01 CA116087, and P30 DK058404 to R.M.P. Department of Veterans Affairs 1IK2BX002126-01 to L.A.C.
Footnotes
Disclosures/Conflict of interest: The authors declare there are no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lydia E. Wroblewski, Email: Lydia.Wroblewski@vanderbilt.edu, Division of Gastroenterology, Department of Medicine; Vanderbilt University School of Medicine; Nashville, TN USA, T: 615-322-4215
Richard M. Peek, Jr, Email: Richard.Peek@vanderbilt.edu, Division of Gastroenterology, Department of Medicine; Vanderbilt University School of Medicine; Nashville, TN USA, T: 615-343-1596
Lori A. Coburn, Email: Lori.coburn@vanderbilt.edu, Veterans Affairs Tennessee Valley Healthcare System; Division of Gastroenterology, Department of Medicine; Vanderbilt University School of Medicine; Nashville, TN USA, T: 615-875-4222, F: 615-343-4229
References
- 1.Blaser MJ. The microbiome revolution. The Journal of Clinical Investigation. 2014;124(10):4162–4165. doi: 10.1172/JCI78366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: A Cancer Journal for Clinicians. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The Lancet Oncology. 2012;13(6):607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 5.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host & Microbe. 2011;10(4):324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs CS, Mayer RJ. Gastric carcinoma. The New England Journal of Medicine. 1995;333(1):32–41. doi: 10.1056/NEJM199507063330107. [DOI] [PubMed] [Google Scholar]
- 8.Howson CP, Hiyama T, Wynder EL. The decline in gastric cancer: epidemiology of an unplanned triumph. Epidemiologic Reviews. 1986;8:1–27. doi: 10.1093/oxfordjournals.epirev.a036288. [DOI] [PubMed] [Google Scholar]
- 9.Blot WJ, Devesa SS, Kneller RW, et al. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA. 1991;265(10):1287–1289. [PubMed] [Google Scholar]
- 10.Pera M, Cameron AJ, Trastek VF, et al. Increasing incidence of adenocarcinoma of the esophagus and esophagogastric junction. Gastroenterology. 1993;104(2):510–513. doi: 10.1016/0016-5085(93)90420-h. [DOI] [PubMed] [Google Scholar]
- 11.Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. International Journal of Cancer. 2015;136(2):487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- 12.Wroblewski LE, Peek RM, Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clinical Microbiology Reviews. 2010;23(4):713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linz B, Balloux F, Moodley Y, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amieva M, Peek RM., Jr Pathobiology of Helicobacter pylori-Induced Gastric Cancer. Gastroenterology. 2016;150(1):64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wroblewski LE, Peek RM., Jr Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterology Clinics of North America. 2013;42(2):285–298. doi: 10.1016/j.gtc.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nature Reviews Microbiology. 2005;3(4):320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 17.Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends in Microbiology. 2012;20(4):165–174. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Atherton JC, Cao P, Peek RM, Jr, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori Association of specific vacA types with cytotoxin production and peptic ulceration. The Journal of Biological Chemistry. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 19.Atherton JC, Peek RM, Jr, Tham KT, et al. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112(1):92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 20.Miehlke S, Kirsch C, Agha-Amiri K, et al. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. International Journal of Cancer. 2000;87(3):322–327. [PubMed] [Google Scholar]
- 21.Ma JL, Zhang L, Brown LM, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. Journal of the National Cancer Institute. 2012;104(6):488–492. doi: 10.1093/jnci/djs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4(6):505–531. doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreu MT, Peek RM., Jr Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146(6):1534–1546. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(3):732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3(7):e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. The ISME Journal. 2011;5(4):574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Research. 1992;52(24):6735–6740. [PubMed] [Google Scholar]
- 28.Jackson MA, Goodrich JK, Maxan ME, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2015 Dec 30; doi: 10.1136/gutjnl-2015-310861. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2015 Dec 9; doi: 10.1136/gutjnl-2015-310376. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dicksved J, Lindberg M, Rosenquist M, et al. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. Journal of Medical Microbiology. 2009;58(Pt 4):509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 31.Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407–416. doi: 10.1111/hel.12145. [DOI] [PubMed] [Google Scholar]
- 32.Yang I, Woltemate S, Piazuelo MB, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Scientific Reports. 2016;6:18594. doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolig AS, Cech C, Ahler E, et al. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infection and Immunity. 2013;81(5):1382–1389. doi: 10.1128/IAI.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigal M, Rothenberg ME, Logan CY, et al. Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology. 2015;148(7):1392–1404. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 35.Aebischer T, Fischer A, Walduck A, et al. Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunology and Medical Microbiology. 2006;46(2):221–229. doi: 10.1111/j.1574-695X.2005.00024.x. [DOI] [PubMed] [Google Scholar]
- 36.Tan MP, Kaparakis M, Galic M, et al. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Applied and Environmental Microbiology. 2007;73(3):1010–1013. doi: 10.1128/AEM.01675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lofgren JL, Whary MT, Ge Z, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140(1):210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson MJ, Pritchard DM, Boxall SA, et al. Gastric Helicobacter infection induces iron deficiency in the INS-GAS mouse. PLoS One. 2012;7(11):e50194. doi: 10.1371/journal.pone.0050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Fan X, Lindholm C, et al. Helicobacter pylori modulates lymphoepithelial cell interactions leading to epithelial cell damage through Fas/Fas ligand interactions. Infection and Immunity. 2000;68(7):4303–4311. doi: 10.1128/iai.68.7.4303-4311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lertpiriyapong K, Whary MT, Muthupalani S, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63(1):54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Reviews Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Lu X, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137(2):588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blackett KL, Siddhi SS, Cleary S, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Alimentary Pharmacology & Therapeutics. 2013;37(11):1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 44.Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Annals of Gastroenterology. 2014;27(1):9–14. [PMC free article] [PubMed] [Google Scholar]
- 45.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends in Genetics: TIG. 1993;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 46.Dejea C, Wick E, Sears CL. Bacterial oncogenesis in the colon. Future Microbiology. 2013;8(4):445–460. doi: 10.2217/fmb.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. International Journal of Cancer. 2007;120(3):656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 48.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2015 doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keku TO, Dulal S, Deveaux A, et al. The gastrointestinal microbiota and colorectal cancer. American Journal of Physiology Gastrointestinal and Liver Physiology. 2015;308(5):G351–363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. The American Journal of Clinical Nutrition. 2013;98(1):111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115(2):281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 54.Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zackular JP, Rogers MA, Ruffin MTt, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prevention Research. 2014;7(11):1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environmental Microbiology. 2008;10(3):789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 61.Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1(3):138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6(5):e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. The ISME Journal. 2012;6(10):1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen HM, Yu YN, Wang JL, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. The American Journal of Clinical Nutrition. 2013;97(5):1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 65.Greer JB, O’Keefe SJ. Microbial induction of immunity, inflammation, and cancer. Frontiers in Physiology. 2011;1:168. doi: 10.3389/fphys.2010.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vargas AJ, Thompson PA. Diet and nutrient factors in colorectal cancer risk. Nutrition in Clinical Practice. 2012;27(5):613–623. doi: 10.1177/0884533612454885. [DOI] [PubMed] [Google Scholar]
- 67.Chang PV, Hao L, Offermanns S, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(6):2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews Microbiology. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 69.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33(6):1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 71.Sussman DA, Santaolalla R, Strobel S, et al. Cancer in inflammatory bowel disease: lessons from animal models. Current Opinion in Gastroenterology. 2012;28(4):327–333. doi: 10.1097/MOG.0b013e328354cc36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uronis JM, Muhlbauer M, Herfarth HH, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. The Journal of Clinical Investigation. 2013;123(2):700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]