Gearhart and collaborators address the long-standing question of the roles of error-prone DNA polymerases in somatic hypermutation of antibody genes.
Abstract
DNA polymerase ι (Pol ι) is an attractive candidate for somatic hypermutation in antibody genes because of its low fidelity. To identify a role for Pol ι, we analyzed mutations in two strains of mice with deficiencies in the enzyme: 129 mice with negligible expression of truncated Pol ι, and knock-in mice that express full-length Pol ι that is catalytically inactive. Both strains had normal frequencies and spectra of mutations in the variable region, indicating that loss of Pol ι did not change overall mutagenesis. We next examined if Pol ι affected tandem mutations generated by another error-prone polymerase, Pol ζ. The frequency of contiguous mutations was analyzed using a novel computational model to determine if they occur during a single DNA transaction or during two independent events. Analyses of 2,000 mutations from both strains indicated that Pol ι–compromised mice lost the tandem signature, whereas C57BL/6 mice accumulated significant amounts of double mutations. The results support a model where Pol ι occasionally accesses the replication fork to generate a first mutation, and Pol ζ extends the mismatch with a second mutation.
INTRODUCTION
Upon encounter with antigen, B cells express activation-induced deaminase (AID), which deaminates cytosine to uracil in DNA (Maul et al., 2011). The uracil base is then used to induce a vast array of mutations and DNA breaks to promote somatic hypermutation and class switch recombination. During somatic hypermutation, uracils are detected by either the mismatch repair protein complex, MSH2-MSH6 (Wiesendanger et al., 2000), or the base excision repair protein, uracil DNA glycosylase (UNG; Rada et al., 2002). However, these proteins do not function in the canonical repair pathways of removing base damage and allowing faithful DNA synthesis by high fidelity DNA polymerases (Pols) β, δ, and ε. Instead, low fidelity Pols η, ζ, and Rev1 are recruited to synthesize multiple mutations. Pols η and ζ function mainly during synthesis in gaps created by MSH2–MSH6 and exonuclease 1 (Bardwell et al., 2004; Martomo et al., 2005). Pol η is responsible for the majority of mutations of A:T bp (Zeng et al., 2001; Delbos et al., 2007), and Pol ζ contributes to synthesis of tandem double mutations (Daly et al., 2012; Saribasak et al., 2012). Rev1, a deoxycytidyl transferase, inserts C opposite abasic sites generated after removal of uracils by UNG to produce transversions of C:G bp (Jansen et al., 2006). The abasic site could also be nicked by an apurinic/apyrimidinic endonuclease to create a single strand break (Stavnezer et al., 2014), which can allow entry by Pol η to generate mutations of A:T bp (Delbos et al., 2007).
Pol ι would also appear to be an attractive candidate for somatic hypermutation because of its very high misincorporation rate (Tissier et al., 2000). Indeed, Pol ι may be present during gap synthesis because it physically interacts with Pol η through ubiquitination (McIntyre et al., 2013), and both polymerases are recruited to DNA damage foci (Kannouche et al., 2003). However, there was no alteration in mutation frequency or spectra in the 129 strain of mice (McDonald et al., 2003; Martomo et al., 2006), which does not express full length Pol ι due to a naturally occurring point mutation in exon 2 that produces a nonsense codon. It has recently been reported that there is a high incidence of exon 2 skipping in 129-derived strains, and the truncated protein has residual polymerase activity (Aoufouchi et al., 2015). However, another study demonstrated that human Pol ι lacking exon 2 is inactive (Makarova et al., 2011), likely because it is missing critical active-site contacts required for polymerase function. Thus, it remains unclear whether Pol ι does, or does not, participate in somatic hypermutation.
Because of this controversy, we generated a knock-in mouse with catalytically inactive Pol ι. We also considered a new role for Pol ι to act together with Pol ζ during mutagenesis. Multiple polymerases can work sequentially when bypassing DNA lesions, and this includes Pol ι and Pol ζ (Johnson et al., 2000; Ziv et al., 2009). Using a novel computational model to statistically analyze the frequency of tandem mutations, we demonstrate that Pol ι cooperates with Pol ζ to produce contiguous mutations.
RESULTS AND DISCUSSION
Catalytically inactive Pol ι binds DNA and is expressed in knock-in mice
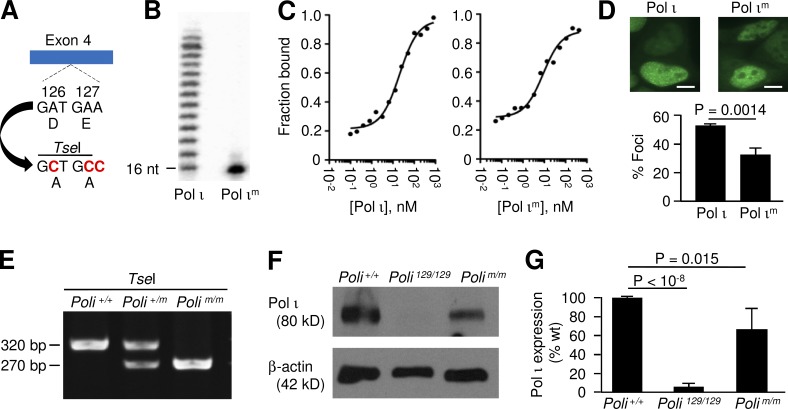
To generate the mutant protein, active site residues D126 and E127 were both changed to alanine (Pol ιm; Fig. 1 A), which should disrupt the ability of Pol ι to chelate Mg2+ required for DNA synthesis. A similar alteration of the corresponding residues in Pol η abolished its catalytic activity (Tissier et al., 2000). To confirm inactivation of Pol ι activity, primer extension assays were performed using Pol ι and Pol ιm proteins expressed in E. coli. As expected, Pol ιm displayed no extension, whereas an identical concentration of wild-type Pol ι had robust synthesis (Fig. 1 B). To see if the lack of synthesis by Pol ιm was caused by an inability to bind to a primer terminus, we compared the mutant enzyme to wild-type enzyme in an electrophoretic mobility shift assay (EMSA; Fig. 1 C). The analysis revealed that Pol ι bound the primer-template with a Kd of 21 ± 3 nM, whereas Pol ιm bound with a comparable Kd of 8.8 ± 1.7 nM. To test the in vivo properties of the mutant protein, we examined if it could accumulate in DNA replication foci analogous to wild-type protein (Kannouche et al., 2003). EGFP-Pol ι and EGFP-Pol ιm constructs were transfected into proliferating HEK293T cells, and fluorescence images of cell nuclei were obtained. Spontaneous replication foci from 600 nuclei in each group were then analyzed. As shown in Fig. 1 D, Pol ι foci were observed in approximately half of the nuclei, and Pol ιm foci occurred in approximately one-third of the nuclei. This shows that Pol ιm is present, albeit in a lesser amount, in physiological transactions during DNA replication, similar to wild-type Pol ι.
Figure 1.
Pol ιm activity and expression in knock-in mice. (A) Nucleotide substitutions introduced into exon 4 produced D126A-E127A altered codons, and created a TseI restriction site for PCR genotyping. (B) DNA primer extension assay comparing wild-type Pol ι and Pol ιm. (C) Binding affinity of Pol ι and Pol ιm for DNA by EMSA analysis. The dissociation constant KD(DNA) was obtained by plotting the fraction of bound DNA as a function of enzyme concentration. (D) Replication foci formation. Representative images are shown for HEK293T cells transformed with either EGFP-Pol ι or EGFP-Pol ιm constructs. The protein is seen as multiple bright foci throughout the green nucleus at the bottom of each picture. Bars, 5 µm. Graph shows the results from three independent experiments, with 200 nuclei each counted for the constructs. Error bars represent the standard deviation; p-value was calculated from a two-tailed Student’s t test. (E) Genotyping analysis of Poli alleles. PCR amplification produced a 320-bp amplicon, which was digested with TseI. (F) Pol ι expression in mouse testis from three strains of mice, comparing Pol ι and β-actin by Western blot. A representative image is shown. (G) Quantification of Western blot signals normalized to β-actin. Error bars signify the standard deviation of values from three independent experiments with two to four mice per experiment. The p-value was determined from a two-tailed Student’s t test.
We next generated a mouse strain expressing Pol ιm. Knock-in mice were made from C57BL/6 embryonic cells using standard genetic techniques. To confirm the existence of the D126A-E127A allele, PCR was used to amplify a 320-bp region surrounding exon 4, and the mutant allele was confirmed by digestion with TseI (Fig. 1, A and E). Homozygous Polim/m mice were then bred and tested for expression of the mutant Pol ι protein. Testis extracts, which contain abundant quantities of the protein, were prepared from Poli/+(C57BL/6), Poli129/129, and Polim/m mice, and analyzed by Western blotting (McDonald et al., 2003). Pol ι protein was expressed at high levels in extracts from Poli+/+ and Polim/m mice, compared with Poli129/129 extracts (Fig. 1 F). Quantitation of the blots revealed that steady-state levels of mutant Pol ι were approximately two-thirds of that observed for wild type (Fig. 1 G). The lower levels of Pol ιm may be a result of either reduced expression or increased turn-over. In contrast, Pol ι was virtually absent in extracts from 129 mice, as previously reported (McDonald et al., 2003).
Mice with defective Pol ι have normal frequency and spectra of mutations
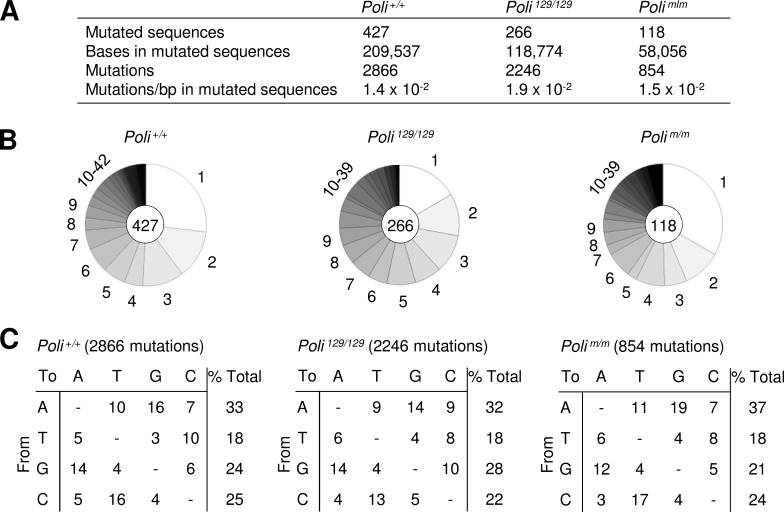
The frequency and types of mutation were measured in B cells from Peyer’s patches, which undergo constitutive activation from gut flora in the small intestine. Germinal center B cells (GL7+ B220+) were isolated by flow cytometry, and DNA was prepared and PCR amplified to produce a 492-bp region downstream of the rearranged JH4 gene segment on the Igh locus. Mutations in this intron region reflect the direct effects of polymerase mutagenesis in the absence of selection. The frequencies in mutations/base pairs from Poli+/+ (1.4 × 10−2), Poli129/129 (1.9 × 10−2), and Polim/m (1.5 × 10−2) sequences (Fig. 2 A), and the number of mutations per sequence (Fig. 2 B) were similar. The types of mutations in the two strains expressing defective Pol ι were also analogous to those from Poli+/+ mice (Fig. 2 C). Thus, the absence of Pol ι in the Poli129/129 strain, or the presence of catalytically inactive protein in the Polim/m strain, did not affect the frequency or spectra of mutations, perhaps because it quickly falls off the template after partial synthesis.
Figure 2.
Somatic hypermutation analyses in JH4 intron of germinal center B cells. (A) Frequency of mutation in the JH4 intron from Peyer’s patch B cells for Poli+/+, Poli129/129, and Polim/m mice. Poli+/+ data are previously published; Poli129/129 data are from McDonald et al. (2003), and four additional independent experiments, with one to three mice per experiment; and Polim/m data are from three independent experiments, with two to five mice per experiment. (B) Number of mutations per sequence. Each pie segment depicts the percentage of sequences with one or more mutations. Center circle shows number of mutated sequences analyzed. (C) Spectra of substitutions. The number of mutations is shown in parentheses. Mutations were recorded from the nontranscribed strand and corrected for base composition of the sequence. Data are expressed as a percentage of mutations.
Pol ι acts sequentially with Pol ζ to generate tandem mutations
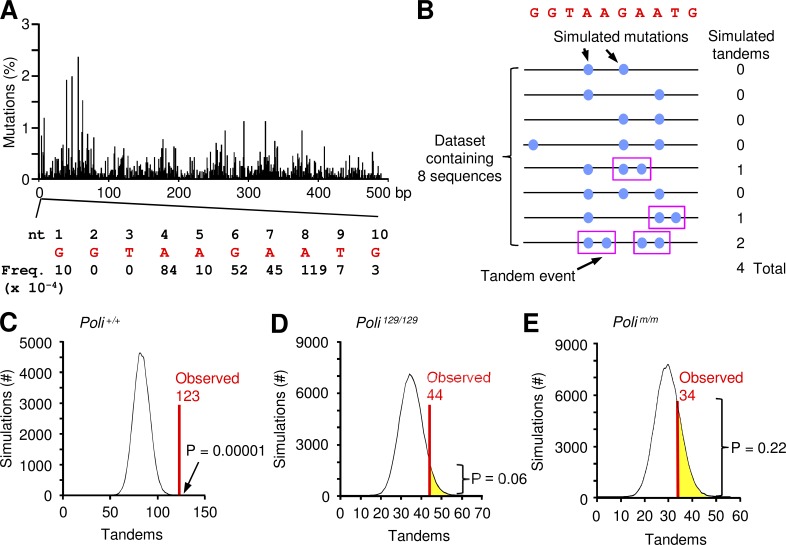
Pol ζ generates a unique error signature in variable regions: tandem or adjacent double mutations (Daly et al., 2012; Saribasak et al., 2012). Because the error frequency of Pol ζ is only 10−3 (Zhong et al., 2006), it would be inefficient at generating the first mutation. However, Pol ζ excels at extending from an initial mismatch with a second mutation to generate a tandem double mutation (Stone et al., 2012). What is the polymerase that puts in the first mutation? In somatic hypermutation, we reported that tandem mutations were elevated in the absence of Pol η (Saribasak et al., 2012), indicating that Pol η is not involved. We propose that the very error-prone Pol ι, with an error frequency as high as 10−1 when copying some bases (Tissier et al., 2000), generates the first mismatch. To address this hypothesis, we developed a sensitive computational model to statistically determine if tandem substitutions were generated during one synthesis event or during two independent events. The model was based on two parameters: (1) the frequency of mutations per nucleotide and (2) the number of mutations per sequence. Frequencies were calculated from 2866 mutations in C57BL/6 sequences (Fig. 3 A). A list of the frequencies for the first 10 bases is displayed in Fig. 3 A; a complete listing for 492 bases in the JH4 intron is shown in Table S1. The data demonstrate that each nucleotide accumulates mutations at a different frequency, which might be caused by hot spot locations and sequence environment (MacCarthy et al., 2009). Sequences with more mutations will have an increased probability of two independent events occurring adjacently during clonal expansion. However, a tandem event in a sequence with only two mutations would be highly significant, and likely result from a single synthesis event.
Figure 3.
Computational model to determine significance of tandem mutations. (A) Frequency of mutation at each nucleotide in the JH4 intron (492 bp). Data are from 2866 mutations observed in C57BL/6 sequences. Expanded view shows the frequency per base of the first 10 nucleotides (red). (B) Schematic description of the computational model. A dataset of eight sequences is constructed with simulated mutations (blue circles) inserted into the red germline sequence of the first 10 nucleotides. Mutations correspond to the observed mutation frequencies shown in A. The number of simulated tandems (circles in pink boxes) that are generated randomly is calculated for each sequence. (C) Plot of simulations versus tandems. For Poli+/+ sequences, the total number of expected tandems in each simulated dataset is shown as a histogram. The observed number of tandems, 123, is marked by a red line. The p-value, 0.00001, was calculated as the fraction of simulations (out of 100,000) in which the number of expected tandems was greater than, plus half of the number equal, to the observed number. (D and E) Values for Poli29/129 and Polim/m sequences. The observed number of tandems is shown in red, and the number of simulations with expected tandems greater than the observed number is highlighted in yellow. P-values were determined as in C.
Starting with the nonmutated germline sequence, the program predicts the location of mutations depending on these two parameters. For example, Fig. 3 B shows eight sequences, with three sequences containing two mutations, four sequences with three mutations, and one sequence with four mutations. There are four predicted tandem events in this simulated dataset. The process is repeated 100,000 times to calculate the number of expected tandems. Their distribution is represented as a histogram, and the area under the histogram illustrates the number of simulations that would yield the expected tandems. We then analyzed data from Peyer’s patches to count the number of actual tandem mutations. In Poli+/+ mice, 425 sequences were examined, and 123 tandems were observed (Table 1). This was compared against the expected distribution of tandems given by the computational program (Fig. 3 C); a p-value of 10−5 indicates that the observed number of tandems (red line) was significantly higher than the simulated number. Tandem mutations were then examined in datasets from Poli129/129 and Polim/m mice. The observed number of tandems was counted for each of these (Table 1), and compared against the expected distribution given by the computational model. Fig. 3 (D and E) show Poli129/129 mice and Polim/m mice had no increase in observed tandems beyond what was expected by random chance (P = 0.06 and 0.22, respectively). Thus, they did not accrue a surplus of contiguous mutations, suggesting that loss of functional Pol ι prevented tandem generation.
Table 1. Sequence data used for tandem analyses.
| Mutations per sequence | Poli+/+ | Poli129/129 | Polim/m | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences | Mutations | Tandems | Sequences | Mutations | Tandems | Sequences | Mutations | Tandems | ||||||
| 1 | 114 | 114 | 0 | 32 | 32 | 0 | 38 | 38 | 0 | |||||
| 2 | 55 | 110 | 3 | 22 | 44 | 0 | 12 | 24 | 0 | |||||
| 3 | 47 | 141 | 1 | 20 | 60 | 3 | 7 | 21 | 0 | |||||
| 4 | 21 | 84 | 1 | 14 | 56 | 1 | 8 | 32 | 1 | |||||
| 5 | 26 | 130 | 2 | 15 | 75 | 2 | 4 | 20 | 0 | |||||
| 6 | 28 | 168 | 7 | 10 | 60 | 1 | 8 | 48 | 0 | |||||
| 7 | 22 | 154 | 6 | 11 | 77 | 4 | 3 | 21 | 0 | |||||
| 8 | 14 | 112 | 5 | 10 | 80 | 3 | 4 | 32 | 1 | |||||
| 9 | 14 | 126 | 3 | 9 | 81 | 5 | 4 | 36 | 1 | |||||
| 10 | 11 | 110 | 3 | 9 | 90 | 1 | 1 | 10 | 0 | |||||
| 11 | 10 | 110 | 8 | 7 | 77 | 1 | 1 | 11 | 1 | |||||
| 12 | 9 | 108 | 3 | 2 | 24 | 0 | 4 | 48 | 0 | |||||
| 13 | 9 | 117 | 10 | 8 | 104 | 3 | 0 | 0 | 0 | |||||
| 14 | 6 | 84 | 6 | 5 | 70 | 5 | 1 | 14 | 0 | |||||
| 15 | 6 | 90 | 5 | 3 | 45 | 0 | 2 | 30 | 2 | |||||
| 16 | 0 | 0 | 0 | 4 | 64 | 2 | 2 | 32 | 1 | |||||
| 17 | 3 | 51 | 3 | 4 | 68 | 3 | 1 | 17 | 1 | |||||
| 18 | 2 | 36 | 5 | 0 | 0 | 0 | 2 | 36 | 2 | |||||
| 19 | 2 | 38 | 5 | 0 | 0 | 0 | 3 | 57 | 4 | |||||
| 20 | 0 | 0 | 0 | 3 | 60 | 2 | 0 | 0 | 0 | |||||
| 21 | 4 | 84 | 7 | 0 | 0 | 0 | 1 | 21 | 2 | |||||
| 22 | 6 | 132 | 8 | 2 | 44 | 4 | 1 | 22 | 2 | |||||
| 23 | 2 | 46 | 2 | 1 | 23 | 0 | 0 | 0 | 0 | |||||
| 24 | 3 | 72 | 3 | 0 | 0 | 0 | 2 | 48 | 4 | |||||
| 25 | 1 | 25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 26 | 2 | 52 | 5 | 1 | 26 | 1 | 0 | 0 | 0 | |||||
| 27 | 1 | 27 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 28 | 3 | |||||
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 29 | 1 | |||||
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 32 | 1 | 32 | 0 | 0 | 0 | 0 | 1 | 32 | 3 | |||||
| 33 | 1 | 33 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 35 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 35 | 1 | |||||
| 36 | 1 | 36 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 37 | 1 | 37 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 38 | 1 | 38 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 39 | 0 | 0 | 0 | 1 | 39 | 3 | 1 | 39 | 4 | |||||
| 40 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| 42 | 2 | 84 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Total | 425 | 2,581 | 123 | 193 | 1,299 | 44 | 114 | 781 | 34 | |||||
Datasets are restricted to sequences containing the full-length 492 bp.
Pols ι and ζ cooperate at the Ig loci
Although the absence of functional Pol ι had no effect on the frequency or spectra of mutation, it did affect the signature of Pol ζ. In yeast and mice, adjacent mutations were abolished in the absence of Pol ζ, and elevated in the presence of a mutagenic form of the polymerase (Daly et al., 2012; Stone et al., 2012). Using a computational model to assess if tandem mutations are produced during a single DNA transaction or during two independent events, we show that Poli+/+ mice have a highly significant excess of tandem substitutions, implying that many of them are synthesized simultaneously. To estimate the frequency of tandem mutations that result from a single synthesis event, the mean number of expected tandems (82) was subtracted from the observed number (123) to acquire the number of single transaction events. This number was then compared with the total mutations (2,581), to show that tandem events make up 1.6% of mutations. This contribution to somatic hypermutation is small, which explains why it was missed before (McDonald et al., 2003; Martomo et al., 2006). Nonetheless, the value is reduced in Poli129/129 and Polim/m mice to 0.7 and 0.5%, respectively. Thus, Pol ι-compromised mice lost most of the tandem signature, suggesting that this polymerase plays a role with Pol ζ to produce the double substitutions.
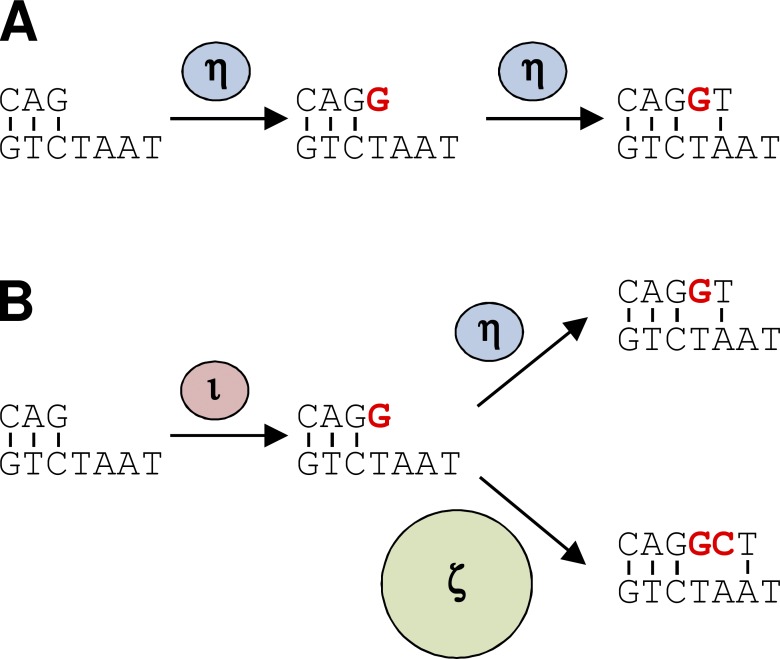
To model this in Fig. 4 A, Pol η is the dominant polymerase in mutation synthesis, and generates mostly single mutations. Pol η does not convert these to tandems because Pol η–deficient mice and yeast have an abundance of tandems (Saribasak et al., 2012; Stone et al., 2012). In Fig. 4 B, we propose that occasionally Pol ι accesses a repair gap, and due to its low fidelity and distributive synthesis, it fills in one or two bases and then dissociates from the template after introducing a mismatch. This predicts that the first mutation in a tandem pair should have the error signature of Pol ι. To enrich for tandems that have likely resulted from one synthesis event, we examined sequences with ≤5 total mutations. The first mutation in five of seven sequences containing a tandem pair had an A to G or A to T substitution. This error signature corresponds to the two most probable mutations generated by Pol ι in vitro (Tissier et al., 2000), supporting a role for Pol ι in incorporating the first mismatch. This mispair can be extended by either Pol η, which faithfully inserts the next nucleotide, or Pol ζ, which has a unique catalytic property that allows it to synthesize a second misinsertion, and then extend from the double mismatch. Could Pol ι insert the second mismatch? Analysis of mismatch extension by Pol ι suggests that it can generate a second mismatch only in a specific sequence context (Vaisman et al., 2001). However, Pol ζ can generate the second mismatch and extend from that mismatch without an apparent sequence bias (Zhong et al., 2006).
Figure 4.
Two-step model of Pol ι and Pol ζ in tandem genesis. (A) Pol η generates mutations (red) when copying A:T bp and extends the single mismatch with the correct base. (B) Occasionally, Pol ι synthesizes a mutation, falls off, and Pol η extends the single mispair. Alternatively, Pol ζ synthesizes a second mutation, and extends the double mismatch with the correct base, producing a tandem mutation.
A role for Pol ι in somatic hypermutation has been proposed in a BL2 human cell line that was deficient for the polymerase, and had decreased mutation frequency (Faili et al., 2002), and it was suggested that Pol ι is involved in the UNG pathway in these cells (Weill and Reynaud, 2008). However, in mice deficient for the polymerase, there was no decrease in the frequency of mutation. Further, our data indicate a role for Pol ι in the MSH2–MSH6 pathway, as tandem mutations are eliminated in Msh2−/− or Msh6−/− mice (Saribasak et al., 2012). It is unclear if these conflicting results are caused by a species difference or by a unique feature of the BL2 cell line.
A biological role for the three translesion polymerases is evident in mice deficient for the enzymes. Pol η–deficient mice have increased levels of sensitivity to ultraviolet irradiation (Lin et al., 2006), Pol ζ–deficient mice sustain developmental defects (Wittschieben et al., 2000), and Pol ι–deficient mice accumulate mesenchymal tumors (Ohkumo et al., 2006). An immunological role for Pol ι may reside in its ability to participate in changing two bases in a codon during one mutagenic DNA synthesis reaction, to rapidly increase the pool of diverse antibodies for selection by antigen. The interplay of these three polymerases in the minefield of AID-induced lesions in variable regions raises interesting questions about hierarchy, repair, and mutagenesis.
MATERIALS AND METHODS
Protein expression and purification
Full length N-terminal His-tagged human Pol ι was expressed using the E. coli strain RW644 and expression plasmid pJM868, as previously described (Frank et al., 2012). The Pol ι mutant was generated by chemically synthesizing a 615-bp BglII–HindIII fragment encoding the N terminus of the E. coli codon-optimized Poli gene with the D126A-E127A substitution (Genscript). The fragment was subsequently subcloned as a BglII–HindIII fragment into plasmid pCT14 (Donigan et al., 2014), and the resulting plasmid, pKD005, was grown in RW644. Pol ι and Pol ιm proteins were purified and determined to be >95% pure by SDS-PAGE analysis.
Primer extension and EMSA
Primer extension reactions (10 µl) contained 2 nM polymerase, 10 nM 32P-labeled 16-mer primer annealed to a 28-mer template, T10AGC (5′-GCAAAAAAAAAAAAGCACGTCCGTACCA-3′; the position of the primer is underlined), and 100 µM nucleotides in reaction buffer (0.5 mM MnCl2, 40 mM Tris-HCl, pH 8.0, 10 mM dithiothreitol, 250 µg/ml bovine serum albumin, 2.5% glycerol, and 10 mM 2-mercaptoethanol). DNA substrates were confirmed to be >95% annealed by incubating with DNA Pol I Klenow fragment exo− (New England Biolabs). The reactions were then incubated at 37°C for 20 min, and quenched by the addition of 10 µl of 95% formamide and 10 mM EDTA. Samples were heated to 100°C for 5 min, and resolved on an 18% polyacrylamide-urea gel. Reaction products were visualized and quantified using a Fuji FLA-5100 Phosphorimager and ImageGauge software. EMSA gels were used to determine the DNA binding constants (KD(DNA)) for Pol ι and Pol ιm (Donigan et al., 2014). Serial dilutions of the enzymes (0.2–819 nM for Pol ι and 0.2–429 nM for Pol ιm) were incubated with radiolabeled DNA substrate in reaction buffer and separated by native gel electrophoresis. The fraction of bound DNA was quantified relative to protein concentration, as previously described (Donigan et al., 2014).
Replication foci
The Poli gene was codon optimized for maximal expression in human cells. The ∼2.2-kb gene was synthesized (Genscript) and subcloned as a XhoI–BamHI fragment into pEGFP-C1 (Invitrogen), to generate EGFP-Pol ι (pDH24). The Polim gene was constructed by synthesizing an ∼500-bp XhoI–PmlI fragment containing D126A and E127A substitutions, as well as silent mutations that generated a unique BstXI restriction enzyme site for subsequent subclone identification. The fragment was cloned as pEGFP-Pol ιm (pDH33), and the insert was confirmed by digestion with BstXI. HEK293T cells (ATCC) were plated onto Number 1 coverslips to be fixed and mounted onto slides after treatments. These plasmids were then transfected into HEK293T cells using TurboFectin 8.0, according to the manufacturer's protocol (OriGene). Cells were fixed using 2% formaldehyde (16% methanol-free formaldehyde; Polysciences, Inc.) in PBS, and mounted onto slides using Mowiol mounting medium. Fluorescence images of cell nuclei were acquired on an Axiovert 40 CFL (Zeiss) with an X-cite Series 120Q light source using Zen 2 (blue edition) software.
Polim/m mouse generation and characterization
The Polim allele was generated in a C57BL/6 background (Ozgene Pty Ltd.) using standard genetic manipulation techniques. The mutant allele was distinguished from the wild-type allele by PCR amplification and subsequent digestion with TseI restriction enzyme (New England Biolabs). To this end, genomic tail DNA was amplified with forward 5′-ACACACACAATACTACACA-3′ and reverse 5′-AAGCTGCTGGAGTCTTTT-3′ primers to generate a 320-bp fragment. The product was then digested with TseI, and separated on an agarose gel to visualize products. The 320-bp wild-type band was not cut, whereas the mutant band generated 270- and 50-bp fragments. For Western blots, protein extracts from mouse testes were prepared and analyzed with affinity-purified rabbit anti-Pol ι (McDonald et al., 2003) and mouse anti–β-actin (clone AC-15; Sigma-Aldrich), followed by goat anti–rabbit or goat anti–mouse IgG peroxidase antibodies (Sigma-Aldrich), respectively. Chemiluminescent detection was performed using ECL plus Western blotting substrate (Thermo Fisher Scientific).
Homozygous Polim/m and C57BL/6 mice were bred in the NIA mouse facility. 129X1/SvJ mice were purchased from The Jackson Laboratory. Mice of both genders were used at 2–6 mo of age. All animal procedures were reviewed and approved by the Animal Care and Use Committee of the National Institute on Aging.
Somatic hypermutation
Lymphocytes were isolated from Peyer’s patches and stained with FITC-labeled anti-B220 (clone RA3-6B2; eBioscience) and Alexa Fluor 647–labeled anti-GL7 (clone GL7; BioLegend) antibodies, followed by cell sorting for the B220+ GL7+ germinal center B cell population. Cells were then lysed in digestion buffer (10 mM Tris, pH 8.0, 25 mM EDTA, 100 mM NaCl, 1% SDS, and 0.1 mg/ml proteinase K) at 55°C overnight. Genomic DNA was isolated by phenol/chloroform extraction and ethanol precipitation. The JH4 intronic region was amplified and analyzed as previously described (Saribasak et al., 2012). For mutation frequency and spectra, mutations were analyzed from previously published data (2,866 mutations) from C57BL/6 mice (Rada et al., 2004; Martomo et al., 2008; Schenten et al., 2009; Saribasak et al., 2011; Zanotti et al., 2015). Poli129/129 data were from McDonald et al. (2003) and data generated in this study, and Polim/m data were generated in-house.
Tandem modeling
For analysis of tandems, a subset of sequences containing only full-length clones was used, and is listed in Table 1. The method starts with the nonmutated germline sequence, together with the mutation frequencies observed at each site in the C57BL/6 dataset (Table S1). The general idea was to use the expected mutation frequencies to generate simulated datasets that represent a null model, which could then be compared with a target dataset, usually from a mutant. In preparation for the simulation, probabilities for mutating each site were calculated such that they were proportional to the mutation frequency at that site. The R function “sample” was then used to generate the simulated mutations at these sites according to the assigned probabilities. When we compared simulated data with a target dataset, we ensured that the simulated dataset contained the same number of sequences and equal numbers of mutations per sequence as the target dataset, to avoid any bias in the simulated datasets. Simulated datasets that, for example, contain higher numbers of mutations per sequence would be biased toward having more tandem mutations. Because we were interested specifically in measuring the number of tandem mutations, only sequences containing at least two mutations in the target dataset were considered. As shown schematically in Fig. 3 B, the number of tandem mutations in each simulated dataset was counted. For each target dataset we generated a total of 100,000 simulated datasets, and the number of tandem mutations gave us an expected, or null model, distribution of tandem mutations, after normalization. We then considered how the number of tandem mutations observed in the target dataset (for example, observed, in Fig. 3 C) compared with this null model distribution. The p-value represents the fraction of simulations (out of 100,000) in which the number of expected tandems was greater than, plus half of the number equal, to the observed value.
Online supplemental material
Table S1 lists the mutation frequency at individual nucleotides in the JH4 intron. Data were accumulated from published C57BL/6 sequences containing 2,866 mutations within the 492-bp region. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151227/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Cao for technical support, and R. Wersto, C. Nguyen, and T. Wallace of the National Institute on Aging flow cytometry unit for sorting. We gratefully acknowledge Ranjan Sen for critical reading of the manuscript.
This work was supported in part by National Institutes of Health (NIH) grant GM111741 to T. MacCarthy, and the Intramural Research Programs of the National Institute of Child Health and Human Development to R. Woodgate, and National Institute on Aging to P.J. Gearhart.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- Pol
- DNA polymerase
- UNG
- uracil DNA glycosylase
References
- Aoufouchi S., De Smet A., Delbos F., Gelot C., Guerrera I.C., Weill J.C., and Reynaud C.A.. 2015. 129-Derived mouse strains express an unstable but catalytically active DNA polymerase iota variant. Mol. Cell. Biol. 35:3059–3070. 10.1128/MCB.00371-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell P.D., Woo C.J., Wei K., Li Z., Martin A., Sack S.Z., Parris T., Edelmann W., and Scharff M.D.. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5:224–229. 10.1038/ni1031 [DOI] [PubMed] [Google Scholar]
- Daly J., Bebenek K., Watt D.L., Richter K., Jiang C., Zhao M.L., Ray M., McGregor W.G., Kunkel T.A., and Diaz M.. 2012. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity. J. Immunol. 188:5528–5537. 10.4049/jimmunol.1102629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F., Aoufouchi S., Faili A., Weill J.C., and Reynaud C.A.. 2007. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 204:17–23. 10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan K.A., McLenigan M.P., Yang W., Goodman M.F., and Woodgate R.. 2014. The steric gate of DNA polymerase ι regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J. Biol. Chem. 289:9136–9145. 10.1074/jbc.M113.545442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A., Aoufouchi S., Flatter E., Guéranger Q., Reynaud C.A., and Weill J.C.. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 419:944–947. 10.1038/nature01117 [DOI] [PubMed] [Google Scholar]
- Frank E.G., McDonald J.P., Karata K., Huston D., and Woodgate R.. 2012. A strategy for the expression of recombinant proteins traditionally hard to purify. Anal. Biochem. 429:132–139. 10.1016/j.ab.2012.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., and de Wind N.. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203:319–323. 10.1084/jem.20052227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Washington M.T., Haracska L., Prakash S., and Prakash L.. 2000. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 406:1015–1019. 10.1038/35023030 [DOI] [PubMed] [Google Scholar]
- Kannouche P., Fernández de Henestrosa A.R., Coull B., Vidal A.E., Gray C., Zicha D., Woodgate R., and Lehmann A.R.. 2003. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 22:1223–1233. 10.1093/emboj/7595006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Clark A.B., McCulloch S.D., Yuan T., Bronson R.T., Kunkel T.A., and Kucherlapati R.. 2006. Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res. 66:87–94. 10.1158/0008-5472.CAN-05-1862 [DOI] [PubMed] [Google Scholar]
- MacCarthy T., Kalis S.L., Roa S., Pham P., Goodman M.F., Scharff M.D., and Bergman A.. 2009. V-region mutation in vitro, in vivo, and in silico reveal the importance of the enzymatic properties of AID and the sequence environment. Proc. Natl. Acad. Sci. USA. 106:8629–8634. 10.1073/pnas.0903803106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova A.V., Grabow C., Gening L.V., Tarantul V.Z., Tahirov T.H., Bessho T., and Pavlov Y.I.. 2011. Inaccurate DNA synthesis in cell extracts of yeast producing active human DNA polymerase iota. PLoS One. 6:e16612 10.1371/journal.pone.0016612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Wersto R.P., Ohkumo T., Kondo Y., Yokoi M., Masutani C., Hanaoka F., and Gearhart P.J.. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA. 102:8656–8661. 10.1073/pnas.0501852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Vaisman A., Maas A., Yokoi M., Hoeijmakers J.H., Hanaoka F., Woodgate R., and Gearhart P.J.. 2006. Normal hypermutation in antibody genes from congenic mice defective for DNA polymerase iota. DNA Repair (Amst.). 5:392–398. 10.1016/j.dnarep.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Martomo S.A., Saribasak H., Yokoi M., Hanaoka F., and Gearhart P.J.. 2008. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst.). 7:1603–1608. 10.1016/j.dnarep.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul R.W., Saribasak H., Martomo S.A., McClure R.L., Yang W., Vaisman A., Gramlich H.S., Schatz D.G., Woodgate R., Wilson D.M. III, and Gearhart P.J.. 2011. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat. Immunol. 12:70–76. 10.1038/ni.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Frank E.G., Plosky B.S., Rogozin I.B., Masutani C., Hanaoka F., Woodgate R., and Gearhart P.J.. 2003. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J. Exp. Med. 198:635–643. 10.1084/jem.20030767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre J., Vidal A.E., McLenigan M.P., Bomar M.G., Curti E., McDonald J.P., Plosky B.S., Ohashi E., and Woodgate R.. 2013. Ubiquitin mediates the physical and functional interaction between human DNA polymerases η and ι. Nucleic Acids Res. 41:1649–1660. 10.1093/nar/gks1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkumo T., Kondo Y., Yokoi M., Tsukamoto T., Yamada A., Sugimoto T., Kanao R., Higashi Y., Kondoh H., Tatematsu M., et al. 2006. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol. Cell. Biol. 26:7696–7706. 10.1128/MCB.01076-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., and Neuberger M.S.. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. 10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., and Neuberger M.S.. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Saribasak H., Maul R.W., Cao Z., McClure R.L., Yang W., McNeill D.R., Wilson D.M. III, and Gearhart P.J.. 2011. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J. Exp. Med. 208:2209–2216. 10.1084/jem.20111135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribasak H., Maul R.W., Cao Z., Yang W.W., Schenten D., Kracker S., and Gearhart P.J.. 2012. DNA polymerase ζ generates tandem mutations in immunoglobulin variable regions. J. Exp. Med. 209:1075–1081. 10.1084/jem.20112234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D., Kracker S., Esposito G., Franco S., Klein U., Murphy M., Alt F.W., and Rajewsky K.. 2009. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 206:477–490. 10.1084/jem.20080669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Linehan E.K., Thompson M.R., Habboub G., Ucher A.J., Kadungure T., Tsuchimoto D., Nakabeppu Y., and Schrader C.E.. 2014. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc. Natl. Acad. Sci. USA. 111:9217–9222. 10.1073/pnas.1405590111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.E., Lujan S.A., Kunkel T.A., and Kunkel T.A.. 2012. DNA polymerase zeta generates clustered mutations during bypass of endogenous DNA lesions in Saccharomyces cerevisiae. Environ. Mol. Mutagen. 53:777–786. 10.1002/em.21728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A., McDonald J.P., Frank E.G., and Woodgate R.. 2000. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Vaisman A., Tissier A., Frank E.G., Goodman M.F., and Woodgate R.. 2001. Human DNA polymerase iota promiscuous mismatch extension. J. Biol. Chem. 276:30615–30622. 10.1074/jbc.M102694200 [DOI] [PubMed] [Google Scholar]
- Weill J.C., and Reynaud C.A.. 2008. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 8:302–312. 10.1038/nri2281 [DOI] [PubMed] [Google Scholar]
- Wiesendanger M., Kneitz B., Edelmann W., and Scharff M.D.. 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 191:579–584. 10.1084/jem.191.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J., Shivji M.K., Lalani E., Jacobs M.A., Marini F., Gearhart P.J., Rosewell I., Stamp G., and Wood R.D.. 2000. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 10:1217–1220. 10.1016/S0960-9822(00)00725-9 [DOI] [PubMed] [Google Scholar]
- Zanotti K.J., Maul R.W., Castiblanco D.P., Yang W., Choi Y.J., Fox J.T., Myung K., Saribasak H., and Gearhart P.J.. 2015. ATAD5 deficiency decreases B cell division and Igh recombination. J. Immunol. 194:35–42. 10.4049/jimmunol.1401158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Winter D.B., Kasmer C., Kraemer K.H., Lehmann A.R., and Gearhart P.J.. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. 10.1038/88740 [DOI] [PubMed] [Google Scholar]
- Zhong X., Garg P., Stith C.M., Nick McElhinny S.A., Kissling G.E., Burgers P.M., and Kunkel T.A.. 2006. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 34:4731–4742. 10.1093/nar/gkl465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O., Geacintov N., Nakajima S., Yasui A., and Livneh Z.. 2009. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc. Natl. Acad. Sci. USA. 106:11552–11557. 10.1073/pnas.0812548106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.