Xiao and collaborators show that miR-155 regulates T follicular helper cell development and function by suppressing the E3 ubiquitin ligase Peli1.
Abstract
MicroRNA (miRNA) deficiency impairs the generation of T follicular helper (Tfh) cells, but the contribution of individual miRNAs to this phenotype remains poorly understood. In this study, we performed deep sequencing analysis of miRNAs expressed in Tfh cells and identified a five-miRNA signature. Analyses of mutant mice deficient of these miRNAs revealed that miR-22 and miR-183/96/182 are dispensable, but miR-155 is essential for the generation and function of Tfh cells. miR-155 deficiency led to decreased proliferation specifically at the late stage of Tfh cell differentiation and reduced CD40 ligand (CD40L) expression on antigen-specific CD4+ T cells. Mechanistically, miR-155 repressed the expression of Peli1, a ubiquitin ligase that promotes the degradation of the NF-κB family transcription factor c-Rel, which controls cellular proliferation and CD40L expression. Therefore, our study identifies a novel miR-155–Peli1–c-Rel pathway that specifically regulates Tfh cell generation and function.
INTRODUCTION
Germinal center (GC) response is one of the defining features of adaptive immunity. T and B cells interact in specialized structures called GCs that form within secondary lymphoid organs after encountering pathogens or vaccination. GC B cells undergo class switch recombination, somatic hypermutation, affinity maturation, and differentiation into plasma cells and memory B cells. A distinct CD4+ effector T cell subset, T follicular helper cells (Tfh cells), provides critical help to B cells in the GC response (Crotty, 2011). Recent studies suggest that Tfh cell differentiation and function are essential in the control of chronic virus infections (Fahey et al., 2011; Harker et al., 2011; Kang et al., 2013), whereas Tfh cell expansion has been observed in a subset of patients with autoimmune diseases and several mouse models of autoimmunity and was shown to play a causative role in disease pathogenesis in some models (Linterman et al., 2009; Simpson et al., 2010; Zhang et al., 2013). Therefore, elucidating the cellular and molecular mechanisms underlying Tfh cell differentiation and function is of fundamental importance for the design of better vaccines and therapies aimed to boost antibody production in infectious settings or to mute autoantibody production in autoimmune diseases.
MicroRNAs (miRNAs) are endogenously encoded small RNAs that regulate the expression of protein-coding genes by pairing with their target mRNAs and promoting their degradation or translational repression (Bartel, 2009). Hundreds of miRNAs are expressed in the immune system (Kuchen et al., 2010). Genetic studies have demonstrated that miRNAs are critical regulators of the GC response (Baumjohann and Ansel, 2014). Thus, T cell–specific ablation of DiGeorge syndrome critical region gene 8 (Dgcr8), a critical subunit of the microprocessor complex that mediates the biogenesis of pre-miRNAs from the primary miRNA transcripts, results in severe defects in Tfh cell differentiation (Baumjohann et al., 2013). Antigen-specific Dgcr8-deficient CD4+ T cells exhibited impaired proliferation, failed to up-regulate Bcl6 and CXCR5, and were unable to enter B cell follicles during immune responses (Baumjohann et al., 2013). Recent studies of individual miRNA genes that regulate Tfh cell differentiation have been focused on the miR-17∼92 family miRNAs, which control the expression of Bcl6 and CXCR5 in antigen-specific CD4+ T cells, as well as their migration into B cell follicles. However, miR-17∼92 family miRNA-deficient antigen-specific CD4+ T cells exhibited normal in vivo proliferation, suggesting that other miRNA genes are responsible for controlling cellular proliferation during Tfh cell differentiation (Baumjohann et al., 2013; Kang et al., 2013).
In this study, we performed deep sequencing analysis of miRNAs expressed in Tfh cells and identified a five-miRNA signature that is highly induced upon Tfh cell differentiation. Functional analyses of mutant mice deficient of these Tfh-specific miRNAs revealed that miR-155 promotes the generation and function of Tfh cells. Further mechanistic studies identified a miR-155–Peli1–c-Rel pathway that specifically controls the late stage proliferation of and CD40 ligand (CD40L) expression on Tfh cells.
RESULTS
Deep sequencing identifies a miRNA signature of Tfh cells
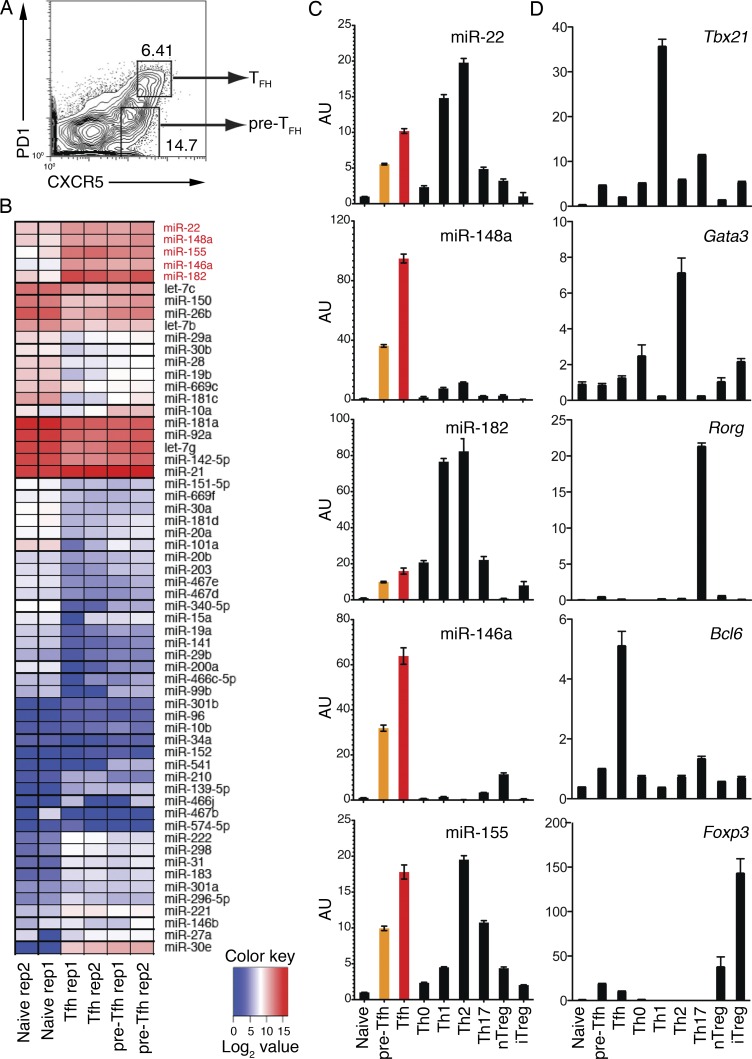
We hypothesized that, in addition to the miR-17∼92 family miRNAs, there are other miRNAs that play important roles in regulating the generation and function of Tfh cells. To identify these miRNAs, we performed small RNA deep sequencing analysis of pre-Tfh and Tfh cells sorted from mice immunized with OVA precipitated in alum together with LPS (OVA/alum/LPS; Fig. 1 A and Table S1). Clustering of the top 60 most variable miRNAs revealed that pre-Tfh and Tfh cells exhibited similar miRNA expression profiles, which are distinct from that of naive CD4+ T (CD4+CD44−CXCR5−PD-1−) cells sorted from the same immunized mice (Fig. 1 B). Five miRNAs were found highly induced in pre-Tfh and Tfh cells, including miR-22, miR-148a, miR-182, miR-146a, and miR-155. To validate the small RNA deep sequencing results, we prepared naive CD4+ T cells, various T helper cell subsets, and regulatory T cells (T reg cells) in independent experiments and examined the expression of these five miRNAs, as well as signature transcription factors for these cell subsets, by quantitative RT-PCR (Fig. 1, C and D). Indeed, all of these five miRNAs were highly induced upon differentiation into pre-Tfh cells, and their expression levels were further increased in Tfh cells. We termed these five miRNAs the miRNA signature of Tfh cells.
Figure 1.
Deep sequencing analysis of miRNA expression profiles of Tfh cells. (A) FACS sorting strategy to purify CXCR5intPD1lo pre-Tfh and CXCR5hiPD1hi Tfh cells. WT C57BL/6 mice were immunized with OVA/alum/LPS, and cell subsets were sorted from splenocytes on day 7 after immunization. (B) Heat map of miRNA signatures for naive CD4+ T (CD44−CXCR5−PD1−), pre-Tfh, and Tfh cells. The color scale represents the log2 value of miRNA read counts in tags per million. The Tfh cell signature miRNAs are marked in red. See details in Table S1. (C and D) Quantitative RT-PCR analysis of individual miRNAs (C) and transcription factors (D) in naive CD4+ T, pre-Tfh, Tfh, and in vitro generated Th cell subsets and natural T reg (nTreg) cells. AU, arbitrary units. Error bars represent SD. Data are one representative result of two independent experiments and triplicate for each RT-PCR reaction.
miR-22 and miR-183/96/182 are dispensable for Tfh cell generation and function
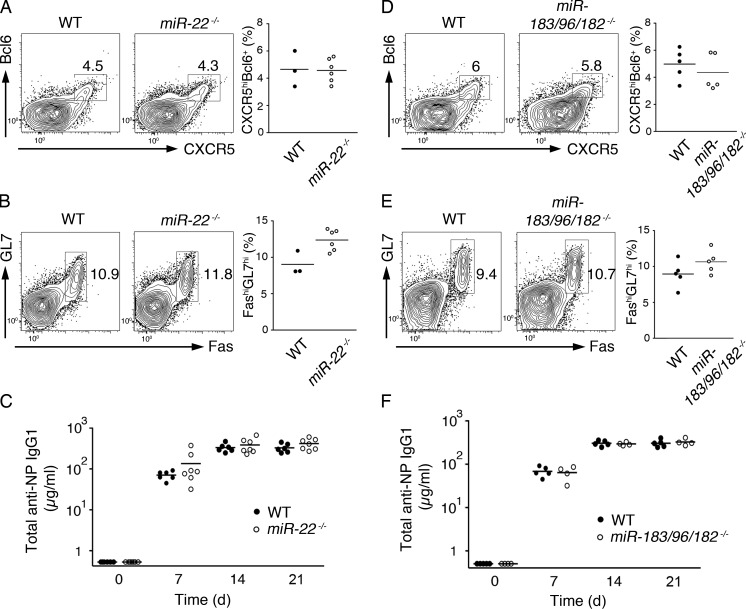
Among the five miRNAs, KO mice were previously generated and reported for miR-22, miR-146a, miR-155, and the miR-183/96/182 miRNA cluster, which encodes for miR-183, miR-96, and miR-182 (Rodriguez et al., 2007; Thai et al., 2007; Boldin et al., 2011; Gurha et al., 2012; Lumayag et al., 2013). miR-148a–deficient mice are currently not available. Recent studies of miR-146a–deficient mice showed that miR-146a played a negative role in the generation and accumulation of Tfh cells and GC B cells and that miR-146a repressed several Tfh cell–expressed genes (Hu et al., 2014; Pratama et al., 2015). To examine the functions of other Tfh cell signature miRNAs, we immunized mice deficient of miR-22 or the miR-183/96/182 cluster. It was previously reported that miR-182 is induced by IL-2 and plays a potent role in promoting clonal expansion of activated helper T cells (Stittrich et al., 2010), suggesting that it may have an important function in Tfh cell generation. Surprisingly, the generation of Tfh cells was normal in both miR-22– and miR-183/96/182–deficient mice (Fig. 2, A and D). In addition, GC B cell formation and antigen-specific antibody production were not affected by the loss of miR-22 or miR-183/96/182 (Fig. 2, B, C, E, and F). Therefore, miR-22 and the miR-183/96/182 cluster are dispensable for Tfh cell generation and function.
Figure 2.
miR-22 and miR-183/96/182 are dispensable for Tfh cell generation and function. (A, B, D, and E) WT, miR-22−/−, and miR-183/96/182−/− mice were immunized with OVA/alum/LPS, and CXCR5hiBcl6+ Tfh cells (A and D) and FAShiGL7hi GC B cells (B and E) were analyzed on day 7 after immunization by flow cytometry. (Left) Representative FACS plots. (Right) Summary of percentages of Tfh and GC B cells. (C and F) In independent experiments, NP-OVA/alum–immunized mice were bled at the indicated time points, and the amounts of NP-specific IgG1 were determined by ELISA. Each symbol represents an individual mouse. Horizontal lines indicate the mean. n = 3–7.
miR-155 deficiency in T cells leads to impaired generation of Tfh cells
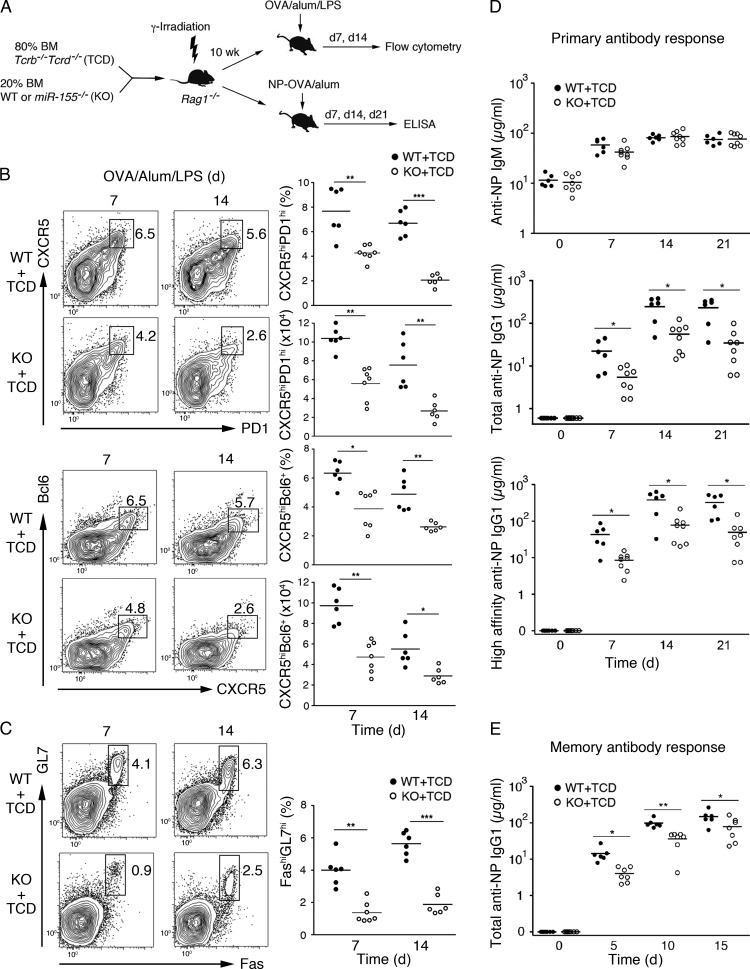
Previous studies showed that miR-155 plays a critical role in controlling GC responses and Tfh cell generation, but the underlying cellular and molecular mechanisms remain poorly understood (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007; Hu et al., 2014). To investigate the T cell–intrinsic role of miR-155 in Tfh cell generation, we generated mixed BM chimeras with T cell–specific loss of miR-155, which were created by reconstituting sublethally irradiated Rag1−/− mice with 80% of Tcrb−/−Tcrd−/− (TCD) plus 20% of WT or miR-155−/− (KO) BM cells (Fig. 3 A). The Tcrb−/−Tcrd−/− mutation prevents the generation of T cells; therefore, T cells in the animals receiving miR-155−/− BM will be miR-155 deficient (KO + TCD), whereas the recipients of WT BM will have WT T cells (WT + TCD). The 80:20 ratio favors reconstitution of all the other hematopoietic lineages from WT precursors. Both groups of chimeras have similar percentages and numbers of B and T cells (unpublished data). We immunized these mice with OVA/alum/LPS or 4-hydroxy-3-nitrophenyl (NP) linked to OVA precipitated in alum (NP-OVA/alum) to study the effect of T cell–specific miR-155 deficiency on GC reaction and antibody responses (Fig. 3 A). The KO + TCD chimeras showed severe defects in the generation of GC B and Tfh cells and produced decreased amounts of total and high-affinity NP-specific IgG1 antibody (Fig. 3, B–D). The KO + TCD chimeras also had a defective secondary antibody response after reimmunization (Fig. 3 E), suggesting that miR-155 expression in T cells is required not only for the initial generation and function of Tfh cells, but also for the establishment of long-lived protective CD4+ T cell–dependent B cell responses. These mice produced NP-specific IgM antibody in amounts comparable with that produced by WT + TCD chimeras (Fig. 3 D), suggesting that general B cell activation and plasma cell differentiation were not affected by the absence of miR-155 expression in T cells. These results demonstrate a T cell–intrinsic requirement of miR-155 for the generation of Tfh cells and GC B cells, as well as the production of high-affinity and class-switched antibodies.
Figure 3.
T cell–intrinsic requirement of miR-155 for generating GC B and Tfh cells. (A–D) Generation and immunization of mixed BM chimera mice with T cell–specific loss of miR-155. Sublethally irradiated Rag1−/− mice were reconstituted with BM cells from miR-155−/− and Tcrb−/−Tcrd−/− (TCD) or WT and TCD mice at a 1:4 ratio and subsequently immunized with OVA/alum/LPS (B and C) or NP-OVA/alum (D). (B, left) Flow cytometry analysis of Tfh cells. Numbers adjacent to the outlined areas indicate percentages of CXCR5hiPD1hi (top) and CXCR5hiBcl-6+ (bottom) Tfh cells among total CD4+ T cells. (Right) Summary of the percentage and cell number of CXCR5hiPD1hi and CXCR5hiBcl-6+ Tfh cells. (C, left) Flow cytometry analysis of GC B cells. Numbers adjacent to the outlined areas indicate the percentage of GC B cells (FashiGL7hi) among total B cells (B220+). (Right) Summary of the percentage of GC B cells. (D) ELISA analysis of NP-specific IgM (top), total NP-specific IgG1 (middle), and high-affinity anti-NP IgG1 (bottom) antibodies in the sera from NP-OVA/alum–immunized mice. (E) Mixed BM chimera mice in Fig. 3 A were reimmunized with NP-OVA 3 mo after primary immunization. Total anti-NP IgG1 in the sera was determined by ELISA. Each dot represents an individual mouse. Horizontal lines indicate the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data were combined from two independent experiments (B and C). n = 6–8.
miR-155 is specifically required for the late stage of proliferation during Tfh cell differentiation
We introduced the transgene encoding the OVA-specific OT-II TCR into miR-155−/− mice to study the effect of miR-155 on antigen-specific CD4+ T cells during Tfh cell differentiation (Barnden et al., 1998). We labeled naive CD45.2+ WT or miR-155−/− OT-II CD4+ T cells with the cytosolic dye CFSE and adoptively transferred them into CD45.1+ WT recipients, immunized the recipients with OVA/alum, and analyzed the transferred OT-II CD4+ T cells in draining lymph nodes at various time points. The percentage of adoptively transferred miR-155−/− OT-II cells among total CD4+ T cells was comparable with that of their WT counterparts on days 2.5 and 3.5 after immunization but was drastically decreased by day 6.5, when both the percentage and absolute number of miR-155−/− OT-II and CXCR5hiBcl6+ Tfh cells were threefold less than that of WT OT-II cells (Fig. 4, A and B). Among miR-155−/− OT-II cells, the frequency of CXCR5hiBcl6+ Tfh cells was comparable with that among WT OT-II cells (Fig. 4 C). These data suggest that much fewer Tfh cells were generated from miR-155−/− OT-II cells than from WT OT-II cells because of the lower numbers of miR-155−/− OT-II cells by day 6.5 (Fig. 4 B). The expression of Tfh cell markers, such as CXCR5, PD-1, and Bcl6, and activation markers, such as CD44 and inducible T cell co-stimulator (ICOS), was marginally affected by miR-155 deficiency (unpublished data). The reduction in miR-155−/− OT-II cells on day 6.5 could be caused by a decrease of cell proliferation, an increase of cell death, or both. Examination of CFSE dilution showed that miR-155−/− OT-II cells proliferated less than WT OT-II cells between days 3.5 and 6.5 (Fig. 4, D and E), whereas cell death was not significantly affected by miR-155 deficiency (Fig. 4 E, bottom). These results prompted us to examine the de novo expansion of antigen-specific CD4+ T cells at different stages of Tfh cell differentiation. In the same experimental setting as in Fig. 4 (A–D), BrdU was injected into immunized recipient mice 1 d before analysis to label cells undergoing proliferation, and OT-II cells were analyzed on day 3 or 4.5 for BrdU incorporation. Consistent with the CFSE dilution results (Fig. 4 D), miR-155−/− OT-II cells exhibited impaired proliferation between days 3.5 and 4.5 but not between days 2 and 3 after immunization (Fig. 4 F). We also examined the location of miR-155−/− OT-II CD4+ cells in draining lymph nodes during immune responses. Consistent with their reduced frequency, the density of miR-155−/− OT-II CD4+ cells (identified by CD45.2 staining) in draining lymph nodes was much lower than that of WT OT-II CD4+ cells (Fig. 4, G and H). The reduction in miR-155−/− OT-II CD4+ cell density was found in both B and T cell zones, suggesting that the migration of CD4+ cells from the T cell zone into B cell follicles is not affected by miR-155 deficiency (Fig. 4, G and H). Together, these results reveal a critical role of miR-155 in controlling the proliferation of antigen-specific CD4+ T cells, specifically at the late stage of Tfh cell differentiation, but not their survival or migration.
Figure 4.
miR-155 controls the optimal expansion of antigen-specific CD4+ T cells. (A–H) Naive CD4+CD45.2+ T cells from WT or miR-155−/− OT-II (KO) mice were adoptively transferred into WT CD45.1+ mice. Recipient mice were immunized with OVA/alum subcutaneously into the flank 1 d after cell transfer. Cells in the draining lymph nodes were analyzed by flow cytometry at the indicated time points. (A) Flow cytometry analysis of CD45.2+ OT-II cells among total CD4+ T cells. (B) Absolute number of transferred OT-II CD4+ T and CXCR5hiBcl-6+ Tfh cells in draining lymph nodes at 6.5 d after immunization. (C) Flow cytometry analysis of CXCR5hiBcl6+ Tfh cells among CD4+CD45.2+ OT-II cells. (D) Flow cytometry analysis of cell proliferation of CD4+CD45.2+ OT-II cells. The proliferation index was calculated by using FlowJo analysis. (E) Percentage of CD45.2+ OT-II cells with five or more cell divisions (bottom) and active caspase 3+ (apoptotic) cells among CD4+CD45.2+ OT-II cells on day 6.5 after immunization (top). Representative figures were adapted from the experiment in D to indicate that the cells underwent five or more cell divisions. Error bars represent SD. (F) BrdU was injected into mice i.p. on day 2 or day 3.5 after immunization and euthanized for analysis 24 h later (day 3 or day 4.5, respectively). CD4+CD45.2+ OT-II cells were analyzed by flow cytometry for BrdU incorporation (BrdU+). (G) Immunohistochemistry of WT or miR-155−/− OT-II cells in draining lymph nodes. Cryosections of the draining lymph nodes from the immunized recipient mice were stained with CD45.2 (white, adoptively transferred OT-II cells) and IgD (red, naive B cells) and analyzed by fluorescence microscopy. Bar, 70 µm. (H) Cell density of CD45.2+ OT-II cells in the whole area and B cell zone. Each dot represents an individual mouse. Horizontal lines indicate the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). ns, not significant. n = 3–14.
miR-155 controls c-Rel and CD40L expression by targeting Peli1
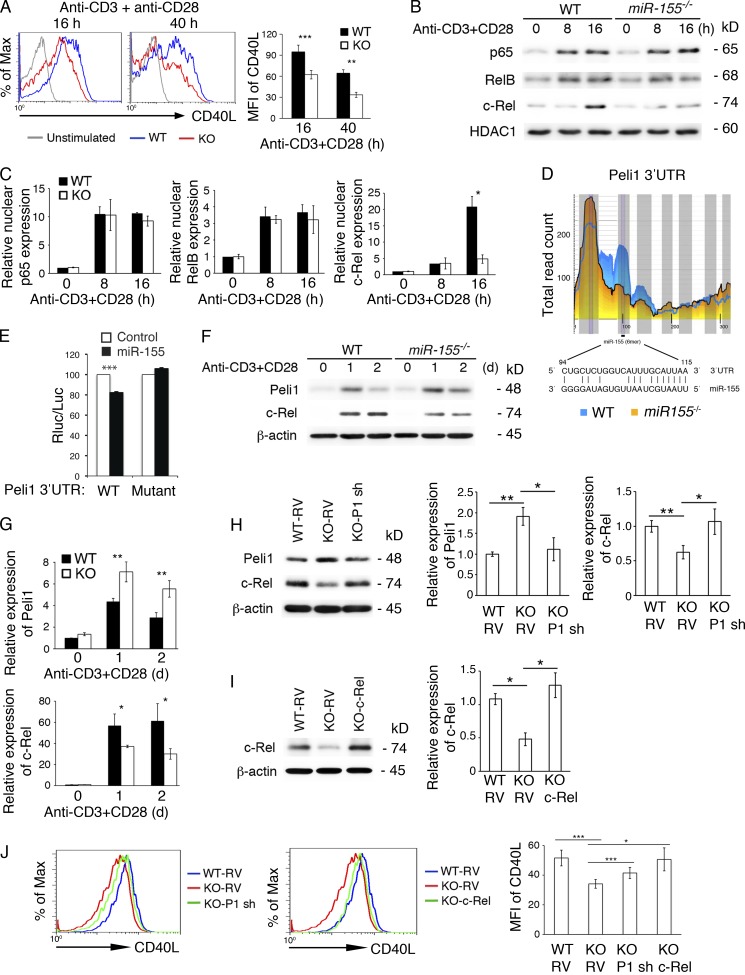
Previous studies showed that the strength of TCR binding plays an important role in the Tfh cell differentiation program, suggesting the functional importance of initial T cell activation in the generation of Tfh cells (Fazilleau et al., 2009; Tubo et al., 2013). To elucidate the molecular mechanisms through which miR-155 controls the generation of Tfh cells, we first investigated whether miR-155 deficiency affects T cell activation by determining IL-2 production and expression of activation markers. TCR- and CD28-mediated IL-2 production by miR-155−/− CD4+ T cells was largely normal (unpublished data). The expression of activation markers such as CD25, CD44, CD69, and ICOS was comparable between miR-155−/− and WT CD4+ T cells (unpublished data). Surface expression of CD71 (transferrin receptor) on miR-155−/− CD4+ T cells showed a slight reduction, which did not reach statistical significance (unpublished data). In contrast, surface expression of CD40L, which is essential for T cell–B cell interaction during GC response (Crotty, 2011), on miR-155−/− CD4+ T cells was reduced to 64.5% (16 h) and 50% (40 h) of WT levels (Fig. 5 A). Therefore, miR-155 specifically regulates CD40L expression during T cell activation.
Figure 5.
miR-155 controls c-Rel and CD40L expression by targeting Peli1 in T cells. (A–J) Naive CD4+ T cells from miR-155−/− (KO) and WT mice were stimulated with 5 µg/ml anti-CD3 and 2 µg/ml anti-CD28 antibodies for the indicated amounts of time. (A) Surface expression of CD40L was analyzed by flow cytometry. The bar graph summarizes mean fluorescence intensity (MFI) of CD40L. n = 4–7 per group. (B and C) Immunoblot analysis of p65, RelB, and c-Rel in the nuclear extracts of naive CD4+ T cells from miR-155−/− (KO) and WT mice stimulated with 5 µg/ml anti-CD3 and 2 µg/ml anti-CD28 antibodies for the indicated amounts of time. Two independent immunoblot experiments were normalized by β-actin and summarized in C. (D) Schematic representation of the Peli1 3′UTR and the miR-155–binding region. The figure was adapted from CLIP base (Loeb et al., 2012). Note that Ago2 binding to the miR-155 site is much reduced in in vitro activated miR-155−/− CD4+ T cells. (E) Reporter assay of the Peli1 3′UTR fragment containing the WT or mutated miR-155–binding site. (F and G) Immunoblot analysis of Peli1, c-Rel, and β-actin expression in naive CD4+ T cells stimulated with 5 µg/ml anti-CD3 and 2 µg/ml anti-CD28 antibodies for the indicated amounts of time. Four independent immunoblot experiments were normalized by β-actin and summarized in G. (H) Immunoblot analysis of Peli1, c-Rel, and β-actin expression in WT and miR-155−/− (KO) CD4+ T cells transduced with control or Peli1 shRNA (P1 sh)–encoding RVs. (I) Immunoblot analysis of c-Rel and β-actin expression in WT and miR-155−/− (KO) CD4+ T cells transduced with control or c-Rel–encoding RVs. (H and I, left) Representative immunoblots. (Right) Bar graphs showing quantification results. (J) Flow cytometry analysis of CD40L expression on RV-transduced CD4+ T cells 1 d after retroviral transduction. The bar graph summarizes mean fluorescence intensity of CD40L. n = 5 per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data represent two (B, C, I, and J), three (H), and four (A, E, F, and G) independent experiments. Error bars represent SD.
It was previously reported that NF-κB binds to the distal end of the CD40L promoter and up-regulates CD40L transcription (Srahna et al., 2001; Schubert et al., 2002; Pham et al., 2005). miR-155 may control CD40L expression by regulating the activity of NF-κB transcription factors. To test this hypothesis, we examined nuclear accumulation of NF-κB family members during T cell activation. The nuclear protein levels of RelA (p65) and RelB were normal in activated miR-155−/− CD4+ T cells, whereas nuclear accumulation of c-Rel was much lower in these cells than in WT cells (Fig. 5, B and C). As c-Rel also regulates T cell proliferation in a dosage-dependent manner (Köntgen et al., 1995), it is very likely that miR-155 controls both CD40L expression and the proliferation of antigen-specific CD4+ T cells by regulating nuclear accumulation of c-Rel.
To gain insights into the molecular pathway through which miR-155 controls c-Rel and CD40L expression, we analyzed the list of miR-155 target genes experimentally identified by using differential high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation (CLIP) of Argonaute 2 (Ago2) in activated CD4+ T cells (Loeb et al., 2012). We looked for potential miR-155 target genes implicated in the regulation of NF-κB activation in T cells. Peli1 was selected because it acts as an E3 ubiquitin ligase of c-Rel and negatively regulates nuclear accumulation of c-Rel by promoting its degradation (Chang et al., 2011). There is one evolutionarily conserved miR-155 binding site in the 3′ untranslated region (UTR) of the Peli1 mRNA. Ago2 binding to this site was significantly reduced in miR-155−/− CD4+ T cells (Fig. 5 D), suggesting that it is a bona fide miR-155 target site. To demonstrate direct regulation of Peli1 by miR-155, we performed a dual-luciferase reporter assay. A Peli1 3′UTR fragment containing the miR-155 binding site was cloned into the expression vector psiCHECK2, which encodes the Renilla luciferase controlled by the cloned 3′UTR. The resulting construct was cotransfected in HEK293 cells with an expression vector encoding miR-155, and regulation of protein expression by miR-155 was measured by luciferase activity. As shown in Fig. 5 E, miR-155 significantly reduced protein expression through the Peli1 3′UTR. When the miR-155 binding site was mutated, the reduction in protein expression was abolished, indicating that miR-155 suppresses the expression of Peli1 through its cognate binding site in the Peli1 3′UTR.
Consistent with miR-155 suppression of Peli1 expression and the promotion of c-Rel degradation by Peli1, in vitro activated miR-155−/− CD4+ T cells expressed more Peli1 and less c-Rel proteins than their WT counterparts (Fig. 5, F and G). We also examined the protein levels of a small number of other miR-155 target genes identified by differential high-throughput sequencing of CLIP (Loeb et al., 2012) and implicated in various aspects of T cell biology. The protein levels of Fbxo11 and SHIP1 were not affected by miR-155 deficiency (unpublished data). Therefore, Peli1 represents an authentic target gene of miR-155 in this cellular context.
To further test the hypothesis that miR-155 regulates c-Rel and CD40L expression by suppressing the expression of Peli1, we used retrovirus (RV)-encoded shRNA to knock down the expression of Peli1 and RV-encoded c-Rel to overexpress c-Rel in miR-155−/− CD4+ T cells. RV-Peli1 shRNA restored Peli1 and c-Rel expression close to WT levels (Fig. 5 H), whereas RV–c-Rel increased c-Rel expression to a level that was slightly higher than the WT level (Fig. 5 I). Both Peli1 knockdown and c-Rel reexpression substantially restored CD40L expression in miR-155−/− CD4+ T cells (Fig. 5 J). Collectively, these results demonstrate that miR-155 controls c-Rel and CD40L expression by suppressing the expression of its target gene, Peli1.
Restoration of Tfh cell generation and function by reducing Peli1 in miR-155−/− T cells
c-Rel and CD40L are critical regulators of the GC response (Crotty, 2011; Gilmore and Gerondakis, 2011). We next asked whether correcting Peli1 expression in miR-155−/− CD4+ T cells would rescue the GC response defects caused by miR-155 deficiency in T cells. miR-155−/− OT-II CD4+ T cells were transduced with RV-Peli1 shRNA and transferred into TCD recipient mice, which were subsequently immunized with OVA/alum. Although WT OT-II CD4+ T cells support the GC response in TCD mice, miR-155−/− OT-II CD4+ T cells exhibited a severe reduction in cell numbers and an inability to provide help to B cells (Fig. 6 A). Strikingly, knocking down Peli1 expression rescued the B cell help ability of miR-155−/− OT-II CD4+ T cells by ∼60–70%, as indicated by increased formation of GC B and Tfh cells, as well as by elevated miR-155−/− OT-II CD4+ T and Tfh cell numbers (Fig. 6, A–C). Accordingly, the production of total and high-affinity NP-specific antibodies was largely restored (Fig. 6 D).
Figure 6.
Peli1 knockdown in miR-155−/− T cells substantially restores the GC response. (A–D) WT and miR-155−/− (KO) OT-II CD4+ T cells transduced with control or Peli1 shRNA (P1 sh)–encoding RVs were adoptively transferred into TCD mice and subsequently immunized with OVA/alum. Splenocytes were prepared from recipient mice on day 10 after immunization. (A) Flow cytometry analysis of GC B cells among total B cells. (B and C) Percentage (B) and total cell numbers (C) of CXCR5hiBcl6+ Tfh cells among CD4+ T cells (left) and OT-II CD4+ T cells among total splenocytes (right) are shown. (D) ELISA analysis of NP-specific IgG1 antibodies in the sera after NP-OVA/alum immunization. Serum titers of total (left) and high-affinity (right) NP-specific IgG1 antibodies are shown. Each symbol represents an individual mouse. Horizontal lines indicate the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data were combined from two independent experiments. ns, not significant. n = 5–9.
We next used a genetic approach to modulate Peli1 expression in miR-155−/− mice. As shown in Fig. 7 (A and B), miR-155−/−;Peli1+/− mice exhibited largely restored generation of Tfh and GC B cells on day 8 after OVA/alum/LPS immunization. To determine whether the effect of genetic rescue on Tfh generation is T cell intrinsic, we generated mixed BM chimera mice with mixed BM cells (TCD BM + WT, miR-155−/−, or miR-155;Peli1+/− BM) in a Rag1−/− background and immunized the chimera mice with NP-OVA/alum/LPS (Fig. 7 C). Heterozygous deletion of Peli1 in miR-155−/− T cells substantially rescued the impaired Tfh cell generation caused by miR-155 deficiency (Fig. 7 D). Peli1 heterozygosity also restored Tfh function, as indicated by close to WT levels of GC B cell numbers (Fig. 7 E) and class-switched and affinity-matured antigen-specific antibodies (Fig. 7 F). These results demonstrate that Peli1 is a major mediator of miR-155 function in CD4+ T cells to control Tfh cell generation, GC response, and antibody production.
Figure 7.
Heterozygous Peli1 deletion restores GC response in miR-155−/− mice. (A and B) Splenocytes were prepared from WT, miR-155−/−, Peli1+/−, and miR-155−/−;Peli1+/− mice on day 8 after immunization with OVA/alum/LPS. CXCR5hiBcl-6+ Tfh cells (top) among total CD4+ T cells and FAS+GL7+ GC B cells (bottom) were analyzed by flow cytometry (A) and represented as dot plots (B). (C) Chimera mice were generated by reconstitution of irradiated Rag1−/− mice with BM cells from WT and TCD, KO and TCD, or KO;Peli+/− and TCD at a 1:4 ratio. The chimera mice were subsequently immunized with NP-OVA/alum/LPS. (D and E) CXCR5hiBcl-6+ Tfh cells (D) and FAShiGL7hi GC B cells (E) were analyzed from spleen after 8 d of immunization by flow cytometry, and combined results are exhibited as dot plots. (F) Amounts of NP-specific total and affinity-matured antibodies were determined by ELISA on day 10 after immunization. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). ns, not significant. n = 4–10.
Reexpression of c-Rel restores Tfh cell generation and function of miR-155–deficient T cells
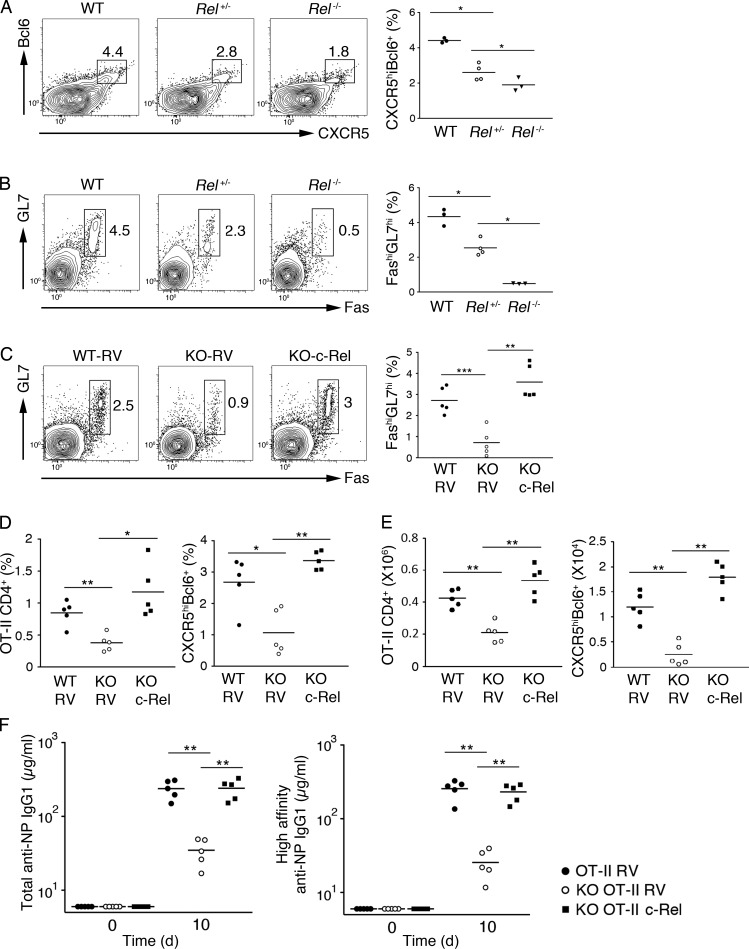
It was previously reported that germline deletion of the Rel gene (encoding c-Rel) caused severely compromised B and T cell proliferation and antibody production and that conditional deletion of c-Rel in activated B cells led to impaired production of high-affinity antibodies because of the collapse of established GCs (Köntgen et al., 1995; Heise et al., 2014). However, the function of c-Rel in the generation and function of Tfh cells remains unclear. As we found that the c-Rel protein levels in CD4+ T cells is controlled by miR-155 via Peli1 and that c-Rel modulates CD4+ T cell expression of CD40L, which is critical for the function of Tfh cells during the GC response, we speculated that the c-Rel protein level in CD4+ T cells is critical for Tfh cell generation and function. To explore the functional importance of c-Rel in T cells, we immunized WT, Rel+/−, and Rel−/− mice with NP-OVA/alum/LPS. As shown in Fig. 8 (A and B), Rel−/− mice exhibited drastically reduced numbers of Tfh and GC B cells, whereas Rel+/− mice had similar but milder phenotypes. These results suggest that c-Rel controls Tfh cell generation and function in a dose-dependent manner.
Figure 8.
Reexpression of c-Rel restores Tfh cell generation and function of miR-155−/− T cells. (A and B) WT, Rel+/−, and Rel−/− mice were immunized with OVA/alum/LPS i.p., and the spleen was analyzed for CXCR5hiBcl6+ Tfh (A) and FAS+GL7+ GC B (B) cells on day 8 after immunization by flow cytometry. (C–F) c-Rel–encoding or control RVs were transduced into WT and miR-155−/− (KO) OT-II CD4+ T cells, adoptively transferred into TCD mice, and immunized subsequently with NP-OVA/alum/LPS. Spleens were analyzed for GC B (C) and Tfh (D) cells by flow cytometry, and amounts of antigen-specific antibodies (F) were determined by ELISA on day 10 after immunization. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). n = 3–5.
We hypothesized that impaired Tfh cell generation and function in miR-155−/− mice is mainly caused by reduced c-Rel protein levels in CD4+ T cells. To test this hypothesis, we attempted to rescue the Tfh cell defects by restoring c-Rel protein levels in miR-155−/− CD4+ T cells. We transduced miR-155−/− OT-II CD4+ T cells with a c-Rel–expressing RV, adoptively transferred them into TCD mice, and immunized the recipient mice with NP-OVA/alum. Strikingly, retroviral expression of c-Rel completely restored the generation and function of Tfh cells from miR-155−/− OT-II cells (Fig. 8, C–E), as well as the production of class-switched and affinity-matured antigen-specific antibodies (Fig. 8 F). Collectively, these results demonstrated that the miR-155–Peli1–c-Rel pathway plays a critical role in controlling the generation and function of Tfh cells.
The miR-155–Peli1–c-Rel pathway is dispensable for the generation of cytotoxic CD8, Th1, and Th17 cells
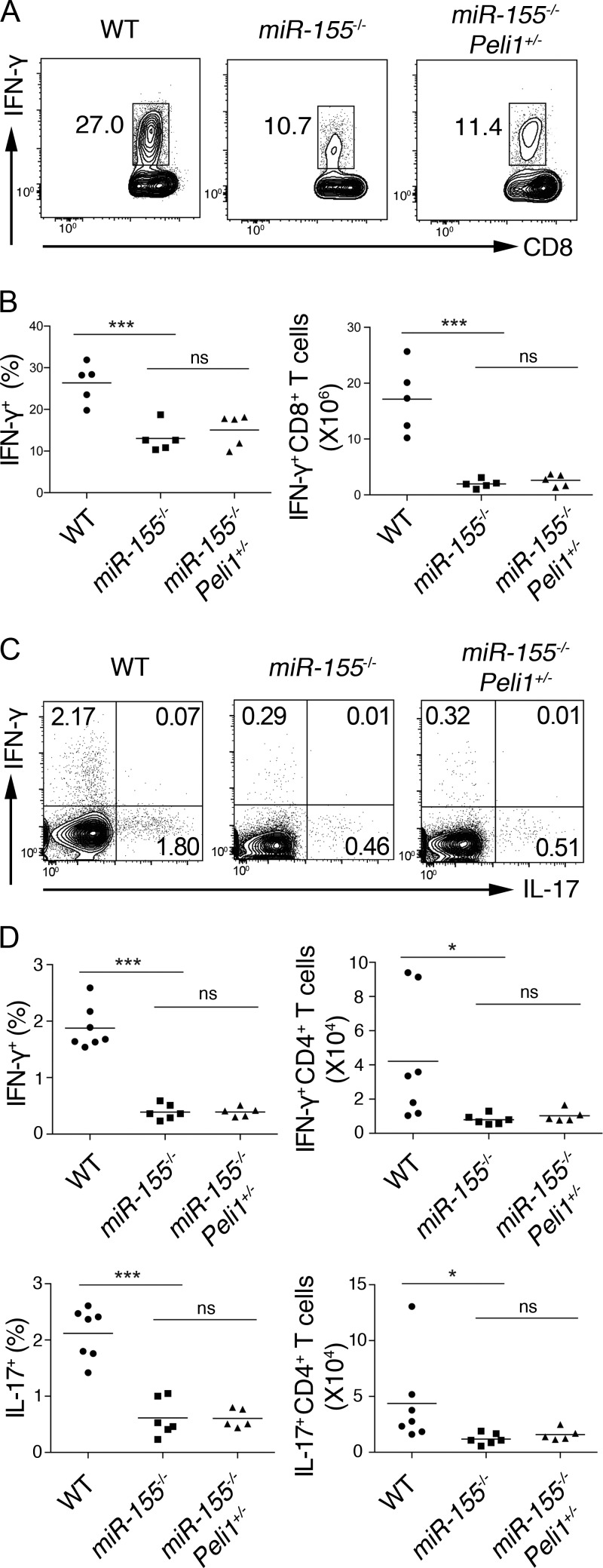
Previous studies showed that miR-155 plays critical roles in other immune cells, such as cytotoxic CD8+ T cells, Th1 cells, and Th17 cells (O’Connell et al., 2010; Dudda et al., 2013; Gracias et al., 2013; Escobar et al., 2014). To examine whether Peli1 functions as a key miR-155 target gene in these cell contexts, WT, miR-155−/−, and miR155−/−;Peli1+/− mice were challenged with lymphocytic choriomeningitis virus (LCMV) Armstrong, whose clearance requires intact cytotoxic CD8+ T cell function. As shown in Fig. 9 (A and B), miR-155−/− mice showed compromised generation of IFN-γ–producing effector CD8+ T cells, but this defect was not rescued in miR-155−/−;Peli1+/− mice. These mice were also examined in the experimental autoimmune encephalomyelitis (EAE) model to study whether the miR-155–Peli1 axis functions in the generation of Th1 and Th17 cells. Consistent with previous studies, miR-155−/− mice were defective in the generation of both Th1 and Th17 cells (O’Connell et al., 2010; Escobar et al., 2014). Heterozygous deletion of Peli1 cannot rescue these defects (Fig. 9, C and D). Collectively, these results showed that miR-155 regulation of Peli1 is dispensable for the generation of cytotoxic CD8+ T, Th1, and Th17 cells, thereby highlighting the functional specificity of the miR-155–Peli1 pathway in regulating the generation of Tfh cells.
Figure 9.
miR-155 regulation of Peli1 is dispensable for the generation of cytotoxic CD8+ T, Th1, and Th17 cells. (A and B) Flow cytometry analysis of IFN-γ–producing effector CD8+ T cells from the spleen of WT, miR-155−/−, and miR155−/−;Peli1+/− mice at day 8 after LCMV Armstrong infection (gated on CD8+ T cells). (C and D) Flow cytometry analysis of Th1 and Th17 cells from draining lymph nodes of WT, miR-155−/−, and miR155−/−;Peli1+/− mice at day 14 after the induction of EAE (gated on CD4+ T cells). *, P < 0.05; ***, P < 0.001 (Student’s t test). ns, not significant. n = 5–7.
DISCUSSION
A previous study has shown that miRNAs control several facets of Tfh cell differentiation, including cellular proliferation, up-regulation of CXCR5 and Bcl6, and migration of CD4+ T cells into B cell follicles (Baumjohann et al., 2013). Recent studies from us and other investigators found that the miR-17∼92 family miRNAs regulate the expression of CXCR5 and Bcl6, as well as CD4+ T cell migration into B cell follicles, but not proliferation of antigen-specific CD4+ T cells (Baumjohann et al., 2013; Kang et al., 2013), suggesting that additional miRNAs regulate the generation and function of Tfh cells. In the present study, we performed the first deep sequencing analysis of miRNA expression profiles of pre-Tfh and Tfh cells and identified five miRNAs that are highly induced upon Tfh cell differentiation. They include miR-22, miR-148a, miR-182, miR-146a, and miR-155.
By analyzing Tfh cell generation, GC B cell formation, and production of class-switched antibodies in mutant mice deficient of these individual miRNA genes, we found that miR-22 and miR-183/96/182 are dispensable, whereas miR-155 is essential for Tfh cell generation and function. This result is surprising because miR-182 was previously reported to play a potent role in promoting clonal expansion of activated Th cells (Stittrich et al., 2010), an integral component of the Tfh cell differentiation program. As miRNAs encoded by this cluster are highly induced in several Th cell subsets (Fig. 1 C; Kuchen et al., 2010), it is possible that they play critical roles in some of these cellular contexts. Indeed, results from our collaborators showed that miR-183/96/182 regulates pathogenic cytokine expression during Th17 cell development (Ichiyama et al., 2016). Studies from other investigators showed that miR-146a plays a negative role in Tfh cell generation and accumulation, likely through suppressing several Tfh cell–expressing genes (Hu et al., 2014; Pratama et al., 2015). The potential role of miR-148a, another Tfh-specific miRNA, in Tfh cell generation and function remains to be explored. This awaits the creation and analysis of mutant mice deficient of this gene.
This study identified the miR-155–Peli1–c-Rel axis as a novel pathway that plays critical roles in CD4+ T cells to control the generation and function of Tfh cells by regulating their proliferation specifically at the late stage of Tfh cell differentiation and CD40L expression on these cells. The nexus point of this pathway is the control of c-Rel protein levels by miR-155 and Peli1. Unlike other NF-κB family members that are broadly expressed, high levels of c-Rel are found primarily in B and T cells (Gilmore and Gerondakis, 2011). c-Rel is not essential for normal hematopoiesis and lymphopoiesis. Instead, it is required for several functions in mature T and B cells. c-Rel–deficient T and B cells are severely impaired in cellular proliferation in response to various stimuli (Köntgen et al., 1995), whereas c-Rel−/− mice exhibit defective GC response and production of class-switched antibodies (Chen et al., 2010). The functions of c-Rel are very sensitive to its gene dosage. c-Rel heterozygosity leads to defects at the cellular and animal levels that are similar to, but milder than, that caused by a complete loss of c-Rel (Köntgen et al., 1995). The critical functions of c-Rel in lymphocytes and its haploinsufficiency make it an ideal target for miR-155 and Peli1. miR-155, Peli1, and c-Rel are all strongly induced upon T cell activation by the engagement of TCR and co-stimulation molecules (Haasch et al., 2002; Thai et al., 2007; Chang et al., 2011). The results presented in this study show that miR-155 sets the Peli1 expression level in a proper range to allow the optimal induction of c-Rel, whose expression must be tightly controlled. Inadequate expression of c-Rel results in impaired B and T cell activation, as well as diminished GC response and antibody production (Köntgen et al., 1995; Chen et al., 2010), whereas exaggerated c-Rel expression is associated with autoimmune diseases and lymphomas (Gilmore and Gerondakis, 2011). In the present study, we showed that c-Rel is a dose-dependent regulator of Tfh cell generation and function. c-Rel controls two critical aspects of the Tfh cell program: proliferation of antigen-specific CD4+ T cells and expression of CD40L on these cells. In the absence of miR-155, CD4+ T cells express increased amounts of Peli1, which leads to reduced nuclear accumulation of c-Rel and, therefore, impaired proliferation of antigen-specific CD4+ T cells at the late stage of Tfh cell differentiation and compromised expression of CD40L on these cells.
The CD40L–CD40 interaction is essential for CD4+ T cells to provide critical help to B cells during the GC response (Crotty, 2011). Although CD40 is constitutively expressed on B cells and other antigen-presenting cells, CD40L is transiently expressed on CD4+ T cells upon activation (Crow and Kirou, 2001). Engagement of CD40 leads to B cell clonal expansion, GC formation, isotype switching, affinity maturation, and generation of long-lived plasma cells (Quezada et al., 2004). CD40L expression is a limiting factor in the GC response (Pérez-Melgosa et al., 1999). Both the magnitude and duration of CD40L expression are subjected to tight regulation (Crow and Kirou, 2001). CD40L-deficient mice exhibit impaired T cell–dependent immune response and GC formation (Grewal et al., 1995; van Essen et al., 1995). CD40L deficiency in humans results in immunodeficiency associated with a severe defect in humoral immune response and a Tfh cell defect. In the absence of CD40L, T cells differentiate poorly into Tfh cells, and B cell–T cell interactions in the GC are impaired (Durandy et al., 2013). Conversely, in human systemic lupus erythematosus (SLE) patient and mouse lupus models, CD40L expression on the T cell surface is much prolonged after activation. In addition, high levels of the soluble form of CD40L are found in SLE patient sera. Both the surface and soluble forms of CD40L can activate autoimmune B cells and initiate autoantibody production in SLE (Crow and Kirou, 2001). Our study found that miR-155 specifically controls CD40L expression on CD4+ T cells. In the absence of miR-155, activated CD4+ T cells express ∼50–65% of WT levels of CD40L. A previous study showed that CD40Ltg+ mice exhibited a 1.1–2-fold increase in CD40L expression on activated T cells, and this led to a twofold increase in the production of antigen-specific high-affinity IgG1 antibody (Pérez-Melgosa et al., 1999). Conversely, mutant mice with T cell–specific transgenic expression of a dominant-negative TFE3 protein exhibited reduced CD40L expression on activated CD4+ T cells, which resulted in impaired GC formation and a drastic decrease in the production of class-switched antibodies (Huan et al., 2006). Considering the high sensitivity of GC response to CD40L expression levels on CD4+ T cells, it is likely that the reduction in CD40L expression on miR-155−/− CD4+ T cells contributes to impaired GC response in miR-155−/− mice.
Our results demonstrate that Peli1 is a key functional target gene of miR-155 in CD4+ T cells to regulate Tfh cell generation and function. A previous study has shown that deletion of Peli1 causes hyperactivation of T cells, and Peli1−/− mice spontaneously develop autoimmune diseases characterized by multiorgan inflammation and autoantibody production (Chang et al., 2011). Peli1 exerts its functions in T cells mainly through promoting the degradation of c-Rel protein and thereby preventing exaggerated accumulation of c-Rel in the nucleus. miR-155−/− CD4+ T cells express increased amounts of Peli1 protein, and correcting Peli1 expression to WT levels by shRNA knockdown substantially rescued GC B cell formation and antibody responses. Furthermore, genetic ablation of one copy of the Peli1 gene in miR-155−/− mice substantially restored the generation and function of Tfh cells. These data demonstrate that miR-155 regulation of Peli1 plays a major role in mediating miR-155 control of Tfh cell generation and function.
miR-155 is highly induced upon the activation of T and B cells and plays critical roles in the immune system (Vigorito et al., 2013). Importantly, miR-155 does not control the general lymphocyte activation and proliferation program. It affects immune responses in a cell type– and context-specific manner, likely through regulating different downstream target genes and pathways in different cell subsets and biological processes (Lu et al., 2015). Thus, miR-155 promotes the differentiation and accumulation of Th17 cells in EAE through regulating Jarid2 and Socs1 (O’Connell et al., 2010; Escobar et al., 2014; Lu et al., 2015), inhibits Th2 cell differentiation through c-Maf (Rodriguez et al., 2007; Thai et al., 2007), controls the competitive fitness of T reg cells through Socs1 (Lu et al., 2009, 2015), and regulates B cell differentiation into plasma cells through PU.1 (Vigorito et al., 2007; Lu et al., 2014). In this study, we found that miR-155 is highly expressed in pre-Tfh and Tfh cells and plays an essential role in the generation and function of Tfh cells. Mechanistically, it controls cellular proliferation at a late stage of Tfh cell differentiation, as well as CD40L expression, through a novel miR-155–Peli1–c-Rel pathway. Importantly, this pathway is dispensable for the generation of cytotoxic CD8+ T, Th1, and Th17 cells, highlighting its specificity in regulating the generation and function of Tfh cells.
In summary, our study identified a five-miRNA signature of Tfh cells, found that miR-22 and miR-183/96/182 are dispensable in Tfh cells, and established the miR-155–Peli1–c-Rel axis as an important pathway that regulates the generation and function of Tfh cells. In the long run, manipulating this pathway may help us to design better ways to boost immune responses to fight pathogen infections and to attenuate autoantibody production to treat autoimmune diseases.
MATERIALS AND METHODS
Mice
miR-22−/− (Jax stock 018155 on a 129S7/SvEvBrd background), miR-155−/− (Jax stock 007745 on a 129S6/SvEvTac × C57BL/6NTac mixed background), Tcrb−/−Tcrd−/− (Jax stock 002122 on a C57BL/6J background), OT-II (Jax stock 004194 on a C57BL/6 background), and Rag1−/− (Jax stock 002216 on a C57BL/6 background) mice were from The Jackson Laboratory. miR-183/96/182 (on a 129 × C57BL/6 background) and Peli1-deficient (on a C57BL/6 background; provided by S. Sun, The University of Texas M.D. Anderson Cancer Center, Houston, TX) mice were previously described (Chang et al., 2011; Lumayag et al., 2013). miR-155−/−;Peli1+/− mice were generated by breeding miR-155−/− and Peli1−/− mice. All mice were bred and housed under specific pathogen-free conditions. Both male and female mice were used for all experiments, and no sex differences were observed. For BM reconstitution, BM cells isolated from C57BL/6, miR-155−/−, and miR-155−/−;Peli1+/− mice were subjected to red blood cell lysis and T cell depletion by magnetic cell separation, mixed with BM cells from Tcrb−/−Tcrd−/− mice at a 1:4 ratio (5 × 106 cells in total), and transferred by tail vein injection into Rag1−/− mice irradiated with two rounds of 400 Rad with a 3-h interval (Vigorito et al., 2007). 10 wk after reconstitution, recipient mice were immunized with OVA/alum/LPS i.p. For LCMV infection, mice were injected i.p. with 2 × 105 PFU LCMV Armstrong and analyzed on day 8 after infection. All mice were used in accordance with guidelines from the Institutional Animal Care and Use Committees of The Scripps Research Institute and Xiamen University.
Antibodies and reagents
Anti-CD4 (GK1.5), anti-CD25 (PC61), anti-CD44 (IM7), anti-CD69 (H1.2F3), anti-CD71 (RI7217), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD40L (MR1), anti-CD90.1 (Thy1.1), anti–PD-1 (RMP1-30), anti-ICOS (15F9), and biotin anti–rat IgG2a (MRG2a-83) were from BioLegend. Anti-CXCR5 (2G8), anti–Bcl-6 (K112-91), anti-CD95 (Jo2), anti-IgD (11-26c.2a), FITC-conjugated anti-CD3 (2C11), and Alexa Fluor 647–conjugated anti-GL7 (GL7) were from BD. Anti-SHIP1 and anti-Fbxo11 were from Cell Signaling Technology. Anti-pellino1/2 (F7), c-Rel (N), p65 (C-20), and RelB (C-19) were from Santa Cruz Biotechnology, Inc. Anti–β-actin (AC-74) was from Sigma-Aldrich.
RNA isolation and miRNA expression profiling of Tfh cells
2–3-mo-old C57BL/6 mice were immunized with OVA/alum/LPS i.p. On day 7 after immunization, naive (CD4+CD44−CXCR5−PD1−), pre-Tfh (CD4+CXCR5intPD1lo), and Tfh (CD4+CXCR5hiPD1hi) cells were FACS sorted from splenocytes. Total RNA was extracted using TRIzol reagent and subjected to standard small RNA deep sequencing analysis. In brief, small RNA in the range of 18–35 nucleotides was isolated by denaturing PAGE. 5′ and 3′ adapters were added to the small RNAs sequentially, followed by RT and PCR amplification. The amplified cDNA was gel purified and loaded on the Genome Analyzer II (Illumina) for deep sequencing according to the manufacturer’s instructions. The miRDeep software package was used to identify known miRNAs, predict novel miRNAs, and establish a miRNA expression profile for each sample (Friedländer et al., 2008).
To identify differentially expressed miRNAs, variances and SD among samples were first calculated using the rowVars function from the gene filter library in R. Then, the miRNA expression levels of the selected most differentially regulated 60 miRNAs were subjected to heat map analysis. In the heat map, these miRNAs are ordered and clustered together when the expression patterns are similar based on row dendrogram. The small RNA deep sequencing data have been deposited on the NCBI Gene Expression Omnibus database (accession no. GSE72975).
Generation of Th cell subsets
CD62L+CD25− naive CD4+ T cells in the spleen were sorted by flow cytometers (FACSAria; BD) from Foxp3-GFP (for Th0, Th1, and induced T reg [iT reg] cells; Fontenot et al., 2005), IL-4–GFP (for Th2 cells; Mohrs et al., 2001), and IL-17A–GFP mice (for Th17 cells; Jax stock 018472) and were stimulated for 4 d with 1 µg/ml anti-CD3 and mitomycin C (Sigma-Aldrich)–treated congenic allophycocyanin (APC) and 50 U/ml recombinant human IL-2 under Th0-, Th1-, iT reg–, Th2-, or Th17-polarizing conditions. Th cell polarization mediums were supplemented as follows: for Th0 cell differentiation, 10 µg/ml anti–IL-4, 10 µg/ml anti–IL-12, and 10 µg/ml anti–IFN-γ; for Th1 cell differentiation, 2 U/ml IL-12 and 10 µg/ml anti–IL-4; for Th2 cell differentiation, 20 ng/ml IL-4, 10 µg/ml anti–IL-12, and 10 µg/ml anti–IFN-γ; for iT reg cell differentiation, 1 ng/ml human TGF-β; and for Th17 cell differentiation, 2 ng/ml human TGF-β and 20 ng/ml IL-6. Th cells were sorted by the expression of CD45 congenic marker and GFP reporter. Natural T reg cells were sorted directly from Foxp3-IRES (internal ribosomal entry site)-Thy1.1 mice (Liston et al., 2008). All antibodies were purchased from Bio X Cell, and recombinant cytokines were from PeproTech.
Flow cytometry and intracellular staining
Single-cell suspensions were prepared from spleen or draining lymph nodes. Antibodies for cell surface markers were added, followed by incubation for 30 min at 4°C. For staining of CXCR5, single cells were stained with purified antibody to mouse CXCR5 (2G8; BD), followed by biotin-conjugated antibody to rat IgG2a (MRG2a-83; BioLegend) and streptavidin-APC. For staining of IL-2, cell surface markers were stained, and then cells were fixed for 30 min at 4°C with 1% paraformaldehyde, permeabilized with FACS buffer (2% FBS in PBS) containing 0.1% saponin (Sigma-Aldrich), and incubated for 30 min at 4°C with APC-conjugated anti–IL-2. A Foxp3 staining kit was used to stain Bcl-6 according to the manufacturer’s instructions (eBioscience). Cells on which surface markers had been stained were fixed for 1 h at 4°C with fixation/permeabilization buffer, washed with 1× permeabilization buffer, and incubated for 1 h at room temperature with anti–Bcl-6 in 1× permeabilization buffer. Apoptotic cells were assessed by using a CaspGLOWFluorescein Active Caspase-3 Staining kit according to the manufacturer’s instructions (eBioscience).
Immunofluorescence microscopy
Mouse lymph nodes were washed with PBS and placed in optimal cutting temperature compound and snap frozen in dry ice. Cryosections of 6 µm in thickness were fixed with acetone and then stained with anti–mouse IgD (BioLegend), anti–rat Alexa Fluor 488 (Thermo Fisher Scientific), anti-CD45.2–APC (BioLegend), and Hoechst 33342 (Thermo Fisher Scientific). Images were acquired using a confocal microscope (780; Leica Biosystems) and analyzed with Imaris software (Bitplane).
T cell stimulation and retroviral transduction
Annealed oligonucleotides encoding Peli1-specific shRNA (5′-GATCCGGTCAACTGAAAGTCCTATTCTCGAGAATAGGACTTTCAGTTGACCGTTTTTG-3′) or mouse c-Rel cDNA from c-Rel Flag pCDNA3 (plasmid 20013; Addgene) were inserted into a Thy-1.1–expressing retroviral vector provided by A. Rao (La Jolla Institute for Allergy and Immunology, La Jolla, CA; plasmid 17442; Addgene). S-Eco packaging cells were transfected by Fu-GENE HD reagent (Promega), and retroviral supernatants were collected 48 h after transfection. Naive CD4+ (CD25−CD44−CD69−) T cells were purified as previously described (Kang et al., 2013), stimulated with 5 µg/ml plated-bound anti-CD3 (2C11; BioLegend) and 2 µg/ml anti-CD28 (37.51; BioLegend) for 1 d, and then transduced by spin infection for 1 h at 1,800 rpm with retroviral supernatants in the presence of 6 µg/ml polybrene (Sigma-Aldrich).
Adoptive transfer of OT-II CD4+ T cells
106 naive WT or miR-155−/− CD4+CD45.2+ OT-II CD4+ T cells were transferred into CD45.1+ C57BL/6 recipient mice by tail vein injection, followed by subcutaneous immunization with 25 µg OVA/alum into the flank 1 d after cell transfer. For the transfer of RV-infected WT or miR-155−/− OT-II CD4+ T cells, 106 Thy-1.1+ cells were purified by magnetic cell separation and transferred into Tcrb−/−Tcrd−/− mice. 1 d after cell transfer, recipient mice were immunized with 25 µg OVA/alum or NP-OVA/alum i.p.
ELISA
Microtiter plates were coated with NP30-BSA or NP5-BSA (10 µg/ml in PBS; LGC Biosearch Technologies) for measurement of total or high-affinity NP-specific antibody, respectively. Nonspecific binding was blocked with 0.5% BSA in PBS. Serum samples were serially diluted in 0.5% BSA in PBS and were incubated in blocked plates at room temperature for 2 h. Plates were incubated for 2 h with biotin-conjugated anti-IgM (1020-08) or anti-IgG1 (1070-08; SouthernBiotech) for 1 h with streptavidin–alkaline phosphatase (Roche) and then with alkaline phosphatase substrate solution containing 4-nitro-phenyl phosphate (Sigma-Aldrich) for color development, followed by quantification on a microplate reader (VERSAmax; Molecular Devices).
Immunoblot analysis
Naive and stimulated CD4+ T cells were washed with PBS twice and lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% SDS) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Cell lysates were resolved on SDS-PAGE. Primary antibodies were diluted in 5% (wt/vol) BSA or nonfat milk in 1× Tris-buffered saline buffer (10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) followed by incubation overnight at 4°C.
Preparation of nuclear extracts
10 × 106 naive CD4+ cells before or after stimulation were harvested and resuspended in 100 µl of 0.1% NP-40 lysis buffer (10 mM Tris-Cl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40) and allowed to swell at 4°C for 5 min. The homogenates were centrifuged at 800 g for 5 min, and the supernatant was stored at −80°C as cytoplasmic extracts. The nuclear pellet was washed twice with washing buffer (10 mM Tris-Cl, pH 7.5, 10 mM NaCl, and 3 mM MgCl2) and resuspended in 20 µl radioimmunoprecipitation assay buffer for Western blot analysis. After incubation at 4°C for 30 min with occasional vigorous vortex mixing, the nuclear extracts were centrifuged at 16,000 g for 10 min at 4°C, and the supernatant was frozen in aliquots at −80°C.
Luciferase reporter assay
Reporter constructs for the assay were prepared by cloning of a fragment of the Peli1 3′UTR (nucleotide position 1541–2033) containing the putative miR-155 binding site into psiCheck2 (Promega). 11 nucleotides of the miR-155 binding site spanning the miR-155 seed sequence (5′-ATTTGCATTAA-3′) were removed to generate the mutated reporter construct. The reporter constructs were cotransfected with empty or miR-155 expression plasmid into HEK293 cells. 24 h after transfection, luciferase activity was analyzed by using the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer’s guide.
EAE
Mice were injected subcutaneously with 100 µl of an emulsion containing 100 µg MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK) in complete Freund adjuvant. Mice were also injected i.p. with 200 ng pertussis toxin in 200 µl PBS on days 0 and 2 (Li et al., 2014).
Intracellular cytokine staining
For LCMV Armstrong–infected mice, splenocytes were harvested on day 8 after infection and stimulated for 5 h with 2 µg/ml MHC class I–restricted peptides of LCMV glycoprotein amino acids 33–41 (>99% pure; American Peptide and The Scripps Research Institute) in the presence of 4 µg/ml brefeldin A (Sigma-Aldrich). Cells were fixed, permeabilized with 2% saponin, and stained intracellularly with anti–IFN-γ (XMG1.2; BioLegend), anti-TNF (MP6-XT22), and anti–IL-2 (JES6-5H4; BioLegend). For EAE mice, draining lymph node cells were stimulated for 5 h with 50 ng/ml PMA (eBioscience) and ionomycin (eBioscience) in the presence of brefeldin A and monensin (eBioscience). Cells were fixed and permeabilized with 2% saponin and stained with anti–IFN-γ (XMG1.2) for Th1 cell differentiation or IL-17A (TC11-18H10.1; BioLegend) for Th17 cell differentiation.
Statistics
P-values were determined by using two-tailed Student’s t tests. Statistical significance is displayed as *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Table S1 contains small RNA deep sequencing analysis of Tfh cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20160204/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shao-Cong Sun for providing Peli1−/− mice, members of the Xiao laboratory for discussion and critical reading of the manuscript, and Yuqiong Liang at the Ye Zheng laboratory for technical assistance.
C. Xiao is a Pew Scholar in Biomedical Sciences. This study is supported by the PEW Charitable Trusts, Cancer Research Institute, Lupus Research Institute, National Institutes of Health (grants R01AI087634, R01AI089854, R56AI110403, and R56AI121155 to C. Xiao and grants R01AI103646 and R01AI108651 to L.-F. Lu), National Natural Science Foundation of China (grant 31570882 to W.-H. Liu, grant 31570883 to N. Xiao, and grant 31570911 to G. Fu), 1000 Young Talents Program of China (grant K08008 to N. Xiao), the Fundamental Research Funds for the Central Universities of the People’s Republic of China (grant 20720150065 to N. Xiao and G. Fu), the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, Information and Communications Technology, and Future Planning (grant NRF-2015R1C1A1A01052387 to S.G. Kang), and a 2016 research grant from Kangwon National University (to S.G. Kang).
The authors declare no competing financial interests.
Author contributions: W.-H. Liu, S.G. Kang, Z. Huang, and C. Xiao conceived and designed the project. C.-J. Wu generated various Th cell subsets for miRNA expression validation under the supervision of L.-F. Lu and Y. Zheng. C.J. Maine performed immune staining. Y. Liu and A. Hoffmann bred and provided Rel-deficient mice. M. Sabouri-Ghomi and H.Y. Jin analyzed the small RNA deep sequencing data. S. Xu provided miR-183/96/182–deficient mice. W.-H. Liu, S.G. Kang, Z. Huang, J. Shepherd, and A. Gonzalez-Martin performed all other experiments under the supervision of C. Xiao. W.-H. Liu, S.G. Kang, Z. Huang, and C. Xiao wrote the manuscript with input from other authors. C. Xiao, W.-H. Liu, G. Fu, N. Xiao, and L.-F. Lu provided funding and facility support and helped with data analysis and manuscript preparation.
Footnotes
Abbreviations used:
- APC
- allophycocyanin
- CD40L
- CD40 ligand
- CLIP
- cross-linking and immunoprecipitation
- EAE
- experimental autoimmune encephalomyelitis
- GC
- germinal center
- ICOS
- inducible T cell co-stimulator
- LCMV
- lymphocytic choriomeningitis virus
- miRNA
- microRNA
- RV
- retrovirus
- SLE
- systemic lupus erythematosus
- UTR
- untranslated region
References
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., and Ansel K.M.. 2014. MicroRNA regulation of the germinal center response. Curr. Opin. Immunol. 28:6–11. 10.1016/j.coi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Kageyama R., Clingan J.M., Morar M.M., Patel S., de Kouchkovsky D., Bannard O., Bluestone J.A., Matloubian M., Ansel K.M., and Jeker L.T.. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 14:840–848. 10.1038/ni.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., et al. 2011. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208:1189–1201. 10.1084/jem.20101823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Jin W., Chang J.H., Xiao Y., Brittain G.C., Yu J., Zhou X., Wang Y.H., Cheng X., Li P., et al. 2011. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 12:1002–1009. 10.1038/ni.2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hardy K., Bunting K., Daley S., Ma L., and Shannon M.F.. 2010. Regulation of the IL-21 gene by the NF-κB transcription factor c-Rel. J. Immunol. 185:2350–2359. 10.4049/jimmunol.1000317 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crow M.K., and Kirou K.A.. 2001. Regulation of CD40 ligand expression in systemic lupus erythematosus. Curr. Opin. Rheumatol. 13:361–369. 10.1097/00002281-200109000-00004 [DOI] [PubMed] [Google Scholar]
- Dudda J.C., Salaun B., Ji Y., Palmer D.C., Monnot G.C., Merck E., Boudousquie C., Utzschneider D.T., Escobar T.M., Perret R., et al. 2013. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 38:742–753. 10.1016/j.immuni.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy A., Kracker S., and Fischer A.. 2013. Primary antibody deficiencies. Nat. Rev. Immunol. 13:519–533. 10.1038/nri3466 [DOI] [PubMed] [Google Scholar]
- Escobar T.M., Kanellopoulou C., Kugler D.G., Kilaru G., Nguyen C.K., Nagarajan V., Bhairavabhotla R.K., Northrup D., Zahr R., Burr P., et al. 2014. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 40:865–879. 10.1016/j.immuni.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., and Brooks D.G.. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999. 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N., McHeyzer-Williams L.J., Rosen H., and McHeyzer-Williams M.G.. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 10:375–384. 10.1038/ni.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., and Rudensky A.Y.. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Friedländer M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., and Rajewsky N.. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26:407–415. 10.1038/nbt1394 [DOI] [PubMed] [Google Scholar]
- Gilmore T.D., and Gerondakis S.. 2011. The c-Rel transcription factor in development and disease. Genes Cancer. 2:695–711. 10.1177/1947601911421925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracias D.T., Stelekati E., Hope J.L., Boesteanu A.C., Doering T.A., Norton J., Mueller Y.M., Fraietta J.A., Wherry E.J., Turner M., and Katsikis P.D.. 2013. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat. Immunol. 14:593–602. 10.1038/ni.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal I.S., Xu J., and Flavell R.A.. 1995. Impairment of antigen-specific T-cell priming in mice lacking CD40 ligand. Nature. 378:617–620. 10.1038/378617a0 [DOI] [PubMed] [Google Scholar]
- Gurha P., Abreu-Goodger C., Wang T., Ramirez M.O., Drumond A.L., van Dongen S., Chen Y., Bartonicek N., Enright A.J., Lee B., et al. 2012. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 125:2751–2761. 10.1161/CIRCULATIONAHA.111.044354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasch D., Chen Y.W., Reilly R.M., Chiou X.G., Koterski S., Smith M.L., Kroeger P., McWeeny K., Halbert D.N., Mollison K.W., et al. 2002. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217:78–86. 10.1016/S0008-8749(02)00506-3 [DOI] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., and Zuniga E.I.. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829. 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N., De Silva N.S., Silva K., Carette A., Simonetti G., Pasparakis M., and Klein U.. 2014. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J. Exp. Med. 211:2103–2118. 10.1084/jem.20132613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Kagele D.A., Huffaker T.B., Runtsch M.C., Alexander M., Liu J., Bake E., Su W., Williams M.A., Rao D.S., et al. 2014. miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity. 41:605–619. 10.1016/j.immuni.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C., Kelly M.L., Steele R., Shapira I., Gottesman S.R., and Roman C.A.. 2006. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat. Immunol. 7:1082–1091. 10.1038/ni1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K., Gonzalez-Martin A., Kim B.S., Jin H.Y., Jin W., Xu W., Sabouri-Ghomi M., Xu S., Zheng P., Xiao C., and Dong C.. 2016. The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity. 44:1284–1298. 10.1016/j.immuni.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.G., Liu W.H., Lu P., Jin H.Y., Lim H.W., Shepherd J., Fremgen D., Verdin E., Oldstone M.B., Qi H., et al. 2013. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat. Immunol. 14:849–857. 10.1038/ni.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köntgen F., Grumont R.J., Strasser A., Metcalf D., Li R., Tarlinton D., and Gerondakis S.. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977. 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- Kuchen S., Resch W., Yamane A., Kuo N., Li Z., Chakraborty T., Wei L., Laurence A., Yasuda T., Peng S., et al. 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 32:828–839. 10.1016/j.immuni.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liang Y., LeBlanc M., Benner C., and Zheng Y.. 2014. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 158:734–748. 10.1016/j.cell.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Rigby R.J., Wong R.K., Yu D., Brink R., Cannons J.L., Schwartzberg P.L., Cook M.C., Walters G.D., and Vinuesa C.G.. 2009. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 206:561–576. 10.1084/jem.20081886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Nutsch K.M., Farr A.G., Lund J.M., Rasmussen J.P., Koni P.A., and Rudensky A.Y.. 2008. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl. Acad. Sci. USA. 105:11903–11908. 10.1073/pnas.0801506105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., and Rudensky A.Y.. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 48:760–770. 10.1016/j.molcel.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Nakagawa R., Lazzaro S., Staudacher P., Abreu-Goodger C., Henley T., Boiani S., Leyland R., Galloway A., Andrews S., et al. 2014. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J. Exp. Med. 211:2183–2198. 10.1084/jem.20140338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.F., Thai T.H., Calado D.P., Chaudhry A., Kubo M., Tanaka K., Loeb G.B., Lee H., Yoshimura A., Rajewsky K., and Rudensky A.Y.. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 30:80–91. 10.1016/j.immuni.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.F., Gasteiger G., Yu I.S., Chaudhry A., Hsin J.P., Lu Y., Bos P.D., Lin L.L., Zawislak C.L., Cho S., et al. 2015. A single miRNA-mRNA interaction affects the immune response in a context- and cell-type-specific manner. Immunity. 43:52–64. 10.1016/j.immuni.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumayag S., Haldin C.E., Corbett N.J., Wahlin K.J., Cowan C., Turturro S., Larsen P.E., Kovacs B., Witmer P.D., Valle D., et al. 2013. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. USA. 110:E507–E516. 10.1073/pnas.1212655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrs M., Shinkai K., Mohrs K., and Locksley R.M.. 2001. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 15:303–311. 10.1016/S1074-7613(01)00186-8 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Kahn D., Gibson W.S., Round J.L., Scholz R.L., Chaudhuri A.A., Kahn M.E., Rao D.S., and Baltimore D.. 2010. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 33:607–619. 10.1016/j.immuni.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Melgosa M., Hollenbaugh D., and Wilson C.B.. 1999. Cutting edge: CD40 ligand is a limiting factor in the humoral response to T cell-dependent antigens. J. Immunol. 163:1123–1127. [PubMed] [Google Scholar]
- Pham L.V., Tamayo A.T., Yoshimura L.C., Lin-Lee Y.C., and Ford R.J.. 2005. Constitutive NF-κB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood. 106:3940–3947. 10.1182/blood-2005-03-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama A., Srivastava M., Williams N.J., Papa I., Lee S.K., Dinh X.T., Hutloff A., Jordan M.A., Zhao J.L., Casellas R., et al. 2015. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nat. Commun. 6:6436 10.1038/ncomms7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada S.A., Jarvinen L.Z., Lind E.F., and Noelle R.J.. 2004. CD40/CD154 interactions at the interface of tolerance and immunity. Annu. Rev. Immunol. 22:307–328. 10.1146/annurev.immunol.22.012703.104533 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611. 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert L.A., Cron R.Q., Cleary A.M., Brunner M., Song A., Lu L.S., Jullien P., Krensky A.M., and Lewis D.B.. 2002. A T cell-specific enhancer of the human CD40 ligand gene. J. Biol. Chem. 277:7386–7395. 10.1074/jbc.M110350200 [DOI] [PubMed] [Google Scholar]
- Simpson N., Gatenby P.A., Wilson A., Malik S., Fulcher D.A., Tangye S.G., Manku H., Vyse T.J., Roncador G., Huttley G.A., et al. 2010. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 62:234–244. 10.1002/art.25032 [DOI] [PubMed] [Google Scholar]
- Srahna M., Remacle J.E., Annamalai K., Pype S., Huylebroeck D., Boogaerts M.A., and Vandenberghe P.. 2001. NF-κB is involved in the regulation of CD154 (CD40 ligand) expression in primary human T cells. Clin. Exp. Immunol. 125:229–236. 10.1046/j.1365-2249.2001.01601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittrich A.B., Haftmann C., Sgouroudis E., Kühl A.A., Hegazy A.N., Panse I., Riedel R., Flossdorf M., Dong J., Fuhrmann F., et al. 2010. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 11:1057–1062. 10.1038/ni.1945 [DOI] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608. 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- Tubo N.J., Pagán A.J., Taylor J.J., Nelson R.W., Linehan J.L., Ertelt J.M., Huseby E.S., Way S.S., and Jenkins M.K.. 2013. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 153:785–796. 10.1016/j.cell.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essen D., Kikutani H., and Gray D.. 1995. CD40 ligand-transduced co-stimulation of T cells in the development of helper function. Nature. 378:620–623. 10.1038/378620a0 [DOI] [PubMed] [Google Scholar]
- Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 27:847–859. 10.1016/j.immuni.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E., Kohlhaas S., Lu D., and Leyland R.. 2013. miR-155: an ancient regulator of the immune system. Immunol. Rev. 253:146–157. 10.1111/imr.12057 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ing S., Fraser A., Chen M., Khan O., Zakem J., Davis W., and Quinet R.. 2013. Follicular helper T cells: new insights into mechanisms of autoimmune diseases. Ochsner J. 13:131–139. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.