Car enzyme inhibition prevents mast cell responses and inflammation following Trichinella spiralis infection or the induction of food allergy–like disease.
Abstract
Type 2 cytokine responses are necessary for the development of protective immunity to helminth parasites but also cause the inflammation associated with allergies and asthma. Recent studies have found that peripheral hematopoietic progenitor cells contribute to type 2 cytokine–mediated inflammation through their enhanced ability to develop into mast cells. In this study, we show that carbonic anhydrase (Car) enzymes are up-regulated in type 2–associated progenitor cells and demonstrate that Car enzyme inhibition is sufficient to prevent mouse mast cell responses and inflammation after Trichinella spiralis infection or the induction of food allergy–like disease. Further, we used CRISPR/Cas9 technology and illustrate that genetically editing Car1 is sufficient to selectively reduce mast cell development. Finally, we demonstrate that Car enzymes can be targeted to prevent human mast cell development. Collectively, these experiments identify a previously unrecognized role for Car enzymes in regulating mast cell lineage commitment and suggest that Car enzyme inhibitors may possess therapeutic potential that can be used to treat mast cell–mediated inflammation.
INTRODUCTION
Type 2 cytokine responses are characterized by the development of T helper type 2 cells (Th2 cells), IL-4, -5, -9, and -13 expression, basophil and mast cell responses, and increased IgE production. Type 2 cytokines are necessary for the development of protective immunity to helminth parasites and also promote the pathology associated with allergies and asthma (Allen and Maizels, 2011; Pulendran and Artis, 2012). Helminth parasites infect an estimated 2 billion people and cause anemia, retarded growth, and exert enormous economic burdens on heavily infected areas (Allen and Maizels, 2011). Allergic diseases including food allergies have risen to epidemic proportions in developed areas of the world and result in significant morbidity and even death (Pulendran and Artis, 2012). Current therapeutic strategies to treat helminth infections and allergic inflammation are limited by our incomplete understanding of the events that promote type 2 inflammation (Pulendran and Artis, 2012).
An emerging body of literature has identified that type 2 inflammation can be promoted by specialized progenitor cells that enter peripheral tissues and undergo in situ hematopoiesis. These studies demonstrate that lineage negative, CD34+, c-Kit+ hematopoietic progenitors accumulate in peripheral tissues after stimulation with cytokine alarmins, exposure to helminths, or the initiation of allergic inflammation (Saenz et al., 2010; Siracusa et al., 2013). The conserved presence of these progenitors in the context of type 2 responses allows them to be characterized as type 2 associated. It has been shown that type 2 progenitors promote inflammation via their enhanced ability to develop into mast cells compared with phenotypically similar BM-resident progenitors (Siracusa et al., 2013). Collectively, these studies suggest that targeting progenitors to prevent mast cell differentiation may be sufficient to regulate type 2 inflammation.
Here, we show that type 2 progenitors express elevated levels of carbonic anhydrase (Car) enzymes. Car enzymes are a family of metabolic enzymes that regulate pH and CO2 homeostasis and have been targeted to treat diseases including glaucoma and gastric ulcers (Supuran, 2008). Car enzymes have also been associated with type 2 cytokine responses (Kamsteeg et al., 2007; Wen et al., 2014). Our experiments demonstrate that pan-inhibition of Car enzymes with an FDA-approved inhibitor was sufficient to prevent inflammatory mast cell responses in the context of helminth infection and models of allergic inflammation. In contrast, Car enzyme inhibition promoted immunity to Aspergillus fumigatus, suggesting that Car enzymes can selectively affect specific immune modules. Furthermore, selectively editing Car1 via CRISPR/Cas9 was sufficient to reduce mast cell development from stem cells but had no effect on macrophage commitment. Finally, we demonstrate that Car enzyme inhibition was also sufficient to prevent human mast cell development. Collectively, these studies identify a previously unappreciated mechanism through which mammalian immune cells are instructed by inflammatory cues and provide insight into the therapeutic potential of targeting Car1 to treat mast cell–mediated inflammation.
RESULTS AND DISCUSSION
Mast cells express elevated levels of Car enzymes
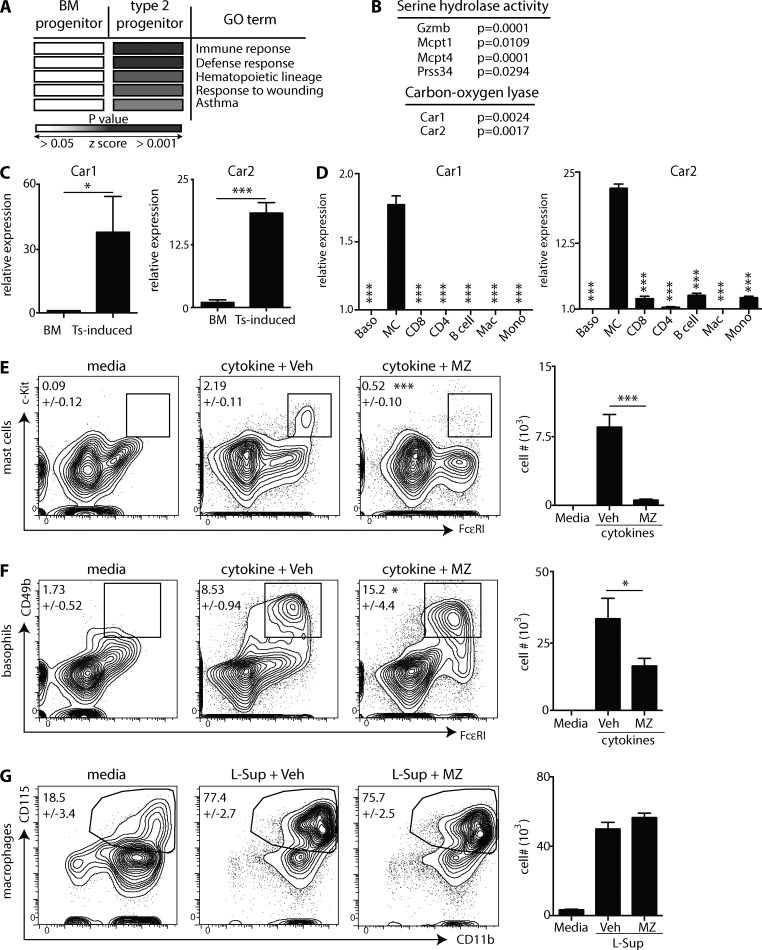
We first compared the transcriptional profiles of naive BM-resident progenitors to those of type 2 progenitors (Siracusa et al., 2013) to identify fundamental pathways associated with mast cell development. The top 200 genes expressed at higher levels in type 2 progenitors were run through pathway analysis (Dennis et al., 2003). Type 2 progenitors were enriched for genes associated with immune and defense responses, hematopoietic lineages, responses to wounding, and asthma (Fig. 1 A). Type 2 progenitors also expressed genes associated with serine hydrolases, known to be present in mast cells (Long and Cravatt, 2011), and carbon–oxygen lyase activity (Fig. 1 B). The carbon–oxygen lyase pathway was comprised of the genes encoding Car enzymes 1 and 2. To confirm these data, we sort purified Trichinella spiralis–induced progenitors from infected mice (Siracusa et al., 2013) and compared Car1 and 2 expression to that of naive BM progenitors. The genes encoding Car1 and 2 were expressed at significantly higher levels in T. spiralis–induced progenitors (Fig. 1 C). Collectively, these data demonstrate that Car1 and 2 are highly expressed in mast cell precursors. Next, we sort purified BM-derived basophils and mast cells, CD8 T cells, CD4 T cells, B cells, macrophages, and monocytes and evaluated their expression levels of Car1 and 2. Car1 and 2 were expressed at significantly higher levels in mature mast cells compared with other populations (Fig. 1 D). These data provoke the hypothesis that Car1 and 2 may regulate mast cell development.
Figure 1.
Mast cell development is associated with increased expression of Car enzymes. Transcriptional profiles of BM-resident or type 2 progenitors were compared. (A and B) Enriched pathways in type 2 progenitors were identified, and gene ontology (GO) terms were listed. (C) Progenitors were purified from the BM of naive mice or the spleens of T. spiralis (Ts)-infected mice, and gene expression was determined. (D) Cell populations were purified from naive mice or cell cultures, and gene expression (relative to basophils [Baso]) was determined. Mac, macrophage; MC, mast cell; Mono, monocyte. (E–G) BM-resident progenitors were isolated and cultured with IL-3 or L cell culture supernatant (L-sup) in the presence of vehicle (Veh) or MZ, and mast cells (E), basophils (F), and macrophages (G) were quantified. (C) Results are representative of three to five pooled biological replicates. (D) Results are representative of four biological replicates. (E-G) Results are representative of at least three individual experiments. Numbers in cytometry plots represent the percentage of cells gated. Statistical analysis was performed using a Student’s t test. *, P < 0.05; ***, P < 0.001. Error bars represent SD.
Car enzyme inhibitors prevent in vitro mast cell development
The Car enzyme inhibitor methazolamide (MZ) has been shown to inhibit Car1 and 2 (Supuran, 2008). Therefore, we used mast cell developmental assays (Saenz et al., 2010; Siracusa et al., 2011, 2013) to determine whether Car enzyme inhibition affected mast cell development. Critically, cultures treated with MZ exhibited significantly reduced mast cell populations compared with controls (Fig. 1 E). In addition to mast cells, type 2 progenitors are also capable of developing into basophils (Siracusa et al., 2013). Therefore, we sought to determine whether Car enzyme inhibition altered basophil development. Although treatment of cultures with MZ resulted in significantly increased percentages of basophils (Fig. 1 F), Car enzyme inhibition resulted in decreased basophil numbers. Next, we sought to determine whether Car enzyme inhibition alters the development of other myeloid cell populations (Marim et al., 2010). Importantly, treatment with MZ had no effect on macrophage development (Fig. 1 G).
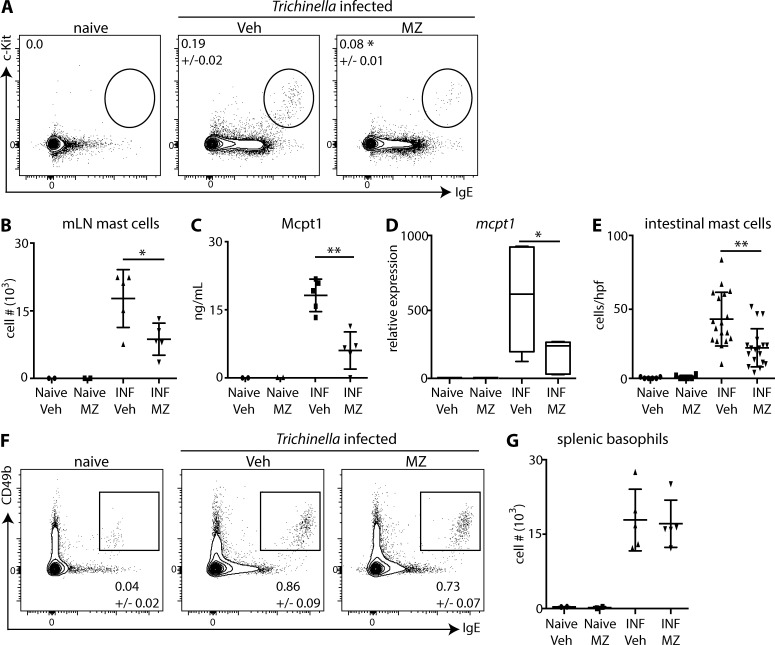
Inhibition of Car enzymes is sufficient to prevent infection-induced mastocytosis
Protective immunity to T. spiralis is dependent on type 2 cytokine responses and is associated with the population expansion of mast cells and basophils (Urban et al., 2000; Giacomin et al., 2012). Therefore, T. spiralis–infected mice were treated with vehicle or MZ, and mast cells and basophils were monitored. T. spiralis infection resulted in increased percentages and numbers of mast cells in the mesenteric LNs (mLNs; Fig. 2, A and B), increased serum levels of mast cell protease 1 (Mcpt1; Fig. 2 C), increased expression of intestinal mcpt1 (Fig. 2 D), and increased intestinal mast cells identified by histological analysis (Fig. 2 E). Critically, treatment of mice with MZ resulted in significantly reduced mast cell responses across these parameters (Fig. 2, A–E). In contrast to mast cells, treatment of T. spiralis–infected mice with MZ had no effect on infection-induced basophilia (Fig. 2, F and G). These experiments suggest that the moderate effects of Car enzyme inhibition on in vitro basophil development (Fig. 1 F) may be a result of its actions on a limited pool of common progenitors shared between basophils and mast cells (Voehringer, 2013).
Figure 2.
Car enzymes regulate infection-induced mastocytosis. T. spiralis–infected (INF) mice were treated with Veh or MZ, and mast cells in the mLNs (A and B), serum levels of Mcpt1 (C), intestinal expression of mcpt1 (D), intestinal mast cells per high-power field (hpf) in histological sections (E), and splenic basophils (F and G) were evaluated. Results are representative of at least three individual experiments with at least two mice per naive group and at least three mice per T. spiralis–infected group. Numbers in cytometry plots represent the percentage of cells gated. Statistical analysis was performed using a Student’s t test. *, P < 0.05; **, P < 0.01. Error bars represent SD.
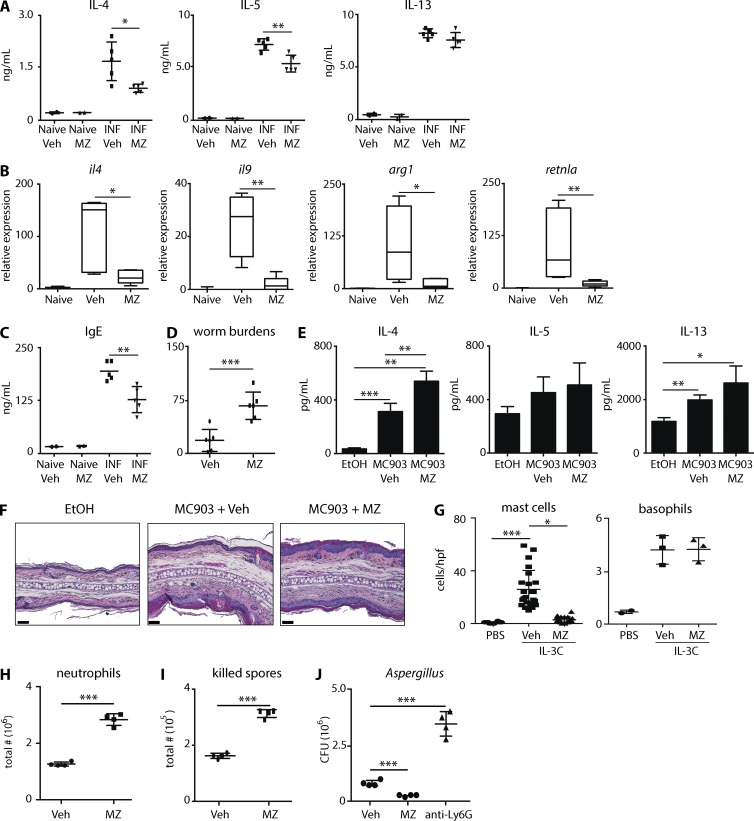
Mast cells have been shown to play critical roles in the development of protective immunity to T. spiralis (Urban et al., 2000; Voehringer, 2013). Therefore, we sought to determine whether the inhibition of T. spiralis–induced mast cell responses correlated with reduced inflammation and immunity. As expected (Urban et al., 2000), T. spiralis–infected mice exhibited increased levels of IL-4, IL-5, and IL-13 production from mLN cells (Fig. 3 A), increased intestinal expression of il4, il9, arg1, and retnla (Fig. 3 B), and elevated serum levels of IgE (Fig. 3 C). Critically, T. spiralis–infected mice treated with MZ exhibited significantly reduced type 2 responses as measured by these parameters (Fig. 3, A–C). Moreover, significantly more worms were recovered from the intestines of mice treated with MZ (Fig. 3 D). These data demonstrate that Car enzyme–mediated inhibition of mast cell responses is associated with reduced inflammation and a loss of protective immunity.
Figure 3.
Inhibition of Car enzymes is sufficient to inhibit mast cell–mediated inflammation. T. spiralis–infected mice (INF) were treated with Veh or MZ. (A–D) Isolated mLN cells were stimulated and levels of IL-4, IL-5, and IL-13 (A), intestinal expression of il4, il9, arg1, and retnla (B), serum IgE levels (C), and worm burdens (D) were evaluated. (E and F) Mice were treated with ethanol (EtOH) or MC903 and Veh or MZ. Skin-draining LN cells were stimulated, and levels of IL-4, IL-5, and IL-13 (E) and skin pathology (F) were evaluated. Error bars represent SD. Bars, 50 µm. (G) Rag1−/− mice were treated with PBS or IL-3C and Veh or MZ. Intestinal mast cells and splenic basophils were evaluated. hpf, high-power field. (H-J) A. fumigatus–infected mice were treated with Veh, MZ, or anti-Ly6G, and neutrophil responses and lung fungal burdens were determined. Results are representative of at least three independent experiments with at least two mice per naive group and at least three mice per experimental group. Statistical analysis was performed using a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The ability of MZ to regulate inflammation could be a result of its effects on mast cells, lymphocytes, or global immunosuppressive qualities. Therefore, we first used a model of atopic dermatitis–like disease in which type 2 cytokine responses and pathology are dependent on group 2 innate lymphoid cells (ILC2) and basophils (Kim et al., 2013, 2014a). Critically, mice treated with MC903 in the presence of MZ exhibited equivalent or significantly increased type 2 cytokine production and similar atopic dermatitis–like pathology as controls (Fig. 3, E and F). These data demonstrate that Car enzyme inhibition does not result in reduced ILC2- and basophil-dependent type 2 cytokine responses.
Mast cells are reported to operate as initiators of T. spiralis–induced Th2 cell development (Knight et al., 2000; Urban et al., 2000) but are also supported by T cell activation (Voehringer, 2013). Therefore, we treated Rag1–deficient mice with IL-3–anti–IL-3 complexes (IL-3C) and monitored intestinal mast cell responses and peripheral basophilia (Finkelman et al., 1993; Ohmori et al., 2009). MZ treatment resulted in significantly reduced mast cell responses but had no effect on IL-3C–induced basophilia in Rag1-deficient animals (Fig. 3 G). These data demonstrate that Car enzyme inhibition is sufficient to selectively inhibit mast cell responses in lymphocyte-sufficient and -deficient environments.
Next, we sought to determine whether Car enzyme inhibition promotes immunosuppressive qualities. A. fumigatus is an opportunistic pathogen that thrives in immunosuppressed hosts (Espinosa et al., 2014). Infecting mice with fluorescent A. fumigatus reporter conidia (Espinosa et al., 2014) allows live and dead spores to be identified in vivo. Neutrophil responses and fungal clearance were increased in MZ-treated mice (Fig. 3, H–J). Collectively, these data demonstrate that Car enzyme inhibition does not promote broad immunosuppressive effects but rather enhances antifungal immunity.
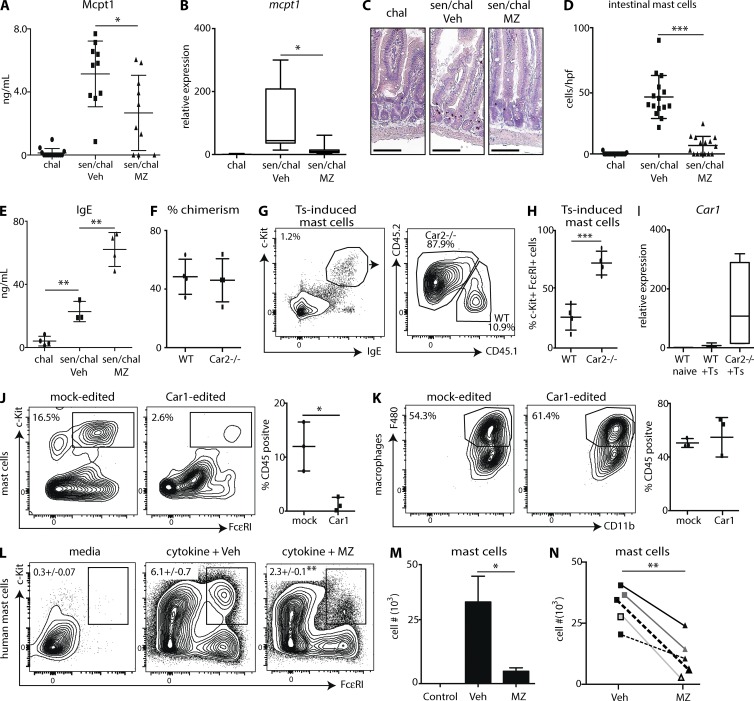
Inhibition of Car enzymes is sufficient to prevent food allergy–induced mast cell responses
Food allergies are a significant and growing public health concern (Sicherer and Sampson, 2014). Mast cells are critically important to the development of intestinal inflammation after sensitization and challenge with food antigens (Wang et al., 2010). Therefore, we used a model of food allergy–like disease that promotes intestinal mastocytosis (Noti et al., 2014) and treated mice with vehicle or MZ. Mice sensitized and challenged with chicken ovalbumin, a food allergen found in eggs (Dang et al., 2014), exhibited increased serum levels of Mcpt1 (Fig. 4 A), elevated intestinal expression of mcpt1 (Fig. 4 B), increased intestinal mast cells identified by histological analysis (Fig. 4, C and D), and increased serum levels of IgE (Fig. 4 E). MZ-treated mice exhibited significantly decreased mast cell responses across all of these parameters (Fig. 4, A–D) but had significantly increased IgE levels (Fig. 4 E). Collectively, these experiments demonstrate that pharmacologic inhibition of Car enzymes is sufficient to significantly reduce food allergy–induced mast cell responses. Furthermore, the increased levels of IgE observed in MZ-treated mice suggest this treatment is not inhibiting lymphocyte activation. The increases observed are likely a result of reductions in mast cell populations capable of binding soluble IgE.
Figure 4.
Car enzymes regulate human mast cell development. OVA challenged (chal) or sensitized and challenge (sen/chal) mice were treated with Veh or MZ. Serum Mcpt1 (A), intestinal expression of mcpt1 (B), intestinal mast cell populations in histological sections (C and D), and serum IgE levels (E) were evaluated. hpf, high-power field. Bars, 100 µm. (F) Mice were reconstituted with WT and Car2−/− BM (50/50), and the percent chimerism was determined. (G and H) Chimeric mice were infected with T. spiralis (Ts), and mLN mast cells were evaluated. (I) Mice reconstituted with 100% WT or 100% Car2−/− BM were infected with T. spiralis, and intestinal expression of Car1 was determined. (J and K) Mock-edited or Car1-edited stem cells were cultured under mast cell (J)– or macrophage (K)-promoting conditions. (L and M) Human CD34+ cells were cultured with mast cell–promoting cytokines in the presence of Veh or MZ. Representative analysis is from one donor. Error bars represent SD. (N) Pooled mast cell numbers from five individual donors. (A and B) Results are illustrated as the combined datasets of three independent experiments. (C–E) Results are representative of three independent experiments of at least two mice per challenged group and at least three mice per sensitized and challenged group. (F and H) Results are representative of two independent experiments of nine biological replicates total. (I) Results are representative of 2 independent experiments of 10 biological replicates total. (J and K) Results are representative of three independent experiments with at least three technical replicates per experiment. (L and M) Results illustrate three technical replicates from one human donor. (N) Data points represent individual human donors. n = 5. Numbers in cytometry plots represent the percentage of cells gated. Statistical analysis was performed using a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Car1 positively regulates mast cell development
We show that inhibition of Car enzyme family members via a pan inhibitor is sufficient to prevent helminth- or allergen-induced mastocytosis. However, whether specifically targeting Car1 or 2 is sufficient to regulate mast cell responses remains unknown. To address this, we lethally irradiated mice and reconstituted them with BM cells from WT and Car2−/− mice at a 50:50 ratio. Both WT and Car2−/− cells were present at equal percentages (Fig. 4 F); however, when chimeric mice were infected with T. spiralis, ∼80% of T. spiralis–induced mast cells were derived from Car2−/− cells (Fig. 4, G and H). These data countered our working hypothesis that Car1 and 2 promote mast cell development. It has been shown previously that deletion of one Car enzyme can result in the increased expression of other family members (Pan et al., 2006). Interestingly, we found that chimeric mice reconstituted with 100% Car2−/− cells expressed higher levels of Car1 after T. spiralis infection than mice reconstituted with WT BM (Fig. 4 I). Collectively, these data provoke the hypothesis that Car1 operates as a positive regulator of mast cell development.
Because Car1-deficient mice are not readily available, we used CRISPR/Cas9 technology to genetically edit Car1 in mouse embryonic stem cells (ESCs). Car1 was edited in ∼60% of ESCs electroporated with Car1-specific RNA guides and Cas9 protein (unpublished data). Furthermore, when Car1-edited stem cells were cultured, they exhibited a reduced capacity to develop into mast cells compared with controls (Fig. 4 J) but showed no defect in macrophage development (Fig. 4 K). Collectively, these experiments illustrate that CRISPR/Cas9-mediated editing of Car1 results in a selective reduction in mast cell hematopoietic potential. However, whether Car1 regulates mast cell development via its effects on other Car family members requires further examination.
Car enzyme inhibition is sufficient to prevent human mast cell development
Patients suffering from allergic diseases such as food allergies present with increased mast cell responses, suggesting that targeting mast cell development may represent a viable therapeutic strategy (Reimann and Lewin, 1988; Bischoff and Crowe, 2005; Ramsay et al., 2010; Voehringer, 2013). Here, we have identified that mouse Car1 operates as a positive regulator of mast cell development. However, whether Car enzymes can be targeted to prevent human mast cell development remains unknown. Therefore, we isolated CD34+ progenitor cells from the blood of healthy human donors and used an established protocol to grow human mast cells (Saito et al., 2006). Human cultures treated with MZ exhibited significantly reduced percentages and total numbers of mast cells (Fig. 4, L and M). The ability of MZ to prevent human mast cell development was found to be consistent across five individual donors (Fig. 4 N).
Recent studies have begun to introduce a model through which alterations in metabolic processes can instruct immune cell development and activation (Pearce and Pearce, 2013). Variations in pH often accompany infectious and inflammatory insults and can have potent effects on immune cell function (Lardner, 2001). The experiments presented here suggest that, similar to germline-encoded receptors that recognize pathogen-derived molecules (Tang et al., 2012), Car enzymes may operate as sensory mechanisms that instruct immune cell hematopoiesis under inflammatory conditions. This hypothesis is supported by a recent study identifying Car4 as the signature gene expressed by pulmonary macrophages (Lavin et al., 2014). Here, our data suggest that Car1 operates as a positive regulator of mast cell development and that targeting Car1 may prove to be effective in treating mast cell–mediated inflammation.
MATERIALS AND METHODS
Mice
C57BL(6) WT mice were purchased from The Jackson Laboratory. Mice were maintained in specific pathogen–free facilities at the Rutgers New Jersey Medical School. All protocols were approved by the Rutgers Institutional Animal Care and Use Committee. Car2-deficient mice on a C57BL(6) background were provided by M. Soleimani (University of Cincinnati, Cincinnati, OH).
Flow cytometry and cell sorting
Cells were stained with monoclonal anti–mouse fluorescently conjugated antibodies: Sca-1 (D7), c-Kit (ACK2), CD3 (145-2C11), CD4 (GK1.5), CD8 (SK1), CD5 (53-73), CD19 (1D3), NK1.1 (PK136), CD11b (MI/70), CD11c (N418), CD16(32) (93), IgE (23G3), FcεRI (MAR-1), CD49b (DX5), CD115 (AFS98), F4/80 (BM8), and CD34 (RAM34) from eBioscience; or monoclonal anti–human fluorescently conjugated antibodies: CD19 (HIB19), FcεRI (AER-37), CD34 (4H11), c-Kit (104D2), and TCRαβ (IP26) from eBioscience. Samples were acquired on an LSRII or LSRFORTESSA X-20 flow cytometer (BD) and analyzed using FlowJo software (v10.0.5; Tree Star). Cell sorting was performed using a FACSAria II flow cytometer (BD). BM-resident and splenic progenitors were sort purified as CD3−, CD19−, CD11b−, CD11c−, NK1.1−, Sca-1−, FcεRI−, CD34+, and c-Kit+ cells. Splenic progenitors were isolated on day 7 after T. spiralis infection.
Microarray and gene expression analysis
Genome-wide transcriptional profiles were downloaded from the NCBI Gene Expression Omnibus (GEO) under the accession no. GSE52485. The Affymetrix CEL files from BM progenitor datasets (GEO accession no.s GSM1267747, GSM1267748, and GSM1267749) and type 2 progenitor datasets (GEO accession nos. GSM1267744, GSM1267745, and GSM1267746) were loaded into Genomics Suite software (version 6.6; Partek Inc.). The CEL files were RMA normalized, and a one-way ANOVA analysis was performed. A subset of the top 200 annotated genes, which were up-regulated in type 2 progenitors versus BM progenitors and that passed a p-value cut off of ≤0.05, was then merged with the RMA-normalized signal intensities and exported as a text file. The top 200 enriched genes in type 2 progenitors were analyzed for gene pathways with the Database for Annotation, Visualization and Integrated Discovery.
Isolation and processing of immune cells for Car expression profiling
Indicated cell populations were isolated from four naive CCR2-GFP mice on a C57BL(6) background. B cells and T cells were sorted from spleens. B cells were sort purified as CD45+ CD11c− NK1.1− CCR2GFP− B220+. T cells were sort purified as CD45+ CD11c− NK1.1− CCR2-GFP− B220− Thy1.2+ and either CD4+ or CD8+. Macrophages and monocytes were isolated from collagenase-digested lungs. Macrophages were sort purified as CD45+ F480+ Siglec F+ CD11c+ MHCII+. Monocytes were sort purified as CD45+ CD11b+ CCR2GFP+ NK1.1− CD11c−. BM-derived mast cells were sort purified as CD3− CD19− CD49b− CD200R+ c-Kit+ and BM-derived basophils as CD3− CD19− c-Kit− CD200R+ CD49b+. BM-derived cells were sort purified on day 7 after culture. Purity for all cell populations was determined to be ≥98%. RNA was extracted using an RNA isolation kit (RNeasy; QIAGEN) and amplified using an RNA amplification kit (MessageAmp II; Ambion). Relative mRNA levels were determined by real-time PCR. RNA was reverse transcribed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). For each gene, Taq Man Fast Universal PCR Master Mix (2×) No Amp and TaqMan probes (Applied Biosystems) were used.
Mouse mast cell and basophil cultures
BM cells were isolated from the femur of WT mice and cultured in the presence of 10 ng/ml IL-3 and vehicle (1:10, DMSO/RPMI complete media) or 30 µg/ml MZ. Fresh IL-3 with either vehicle or MZ was added daily, and mast cell and basophil populations were evaluated on day 7 after culture.
Mouse macrophage cultures
BM cells were isolated from the femur of WT mice and cultured in the presence of 30% L cell culture supernatant and vehicle (1:10, DMSO/RPMI complete media) or 30 µg/ml MZ. Fresh macrophage medium with either vehicle or MZ was added daily, and macrophage cell populations were evaluated on day 5 after culture.
T. spiralis infection and IL-3C treatment
Methods for maintenance, recovery, infection, and isolation of T. spiralis larvae were performed as previously described (Urban et al., 2000). Mice were infected with 500 T. spiralis muscle larvae by oral gavage and were treated with vehicle (1:5, DMSO/PBS) or 2–3 mg MZ i.p. daily; mice were sacrificed between 10 and 11 d after infection. At necropsy, single-cell suspensions of mLNs and spleens were prepared as previously described (Giacomin et al., 2012) for basophil and mast cell analysis. Blood was collected, and serum was isolated for analysis of Mcpt1 and IgE. 1-cm sections of small intestine (jejunum) were collected for real-time PCR and histological analysis. The remaining sections of small intestine were slit lengthwise, rinsed in PBS, and placed in HBSS for 4 h at 37°C, and adult worms were quantified by microscopic analysis as previously described (Urban et al., 2000). Rag1−/− mice were injected i.p. with a combination of 1 µg of recombinant IL-3 and 0.5 µg α–IL-3 antibody (clone MP2-8F8; BioLegend) in 200 µl PBS every 3 d for 7–10 d. IL-3C–treated mice were treated with 2 mg MZ or vehicle (1:5, DMSO/PBS) every day.
RNA isolation and quantitative real-time PCR analysis
RNA from sections of small intestine was isolated by homogenization in TRIzol (Invitrogen) followed by phenol-chloroform extraction and isopropanol precipitation. cDNA was generated per standard protocol with Superscript reverse transcription (Invitrogen) and used as an input for real-time PCR. Real-time data were analyzed using the ΔΔ cycle threshold (CT) method using SYBR green chemistry (Applied Biosystems) with β-actin serving as the endogenous housekeeping gene. All reactions were run on a Fast Real-Time PCR system (ABI 7500; Applied Biosystems). Samples were normalized to naive controls. The following QuantiTech primer assays from QIAGEN were used: Mcpt1 (QT00157864), IL-4 (QT00160678), IL-9 (QT00107555), Arg1 (QT00134288), Retnla (QT00254359), Car1 (QT01058113), and Car2 (QT00494592).
T cell stimulations and ELISAs
Single-cell suspensions were made from mLNs or skin draining LNs. LN cells were cultured in RPMI complete medium and treated with and without anti-CD3 (clone 145-2C11; BD) and anti-CD28 antibodies (clone 37.51; BD) for 72 h. Standard sandwich ELISA (eBioscience) was performed to measure IL-4 (clones 11B11 and BVD6-24G2), IL-5 (clones TRFK5 and TRFK4), and IL-13 (clones eBio13A and eBio1316H) levels in the cell-free supernatants. Serum quantities of Mcpt1 and IgE were measured by commercial sandwich ELISA kits (BD; eBioscience).
Atopic dermatitis–like disease
Mice were treated once a day topically with 2 nmol of MC903 (calcipotriol; Tocris Bioscience) in 20 µl ethanol for 6 d as described previously (Kim et al., 2013). MC903-treated mice were treated with vehicle (1:5, DMSO/PBS) or 2–3 mg MZ i.p. daily and sacrificed on day 7 after treatment.
Preparation of A. fumigatus conidia
A. fumigatus strain Af293 was grown on glucose minimal media agar plates for 3 d or sabouraud dextrose agar for 7–10 d at 37°C. Conidia were harvested by adding 0.01% Tween 80 to plates and gently scraping conidia from the plates using a cell scraper. Conidia were then filtered through sterile Miracloth, washed, resuspended in PBS, and counted on a hemocytometer (3520; Hausser Scientific). Fluorescent A. fumigatus reporter conidia were prepared as previously described (Espinosa et al., 2014). Mice were challenged with 5 × 107 A. fumigatus conidia intratracheally as described previously (Espinosa et al., 2014). Mice were treated with vehicle (1:5, DMSO/PBS) or 2–3 mg MZ i.p. daily and sacrificed 72 h after infection. Bronchoalveolar lavage fluid was collected by washing the lungs with 2 ml PBS containing 0.05 M EDTA. After collection of the bronchoalveolar lavage fluid, lung samples were minced in RPMI containing 100 units/ml collagenase (Gibco), incubated at 37°C for 60 min, disrupted, and filtered. Subsequently, red blood cells were lysed using a Tris ammonium chloride solution. Analysis of neutrophils was conducted using a panel of cell surface markers (CD45, CD11b, and Ly6G) as previously described (Espinosa et al., 2014). To deplete lung neutrophils, mice were injected with 500 µg anti-Ly6G antibody (clone 1A8; Bio X Cell) i.p. followed by another dose of 100 µg intratracheally (Espinosa et al., 2014).
Food allergy
Food allergy–like disease was initiated as previously described (Noti et al., 2014). In brief, mice were treated daily with 2 nmol MC903 (calcipotriol; Tocris Bioscience) in 20 µl of 100% ethanol on ears in the presence of 100 µg OVA daily for 14 d. As a vehicle control, the same volume of ethanol and OVA was applied. All mice were challenged intragastrically with 50 mg OVA on days 14–17.5. Mice were sacrificed on day 18. Groups of mice were treated with vehicle (1:5, DMSO/PBS) or 2–3 mg MZ i.p. daily until sacrifice.
BM chimeras
Mice were lethally irradiated (1,050 rad) and reconstituted with 106 WT BM cells, 106 Car2−/− BM cells, or a mix of 5 × 105 WT and 5 × 105 Car2−/− BM cells. Mice were allowed to reconstitute for a period of 8 wk.
Mouse ESC culture
R1 mouse ESCs (Nagy et al., 1993) were provided by A. Gravy (Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada) and cultured as described previously (Lin and Talbot, 2011). In brief, irradiated MEFs (R&D Systems) were cultured according to the manufacturer’s protocol. 1 d later, 106 ESCs were cultured over MEFs for ∼4 d until they reached a confluence of ∼80–90%. The medium was replaced every 24 h. ESCs and MEFs were detached using StemPro Accutase cell dissociation reagent (Thermo Fisher Scientific) following the manufacturer’s instructions until cell suspensions were obtained. Cell suspensions were then plated on gelatin-coated vessels for 45 min to separate MEFs from ESCs.
CRISPR/Cas9 transfections
CRISPR/Cas9 genome editing was performed as described previously (Kim et al., 2014b). 1 µg/µl Cas9 nuclease (Invitrogen) was incubated with 240 ng/µl in vitro transcribed guide RNAs (Invitrogen) targeting the gene encoding Car1 (5′-AGAATATCTAGTTCCATCCA-3′) according to manufacturer’s instructions. 106 ESCs were washed once with 1× PBS and resuspended in 50 µl of resuspension buffer. 5 µl ESC suspension was mixed with 7 µl Cas9/gRNA complexes, and cells were electroporated using the Neon Transfection System (1300V; 10 width and 3 pulses; Invitrogen). Mock-transfected control cells were electroporated with Cas9 protein only. ESCs were cultured over MEFs immediately after transfection and until they reached a confluence of 80–90%. Genome targeting efficiency was determined as previously described (Guschin et al., 2010). In brief, genomic DNA was isolated using the QIAamp DNA mini kit (QIAGEN). Then, a 600-bp gene region containing the targeted sequence for car1 was amplified by PCR (forward primer: 5′-GCTCTGTGATTAAAGTCCAG-3′; reverse primer: 5′-TTCCATCGTGCACAAGGCA-3′). The PCR product was reannealed and incubated with 1 µl T7 endonuclease (New England Biolabs, Inc.). PCR products were run on a 2% agarose gel, and the band’s percentage intensity was analyzed using the Image Lab software (Bio-Rad Laboratories). After genome targeting efficiency assays, aliquots of cells from this parent culture of ∼3 × 106 cell were removed and plated for developmental assays.
Mast cell and macrophage differentiation from ESCs
Mast cell and macrophage differentiation was performed as previously described (Kovarova and Koller, 2012). In brief, ESCs were cultured with 9 ml of differentiation medium (α-MEM supplemented with 15% ESC-qualified FBS, penicillin/streptomycin, 5% of protein-free hybridoma medium, insulin transferrin selenium supplement, 1-thioglycerol, and ascorbic acid) on Petri dishes for 10 d replacing the medium every 3 d to generate embryonic bodies. Embryonic bodies were dissociated as described previously (Kovarova and Koller, 2012) and washed. 5 × 105 cells were plated and treated with 10 ng/ml recombinant IL-3 to generate mast cells or with 30% L cell culture supernatant to induce macrophage differentiation. The media was replaced every 48 h. After 3 wk of culture, mast cell and macrophage populations were analyzed by flow cytometry.
Human mast cell cultures
Blood from healthy human donors was obtained from the New York Blood Center under protocols approved by the Rutgers Institutional Review Boards. Peripheral blood cells were isolated using a Ficoll gradient. CD34+ cells were purified using a CD34 MicroBead isolation kit according to the manufacturer’s protocol (130-048-702; Miltenyi Biotec). 25–50 × 103 CD34+ cells were placed in methocult (H4236) supplemented with stem cell factor, IL-6, and IL-3 as described previously (Saito et al., 2006). Mast cell populations were evaluated on day 14 after culture.
Statistics
Results are shown as mean ± SD. Statistical analysis was performed using Student’s t tests in Prism (version 6; GraphPad Software).
ACKNOWLEDGMENTS
We thank members of the Center for Immunity and Inflammation for discussions and critical reading, the New Jersey Medical School (NJMS) Flow Cytometry and Immunology Core Laboratory for technical assistance, and the NJMS Genomics Research Lab for bioinformatics assistance.
This work was supported by the National Institutes of Health (K22 AI110573 and RO1 AI123224 to M.C. Siracusa) and the New Jersey Health Foundation (PC 17-15).
The authors declare no competing financial interests.
Author contributions: E.K. Henry, C.B. Sy, V. Espinosa, J.M. Inclan-Rico, A. Rivera, and M.C. Siracusa designed and performed the research. S.S. Ghanny, D.F. Dwyer, and P. Soteropoulos analyzed transcriptional profiles. E.K. Henry, C.B. Sy, V. Espinosa, A. Rivera, and M.C. Siracusa analyzed experimental data. E.K. Henry and M.C. Siracusa wrote the paper.
Footnotes
Abbreviations used:
- ESC
- embryonic stem cell
- Mcpt1
- mast cell protease 1
- mLN
- mesenteric LN
- MZ
- methazolamide
References
- Allen J.E., and Maizels R.M.. 2011. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 11:375–388. 10.1038/nri2992 [DOI] [PubMed] [Google Scholar]
- Bischoff S., and Crowe S.E.. 2005. Gastrointestinal food allergy: new insights into pathophysiology and clinical perspectives. Gastroenterology. 128:1089–1113. 10.1053/j.gastro.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Dang T.D., Mills C.E., and Allen K.J.. 2014. Determination of the clinical egg allergy phenotypes using component-resolved diagnostics. Pediatr. Allergy Immunol. 25:639–643. 10.1111/pai.12301 [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., and Lempicki R.A.. 2003. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Espinosa V., Jhingran A., Dutta O., Kasahara S., Donnelly R., Du P., Rosenfeld J., Leiner I., Chen C.C., Ron Y., et al. 2014. Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS Pathog. 10:e1003940 10.1371/journal.ppat.1003940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Madden K.B., Morris S.C., Holmes J.M., Boiani N., Katona I.M., and Maliszewski C.R.. 1993. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151:1235–1244. [PubMed] [Google Scholar]
- Giacomin P.R., Siracusa M.C., Walsh K.P., Grencis R.K., Kubo M., Comeau M.R., and Artis D.. 2012. Thymic stromal lymphopoietin-dependent basophils promote Th2 cytokine responses following intestinal helminth infection. J. Immunol. 189:4371–4378. 10.4049/jimmunol.1200691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., and Rebar E.J.. 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649:247–256. 10.1007/978-1-60761-753-2_15 [DOI] [PubMed] [Google Scholar]
- Kamsteeg M., Zeeuwen P.L., de Jongh G.J., Rodijk-Olthuis D., Zeeuwen-Franssen M.E., van Erp P.E., and Schalkwijk J.. 2007. Increased expression of carbonic anhydrase II (CA II) in lesional skin of atopic dermatitis: regulation by Th2 cytokines. J. Invest. Dermatol. 127:1786–1789. 10.1038/sj.jid.5700752 [DOI] [PubMed] [Google Scholar]
- Kim B.S., Siracusa M.C., Saenz S.A., Noti M., Monticelli L.A., Sonnenberg G.F., Hepworth M.R., Van Voorhees A.S., Comeau M.R., and Artis D.. 2013. TSLP elicits IL-33–independent innate lymphoid cell responses to promote skin inflammation. Sci. Transl. Med. 5:170ra16 10.1126/scitranslmed.3005374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.S., Wang K., Siracusa M.C., Saenz S.A., Brestoff J.R., Monticelli L.A., Noti M., Tait Wojno E.D., Fung T.C., Kubo M., and Artis D.. 2014a Basophils promote innate lymphoid cell responses in inflamed skin. J. Immunol. 193:3717–3725. 10.4049/jimmunol.1401307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S.W., Kim J., and Kim J.S.. 2014b Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24:1012–1019. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P.A., Wright S.H., Lawrence C.E., Paterson Y.Y., and Miller H.R.. 2000. Delayed expulsion of the nematode Trichinella spiralis in mice lacking the mucosal mast cell-specific granule chymase, mouse mast cell protease-1. J. Exp. Med. 192:1849–1856. 10.1084/jem.192.12.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarova M., and Koller B.. 2012. Differentiation of mast cells from embryonic stem cells. Curr. Protoc. Immunol. Chapter 22:10.1–10.16. 10.1002/0471142735.im22f10s97 [DOI] [PubMed] [Google Scholar]
- Lardner A. 2001. The effects of extracellular pH on immune function. J. Leukoc. Biol. 69:522–530. [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., and Talbot P.. 2011. Methods for culturing mouse and human embryonic stem cells. Methods Mol. Biol. 690:31–56. 10.1007/978-1-60761-962-8_2 [DOI] [PubMed] [Google Scholar]
- Long J.Z., and Cravatt B.F.. 2011. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 111:6022–6063. 10.1021/cr200075y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marim F.M., Silveira T.N., Lima D.S. Jr., and Zamboni D.S.. 2010. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One. 5:e15263 10.1371/journal.pone.0015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W., and Roder J.C.. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 90:8424–8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noti M., Kim B.S., Siracusa M.C., Rak G.D., Kubo M., Moghaddam A.E., Sattentau Q.A., Comeau M.R., Spergel J.M., and Artis D.. 2014. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietin–basophil axis. J. Allergy Clin. Immunol. 133:1390–1399.e6. 10.1016/j.jaci.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori K., Luo Y., Jia Y., Nishida J., Wang Z., Bunting K.D., Wang D., and Huang H.. 2009. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J. Immunol. 182:2835–2841. 10.4049/jimmunol.0802870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P., Leppilampi M., Pastorekova S., Pastorek J., Waheed A., Sly W.S., and Parkkila S.. 2006. Carbonic anhydrase gene expression in CA II-deficient (Car2−/−) and CA IX-deficient (Car9−/−) mice. J. Physiol. 571:319–327. 10.1113/jphysiol.2005.102590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., and Pearce E.J.. 2013. Metabolic pathways in immune cell activation and quiescence. Immunity. 38:633–643. 10.1016/j.immuni.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., and Artis D.. 2012. New paradigms in type 2 immunity. Science. 337:431–435. 10.1126/science.1221064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay D.B., Stephen S., Borum M., Voltaggio L., and Doman D.B.. 2010. Mast cells in gastrointestinal disease. Gastroenterol. Hepatol. (N. Y.). 6:772–777. [PMC free article] [PubMed] [Google Scholar]
- Reimann H.J., and Lewin J.. 1988. Gastric mucosal reactions in patients with food allergy. Am. J. Gastroenterol. 83:1212–1219. [PubMed] [Google Scholar]
- Saenz S.A., Siracusa M.C., Perrigoue J.G., Spencer S.P., Urban J.F. Jr., Tocker J.E., Budelsky A.L., Kleinschek M.A., Kastelein R.A., Kambayashi T., et al. 2010. IL25 elicits a multipotent progenitor cell population that promotes TH2 cytokine responses. Nature. 464:1362–1366. 10.1038/nature08901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Kato A., Matsumoto K., and Okayama Y.. 2006. Culture of human mast cells from peripheral blood progenitors. Nat. Protoc. 1:2178–2183. 10.1038/nprot.2006.344 [DOI] [PubMed] [Google Scholar]
- Sicherer S.H., and Sampson H.A.. 2014. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J. Allergy Clin. Immunol. 133:291–307.e5. 10.1016/j.jaci.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Siracusa M.C., Saenz S.A., Hill D.A., Kim B.S., Headley M.B., Doering T.A., Wherry E.J., Jessup H.K., Siegel L.A., Kambayashi T., et al. 2011. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 477:229–233. 10.1038/nature10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa M.C., Saenz S.A., Wojno E.D., Kim B.S., Osborne L.C., Ziegler C.G., Benitez A.J., Ruymann K.R., Farber D.L., Sleiman P.M., et al. 2013. Thymic stromal lymphopoietin-mediated extramedullary hematopoiesis promotes allergic inflammation. Immunity. 39:1158–1170. 10.1016/j.immuni.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran C.T. 2008. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 7:168–181. 10.1038/nrd2467 [DOI] [PubMed] [Google Scholar]
- Tang D., Kang R., Coyne C.B., Zeh H.J., and Lotze M.T.. 2012. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 249:158–175. 10.1111/j.1600-065X.2012.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J.F. Jr., Schopf L., Morris S.C., Orekhova T., Madden K.B., Betts C.J., Gamble H.R., Byrd C., Donaldson D., Else K., and Finkelman F.D.. 2000. Stat6 signaling promotes protective immunity against Trichinella spiralis through a mast cell- and T cell-dependent mechanism. J. Immunol. 164:2046–2052. 10.4049/jimmunol.164.4.2046 [DOI] [PubMed] [Google Scholar]
- Voehringer D. 2013. Protective and pathological roles of mast cells and basophils. Nat. Rev. Immunol. 13:362–375. 10.1038/nri3427 [DOI] [PubMed] [Google Scholar]
- Wang M., Takeda K., Shiraishi Y., Okamoto M., Dakhama A., Joetham A., and Gelfand E.W.. 2010. Peanut-induced intestinal allergy is mediated through a mast cell–IgE–FcεRI–IL-13 pathway. J. Allergy Clin. Immunol. 126:306–316.e12. 10.1016/j.jaci.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T., Mingler M.K., Wahl B., Khorki M.E., Pabst O., Zimmermann N., and Rothenberg M.E.. 2014. Carbonic anhydrase IV is expressed on IL-5–activated murine eosinophils. J. Immunol. 192:5481–5489. 10.4049/jimmunol.1302846 [DOI] [PMC free article] [PubMed] [Google Scholar]