Zhao and collaborators use linear amplification–mediated high-throughput genome-wide translocation sequencing to examine TCRδ VDJ recombination at an unprecedented resolution.
Abstract
T cell antigen receptor δ (Tcrd) variable region exons are assembled by RAG-initiated V(D)J recombination events in developing γδ thymocytes. Here, we use linear amplification–mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) to map hundreds of thousands of RAG-initiated Tcrd D segment (Trdd1 and Trdd2) rearrangements in CD4−CD8− double-negative thymocyte progenitors differentiated in vitro from bone marrow–derived hematopoietic stem cells. We find that Trdd2 joins directly to Trdv, Trdd1, and Trdj segments, whereas Trdd1 joining is ordered with joining to Trdd2, a prerequisite for further rearrangement. We also find frequent, previously unappreciated, Trdd1 and Trdd2 rearrangements that inactivate Tcrd, including sequential rearrangements from V(D)J recombination signal sequence fusions. Moreover, we find dozens of RAG off-target sequences that are generated via RAG tracking both upstream and downstream from the Trdd2 recombination center across the Tcrd loop domain that is bounded by the upstream INT1-2 and downstream TEA elements. Disruption of the upstream INT1-2 boundary of this loop domain allows spreading of RAG on- and off-target activity to the proximal Trdv domain and, correspondingly, shifts the Tcrd V(D)J recombination landscape by leading to predominant V(D)J joining to a proximal Trdv3 pseudogene that lies just upstream of the normal boundary.
INTRODUCTION
The RAG endonuclease (RAG) initiates V(D)J recombination by introducing DSBs between a pair of variable (V), diversity (D), and joining (J) gene segments and flanking recombination signal sequences (RSSs) to generate a pair of blunt signal ends (SEs) and a pair of hairpin-sealed coding ends (CEs; Schatz and Swanson, 2011; Alt et al., 2013). Bona fide RSSs are composed of a conserved heptamer (consensus: 5′-CACAGTG) and an AT-rich nonamer separated by nonconserved 12- (12RSS) or 23-bp (23RSS) spacers (Schatz and Swanson, 2011). Normal RAG targeting and cleavage occurs only at pairs of coding segments flanked, respectively, by 12RSSs and 23RSSs (Alt et al., 2013). After binding to a Y-shaped RAG heterodimer (Kim et al., 2015; Ru et al., 2015) and subsequent cleavage, the CEs and SEs are held in a postcleavage synaptic complex from which SEs are directly joined to each other and hairpin CEs are opened, processed, and joined to each other (Schatz and Swanson, 2011). The joining steps occur via classical nonhomologous end joining (Alt et al., 2013).
V(D)J recombination occurs in early B and T lymphocyte development and is tightly regulated by modulating accessibility of V, D, and J RSSs to RAG (Alt et al., 2013). Prior studies have shown that V(D)J recombination is initiated from a recombination center (RC) where RAG is recruited by epigenetic modifications and other factors (Matthews and Oettinger, 2009; Desiderio, 2010; Ji et al., 2010). In the immunoglobulin heavy chain (IgH) locus, the initial RC appears to form over the proximal D and JH region (Teng et al., 2015). The IgH locus contains a key V(D)J recombination regulatory element, termed intergenic control region 1 (IGCR1), which lies between the IgH VH and DH gene segments (Guo et al., 2011). IGCR1 regulates IgH V(D)J recombination in the context of lineage specificity, order, and proximal VH feedback regulation (Guo et al., 2011; Lin et al., 2015). IGCR1 function in these contexts relies on a pair of divergently oriented CTCF-binding elements (termed CBE1 and CBE2). Mutation of both CBE1 and CBE2 abrogates all of these forms of regulation and results in strongly increased utilization of the most proximal VH (VH81x) coupled with a major reduction in distal VH utilization (Guo et al., 2011). The two IGCR1 CBEs have been suggested to cooperatively regulate IgH V(D)J recombination by limiting the activity of the DJH RC to a chromosomal loop domain containing the D and JH segments (Lin et al., 2015) and insulating the activity from the VHs portion of the locus (Hu et al., 2015; Lin et al., 2015). Such a mechanism would ensure ordered rearrangement of D-to-JH segments before appendage of a VH segment (Lin et al., 2015).
A potentially new mechanistic aspect of RAG function involving directional, linear tracking was recently implicated through the study that used the linear amplification–mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) approach to follow RAG cleavage and joining events (Hu et al., 2015). This study showed that pairs of bona fide RSSs integrated into a variety of chromosomal loop domains at various sites across the genome promote robust RAG off-target activity at flanking cryptic target sites, with cleavage occurring between convergent CAC motifs and associated surrogate CEs. Such joining is directionally oriented with respect to CAC motifs used and confined within the specific convergently oriented CTCF-anchored loop domains containing the bona fide RSSs (Hu et al., 2015). The mechanism that drives such RAG off-target directional- and orientation-specific joining biases has been proposed to involve unidirectional RAG tracking over great linear distances after being activated in the context of paired bona fide RSSs within a RC. In progenitor (pro)–B cells harboring a DJH rearrangement, such RAG tracking is robust within a V(D)J recombination domain that extended from IGCR1 to just downstream of the DJH RC (Hu et al., 2015). Moreover, deletion of IGCR1 allowed this off-target activity to directionally extend from the DJH to the proximal VH81x, resulting in dramatically increased overutilization of VH81x in joining to the downstream DJH (Hu et al., 2015).
The TCRδ gene (Tcrd) lies within the locus encoding TCRα (Tcra) in a contiguous 1.5-Mb region of the 129 mouse strain (Carico and Krangel, 2015). The 3′ portion of Tcrd consists of two Ds (Trdd1 and Trdd2) upstream of two Js (Trdj1 and Trdj2), followed by Cδ. There are 16 Vδs, five of which lie in a proximal unique region upstream of the Dδs, and one of which is in an inverted orientation downstream of Cδ (Carico and Krangel, 2015). Other Vδs, also used by Tcra, lie at greater distances from Dδs (Carico and Krangel, 2015). Unlike ordered IgH rearrangement, which generates D-to-J rearrangements before V-to-DJ rearrangements (Alt et al., 2013), Tcrd rearrangements in mice have been concluded to be disordered, with intermediate V-to-D, D-to-D, D-to-J, V-to-D(D), and D(D)-to-J joins (Chien et al., 1987; Migone et al., 1995; Carico and Krangel, 2015). Based on RAG binding, the Trdd2-Trdj1 region has been indicated to contain the initiating Tcrd RC (Ji et al., 2010; Teng et al., 2015). Tcrd recombination relies on the Eδ enhancer (Monroe et al., 1999) and occurs early in T cell development in CD4−CD8− double-negative (DN) thymocytes at the DN2/DN3 stage (CD44+CD25+/CD44−CD25+; Capone et al., 1998; Livák et al., 1999; Carico and Krangel, 2015). Tcra recombination is dependent on the Eα enhancer downstream of Cα and on the T early α (TEA) promoter region upstream of the Traj cluster and occurs in CD4+CD8+ double-positive thymocytes (Sleckman et al., 1997; Carico and Krangel, 2015). Like IgH, Tcrd also contains a pair of intergenic CBEs (INT1 and INT2) between the Vδs and Dδs, and INT2 makes a loop (termed a chromatin interaction loop [CIL]) with a downstream convergently oriented CBE within the TEA region (Chen et al., 2015). The INT1-2 CBEs have been implicated in regulating Tcrd and Tcra V(D)J recombination (Chen et al., 2015).
To gain additional insight into how RAG orchestrates V(D)J recombination within the Tcra-Tcrd locus, we now use the LAM-HTGTS approach to map genome-wide junctions from RSSs flanking Trdd2 and Trdd1 gene segments during early T cell development.
RESULTS
LAM-HTGTS detection of joining events involving RAG-initiated DSBs at Trdd2
To study mechanisms of RAG activity and V(D)J recombination control in the Tcrd locus, we performed LAM-HTGTS studies with primary DN2/DN3 T cell precursors that represent the developmental stage at which Tcrd V(D)J recombination occurs (Capone et al., 1998; Livák et al., 1999; Carico and Krangel, 2015). To gain further potential insights into the initiating Tcrd RC (Teng et al., 2015), we examined the published RAG ChIP-seq profiles at higher resolution and noted that the major peak of RAG binding lies over Trdd2 (Fig. S1). Therefore, for initial application of LAM-HTGTS for Tcrd V(D)J recombination studies, we performed LAM-HTGTS studies using RAG-initiated DSBs at RSSs flanking one or the other sides of Trdd2 as bait (Fig. 1, A–C; and Table S1). To generate large numbers of DN2/DN3 T cell precursors for individual libraries, we cultured bone marrow–derived WT hematopoietic stem cells on OP9-DL1 cells (Schmitt et al., 2004; Holmes and Zúñiga-Pflücker, 2009) in the presence of IL-7 and Flt3-L for 14 d (Huang et al., 2005; Zakrzewski et al., 2006).
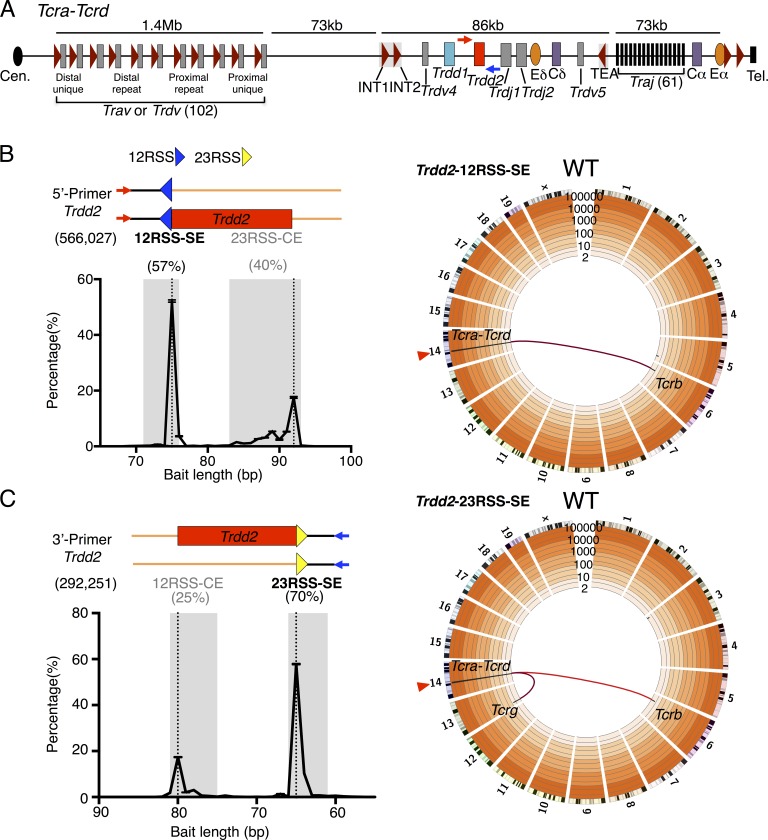
Figure 1.
Genome-wide translocation landscape of DN2/DN3 T cell progenitors. (A) Schematic of murine Tcra-Tcrd locus. Dark red triangles represent position and orientation of CBEs. INT1-2 and TEA CBEs are labeled. Cen., centromere; Tel., telomere. 5′- and 3′-Primers (red and blue arrows, respectively) for generation of Trdd2 LAM-HTGTS libraries are indicated. (B and C, top left) Junction reads are composed of the bait primer (red or blue arrows) and the bait sequence (black lines) leading up to the junction (encompassing bait and prey BE sequences), followed by the prey sequence (orange lines). Relative position of 12RSS (blue triangle), 23RSS (yellow triangle), and Trdd2 gene segment (red rectangle) are indicated. (bottom left) Distribution of bait sequence length, plotted as the percentage of total junctions, corresponds to the relative position of the predicted bait break-sites (dotted lines). Numbers in parentheses denote total junctions analyzed. Ranges of bait sequence lengths of signal end (SE) and coding end (CE) bait libraries are shaded in gray (see Materials and methods for details). Junction percentages of total junctions in indicated ranges are shown above. 5′-Primer (n = 4; B); 3′-Primer (n = 3; C). (right) Circos plots displaying genome-wide prey junction distribution from either Trdd2-12RSS-SE (n = 4; B) or Trdd2-23RSS-SE (n = 3; C) libraries. Bin size is 5 Mb (black bars). Trdd2 bait site (red triangle) and translocations from the bait to Tcr loci hotspots (red links) are indicated. Trdd2-12RSS-SE and Trdd2-12RSS-CE (Fig. S2) are normalized to 76,966 junctions. Trdd2-23RSS-SE and Trdd2-23RSS-CE (Fig. S2) are normalized to 106,193 junctions.
To capture joins involving the 12RSS SE (Trdd2-12RSS-SE) broken ends (BEs), we used a bait primer 75 bp upstream of Trdd2 (5′-Primer; Fig. 1 B). This primer also captures joins to downstream 23RSS CE (Trdd2-23RSS-CE) BEs, but only those on alleles that had not undergone a prior joining event to the upstream Trdd2-12RSS-CE (Fig. 1 B). We used a bait primer 64 bp downstream of Trdd2 (3′-Primer) to capture joins of the 23RSS SE (Trdd2-23RSS-SE) BEs, and also upstream Trdd2-12RSS-CE BEs that occur on alleles that had not undergone a prior joining event to the downstream Trdd2-23RSS-CE (Fig. 1 C). For both the 5′ and 3′ Trdd2 priming strategies, prey junctions with bait sequence lengths that corresponded to either SEs or CEs of the Trdd2 12 or 23 bait RSSs were identified (Fig. 1, B and C; Fig. S2, A and B; and Table S2). As anticipated, more junctions were recovered from primer-proximal SE baits than from primer-distal CE baits (57 vs. 40% for 12RSS and 70 vs. 25% for 23RSS, respectively; Fig. 1, B and C) due to disordered V-to-D, D-to-D, and D-to-J joining of Trdd2 (Chien et al., 1987; Migone et al., 1995; Carico and Krangel, 2015). Given the expected underrepresentation of Trdd2 CE bait junctions, we limit quantitative analyses to SE bait junctions. However, we observed reciprocal junction patterns for Trdd2-12RSS SE and CE baits and also for Trdd2-23RSS SE and CE baits, consistent with normal V(D)J joining (e.g., Figs. 2 and 3).
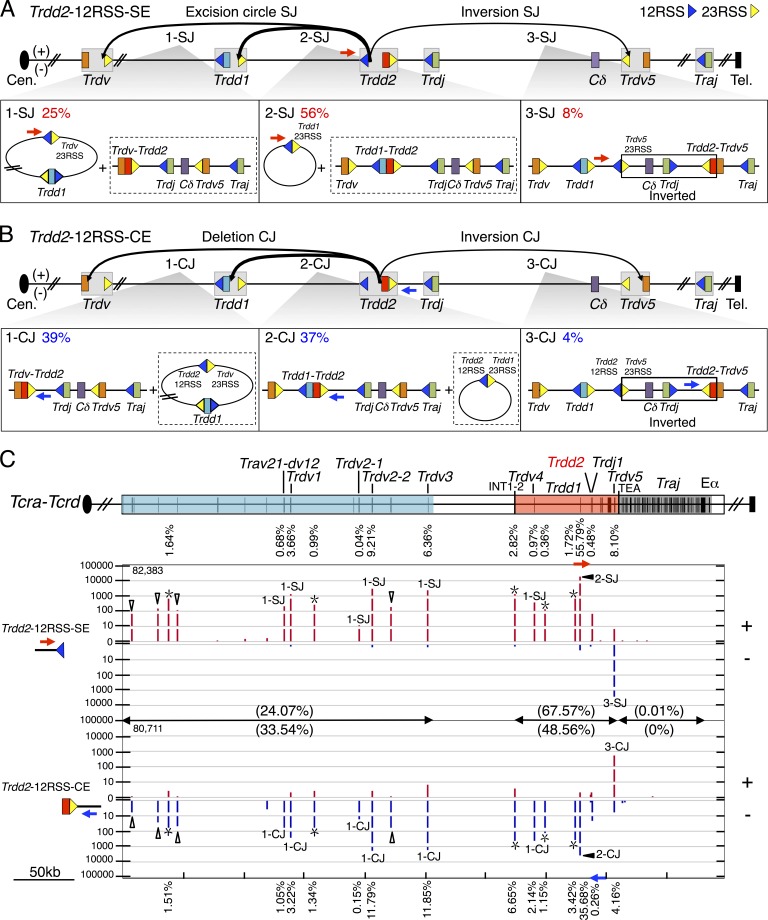
Figure 2.
Joining distribution of Trdd2-12RSS-SE and Trdd2-12RSS-CE libraries across the Tcra-Tcrd locus. (A and B, top) Predicted joining outcomes of Trdd2-12RSS-SE (A) and Trdd2-12RSS-CE (B) baits. Gray boxes indicate gene segments with their flanking RSS(s): 12RSS and 23RSS are represented by blue and yellow triangles, respectively. The used primers are labeled with blue or red arrows. (bottom) Diagram of joining outcomes in excision circles plus deletional joins (1-SJ, 2-SJ, 1-CJ, and 2-CJ); and inversional joins (3-SJ and 3-CJ). The junction percentage of such joining outcomes as of total RAG on-targets are listed in the top left corner. Associated joining outcomes not detected by the primer listed are indicated in dashed box. Inverted regions in 3-SJ and 3-CJ are boxed. (C, top) Gene segment organization of the Tcra-Tcrd locus, organized in a centromere to telomere (+) chromosomal orientation. Proximal unique Travs and Trdvs (blue), the CIL (red), and the downstream Traj cluster (dark gray) are shown. Black vertical bars in the colored regions indicate the position of gene segments. (bottom) IGV plots displaying distribution of RAG on-targets (red in + and blue in – orientations) from pooled Trdd2-12RSS-SE (n = 4) and -CE libraries (n = 3). Junctions are displayed as stacked tracks (i.e., log scale between tracks, linear scale within each track) with the total number of RAG on-targets for the bait used are listed in the top left corner. Junction percentages for each indicated gene segment are labeled above or below their corresponding junction peaks. Open triangles mark Trav junctions and asterisks indicate δREC (δREC1, δREC2, δREC3, δREC4, and δREC5 from centromere to telomere). Junction percentages for each region are denoted in parentheses. Trdd2-12RSS-SE and -CE libraries are normalized to 83,572 total junctions.
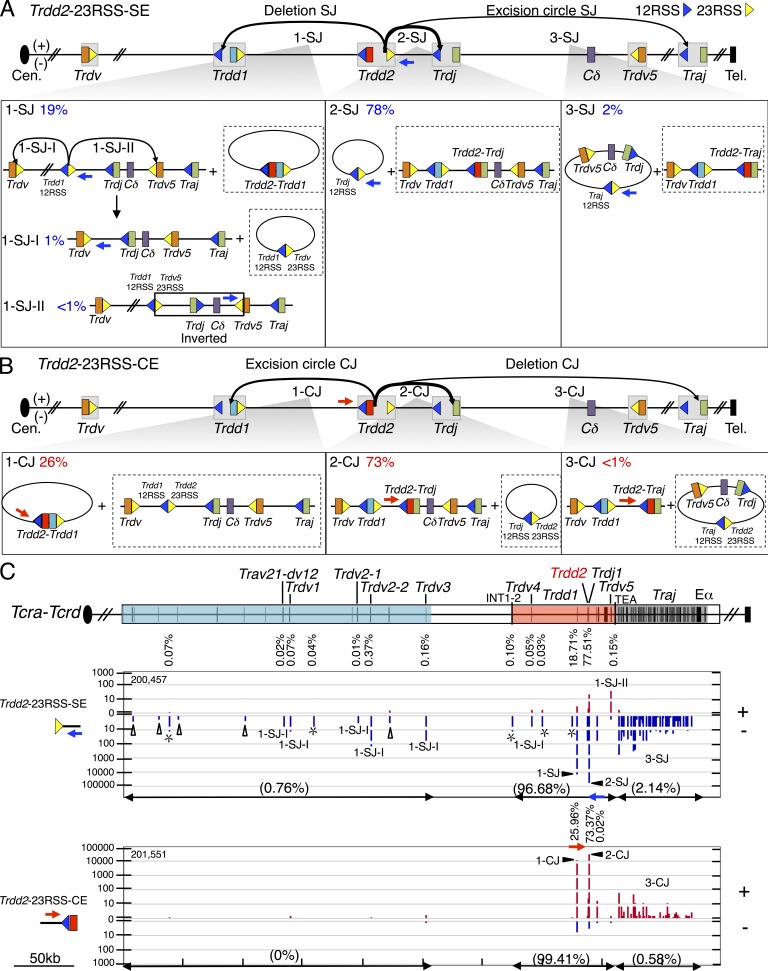
Figure 3.
Joining distribution of Trdd2-23RSS-SE and Trdd2-23RSS-CE libraries across the Tcra-Tcrd locus. (A and B) Schematic of predicted joining outcomes for Trdd2-23RSS-SE (A) and Trdd2-23RSS-CE (B) bait libraries. Each class of joining events links to a lower box illustrating the corresponding joining outcomes in excision circles plus deletional joins (see Fig. 2 legend for other details). 1-SJ panel depicts secondary events from the Trdd1-12RSS-SE/Trdd2-23RSS-SE fusion (i.e., 1-SJ-I and 1-SJ-II). Inverted region in 1-SJ-II is boxed. (C) Gene segment organization of the Tcra-Tcrd locus is displayed in the top panel. IGV plot panels below show junction distribution of pooled Trdd2-23RSS-SE (n = 3) and -CE (n = 4) libraries (see Fig. 2 legend). Trdd2-23RSS-SE and -CE libraries are normalized to 202,196 total junctions.
The highly precise joining of RAG-initiated SE junctions and limited diversity of CE junctions during normal V(D)J recombination tremendously limits junction diversity relative to that recovered, for example, with designer endonucleases. Therefore, we include duplicate junctions in our analysis of LAM-HTGTS libraries of RAG-initiated baits and obtain overall significance by analyzing at least three biological repeats of each experiment with a given bait or genetic background (Hu et al., 2015). We analyzed 323,910 Trdd2-12RSS-SE junctions (four libraries) and 76,966 Trdd2-12RSS-CE junctions (three libraries), as well as 203,485 Trdd2-23RSS-SE junctions (three libraries) and 229,666 Trdd2-23RSS-CE junctions (four libraries; Table S2). For both upstream and downstream Trdd2 baits, >99% of recovered junctions were within the Tcra-Tcrd locus, with very low, but clear-cut, translocation junctions to Tcrb and Tcrg (Fig. 1, B and C; Fig. S2, A and B; and Table S3). Strikingly, though, in contrast to studies performed with designer nuclease bait DSBs in various cell types including DN2/DN3 T cell precursors (Hu et al., 2014; Frock et al., 2015; unpublished data), there were virtually no junctions recovered along the break-site chromosome or from other nonantigen receptor locus genomic sites in Trdd2 bait libraries from WT T cell precursors (Fig. 1, B and C; Fig. S2, A and B; and Table S3).
RAG-initiated Trdd2 SE and CE junctions in WT T cell precursors
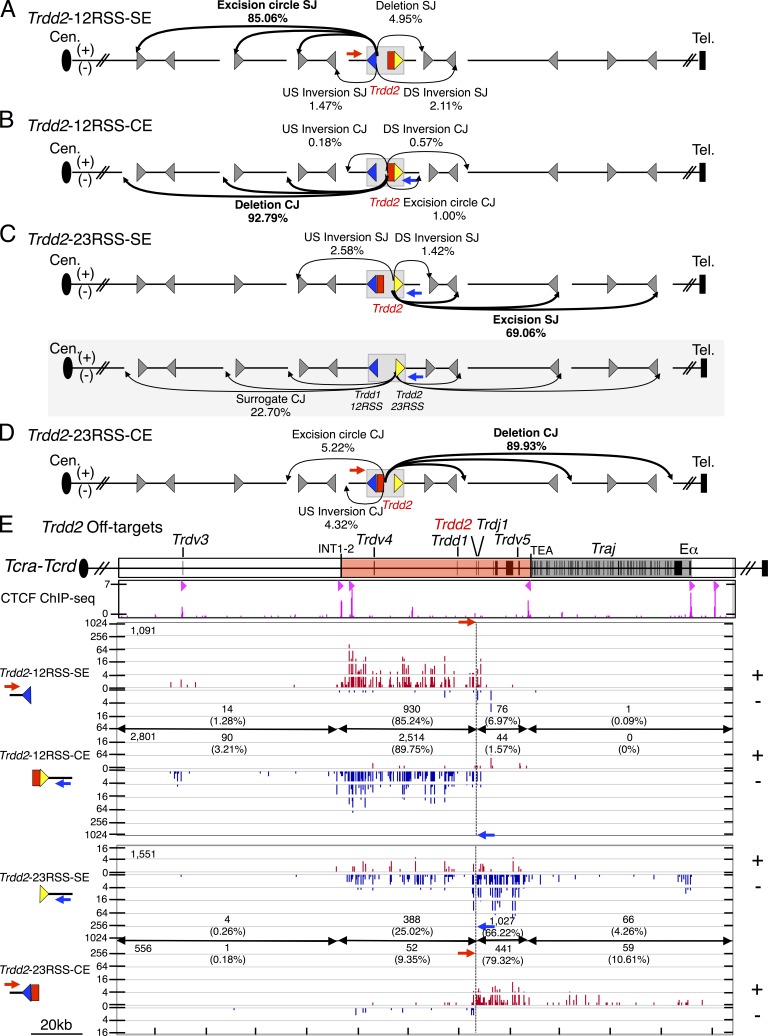
The 12RSS of Trdd2 can pair with bona fide 23RSSs of the Trdd1 or Trdv gene segments (which all lie upstream of Trdd2) resulting in excision circle signal joins (Fig. 2 A) and deletional Trdd1-to-Trdd2 or Trdv-to-Trdd2 coding joins (Fig. 2 B). Both types of coding joins commonly contribute to the normal Tcrd repertoire in mice (Carico and Krangel, 2015). The 12RSS of Trdd2 also can pair with the 23RSS of the inverted Trdv5 that lies downstream of Trdd2, resulting in inversional signal joins and inversional Trdv5-to-Trdd2 coding joins, respectively, which also inverts the intervening sequence between Trdd2 and Trdv5 that contains Trdj1, Trdj2, and Cδ (Fig. 2, A and B). We visualized overall RAG on-target patterns across Tcra-Tcrd via IGV plots (Fig. 2 C; Robinson et al., 2011; Hu et al., 2015). Junctions are displayed in + orientation if the prey sequence aligns in a centromere to telomere direction and in – orientation if prey sequence aligns in the opposite direction (Chiarle et al., 2011; Hu et al., 2015). Trdd2-12RSS-SE and Trdd2-12RSS-CE libraries identified all of the same bona fide RSS sites throughout the Tcrd locus, but, as expected for normal V(D)J recombination, occurred in + and – orientations, respectively (Fig. 2 C).
The most frequent class of joins recovered from the Trdd2-12RSS-SE libraries involved excision circle joining between the Trdd2-12RSS-SEs and the 23RSS of Trdd1 (56%; Fig. 2, A [2-SJ] and C; and Table S4). Trdd2-12RSS-SE libraries contained substantial numbers of excision circle signal joins to 23RSSs of upstream Trdv segments (25%; Fig. 2 A [1-SJ] and C; and Table S4) and also inversional signal joins to the downstream Trdv5 23RSS (8%; Fig. 2, A [3-SJ] and C; and Table S4). The most frequent upstream Trdv-to-Trdd2 joins involved Trdv2-2 (9%) and the Trdv3 pseudogene (6%), which lie at the D-proximal end of the proximal unique region (Fig. 2 C). Trdv2-2 has also been found to be one of the most frequently used Trdvs by repertoire sequencing (Passoni et al., 1997; Weber-Arden et al., 2000). Despite its location in the CIL and close proximity to Trdd2, Trdv4 junctions occurred very rarely (<1%), possibly consistent with its preferential usage in fetal repertoires (Hao and Krangel, 2011). The Trdd2-12RSS-SE also revealed joining to five additional bona fide RSSs not associated with a known coding segment that lie upstream of Trdd2 and therefore qualify as δ-deleting elements (δRECs; Fig. 2 C), termed δREC1, δREC2, δREC3, δREC4, and δREC5 (positioned from centromere to telomere). The strongest of these sites is δREC3, which represents the previously described δREC (de Villartay et al., 1988; Hockett et al., 1989), whereas the other four have not been previously described. δREC2 and δREC3 are conserved across multiple species, whereas δREC1, δREC4, and δREC5 are conserved between mouse and rat (unpublished data). Notably, junctions involving the Trdd2-12RSS-CEs gave the same pattern of joins to all of these Trdvs, Trdd1, and δRECs, but, consistent with normal V(D)J joining, in the opposite orientation (Fig. 2 C).
The 23RSS downstream of Trdd2 can pair with a bona fide 12RSS from the upstream Trdd1 to form deletional signal joins (Fig. 3 A [1-SJ]) and excision circle Trdd2-to-Trdd1 coding joins (Fig. 3 B [1-CJ]). The Trdd2 23RSS can also pair with a 12RSS from the downstream Trdjs leading to excision circle signal joins and normal deletional DJ coding joins (Fig. 3, A [2-SJ] and B [2-CJ]). The Trdd2-23RSS-SE and Trdd2-23RSS-CE libraries again identified all of the same major bona fide RSS sites throughout the Tcrd locus, which, as expected for normal V(D)J recombination, occurred in − and + orientations, respectively (Fig. 3 C). The most frequent class of joins recovered were the excision circle signal joins between the Trdd2-23RSS-SEs and the 12RSS of Trdj1 (78%; Fig. 3, A [2-SJ] and C) and the corresponding normal intra-chromosomal coding joins (Fig. 3, B [2-CJ] and C). However, there were few Trdd2-23RSS-SE joins to the12RSS of Trdj2 (<1%) consistent with earlier findings (Chien et al., 1987; Fig. 3 C). We also found a high frequency of Trdd2-23RSS-SE joins to the 12RSS of Trdd1 (19%; Fig. 3, A [1-SJ] and C) and a high frequency of the reciprocal coding joining of Trdd2-to-Trdd1 (26%) that results in the chromosomal deletion of both Trdd1 and Trdd2 coding gene segments within excision circles (Fig. 3, B [1-CJ] and C). This finding is striking and suggests this class of Trdd recombination could severely diminish repertoire diversity and, given that direct Trdv to Trdj joining appears very infrequent (Table S4), potentially functionally inactivate the Tcrd locus. We also found a small fraction (2%) of excision circle Trdd2-23RSS-SE and Trdd2-23RSS-CE junctions to Traj 12RSS SEs and CE, respectively (Fig. 3, A [3-SJ], B [3-CJ], and C). Finally, we found very low levels of Trdd2-23RSS-SE joins to Trdv-23RSS-CE (Fig. 3, A [1-SJ-I and 1-SJ-II] and C), which appear to arise via a 12/23 restricted intermediate (see the following section).
Profiles of RAG-initiated Trdd1 SE and CE junctions in T cell precursors
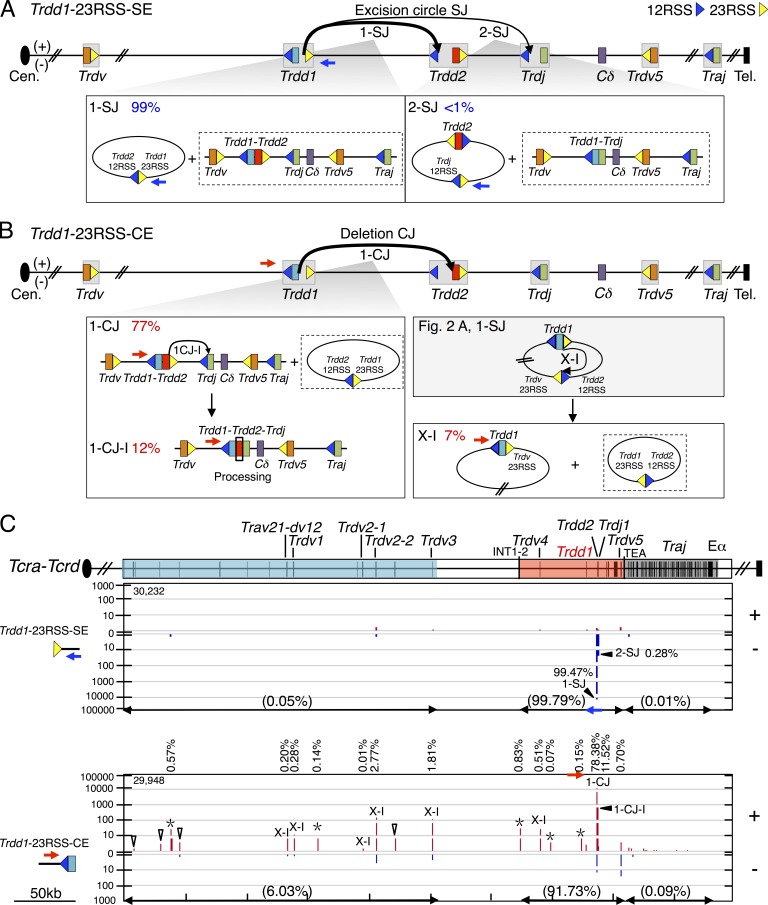
We investigated joining patterns of DSBs generated at Trdd1 bona fide RSSs by using 5′ and 3′ primers flanking Trdd1, as described above for Trdd2 (Fig. S3 A and B; and Table S1; see Materials and methods). We analyzed 36,956 Trdd1-12RSS-SE junctions (three libraries) and 43,699 Trdd1-12RSS-CE junctions (three libraries), as well as 119,344 Trdd1-23RSS-SE junctions (three libraries) and 30,322 Trdd1-23RSS-CE junctions (three libraries; Table S2). Genome-wide joining patterns for all Trdd1 baits used mostly were similar to those described above for Trdd2 baits (Fig. S3, A and B; and Table S3).
Within the Tcra-Tcrd locus, we found a substantial level (15%) of excision circle Trdd1-12RSS-SE joins to 23RSSs associated with upstream proximal Trdv segments, most notably to Trdv2-2 and Trdv3 (Fig. 4, A [1-SJ] and C), with δRECs (4%; Fig. 4 C), and also with the downstream inverted Trdv5 (6%; Fig. 4, A [3-SJ] and C). However, recovery of Trdd1-12RSS-CE joins to Trdv segments were surprisingly rare (4%; Fig. 4 B [1-CJ and 3-CJ]), which in combination with the total Trdd1-12RSS-SE junctions involving Trdv 23RSSs (>20% of junctions; Fig. 4 A [1-SJ and 3-SJ]) raises the possibility that Trdd1 joining is, in fact, ordered. Consistent with this notion, the vast majority of joins recovered from Trdd1-23RSS-SE libraries involved excision circle joining between Trdd1-23RSS-SE and the 12RSS of Trdd2 (99%; Fig. 5, A [1-SJ] and C) with surprisingly few Trdd1-23RSS SE joins to the 12RSS of Trdj (<1%; Fig. 5, A [2-SJ] and C). Although we observe apparent coding joining to Trdj1 from the Trdd1-23RSS-CE, this joining is not direct, as these junctions contain intervening Trdd2 coding sequence (Fig. 5 B [1-CJ-I]). We also addressed this finding by analyzing Trdj1-12RSS-SE bait libraries (404,591 junctions; three libraries; Tables S1 and S2). These studies confirmed that <1% of Trdj1-12RSS-SE junctions joined directly to Trdd1, whereas 99% joined directly to Trdd2 (Table S4). As expected, Trdd1-Trdd2-Trdj1 sequential joining was also detected from Trdj1-12RSS-CE libraries (40,274 junctions; three libraries; Tables S1, S2, and S4). Overall, this set of findings indicates that for Trdd1 segments with an intact 23RSS, Trdd1 12RSS rarely joins to Trdvs; moreover, the Trdd1 23RSS rarely joins to Trdj. Therefore, in contrast to prior expectations, it appears that most mature VDJ junctions involving Trdd1 arise from a Trdd1-to-Trdd2 intermediate via an ordered, as opposed to disordered, joining process.
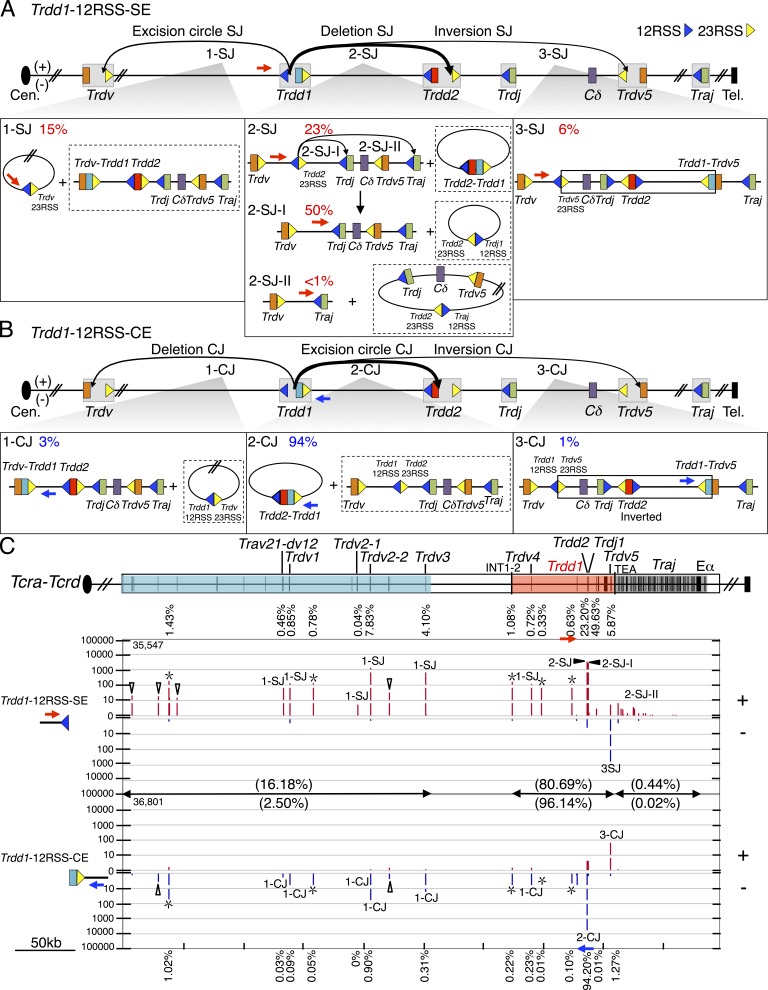
Figure 4.
Joining distribution of Trdd1-12RSS-SE and Trdd1-12RSS-CE libraries across the Tcra-Tcrd locus (A and B) Predicted joining outcomes for Trdd1-12RSS-SE (A) and Trdd1-12RSS-CE (B) bait libraries. Labels (e.g., 1-SJ) for each class of joining events correspond to a lower box displaying joining outcomes and their corresponding percentages as of total RAG on-target joining events (see Fig. 2 legend). 2-SJ panel includes secondary events arising from Trdd1-12RSS-SE/Trdd2-23RSS-SE fusions (i.e., 2-SJ-I and 2-SJ-II). (C, top) Tcra-Tcrd gene segment organization. (C, bottom) Profile of RAG on-targets displayed by IGV from pooled Trdd1-12RSS-SE (n = 3) and -CE libraries (n = 3; see Fig. 2 legend). Trdd1-12RSS-SE and -CE libraries are normalized to 36,596 total junctions.
Figure 5.
Joining distribution of Trdd1-23RSS-SE and Trdd1-23RSS-CE libraries across the Tcra-Tcrd locus. (A and B) Diagram of expected joining outcomes for Trdd1-23RSS-SE (A) and Trdd1-23RSS-CE (B) libraries. Each event links to a lower box showing joining outcomes resulting in excision circles plus deletional joins. (B) Event 1-CJ-I indicates hairpin processing on both sides of Trdd2. Event X-I comes from the excision circle harboring Trdv-23RSS and Trdd2-12RSS, described in Fig. 2 A. (C, top) Organization of gene segments in the Tcra-Tcrd locus. (bottom) IGV plots displaying distribution of RAG on-target junctions (red in + and blue in – orientations) from pooled Trdd1-23RSS-SE (n = 3) and -CE libraries (n = 3; see Fig. 2 legend). Trdd1-23RSS-SE and -CE libraries are normalized to 30,313 total junctions.
We also found several Trdd1 joining patterns, some of which are major, that had not been previously recognized. First, we found that the 12RSS of Trdd1 also joins to the 23RSS of Trdd2 to generate a substantial level (23%) of deletional signal joins, which would leave the two fused 12RSS/23RSS SEs in the chromosome (Fig. 4, A [2-SJ] and C). Remarkably, most (94%) of the recovered Trdd1-12RSS-CE joins are to the 23RSS CE of Trdd2, which resulted in deletion of the fused Trdd1 and Trdd2 segments within excision circles, and thus would severely reduce repertoire diversity (Fig. 4, B [2-CJ] and C). Although these 12RSS SE and CE joins would be expected to be reciprocal products, their differential recovery (23 vs. 94%) is striking. An explanation, however, comes from our finding that 50% of recovered Trdd1-12RSS-SE junctions occur at the CE of Trdj1 (Fig. 4, A [2-SJ-I] and C). and appear to represent secondary rearrangements of the fused Trdd1-12RSS-SE/Trdd2-23RSS-SE junctions that explain the lower than expected recovery of the latter junctions. Although these junctions might first suggest apparent hybrid joins that seem to break the 12/23 rule, they appear to arise as a secondary recombination event of the fused Trdd1-12RSS-SE/Trdd2-23RSS-SE (Fig. 4 A [2-SJ]; and Fig. S4 A) in which the Trdd2-23RSS SE subsequently paired with the Trdj1-12RSS SE and the Trdd1-12RSS SE is used as a surrogate CE to join to Trdj1 (Fig. 4 A [2-SJ-I]; and Fig. S4 A). Consistent with this interpretation, the 12RSS side of most of these junctions shows end-processing expected for CE joining, which would then inactivate the RSS as a further RAG target (Fig. S4 A). We also observe evidence for this type of secondary V(D)J joining activity for Trdd1-23RSS-CEs joining to 23RSSs of Trdv segments (Fig. 5 B, X-I), which most likely occurs within excision circles harboring Trdd1 that are generated via Trdd2-12RSS and Trdv-23RSS joins (Fig. 2 A [1-SJ]). The reciprocal joining outcome of excision circles harboring Trdd1-23RSS/Trdd2-12RSS fusions (Fig. 5 B, X-I) is readily detected from Trdd2-12RSS-SE and Trdd1-23RSS-SE libraries and explains why the observed frequency of Trdd1-to-Trdd2 joining events is slightly higher in the SE library than in the corresponding CE library (Figs. 2, A [2-SJ] and B [2-CJ; and 5, A [1-SJ] and B [1-CJ and 1-CJ-I]).
Direction- and orientation-specific joining of RAG off-target DSBs at the Tcra-Tcrd locus
LAM-HTGTS studies with Trdd2-12RSS and Trdd2-23RSS bait BEs also revealed thousands of lower level, but reproducible, off-target junctions that amounted to ∼1% of the total junctions (Table S2). These off-target junctions were largely generated at convergent CAC motifs (Fig. 6, A-E; Fig. S5, A-D; and Table S5). In this regard, 85% of Trdd2-12RSS-SE junctions were joined perfectly to upstream convergent CAC motifs resulting in excision circle junctions (+ orientation; Fig. 6, A and E). Likewise, ∼93% of the Trdd2-12RSS-CE junctions occur upstream and involve the surrogate CEs associated with convergent CAC motifs, resulting in end-processed deletional junctions (Fig. 6, B and E). Both types of these upstream RAG off-target junctions stop abruptly at the CBE-containing INT1-2 elements (Fig. 6 E). A large fraction (69%) of Trdd2-23RSS SE junctions were perfectly joined to downstream convergent CAC motifs, resulting in excision circle (− orientation) junctions; whereas the majority (90%) of Trdd2-23RSS-CEs joined downstream to surrogate CEs associated with these CACs to form deletional, end-processed junctions (Fig. 6, C-E). The downstream junctions also largely terminate at the CBE-containing TEA element, which forms a convergent loop with INT2 (Fig. 6 E).
Figure 6.
Direction- and orientation-specific off-target joining of RAG-initiated Trdd2 rearrangements. (A–D) Joining outcomes to cRSS (CAC) sites (gray triangle) from Trdd2-12RSS-SE (n = 4; A), -CE (n = 3; B), Trdd2-23RSS-SE (n = 3; C), and -CE (n = 4; D) libraries. Diagram of joining outcomes from Trdd2-23RSS-SE bait in the fused Trdd1-12RSS/Trdd2-23RSS configuration to cRSSs is shaded in gray (C). Junction percentages as of total RAG off-targets in excision circle, deletion, upstream (US) inversion, and downstream (DS) inversion joining events are shown. Red and blue arrows indicate the position and orientation of 5′- and 3′-Primers, respectively. Dotted line indicates the position of bait BEs. (E, top) Gene segment organization of the region between Trdv3 and Eα (3′ portion of the Tcra-Tcrd locus). CIL (red) and Traj cluster (dark gray) are indicated. (middle) ChIP-seq profile of CTCF. CBE orientation is marked by light red triangles. (bottom) IGV plots of RAG off-target joining patterns of Trdd2-12RSS-SE and -CE and Trdd2-23RSS-SE and -CE libraries. The total number of RAG off-targets is displayed on the top left corner of each plot. Junction numbers and percentages as of total RAG off-targets in indicated regions are shown.
The majority (74%) of off-target Trdd1-12RSS-SE junctions occurred to upstream convergent CAC motifs (Fig. S5 E), resulting in excision circles and terminated at INT1-2 (Fig. S6, A and E). A smaller portion of Trdd1-12RSS-SE junctions (20%) occurred to downstream surrogate CEs resulting in deletions (Fig. S6, A and E); characteristics of these junctions, including Trdd1-12RSS end processing, indicate that they occur from the fused Trdd1-12RSS-SE/Trdd2-23RSS-SE junctions with the Trdd1-12RSS-SE acting as a surrogate CE (Fig. S4 A). Trdd1-12RSS CE, Trdd1-23RSS-SE, and Trdd1-23RSS-CE off-target joins were found much less frequently (Fig. S6, B–E), and apparently can be explained by ordered Trdd1 joining to Trdd2.
These analyses also identified certain unanticipated types of off-target junctions. In this regard, we identified a surprisingly high frequency (25%) of deletional (− orientation) Trdd2-23RSS-SE junctions to surrogate CEs associated with upstream CAC motifs (Fig. 6, C and E). Notably, the 23RSS SEs involved in such joins frequently showed end processing (Fig. S4 C), consistent with functioning as surrogate CEs from fused Trdd1-12RSS/Trdd2-23RSS junctions (see above). We also found a low, but reproducible, level of Trdd1-23RSS-CE junctions to upstream CACs that are used as surrogate CEs (Fig. S5 F; and Fig. S6, D and E); these junctions likely occur within Trdd2-12RSS/CAC excision circles harboring unrearranged Trdd1 segments (Fig. S6 D).
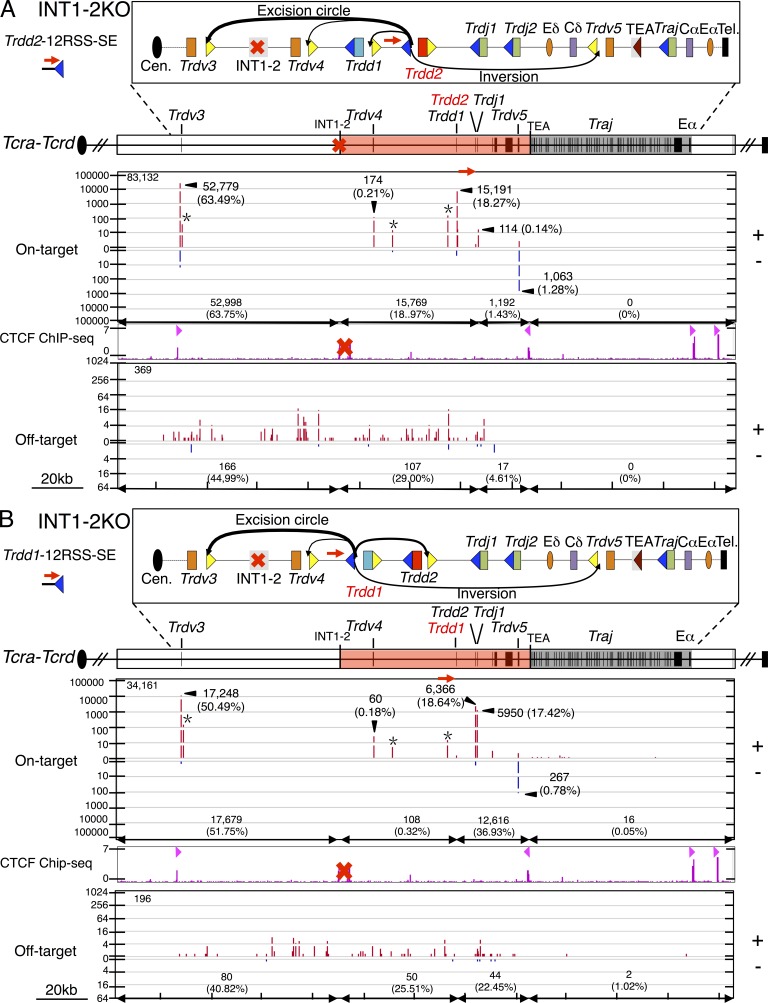
INT1-2 blocks RAG-mediated joining to upstream neighboring domain
To assess the functional role of the INT1-2 CBEs in regulating RAG on- and off-target joining patterns, we performed LAM-HTGTS with Trdd2-12RSS and Trdd2-23RSS bait BEs on DNA from cultured INT1-2–deficient T cell precursors generated from homozygous INT1-2–deficient mice (Chen et al., 2015). The frequency of Trdd2-12RSS-SE joins to bona fide 23RSSs within the Trdd2 to INT1-2 interval decreased in INT1-2–deficient T cell precursors versus those of WT, with the greatest decreases for Trdd1 (threefold; Fig. 2 C; Fig. 7 A; and Table S4). We also observed a large decrease in the frequency of Trdd2-12RSS-SE joins to the downstream bona fide inverted 23RSS of Trdv5 (sixfold; Fig. 2 C; Fig. 7 A; and Table S4). In contrast, the frequency of Trdd2-12RSS-SE junctions with the Trdv3 23RSS, which lies 73 kb upstream of the INT1-2 locale, markedly increased (10-fold) in INT1-2–deficient T cell precursors relative to those of WT (Fig. 2 C; Fig. 7 A; and Table S4). We also found low level, but reproducible, Trdd2-12RSS-SE junctions to an apparently bona fide 23RSS ∼1 kb downstream of Trdv3 in INT1-2–deficient, versus WT, T cell precursors (Fig. 7 A and Table S4). Trdd2-12RSS-SE RAG off-target joins within the Trdd2 to INT1-2 locale interval also decreased in INT1-2–deficient versus WT T cell precursors; however, off-targets then spread 73 kb upstream of the INT1-2 locale to Trdv3 (Fig. 6 E; Fig. 7 A; and Table S4). We also observed similar differences in RAG on- and off-target joining to the 12RSS CEs in INT1-2–deficient versus WT T cell precursors (Table S4). However, Trdd2-23RSS-SE and Trdd2-23RSS-CE RAG on- and off-target joining patterns were similar between INT1-2–deficient and WT, consistent with these joins mainly occurring downstream of Trdd2 and, therefore, not being impacted by the upstream INT1-2 deletion (Table S4).
Figure 7.
INT1-2 CBEs block RAG tracking across the insulated neighboring domain. (A and B, top) Gene segment organization of the 3′ portion of the Tcra-Tcrd locus. Diagram of predicted RAG on-target joining outcomes of INT1-2–deficient Trdd2-12RSS-SE (A) or Trdd1-12RSS-SE (B) libraries is displayed in the box above. (bottom) IGV plots of RAG on- and off-target joining patterns for INT1-2 deficient Trdd2-12RSS-SE (n = 3; A) and Trdd1-12RSS-SE (n = 3; B) bait libraries. Profile of CTCF ChIP-seq is shown and CBE orientation is marked by light red triangles. Red “X” represents the INT1-2 deletion. INT1-2–deficient Trdd2-12RSS-SE and Trdd1-12RSS-SE libraries are normalized to the respective WT libraries described in Figs. 2 and 4. Total RAG on- and off-target numbers of each bait library are listed in the top left corner. RAG on- and off-target numbers and percentages as of the total number of each category in indicated regions are shown.
We also performed LAM-HTGTS with Trdd1-12RSS and Trdd1-23RSS bait BEs on genomic DNA from INT1-2–deficient T cell precursors. In the INT1-2–deficient T cell precursors, we find decreased joining to bona fide 23RSSs in the Trdd1 to the INT1-2 locale interval and a corresponding increase in joining to the Trdv3 23RSS (12-fold) and to the bona fide 23RSS downstream of Trdv3 (Fig. 4 C; Fig. 7 B; and Table S4). Likewise, RAG off-target joining was decreased within the Trdd1 to INT1-2 locale interval in the INT1-2–deficient T cell precursors with off-target activity spreading to Trdv3 (Fig. S6 E; Fig. 7 B; and Table S4). Although we rarely detected Trdd1-12RSS-CE joining to Trdvs in WT precursors, INT1-2 deficiency led to substantial joining of the Trdd1-12RSS-CE to Trdv3 (12.49% of total bona fide joining), indicating increased disordered joining (Table S4). Again, Trdd1-23RSS-SE and CE joining patterns were not substantially altered in INT1-2–deficient T cell precursors (Table S4).
DISCUSSION
Our LAM-HTGTS studies provide strong confirmation of disordered Trdd2 gene segment joining, which comes specifically from the detection of Trdd2-to-Trdj1, Trdd2-to-Trdd1, and Trdd2-to-Trdv as major V(D)J recombination intermediates from Trdd2 CE baits. However, in contrast to Trdd2 disordered joining, primary downstream joining of Trdd1 occurred almost exclusively (99% of junctions) to the upstream CE of Trdd2, either to the germline Trdd2 segment or to a Trdd2-Trdj1 intermediate. These findings indicate that Trdd1 joining is ordered and occurs to Trdd2 before Trdd1-Trdd2 joining to Trdvs via Trdd1-12RSS/Trdv-23RSS pairing. Ordered rearrangement of Trdd1, as opposed to the disordered rearrangement of Trdd2, can be mechanistically understood based on the observation that Trdd2, as opposed to Trdd1, is highly accessible in DN thymocytes (Carabana et al., 2005; Hao and Krangel, 2011) and appears to be the initiating RC (Teng et al., 2015). Thus, RAG bound to the Trdd2-12RSS may capture either Trdd1 or Trdv gene segments, whereas the RAG-poor and weakly accessible Trdd1-12RSS would be much less likely to capture Trdv gene segments. A corollary to this would be that the upstream Trdd1-12RSS must be activated as a RAG substrate once brought into the RC by Trdd1-to-Trdd2 rearrangement. Previous studies have identified germline Tcrd transcripts initiating within Trdd2 and upstream of and within Trdj1, as well as a strong promoter associated with Trdd2 (Carabana et al., 2005). This promoter is likely responsible for Trdd2 accessibility and formation of the Tcrd RC, but it would be disrupted by Trdd1-to-Trdd2 rearrangement. Although analysis of unrearranged alleles identified no germline transcripts mapping to Trdd1, and no substantial promoter activity associated with Trdj1 (Carabana et al., 2005), it is possible that promoter activity associated with one or the other is enhanced by Trdd1-to-Trdd2 rearrangement, allowing RAG-loading at the Trdd1-12RSS to stimulate Trdv-to-Trdd1-Trdd2 rearrangement.
Beyond the well-defined δREC 120 kb upstream of the Trdd gene segments (Janowski et al., 1997; Krangel et al., 1998), we also discovered additional, previously uncharacterized δRECs that between them provide nearly 70% of the deletional activity of δREC and, as a result of their location in the INT1-2-TEA loop or in the proximal unique region, generate variably sized truncations of the Tcrd locus upon joining to Trdd segments. We find that nearly 10% of all such Tcrd V(D)J rearrangements to the upstream 12RSS of Trdd2 occur to this overall set of δRECs, recombination events that would functionally diminish repertoire diversity and potentially severely impair Tcrd V(D)J recombination. Moreover, we also found an additional, even more frequent, mechanism that could lead to Tcrd inactivation. Joins between Trdd1-12RSSs and Trdd2-23RSSs contribute ∼20% of all Trdd2 23RSS rearrangements. This joining event deletes both Trdd1 and Trdd2 segments, leaving in the chromosome the perfectly fused Trdd1-12RSS-SE/Trdd2-23RSS-SE. Such fused 12 and 23 RSSs can be recut and joined as surrogate CEs (Hu et al., 2015). In this regard, we find joining of the Trdd1 12RSSs to Trdj1 and joining Trdd2 23RSS to Trdvs. However, in most of these junctions the Trdd2 or Trdd1 SEs serving as surrogate CEs are processed via normal CE junctional diversification mechanisms and, thus, are inactivated. Together, our findings directly support the notion that Tcrd V(D)J recombination evolved deletional mechanisms to developmentally delete Tcrd and, thereby, to promote appropriate V(D)J recombinational activation of the greater Tcra locus in which Tcrd is embedded (Chen et al., 2015).
Analyses of INT1-2–deficient mice the DN thymoctyes revealed partial inhibition of γδ T cell development and a twofold increase in usage of Trdv2-2 (Vδ4), the most proximal functional Trdv in the unique region upstream of INT1-2 (Chen et al., 2015). Our current LAM-HTGTS analysis of in vitro differentiated DN2/DN3 thymocytes, the stage at which Tcrd rearrangement occurs (Capone et al., 1998; Livák et al., 1999; Carico and Krangel, 2015), confirmed a modest increase in Trdv2-2 usage. However, our current studies further revealed a major 10-fold increase in usage of Trdv3, a pseudogene that is the most proximal upstream Trdv to INT1-2. The markedly increased usage of this Trdv pseudogene in primary rearrangements in the absence of INT1-2 offers a plausible explanation for the γδ T cell developmental defect in INT1-2–deficient thymocytes. Notably, we also find directional joining consistent with RAG tracking upstream from the Trdd2 RC 12RSS to the INT1-2 loop domain boundary in differentiating DN2/DN3 thymocytes, as revealed by junctions to ∼70 different sets of cryptic RSSs and associated surrogate CEs across this domain. This upstream tracking abruptly ends at INT1-2. Correspondingly, INT1-2 deletion, which disrupts the upstream boundary of the INT2-to-TEA loop domain (Chen et al., 2015), allows apparent RAG tracking to continue upstream of the INT1-2 locale into the proximal Trdv domain. Thus, INT1-2 function in sequestering the Trdd2 RC appears similar to that of IGCR1 in sequestering the IgH DJH RC (Guo et al., 2011; Hu et al., 2015), and suggests a potential contribution of RAG tracking to increased Trdv3 utilization. Finally, RAG also potentially tracks downstream from Trdd2 23RSS to TEA, as revealed by directional junctions to 12 convergent cryptic RSSs in this 24-kb region.
Overall, our findings demonstrate that the INT2-to-TEA loop domain largely confines directional RAG joining activity from the initial Trdd2 RC. Correspondingly, major RAG off-target activity is also confined within this domain, helping to suppress RAG activity during Tcrd V(D)J recombination at the huge number of potential off-target sites genome-wide. We do find low-level RAG-initiated Tcrd DSB translocations to DSBs in other TCR loci, consistent with the latter loci undergoing higher levels of DSBs, because they are also RAG targets, than other genomic loci. We also found that the level of such translocations was substantially increased in the absence of ATM (unpublished data), consistent with our prior finding of increased RAG-initiated translocations to Ig loci in ATM-deficient pro–B cells (Hu et al., 2015). Thus, our current findings support the proposal that, in contrast to other types of DSBs, RAG-generated DSBs to two target sites within a chromosomal antigen receptor locus loop are fused in association with the initiating RAG post-cleavage complex by being directly channeled into nonhomologous DNA end-joining (Bredemeyer et al., 2006; Deriano et al., 2011; Hu et al., 2015).
MATERIALS AND METHODS
Mice
The INT1-2KO mice have been previously described (Chen et al., 2015). Mouse work was performed under protocols approved by the Boston Children’s Hospital (Boston, MA) and the Duke University (Durham, NC) Institutional Animal Care and Use Committees.
OP9-DL1 co-culture
The generation of DN2/DN3 T lymphocytes from adult bone marrow cells have been previously described (Huang et al., 2005; Holmes and Zúñiga-Pflücker, 2009). In brief, OP9-DL1 stromal cell lines were maintained in α-MEM (12571–071; Invitrogen), supplemented with 20% FBS (10438–026; Invitrogen). Flt3-L (427FL; R&D Systems) and IL-7 (407-ML; R&D Systems) were used at a concentration of 5 ng/ml each during co-culturing. After 14 d of differentiation, stromal cells and B cells were excluded by MACS-negative selection using CD140a (130–101-547; Miltenyi Biotec) and B220 (130–049-501; Miltenyi Biotec) MicroBeads, respectively. Genomic DNA was collected for LAM-HTGTS library preparation.
LAM-HTGTS and junction mapping
Primers used to generate libraries of Trdd2 and Trdd1 RAG-initiated bait-ends are listed in Table S6. No restriction enzyme blocking was performed for Trdd2 and Trdd1 libraries. All LAM-HTGTS libraries were sequenced by Illumina Miseq and have been described previously (Hu et al., 2016). Junctions were mapped genome-wide using custom circos plots (Krzywinski et al., 2009; Frock et al., 2015) and mapped locally using IGV plots (Robinson et al., 2011; Hu et al., 2015).
Data analysis and normalization
Preprocessing of Miseq reads has been described previously (Frock et al., 2015). Processed reads were aligned using Bowtie2 (Langmead and Salzberg, 2012) to a modified-mm9 genome, in which Tcra-Tcrd locus at positions chr14: 52971974–54848751 was replaced with a 1666365nt-long segment from the 129S1/SvImJ strain (NT_039614.1). Tcra-Tcrd locus annotation was modified accordingly. For libraries from RAG-initiated bait-ends, we included duplicate junctions for analysis (Hu et al., 2015). We compared libraries, including duplicates, with those containing only unique junctions and observed similar patterns between them. We isolated the 12RSS-SE and 23RSS-CE libraries from 5-Primer libraries, as well as 23RSS-SE and 12RSS-CE libraries from 3-Primer libraries according to their coordinated bait sequence length (see the following paragraph). RAG-initiated bait-end libraries prepared from WT and INT1-2–deficient cells were normalized to the isolated junction numbers for comparison.
Isolating SE and CE bait junctions from the library generated by one common primer
We used specific criteria to separate SE and CE bait junctions from the same library. From the predicted SE and CE, we included an additional nucleotide beyond the predicted break site with respect to the primer used, to take into account the small fraction of junctions that coincidentally align beyond the predicted position as a result of nucleotide addition activities. We included several nucleotides for each predicted SE and CE, which represents the end processing of the bait BE sequences. Therefore, junctions with 71–76-bp bait sequence lengths were isolated from 5′-Trdd2 LAM-HTGTS libraries as 12RSS-SE bait-end libraries, and 84–92-bp bait sequence lengths were similarly isolated for 23RSS-CE bait-end libraries, and for 3′-Trdd2 LAM-HTGTS libraries, 61–66-bp bait sequence lengths were separated for 23RSS-SE and 75–81-bp for 12RSS-CE. 5′-Trdd1 LAM-HTGTS libraries were similarly isolated, with 91–103-bp and 106–112-bp representing 12RSS-SE and 23RSS-CE bait sequence lengths, respectively, and for 3′-Trdd1 LAM-HTGTS libraries, 91–98- and 101–107-bp bait sequence lengths for 23RSS-SE and 12RSS-CE, respectively were isolated. Junctions with 46–52-bp bait sequence lengths were isolated from 5′-Trdj1 libraries for Trdj1-12RSS-SE and 45–52-bp bait sequence lengths were isolated from 3′-Trdj1 libraries for Trdj1-12RSS-CE.
Adjusting the sequential joins containing Trdd2 sequence
For Trdd1-23RSS-CE and Trdj1-12RSS-CE libraries, insertion sequences of junctions were screened for Trdd2 consensus sequence by using a sliding window approach with a 1-bp step size and a window size of 5 bp across Trdd2 (12 iterations), and such junctions were manually adjusted back to Trdd2.
Junction hotspots and RAG on- and off-target identification
Junction-enriched regions were identified by MACS2 (Zhang et al., 2008) with custom parameters (extsize, 20 bp; FDR cut-off, 10−9). Enriched regions identified in at least three individual libraries were considered as recurrent hotspots and used in the following study. The bona fide RSS sequence information flanking Tcr gene segments was collected from the IMGT/GENE-DB database (Giudicelli et al., 2005). Hotspots that overlapped with bona fide RSS were recognized as RAG on-targets. Hotspots positioned <10 bp away from the simple CAC motif were defined as associated with RAG off-targets.
ChIP-Seq
We obtained CTCF ChIP-seq data from DN thymocytes from (Shih et al., 2012; available from GEO under accession no. GSM1023416), RAG1, RAG2, and H3K4me3 ChIP-seq data from (Teng et al., 2015; available from GEO under accession nos. GSM1701786, GSM1701790, and GSM30317). Data were reanalyzed with our mm9-modified genome using the ChIP-seq data analysis pipeline Chilin.
Accession code
The GEO accession no. for the datasets reported in this paper is GSE79892.
Online supplemental material
Fig. S1 shows the high-level enrichment of RAG binding and H3K4me3 at Trdd2. Fig. S2 displays genome-wide distribution of prey junctions from Trdd2 12RSS and 23RSS CE libraries. Fig. S3 shows bait length and genome-wide prey junction distributions baited from RAG-initiated Trdd1 DSBs at Trdd1. Fig. S4 includes examples of precise signal joins and processed surrogate coding joins. Fig. S5 shows junctions not corresponding to bona fide RSS sites are associated with RAG off-targets. Fig. S6 displays the profiles of off-targets baited from RAG-initiated DSBs at Trdd1. Table S1 is the summary of all LAM-HTGTS libraries from various bait-ends. Table S2 is the summary of total translocations to RAG on- or off-targets identified by RAG-initiated bait-ends flanking Trdd2, Trdd1, and Trdj1. Table S3 lists the percentage of relative junction distributions of all LAM-HTGTS libraries from RAG-initiated bait-ends flanking Trdd2, Trdd1 and Trdj1. Table S4 lists Tcra-Tcrd gene segments and joining percentages from RAG-initiated bait-ends flanking Trdd2, Trdd1, and Trdj1 in T cell precursors. Table S5 lists of δRECs and RAG off-target sites identified at Tcra-Tcrd via LAM-HTGTS of WT T cell precursors. Table S6 lists LAM-HTGTS oligos used to clone Trdd2, Trdd1, and Trdj1 bait-end junctions. Tables S1–S6 are available as Excel files. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20160670/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alt laboratory members for helpful comments and in-depth discussions.
This work was supported by National Institutes of Health (NIH) grants AI020047 (F.W. Alt) and R37 GM41052 (M.S. Krangel). F.W. Alt is a Howard Hughes Medical Institute Investigator. R.L. Frock was supported by the NIH National Research Service Award T32AI007512.
The authors declare no competing financial interests.
Author contributions: L. Zhao, R.L. Frock, and F.W. Alt designed the study. L. Zhao performed all the experiments. L. Chen and M.S. Krangel provided the INT1-2–deficient mice. Z. Du helped with statistical analyses. J. Hu provided suggestions during manuscript preparation. L. Zhao, R.L. Frock, M.S. Krangel, and F.W. Alt interpreted the data, designed the figures, and wrote the manuscript.
Footnotes
Abbreviations used:
- BE
- broken end
- CBE
- CTCF-binding element
- CE
- coding end
- CIL
- chromatin interaction loop
- CJ
- coding join
- δREC
- δ-deleting element
- DN
- double negative
- IGCR1
- intergenic control region 1
- LAM-HTGTS
- linear amplification–mediated high-throughput genome-wide translocation sequencing
- RAG
- RAG endonuclease
- RC
- recombination center
- RSS
- recombination signal sequence
- SE
- signal end
- SJ
- signal join
- TEA
- T early α
References
- Alt F.W., Zhang Y., Meng F.-L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.-Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., and Sleckman B.P.. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470. 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Capone M., Hockett R.D. Jr., and Zlotnik A.. 1998. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44+CD25+ Pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 95:12522–12527. 10.1073/pnas.95.21.12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabana J., Ortigoza E., and Krangel M.S.. 2005. Regulation of the murine Ddelta2 promoter by upstream stimulatory factor 1, Runx1, and c-Myb. J. Immunol. 174:4144–4152. 10.4049/jimmunol.174.7.4144 [DOI] [PubMed] [Google Scholar]
- Carico Z., and Krangel M.S.. 2015. Chromatin dynamics and the development of the TCRα and TCRδ repertoires. Adv. Immunol. 128:307–361. 10.1016/bs.ai.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Chen L., Carico Z., Shih H.-Y., and Krangel M.S.. 2015. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat. Immunol. 16:1085–1093. 10.1038/ni.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R., Zhang Y., Frock R.L., Lewis S.M., Molinie B., Ho Y.-J., Myers D.R., Choi V.W., Compagno M., Malkin D.J., et al. . 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 147:107–119. 10.1016/j.cell.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y.H., Iwashima M., Wettstein D.A., Kaplan K.B., Elliott J.F., Born W., and Davis M.M.. 1987. T-cell receptor delta gene rearrangements in early thymocytes. Nature. 330:722–727. 10.1038/330722a0 [DOI] [PubMed] [Google Scholar]
- Deriano L., Chaumeil J., Coussens M., Multani A., Chou Y., Alekseyenko A.V., Chang S., Skok J.A., and Roth D.B.. 2011. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 471:119–123. 10.1038/nature09755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S. 2010. Temporal and spatial regulatory functions of the V(D)J recombinase. Semin. Immunol. 22:362–369. 10.1016/j.smim.2010.09.001 [DOI] [PubMed] [Google Scholar]
- de Villartay J.P., Hockett R.D., Coran D., Korsmeyer S.J., and Cohen D.I.. 1988. Deletion of the human T-cell receptor delta-gene by a site-specific recombination. Nature. 335:170–174. 10.1038/335170a0 [DOI] [PubMed] [Google Scholar]
- Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., and Alt F.W.. 2015. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33:179–186. 10.1038/nbt.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V., Chaume D., and Lefranc M.-P.. 2005. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33:D256–D261. 10.1093/nar/gki010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Yoon H.S., Franklin A., Jain S., Ebert A., Cheng H.-L., Hansen E., Despo O., Bossen C., Vettermann C., et al. . 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature. 477:424–430. 10.1038/nature10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., and Krangel M.S.. 2011. Long-distance regulation of fetal V(δ) gene segment TRDV4 by the Tcrd enhancer. J. Immunol. 187:2484–2491. 10.4049/jimmunol.1100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett R.D. Jr., Nuñez G., and Korsmeyer S.J.. 1989. Evolutionary comparison of murine and human delta T-cell receptor deleting elements. New Biol. 1:266–274. [PubMed] [Google Scholar]
- Holmes R., and Zúñiga-Pflücker J.C.. 2009. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb. Protoc. 2009:t5156 10.1101/pdb.prot5156 [DOI] [PubMed] [Google Scholar]
- Hu J., Tepsuporn S., Meyers R.M., Gostissa M., and Alt F.W.. 2014. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc. Natl. Acad. Sci. USA. 111:10269–10274. 10.1073/pnas.1410112111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Zhang Y., Zhao L., Frock R.L., Du Z., Meyers R.M., Meng F.-L., Schatz D.G., and Alt F.W.. 2015. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 163:947–959. 10.1016/j.cell.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Meyers R.M., Dong J., Panchakshari R.A., Alt F.W., and Frock R.L.. 2016. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 11:853–871. 10.1038/nprot.2016.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Garrett K.P., Pelayo R., Zúñiga-Pflücker J.C., Petrie H.T., and Kincade P.W.. 2005. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J. Immunol. 175:4858–4865. 10.4049/jimmunol.175.8.4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski K.M., Ledbetter S., Mayo M.S., and Hockett R.D. Jr. 1997. Identification of a DNA segment exhibiting rearrangement modifying effects upon transgenic delta-deleting elements. J. Exp. Med. 186:91–100. 10.1084/jem.186.1.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Resch W., Corbett E., Yamane A., Casellas R., and Schatz D.G.. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 141:419–431. 10.1016/j.cell.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Lapkouski M., Yang W., and Gellert M.. 2015. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 518:507–511. 10.1038/nature14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S., Hernandez-Munain C., Lauzurica P., McMurry M., Roberts J.L., and Zhong X.P.. 1998. Developmental regulation of V(D)J recombination at the TCR alpha/delta locus. Immunol. Rev. 165:131–147. 10.1111/j.1600-065X.1998.tb01236.x [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., and Marra M.A.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.G., Guo C., Su A., Zhang Y., and Alt F.W.. 2015. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc. Natl. Acad. Sci. USA. 112:1815–1820. 10.1073/pnas.1424936112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livák F., Tourigny M., Schatz D.G., and Petrie H.T.. 1999. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 162:2575–2580. [PubMed] [Google Scholar]
- Matthews A.G.W., and Oettinger M.A.. 2009. RAG: a recombinase diversified. Nat. Immunol. 10:817–821. 10.1038/ni.1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migone N., Padovan S., Zappador C., Giachino C., Bottaro M., Matullo G., Carbonara C., Libero G.D., and Casorati G.. 1995. Restriction of the T-cell receptor V delta gene repertoire is due to preferential rearrangement and is independent of antigen selection. Immunogenetics. 42:323–332. 10.1007/BF00179393 [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Seidl K.J., Gaertner F., Han S., Chen F., Sekiguchi J., Wang J., Ferrini R., Davidson L., Kelsoe G., and Alt F.W.. 1999. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 11:201–212. 10.1016/S1074-7613(00)80095-3 [DOI] [PubMed] [Google Scholar]
- Passoni L., Hoffman E.S., Kim S., Crompton T., Pao W., Dong M.Q., Owen M.J., and Hayday A.C.. 1997. Intrathymic delta selection events in gammadelta cell development. Immunity. 7:83–95. 10.1016/S1074-7613(00)80512-9 [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru H., Chambers M.G., Fu T.-M., Tong A.B., Liao M., and Wu H.. 2015. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 163:1138–1152. 10.1016/j.cell.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., and Swanson P.C.. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202. 10.1146/annurev-genet-110410-132552 [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., de Pooter R.F., Gronski M.A., Cho S.K., Ohashi P.S., and Zúñiga-Pflücker J.C.. 2004. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 5:410–417. 10.1038/ni1055 [DOI] [PubMed] [Google Scholar]
- Shih H.-Y., Verma-Gaur J., Torkamani A., Feeney A.J., Galjart N., and Krangel M.S.. 2012. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc. Natl. Acad. Sci. USA. 109:E3493–E3502. 10.1073/pnas.1214131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., and Alt F.W.. 1997. Function of the TCR alpha enhancer in alphabeta and gammadelta T cells. Immunity. 7:505–515. 10.1016/S1074-7613(00)80372-6 [DOI] [PubMed] [Google Scholar]
- Teng G., Maman Y., Resch W., Kim M., Yamane A., Qian J., Kieffer-Kwon K.-R., Mandal M., Ji Y., Meffre E., et al. . 2015. RAG represents a widespread threat to the lymphocyte genome. Cell. 162:751–765. 10.1016/j.cell.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Arden J., Wilbert O.M., Kabelitz D., and Arden B.. 2000. V delta repertoire during thymic ontogeny suggests three novel waves of gamma delta TCR expression. J. Immunol. 164:1002–1012. 10.4049/jimmunol.164.2.1002 [DOI] [PubMed] [Google Scholar]
- Zakrzewski J.L., Kochman A.A., Lu S.X., Terwey T.H., Kim T.D., Hubbard V.M., Muriglan S.J., Suh D., Smith O.M., Grubin J., et al. . 2006. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nat. Med. 12:1039–1047. 10.1038/nm1463 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.