Abstract
Cytochrome c oxidase (CcO) is a vital enzyme that catalyzes the reduction of molecular oxygen to water and pumps protons across mitochondrial and bacterial membranes. While proton uptake channels as well as water exit channels have been identified for A-type CcOs, the means by which water and protons exit B-type CcOs remain unclear. In this work, we investigate potential mechanisms for proton transport above the dinuclear center (DNC) in ba3-type CcO of Thermus thermophilus. Using long-time scale, all-atom molecular dynamics (MD) simulations for several relevant protonation states, we identify a potential mechanism for proton transport that involves propionate A of the active site heme a3 and residues Asp372, His376 and Glu126II, with residue His376 acting as the proton-loading site. The proposed proton transport process involves a rotation of residue His376 and is in line with experimental findings. We also demonstrate how the strength of the salt bridge between residues Arg225 and Asp287 depends on the protonation state and that this salt bridge is unlikely to act as a simple electrostatic gate that prevents proton backflow. We identify two water exit pathways that connect the water pool above the DNC to the outer P-side of the membrane, which can potentially also act as proton exit transport pathways. Importantly, these water exit pathways can be blocked by narrowing the entrance channel between residues Gln151II and Arg449/Arg450 or by obstructing the entrance through a conformational change of residue Tyr136, respectively, both of which seem to be affected by protonation of residue His376.
Keywords: Cytochrome c oxidase, Proton pumping mechanism, Proton transport, Water exit pathway, Molecular dynamics, Computer simulation
Graphical Abstract
1 Introduction
Cytochrome c oxidase (CcO) is the terminal oxidase of cell respiration in mitochondria and aerobic bacteria and catalyzes the reduction of molecular oxygen to water. Utilizing the resultant energy, this vital enzyme pumps protons across the inner mitochondrial or bacterial cytoplasmic membrane from the negatively charged (N) to the positively charged (P) side, thereby generating an electrochemical proton gradient. [1–4] The energy thus stored is subsequently used to produce adenosine triphosphate in aerobic organisms.
CcO, as one of the heme-copper oxygen reductases, can be classified into A, B and C types. [1, 3, 5–7] A- and B-type CcOs share higher sequence similarity and have a nearly identical three-dimensional structure in the dinuclear reaction center (DNC, or BNC for binuclear center). The DNC, which catalyzes the oxygen reduction, is composed of an Fe-heme (heme a3) and a Cu center (CuB). Additional cofactors are used for the electron transfer from cytochrome c on the P-side of the membrane to the DNC. The cofactors include the homodinuclear copper center (CuA) and a second heme group, which is heme a in aa3-type CcOs and heme b in ba3-type CcOs. A total of four electrons are required to complete a reaction cycle, during which more than four protons are taken up from the N-side of the membrane. Four of these protons, termed scalar protons, are consumed together with the electrons in the DNC to produce water from oxygen, while the remaining protons, called vectorial protons, are transferred to the P-side of the membrane. The proton pumping efficiency is different for A- and B-type CcOs, with 1 H+ pumped/e− and 0.5 H+ pumped/e−, respectively. [8, 9] However, it has been proposed that the pump efficiency of B-type CcO may be underestimated due to experimental limitations. [10, 11]
A number of proton pumping mechanisms have been proposed, [12–20] as well as three plausible entry pathways for proton conduction within the A-type enzyme: the D-, K- and H-channels.[21–24] The H-channel has been identified for bovine oxidase and it is largely conserved among A-type CcOs, however its functional importance remains a subject of debate. [2, 3] The K-channel is used for transfer of some of the scalar protons from the N-side of the membrane to the catalytic site, whereas the D-channel is used both for the transfer of the remaining scalar protons as well as uptake of all the vectorial protons. [25–27] In B-type CcOs, however, there is only one entry channel for the uptake of both scalar and vectorial protons. [28] Although not sharing any significant sequence homology, this channel is located at a similar position as the K-channel in A-type CcO and is therefore referred to as the K-channel analog. While this proton uptake channel has been investigated in detail, [29] the means by which the protons exit the enzyme remain unclear. [30]
Proton pumping requires that residues change their proton affinity during the reaction cycle in order to take up and release protons along the proton transport pathway. Of particular importance are residues within or neighboring the DNC and above (towards the P-side of the membrane). These residues load protons from the N-side and release protons towards the P-side, constituting the so-called proton-loading site (PLS). Although the identity of the PLS of CcO is not known, it seems that this region is common to all members of the same heme-copper oxidase family and that candidates for the PLS are limited. For aa3-type CcOs (e.g. P. denitrificans, R. sphaeroides, and bovine CcO), propionates A and D of the active site heme a3 (PRAa3 and PRDa3), histidine ligands to CuB, and certain residues above the DNC region have all been suggested as the possible PLS, also including surrounding water molecules. [31–36] Recent simulations indicate that the PLS in aa3-type CcOs is in fact a cluster consisting of the propionates PRAa3 and PRDa3 and nearby residues (e.g. Asp A52 and Lys171 in bovine CcO). [37] For ba3-type CcOs, theoretical and experimental data including structural analyses and a set of site-directed mutations suggest that PRAa3 and nearby sites may act as the PLS. [11, 28, 38–41] Among the residues that are thought to participate in proton transport in ba3-type CcO from Thermus thermophilus are Asp372 and His376. [11, 39, 40] Residue Glu126II, which can be hydrogen-bonded to His376, has also been suggested to be involved in proton transport, [40] although recent experimental work does not support this residue as relevant for the proton pumping efficiency. [39]
The driving force for proton pumping resides in the redox reaction of the metal centers and the oxygen binding and reduction chemistry of the DNC. Many features of the chemical mechanism of oxygen reduction and the subsequent mechanistic steps of the reaction cycle are known from spectroscopy and kinetics, but it remains unclear whether proton pumping is directly coupled to the chemistry of oxygen reduction in the DNC or indirectly coupled through conformational and electrostatic changes in the surrounding protein, in particular the nearby homodinuclar CuA and mononuclear Fe-heme redox centers that supply the DNC with electrons. [3, 42–45] Briefly, the catalytic cycle of CcO proceeds through four intermediates denoted as R, F, O, and E. [3, 8, 37, 45, 46] Between each of these intermediate states one electron is transferred into the DNC. When the enzyme is in the fully reduced state R (CuA+-CuA+, Fea2+ or Feb2+, Fea32+, CuB+), oxygen binds to the DNC to form an initial peroxy product state P. After proton and electron transfer (from heme a or heme b) to the DNC, state P progresses to state F in which the oxygen bond is broken. The four electrons that are required for this oxygen reduction thus originated from the oxidation of the DNC (Fea34+, CuB2+) and heme a or heme b (Fea3+ or Feb3+). In the following steps the redox centers are reduced again by four cytochromes c, with one electron added at a time via the CuA pair and heme a or heme b to form the O, E, and R states. In each of these four DNC redox states there must be a specific sequence of intermediate states for the reduction of the CcO cofactors and the transfer of a proton to the DNC and the P-side of the membrane. The consensus model for A-type CcOs is that protons are loaded on the PLS when heme a is reduced and released to the P-side of the membrane when a proton is transferred to the DNC. [3, 37, 44] It is possible that A- and B-type enzymes operate with different mechanisms, using a direct mechanism, an indirect mechanism, or a mixture of both. Because there is only a single input path in B-type enzymes (the K-channel analog), this mechanism must also account for a bifurcation of the proton flow between the scalar and vector pathways at or after the entry point to the DNC. [28, 45] In A-type enzymes, a glutamic acid residue (Glu242 in bovine numbering) is thought to deliver protons both to the PLS cluster and the DNC and its protonation state is assumed to play a role in regulating uptake and release of the proton from the PLS cluster. [37, 47] In B-type enzymes, an isoleucine is present in place of the glutamic acid (Ile235 in ba3-type CcO from Thermus thermophilus), hence the mechanism that regulates proton pumping must differ.
In addition to the vectorial protons, the water that is produced in the chemistry of O2 reduction also needs to leave the active site and enzyme. Several water cavities in the interior of the protein as well as pathways that connect the active site to the protein exterior at the P-side of the membrane have been identified by experiment and computer simulations for aa3-type CcO. [48–52] These water pathways are likely used both for product water and proton pumping out of the protein. [50, 51, 53] However, no clear mechanism on water or proton transport pathways is available for ba3-type CcOs and the experimental kinetic evidence is incomplete.
In the ba3-type CcO from Thermus thermophilus, a salt bridge between Arg225 and Asp287 is located at the interface between protein subunits I and II. Since mutagenesis studies by Chang et al. [39] indicate that Asp287 is important for efficient proton pumping, it has been proposed that this salt bridge operates as a simple electrostatic gate in the proton exit pathway that prevents backflow of protons. [45] The relevance of this salt bridge for proton pumping, however, is not clear.
We have performed unbiased long time-scale molecular dynamics (MD) simulations of the ba3-type CcO from Thermus thermophilus [54] to gain insight into the roles of the relevant residues around the postulated PLS for proton transport. Based on simulations of a variety of different protonation states we present a potential mechanism for proton transport that involves PRAa3, Asp372, His376 and Glu126II. The mechanism is supported by energetics from density functional theory (DFT) calculations for active site cluster models. We furthermore investigate the effect of different protonation states on the strength of the salt bridge Asp287–Arg225 and how this could affect proton transport. Importantly, we also identify two water transport pathways that connect the water pool above the DNC to the P-side of the membrane. These water transport pathways could potentially act also as proton exit pathways.
2 Materials and Methods
2.1 Model preparation
The computational model is based on the high resolution (1.8 Å) X-ray crystal structure of the ba3-type CcO from Thermus thermophilus in the reduced state in lipidic cubic phase (LCP) crystal as obtained from the Protein Data Bank (PDB ID: 3S8F).[54] The crystal structure contains two oxygen atoms between the Fe and Cu centers of the DNC, which have been proposed to be a bridging hydroperoxide. [55] For our calculations, we retained the bridging hydroperoxide and employed oxidation states of the metal centers that correspond to a critical redox intermediate of the reaction cycle, which has been proposed to be a pre-pump state. [40, 45] By a “pre-pump state”, we mean that the reaction chemistry that occurs immediately after this intermediate produces proton pumping. Specifically, the heme b and CuA pair are reduced, that is Feb2+ and CuA+-CuA+, while the DNC (heme a3 and CuB) is oxidized with Fea33+ and CuB2+. The special tyrosine (Tyr237) is deprotonated. This true peroxo state corresponds to state 6 in the reaction cycle presented by Noodleman and coworkers, and is a state that follows the initial adduct state but precedes the P state. [40, 45] The H++ software [56] was used to determine the protonation states of titratable residues for this intermediate state of the reaction cycle, as detailed in the Supporting Information. We denote the lowest energy protonation state as state D in this work.
The protein, including 189 relevant interstitial X-ray crystal water molecules, was inserted into a palmitoyl-oleoyl-phosphatidylcholine (POPC) bilayer containing 180 lipid molecules and solvated in 0.15M KCl solution with a total of 18,866 water molecules by means of the CHARMM-GUI membrane builder. [57] For this purpose, the protein crystal structure without heme groups and metal atoms was employed. VMD [58] was used to combine the lipid and solvent coordinates with the complete enzyme structure including relevant crystal waters via a root-mean-square deviation (RMSD) fit. Subsequently, K+ or Cl− ions were removed to neutralize the system as appropriate for the different protonation states considered in our simulations. The AmberTools [59, 60] charmmlipid2amber.py and LEaP programs were used to convert the atom and residue nomenclature from CHARMM to AMBER format and to assign parameters. The Amber Lipid14 force field [61] was used in combination with the Amber ff12SB force field [62, 63], TIP3P parameters for water [64] and the Joung/Cheatham ion parameters [65]. Non-standard residues were parameterized as detailed below.
2.2 Parameters for CcO cofactors
Force field parameters for the CuA pair, heme b, and the DNC heme a3 and CuB were obtained as follows. Parameters for heme b including bonded parameters to the coordinating histidine residues as well as Lennard-Jones parameters for Cu were taken from the literature. [66, 67] Parameters for heme a3 were derived from these heme b parameters with appropriate modifications for the geranyl-geranyl tail and the formyl group based on the GAFF force field. [68] GAFF parameters were also used for the bridging hydroperoxide in the DNC and the special C-N bond between His233 and Tyr237. Harmonic bond and angle constraints were employed between all metal centers and the ligands, for CuA based on the crystal structure and for the DNC based on geometries optimized for a cluster model with DFT. All parameters are available from Ref [69].
The DFT calculations were performed with the ADF program [70–72] using the OLYP exchange-correlation functional [73, 74] in conjunction with double- and triple-zeta polarized (DZP and TZP) Slater type basis sets (with TZP on the metal atoms and DZP on all other atoms) from the ADF basis set library [75]. The effect of the protein environment was included by means of the COSMO dielectric continuum model [76–78] using a dielectric constant of ε = 18.5. The geometry optimizations of the DNC employed the same 205 atom cluster model used previously by Noodleman et al. [45] consisting of heme a3, the side chain of the axial His384 ligand to heme Fea3, the bridging hydroperoxide, CuB and the side chains of its three coordinating ligands His283, His282 and the special His233 that is covalently linked to Tyr237, as well as the side chains of the three residues Arg449, doubly protonated His376 (positively charged, +1) and protonated Asp372 above the DNC, Gly232 including backbone atoms linked to His233, and 7 water molecules. The low spin Fe3+ and Cu2+ centers of the DNC were antiferromagnetically coupled in our calculations. We constrained the positions of the link atoms to the crystal structure coordinates during the DFT geometry optimizations.
Charges for the cofactors were derived using DFT with the OLYP potential as described above via an electrostatic potential (ESP) fit approach as implemented in the SCRF module [79, 80] of ADF. For the CuA pair we employed a 36 atoms cluster model that includes the two Cu atoms and the side chains of the coordinating histidine and cysteine residues His114II, His157II, Cys149II, and Cys153II. For heme b we included both the heme b and the side chains of its two axial histidine ligands, residues His72 and His386, in the cluster model (97 atoms). For the DNC, we employed the coordinates obtained from the geometry optimization of the cluster model described above, but adding the geranyl-geranyl tail and removing residues Arg449, His376, Asp372 and Gly232 (total 192 atoms). Two different charge sets were generated for the DNC, one with deprotonated PRAa3, and one with protonated PRAa3. The ESP charges derived were used both for the cofactors and the coordinating residues, which are thus modified from the standard Amber ff12SB charges. Charges on symmetry equivalent atoms such as methyl group hydrogens were averaged and all charges were scaled uniformly such that the total charge of our cluster models and the backbone atoms yields the correct integer charge. The charges and the coordinates of our cluster models used to derive the charges are provided in the Supporting Information and in Ref [69].
2.3 Molecular dynamics simulations
All the molecular dynamics (MD) simulations were carried out using the GPU-accelerated version [81–83] of the PMEMD program in the Amber14 software package. [59, 60] The starting structures were minimized for 5,000 steps of steepest descent before the system was gradually heated from 10 to 300 K over 100 ps and then kept at 300 K for 100 ps in the NVT ensemble. The system was subsequently equilibrated for 10 ns in the NPT ensemble at 300 K and 1 bar. The pressure was regulated anisotropically with the Berendsen barostat [84] using a pressure relaxation time of 1.0 ps. The temperature was regulated using Langevin dynamics [85] with a collision frequency of 5.0 ps−1 during heating that was changed to 1.0 ps−1 for equilibration, while the Berendsen weak coupling algorithm [84] with a time constant of 10.0 ps was applied for the production runs. A real-space cutoff of 8.0 Å was applied to non-bonded interactions. Periodic boundary conditions were employed, and the particle mesh Ewald summation method (PME) was used to treat all long-range electrostatics. [86] A long-range dispersion correction was applied to the energy and pressure beyond the cut-off. [87] The SHAKE algorithm was used to constrain bonds involving hydrogen with a relative tolerance of 10−6, [88, 89] and a 2 fs time step was used. Trajectories of 140 ns length were generated for each of the production runs and snapshots for analysis were saved every 2 ps. To ensure that the simulations are well equilibrated, we have monitored the RMSD of the protein backbone carbon atoms and the area per lipid of the lipid bilayer (Supporting Information Figures S1 and S2). Analysis was conducted on the equilibrated portion of the trajectories using the AmberTools CPPTRAJ program. [90]
2.4 Energies of proton transfer and proton uptake
The reaction energies for the proton transfers and proton uptake from the N-side of the membrane were obtained from DFT calculations of DNC cluster models with 204 atoms (205 atoms if protonated) as described above, however, employing both the OLYP and the PW91 [91] semilocal exchange-correlation functionals including dispersion corrections [92] (OLYP-D3, PW91-D3). While OLYP was shown to yield superior spin state splitting energetics, PW91 typically provides good reaction energies. [45, 93] The inner cores of C(1s), N(1s), O(1s), Fe(1s,2s,2p), and Cu(1s,2s,2p) are treated by frozen core approximation in these calculations. We have also performed all-electron single point calculations with the B3LYP-D3 hybrid functional [94, 95] on the PW91-D3 optimized geometries. We account for the free energy of proton uptake from aqueous solution at pH = 7 with values ΔGprot,shift of −3.6 kcal/mol (PW91), −5.2 kcal/mol (OLYP) and 5.6 kcal/mol (B3LYP), which are added to the electronic energies of the protonated clusters. These values are obtained from ΔGprot,shift = ΔZPE - ΔGref(H+) + ΔEcorr(pH = 7). In this equation ΔZPE is the zero point energy difference between the protonated state and the deprotonated state, for which we use 8.0 kcal/mol. The reference energy of the solvated proton is ΔGref(H+) = E(H+) + ΔGsol(H+, 1atm) − TΔSgas(H+) + (5/2)RT. Since ADF computes all energies with respect to a sum of spin-restricted atoms, the gas phase energy of a proton E(H+) is relative to a spin-restricted hydrogen atom and thus not zero and depends on the density functional. For E(H+) we employ the empirically corrected values of 291.5 kcal/mol (PW91), 293.1 kcal/mol (OLYP) and 282.3 kcal/mol (B3LYP) from reference [45]. For the solvation free energy of a proton at 1 atm pressure ΔGsol(H+, 1atm) we use the best available value of −264.0 kcal/mol [96, 97], for the translational entropy contribution to the gas-phase free energy of a proton TΔSgas(H+) at 298 K and 1 atm pressure we use −7.8 kcal/mol, [98] and (5/2)RT =1.5 kcal/mol includes the proton translational energy (3/2)RT and PV = RT. Finally, the correction for the proton concentration for a neutral solvent at pH = 7, ΔEcorr(pH = 7), is 1.3 × 7 kcal/mol = 9.6 kcal/mol. A detailed discussion can be found in the appendix of reference [45].
2.5 Potential of mean force
The potential of mean force (PMF) for the dissociation of the salt bridge between Asp287 and Arg225 was obtained from umbrella sampling [99] simulations. As the reaction coordinate we have chosen the distance between the carbon atom Cγ of the carboxyl group in Asp287 and the carbon atom Cζ of the guanidinium group in Arg225. A harmonic biasing potential V(r) = kumb(r − r0)2 with a force constant of kumb = 10.0 kcal/(mol Å2) was employed to restrain the reaction coordinate in the range of 2.0 Å to 10.8 Å, with windows centered every 0.2 Å. Initial configurations along the reaction coordinate were generated from a snapshot of the neighboring window, followed by 200 ps equilibration. Data was collected for 5 ns at each window and the biased frequency distributions were converted to free energies using the weighted histogram analysis method (WHAM) [100, 101] with a bin size of 0.05 Å and a stringent tolerance of 10−6 kcal/mol on every point of the PMF.
3 Results and Discussion
3.1 Proton transport above the DNC
As mentioned in the introduction, PRAa3 and nearby sites are thought to act as the proton-loading site for proton pumping in ba3-type CcOs. [11, 28, 39–41] Among the residues that site-directed mutagenesis experiments have identified as being important for proton pumping are the residues Asp372 and His376. [11, 39] For instance, the mutants Asp372Ile and His376Asn totally inhibit or severely reduce proton pumping. Residue Glu126II has also been suggested to be of importance based on theoretical considerations. [40] In order to understand potential proton transfer mechanisms that involve these residues, we have analyzed structural features and hydrogen bond networks that were obtained from unbiased MD simulations of a variety of relevant protonation states. In these simulations we focus on a hydroperoxo-bridged intermediate of the reaction cycle that has been postulated to be a pre-pump state. [40, 45] The different protonation states that we have considered are listed in Table 1. Using simulations that are performed for a single redox state precludes drawing direct conclusions about how the proton transport is regulated by the electron transfer processes in CcO and the redox chemistry of oxygen reduction in the DNC. Nevertheless it is possible, as discussed below, to develop a plausible mechanism for proton transfer to/from the PLS that is in line with experimental findings.
Table 1.
Molecular dynamics simulations performed in this work. Protonation state D is the lowest energy protonation state for the investigated true peroxo state of CcO (state 6 in the catalytic cycle of Ref [45]; hydroperoxo-bridged DNC with Fea33+, CuB2+, Feb2+, CuA+-CuA+). See also Figure 1.
| Simulation No. | Protonation states |
|||
|---|---|---|---|---|
| PRAa3 protonated | Asp372 protonated | His376 protonateda | Glu126II protonated | |
| A | no | yes | ε | no |
| B | yes | no | ε | no |
| C | yes | yes | ε | no |
| D | no | yes | ε,δ | no |
| E | no | yes | δ | yes |
| F | no | yes | δ | no |
ε=epsilon-protonated histidine; δ=delta-protonated histidine; ε,δ=doubly protonated histidine.
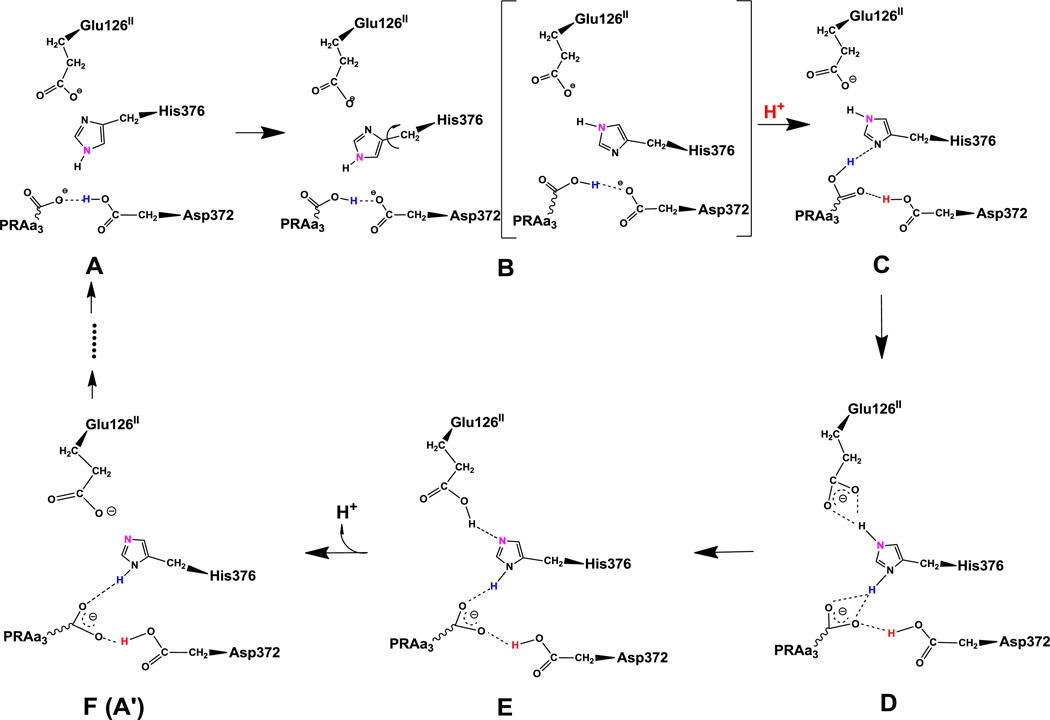
Figure 1 shows the proposed proton transfer pathway. In the redox state of the CcO reaction cycle that forms the basis of our simulations (true peroxo state with hydroperoxo-bridged Fea33+ and CuB2+, reduced Feb2+ and CuA+-CuA+, deprotonated Tyr237), the most favorable protonation state has both PRAa3 and Glu126II deprotonated, while Asp372 is protonated and His376 doubly protonated (state D in Figure 1). This is also what one would expect by analyzing the geometry and bond distances of crystal structure 3S8F. His376 could thus be the PLS and in what follows we demonstrate that proton uptake via protonation states A to C and proton expulsion via protonation states E and F is reasonable. To this end we have performed MD simulations of each of the intermediates A to F and analyzed hydrogen bonding patterns and structural changes.
Figure 1.
Proposed proton transfer pathway. Protonation state D is the lowest energy protonation state for the investigated pre-pump state. Transferred protons and the ε-nitrogen on the imidazole ring of His376 are colored to guide the eye. (A) A strong hydrogen bond exists between protonated Asp372 and PRAa3, facilitating proton transfer. (B) Rotation of His376. (C) Proton uptake by Asp372 results in formation of a strong hydrogen bond between protonated PRAa3 and His376. (D) Proton transfer from PRAa3 to His376 results in a strong hydrogen bond between His376 and Glu126II. (E) Proton transfer from doubly protonated His376 to Glu126II and subsequent release into the water pool. (F) Structure and protonation state equivalent to (A) with exception of the orientation of His376 and protonation site δ-nitrogen.
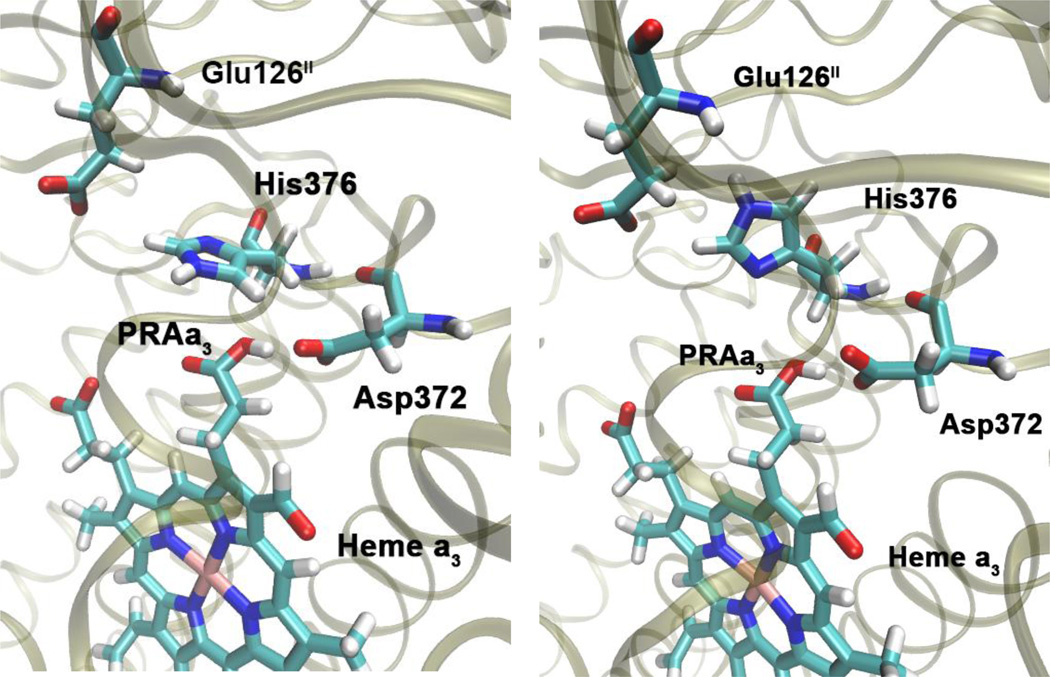
Initially, Glu126II is deprotonated, His376 is ε-protonated, and either Asp372 or PRAa3 is protonated (intermediates A and B). In these protonation states, there is a strong hydrogen bond between Asp372 and PRAa3, which is present throughout our 140 ns simulations. This can be seen from Table 2, which shows that hydrogen bond occupancies between the carboxylate oxygen atoms of PRAa3 (O1A/O2A) and the carboxylate oxygen atoms of Asp372 (OD1/OD2) are very high for states A and B. As a result, it is likely that the proton can easily transfer between Asp372 and PRAa3. In contrast, we could not detect hydrogen bonding between PRAa3 and His376 for these states. This is important, since absence of a strong hydrogen bond allows His376 to rotate and change orientation (Figure 2) such that the δ-nitrogen atom gets positioned to accept a hydrogen bond from PRAa3 at a later stage. In fact, we have observed several spontaneous rotations of the imidazole group of His376 in simulation B.
Table 2.
Hydrogen bond occupancies for simulations of different protonation states. See Table 1 and Figure 1 for the protonation states in the different simulations.
| Simulation | Acceptora | Donora | Occupancyb,c |
|---|---|---|---|
| A | O2A(PRAa3) | OD2(Asp372) | 58.8% |
| O1A(PRAa3) | OD2(Asp372) | 36.2% | |
| O(Asp372) | N(ε-His376) | 4.7% | |
| B | OD2(Asp372) | O2A(PRAa3) | 95.4% |
| OD1(Asp372) | O2A(PRAa3) | 12.0% | |
| O(Asp372) | N(ε-His376) | 51.6% | |
| C | O1A(PRAa3) | OD2(Asp372H) | 68.6% |
| OD1(Asp372H) | O2A(PRAa3) | 9.1% | |
| ND1(ε-His376) | O2A(PRAa3) | 50.5% | |
| O(Asp372H) | N(ε-His376) | 79.2% | |
| OE1(Glu126II) | NE2(ε-His376) | 8.8% | |
| OE2(Glu126II) | NE2(ε-His376) | 7.5% | |
| D | O1A(PRAa3) | OD2(Asp372H) | 99.6% |
| O1A(PRAa3) | ND1(His376H+) | 90.6% | |
| O2A(PRAa3) | ND1(His376H+) | 46.6% | |
| O(Asp372H) | N(His376H+) | 58.8% | |
| OE1(Glu126II) | NE2(His376H+) | 51.4% | |
| OE2(Glu126II) | NE2(His376H+) | 44.3% | |
| E | NE2(δ-His376) | OE2(Glu126HII+) | 29.8% |
| O1A(PRAa3) | OD2(Asp372H) | 46.0% | |
| O2A(PRAa3) | OD2(Asp372H) | 25.2% | |
| O1A(PRAa3) | ND1(δ-His376) | 61.3% | |
| O2A(PRAa3) | ND1(δ-His376) | 46.6% | |
| O(Asp372H) | N(δ-His376) | 82.2% |
Names and residue name of the heavy atoms involved in the hydrogen bond, with the residue name in parentheses. Residues PRAa3, Asp372, His376 and Glu126II are considered.
Only occupancies larger than 3% are listed.
Occupancy is the fraction of simulation frames during which the hydrogen bond is present. Hydrogen bonds are defined through an angle cutoff of 135° and a distance cutoff of 3.0 Å between the hydrogen bonded atoms.
Figure 2.
The side chain of His376 can rotate from its position in the crystal structure (left) such that it is in position to accept a hydrogen bond from PRAa3 (right). Snapshots taken from simulation of protonation state B, see Table 1 and Figure 1.
At this point a proton can be loaded from the K-channel analog onto Asp372, such that both Asp372 and PRAa3 are protonated (state C). This leads to formation of a strong hydrogen bond between the protonated carboxylate group of PRAa3 as donor and the δ-nitrogen of His376 (ND1) as acceptor (Table 2), allowing for efficient proton transfer from PRAa3 to His376 (state D). Alternatively, it may be possible to first transfer a proton from PRAa3 to His376, forming a different intermediate B2 (not shown in Figure 1) in which His376 is doubly protonated and both Asp372 and PRAa3 are deprotonated, followed by reprotonation of Asp372 and formation of state D. DFT calculations (see below) indicate that this pathway via intermediate B2 may actually be energetically favorable.
After proton transfer to His376, there are still strong hydrogen bond interactions between PRAa3 and both protonated Asp372 and protonated His376. Importantly, hydrogen bond interactions between the carboxylate group of Glu126II (OE1/OE2) and the doubly protonated ε-nitrogen of His376 (NE2) strengthen considerably in state D (Table 2). This is to be expected, since a doubly protonated histidine group is a stronger hydrogen bond donor. Formation of such a hydrogen bond indicates that proton transfer from His376 to Glu126II should be facilitated, which results in state E with δ-protonated His376.
Finally, the proton is released from Glu126II to continue the proton transfer through water exit pathways as discussed in the corresponding section below. The resulting state (F) is equivalent to state A with the difference that His376 is now δ-protonated as opposed to ε-protonated. This ring is rotated by 180° (flipped) compared to its orientation in state A. The proton transfer process can now be repeated in an equivalent way as described above and shown in Figure 1, however starting from state F (A’) instead of state A. This is indicated by dots in Figure 1. We have performed MD simulations for the corresponding states and the only significant difference that we could observe was that spontaneous rotation of δ-protonated His376 does not occur in the state that is equivalent to state B. Instead, His376 rotates after proton loading, in the state that is equivalent to state C. The reason is that the protonated δ-nitrogen (ND1) is in a good position to form a hydrogen bond with Asp372 (Table S3). Therefore, the probability to form a hydrogen bond between protonated PRAa3 and the ε-nitrogen of His376 is reduced. Nevertheless, there is considerable flexibility in the spatial arrangement of the functional groups of Asp372, PRAa3, and His376 and we do observe frequent formation of a hydrogen bond network that is equivalent to the depiction of state C in Figure 1. This allows proton transfer from PRAa3 to His376 in state C’. Details of the hydrogen bond occupancies for simulations of states leading from state F to state A are summarized in the Supporting Information.
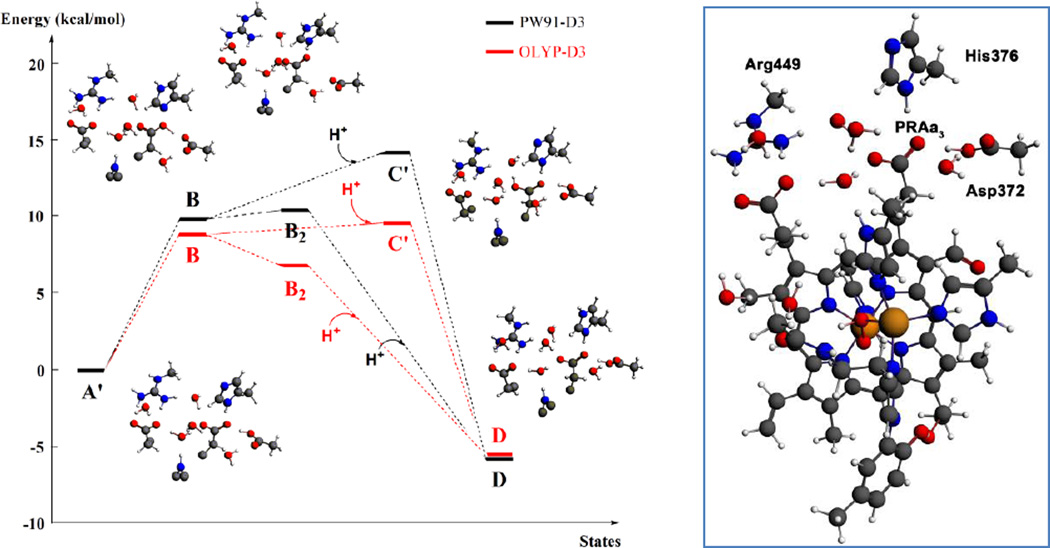
DFT calculations (Table 3 and Figure 3) show that proton uptake from the bulk solution on the N-side of the membrane to His376, the postulated PLS of the reaction cycle, is energetically favorable and that the postulated proton transfer pathway is feasible. For the DFT calculations we have chosen the His376 conformation from the crystal structure, corresponding to state D, in order to be able to preserve the total number of hydrogen bonds in the cluster model and constrain the position of the coordinates of the protein backbone link atoms. We will first discuss results with the semilocal density functionals (PW91-D3/OLYP-D3). The initial proton transfer from Asp372 to PRAa3 (A’→B) is uphill and requires approximately 10 kcal/mol. Reprotonation of Asp372 by uptake of a proton from pH=7 (B→C’) is predicted to require only 4.4 kcal/mol (PW91-D3) or even release some energy (-0.4 kcal/mol, OLYP-D3). The following formation of state D by proton transfer from PRAa3 to His376 (C’→D) is strongly downhill and releases 14.8 (OLYP-D3) to 20.0 kcal/mol (PW91-D3). An alternative pathway, in which proton transfer from PRAa3 to His376 takes place before proton uptake from pH=7 (B→B2 followed by B2→D) is favored by both functionals. PW91-D3 predicts state B2 to be lower in energy than state C’ by 3.7 kcal/mol and OLYP-D3 by 2.5 kcal/mol. The hybrid functional B3LYP-D3 predicts the initial proton transfer to require more energy, but then both states B2 and C’ are downhill from state B. It may thus well be possible that both mechanisms for protonation of His376 operate in tandem. One should keep in mind that the exact values of the computed energies not only depend on the choice of exchange-correlation functional in the DFT calculations but also the hydrogen bonding network of the optimized local minima of the cluster models. Furthermore, explicit electrostatic effects of the larger environment are not taken into account in these DFT calculations. Nevertheless it is clear that the proton will quickly transfer to His376 and not reside on Asp372/PRAa3, which means that His376 can be considered as the real PLS. Once the proton is on His376 and Asp372 reprotonated (state D), it is highly unfavorable for His376 to give back the proton to PRAa3. Therefore, the pair PRAa3/protonated Asp372 could take on the role of a gate that prevents proton back-flow to the DNC.
Table 3.
DFT relative energies (kcal/mol) for proton transfer in Fea33+-(O-OH)−-CuB2+-Y237− DNC models and proton uptake from the N-side of the membrane (at pH = 7), with Q being the total charge of the cluster model. See Table 1 and Figure 1 for the protonation states of Asp372, PRAa3 and His376 in the different intermediates. In state B2, His376 is doubly protonated and both Asp372 and PRAa3 are deprotonated. The prime indicates δ-protonation in place of ε-protonation of His376. See Figure 3 for a representation of the cluster model.
| State | Q | E |
||
|---|---|---|---|---|
| PW91-D3 | OLYP-D3 | B3LYP-D3//PW91-D3 | ||
| A’ + (H+,pH=7) | 0 | 0.0 | 0.0 | 0.0 |
| B + (H+,pH=7) | 0 | 9.8 | 8.8 | 13.4 |
| B2 + (H+,pH=7) | 0 | 10.5 | 6.8 | 8.3 |
| C’ | 1 | 14.2 | 9.3 | 4.9 |
| D | 1 | –5.8 | –5.5 | -13.3 |
Figure 3.
Left: DFT energies for proton transfer and proton uptake from the N-side of the membrane (A’/B/B2 = cluster model + H+ at pH = 7) to His376 (D). The initial step is a proton transfer from Asp372 to PRAa3 (B). Protonation of His376 can occur either before proton uptake (B2) or after proton uptake (C’). The labels correspond to the nomenclature in Table 1 and Figure 1. The prime indicates δ-protonation in place of ε-protonation of His376. Right: complete DNC cluster model for State A’.
It is known that the proton pumping in CcO is coupled to the redox reaction in the DNC and to redox changes in the other cofactors. These redox changes can influence the pKa values of titratable residues through electrostatic interactions and can induce structural changes, for instance conformational changes of histidine residues [42]. In this context, recent studies by Lu and Gunner [37] have demonstrated that the PLS in aa3-type enzymes is likely constituted by several residues and that electrostatic changes in the environment regulate its protonation. In this case the PLS is loaded when heme a is reduced and protons are released from the PLS when protons are transferred into the BNC. Our MD simulations and DFT calculations of the ba3-type CcO from Thermus thermophilus are restricted to a single redox state (state 6 of the catalytic cycle, see ref. [40, 45]) and thus do not capture the driving force for proton pumping. However, the simulations give important insight into the protein dynamics and the dynamics of the hydrogen bond network. They support a plausible mechanism for proton transport that is in line with mutagenesis results that have demonstrated the importance of His376 and in particular Asp372 for proton pumping in ba3-type CcO.[11, 39, 40]
3.2 Role of the salt bridge between Arg225 and Asp287
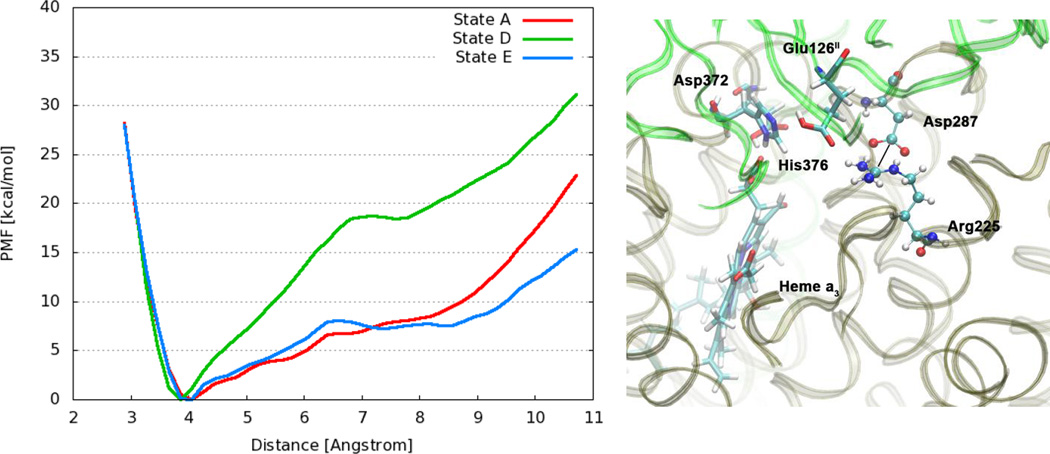
Experimental studies of catalytic turnover and proton pumping efficiency for ba3-type CcO with mutations of Arg225 and Asp287 indicate that these residues are important for efficient proton pumping. [39] Based on these results, it has been speculated that the salt bridge Arg225–Asp287 could act as a simple electrostatic gate, which could operate through changes in the surrounding electrostatic potential, for instance due to the presence or the absence of protons. [45] Details of the role of these residues in the proton transport mechanism are not known, however. To shed some light on the potential role of this salt bridge, we examine its strength in selected protonation states. We have selected protonation states A, D and E (Table 1 and Figure 1), which represent the states at the beginning of the mechanism outlined above (state A), after loading of one proton onto His376 (state D), and after transfer of the proton from His376 to Glu126II (state E). The salt bridge between Arg225 and Asp287 is stable in all protonation states, as can be seen from the salt bridge distance throughout our unbiased simulations, which remains in the range of 3.5 Å to 4.0 Å, which are typical values for a strong salt bridge (Supporting Information Figure S3). Residue Glu126II also interacts with Arg225, both through hydrogen bonds and through electrostatic interactions, forming a somewhat weaker salt bridge. The distance between Glu126II and Arg225 is significantly larger and much more sensitive to the protonation state than the distance between Arg225 and Asp287 (Figure S3).
Figure 4 shows the PMF for the dissociation of the Arg225–Asp287 salt bridge for protonation states A, D and E. The contact ion pair minimum is at a distance of approximately 3.8 Å between the Cζ guanidinium carbon atom of Arg225 and the Cγ carbon atom of the carboxylate group of Asp287. As we can see, in protonation state A the free energy required for dissociating the salt bridge and reaching distances above 7 Å is less than 10 kcal/mol. Upon protonation of the postulated PLS His376 (state D), the strength of the salt bridge increases considerably. This clearly shows how changes in the protonation state, and potentially also other electrostatic changes in the environment, are able to modulate the strength of this salt bridge. In state D, a relatively strong hydrogen bonding interaction is formed between Glu126II and doubly protonated His376 (see Table 2), which increases the distance between Glu126II and Arg225 (see Figure S3). This in turn seems to be the reason for the stronger salt bridge between Arg225 and Asp287 in state D. On the other hand, transfer of the proton from His376 to Glu126II (State E) weakens the salt bridge again, leading to a PMF that is very similar to protonation state A. The simulation of state E shows a non-negligible hydrogen bonding interaction between protonated Glu126II and Arg225, which likely facilitates the dissociation of the Arg225-Asp287 salt bridge relative to state D.
Figure 4.
Left: Free energy profiles for the salt bridge Arg225–Asp287 in different protonation states of CcO. The distance is measured between the carbon atom Cζ of the guanidinium group in Arg225 and the carbon atom Cγ of the carboxylate group in Asp287. (Dotted line) Right: Snapshot from a simulation of state E with protonated Glu126II, highlighting relevant residues.
Since the Arg225–Asp287 salt bridge is stable in the different protonation states that we have investigated, it does not seem to act as a simple electrostatic gate, although its strength can be clearly influenced. Another important observation is that Arg225 is at the entrance of water exit pathway P2 (see section 3.3), and it is thus reasonable to assume that the hydrogen bond interaction between protonated Glu126II and Arg225 in state E could help to facilitate transfer of the proton into this exit pathway P2.
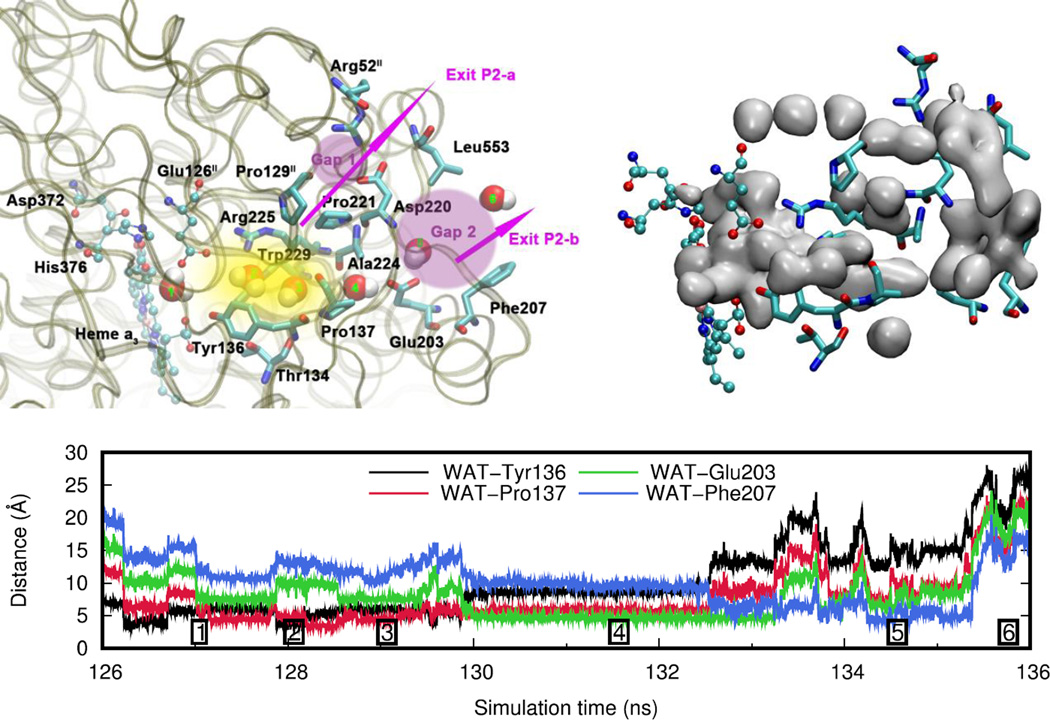
3.3 Water exit pathways
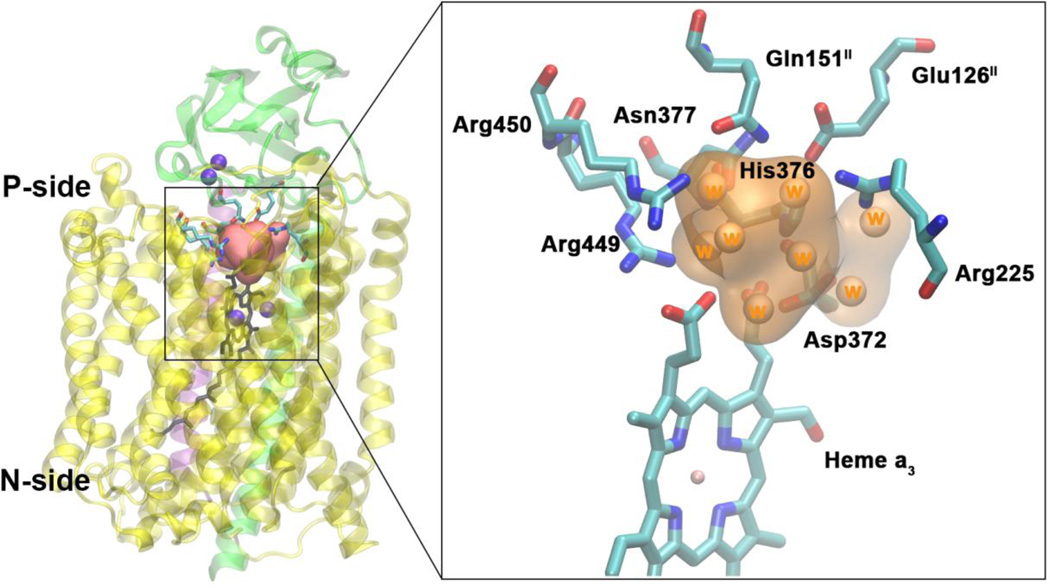
Located just above the DNC is a water pool that is delimited by the heme a3 group and the protein residues Asn377, Asp372, His376, Glu126II, Arg225, Gln151II, Arg449 and Arg450 (Figure 5). In order to identify water transport pathways that connect this water pool to the bulk water on the P-side of the membrane, we have tracked the movement of individual water molecules present in this water pool at the beginning and at the end of the production phase of each of the simulations. Using this approach we have identified two water transport pathways (Figure 6). Proton transfer within the protein requires a path of hydrogen bonds. Although other possibilities for proton exit cannot be excluded, the water pathways presented here are regions in which a chain of hydrogen bonds can be formed via water molecules, making them strong candidates for proton exit pathways.
Figure 5.
Water pool above the DNC in the crystal structure of ba3-type CcO from Thermus thermophilus (PDB ID 3S8F). Left: Location of the water pool within the enzyme with surrounding residues highlighted. Purple spheres are metals in the protein (CuA, CuB, Fea3). Subunits I, II and III are shown in yellow, green and purple. Right: Details of the water pool and surrounding residues. The orange region is the water pool consisting of eight crystal waters.
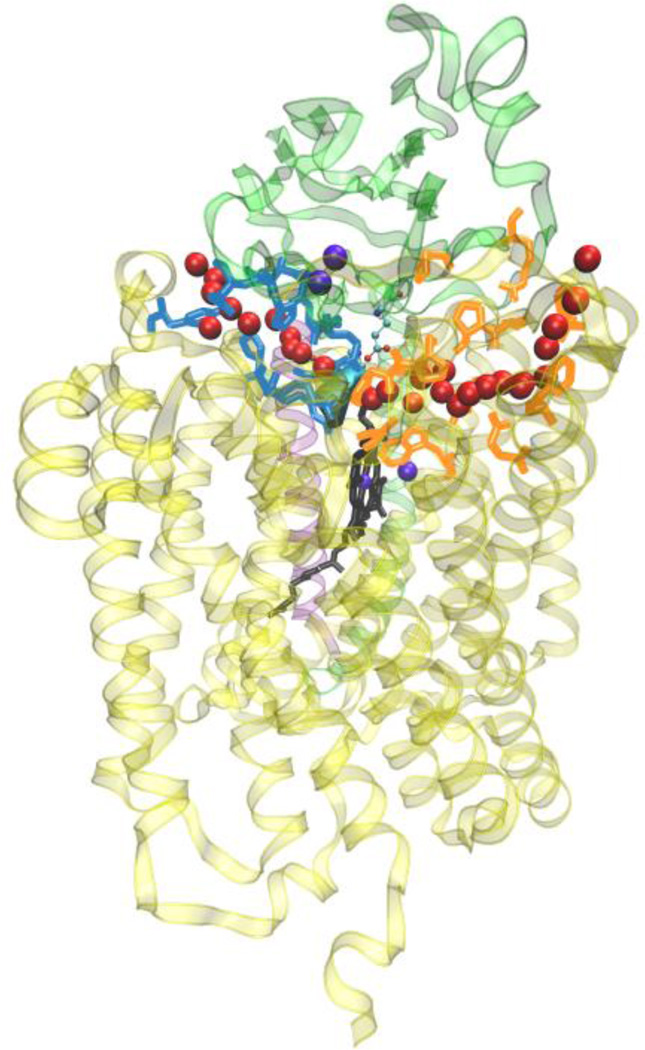
Figure 6.
General view of the two water exit pathways that connect the water pool above the DNC to the P-side of the membrane. Residues in blue color line pathway P1 and residues in orange color line pathway P2. The position of water molecules along one branch of each of the pathways are depicted with red spheres.
Both exit and entry of water molecules through these pathways have been observed. In most simulations we observe a net influx of water molecules, which is consistent with claims that the experimental crystal structures are somewhat dehydrated.[102] The numbers of water molecules that enter and leave the water pool along the two pathways are summarized in Table 4 for each simulation. The table shows that the rate of exchange between the water pool and bulk water differs between the simulations and thus depends on the protonation state of the various residues. Some of this observed difference might be attributed to the finite simulation time (140 ns) and the fact that we have used a simple approach for identifying the water pathways, which is not quantitative with respect to water exchange between protein interior and bulk water. The data presented in Table 4 considers only the first and last frame of the simulations and only water molecules in the water pool, but not water molecules in the water channels and interstitial space that could enter the water pool and subsequently exit through the water pathways. On the other hand, the data in Table 4 gives a clear indication that subtle changes in the protein environment (such as changes in protonation state or redox state) are able to induce structural changes that block the pathways and thus could act as valves that prevent proton backflow. For instance, no water exchange was observed for state D in which the postulated PLS His376 is doubly protonated.
Table 4.
Number of water molecules observed to exchange between the water pool above the DNC and bulk water outside of the enzyme via pathways P1 and P2 for simulations A to F. Exchanging water molecules were identified by comparison of the first and last simulation frames. Water molecules in the water pool were identified through distance to the surrounding residues and visual inspection.
| Simulations | A | B | C | D | E | F | |
|---|---|---|---|---|---|---|---|
| Water molecules in water pool | |||||||
| First frame | 8 | 8 | 8 | 8 | 8 | 8 | |
| Last frame | 14 | 10 | 12 | 8 | 10 | 12 | |
| Water molecules exchanging with bulk | |||||||
| Entera – pathway P1 | 11 | 1 | 3 | 0 | 2 | 7 | |
| Entera – pathway P2 | 0 | 0 | 1 | 0 | 1 | 2 | |
| Exitb – pathway P1 | 4 | 1 | 1 | 0 | 1 | 5 | |
| Exitb – pathway P2 | 0 | 0 | 0 | 0 | 1 | 2 | |
| Total – pathway P1 | 15 | 2 | 4 | 0 | 3 | 12 | |
| Total – pathway P2 | 0 | 0 | 1 | 0 | 2 | 4 | |
Water molecules that moved from the bulk water outside of the protein into the water pool.
Water molecules that moved from the water pool to the bulk water outside of the protein.
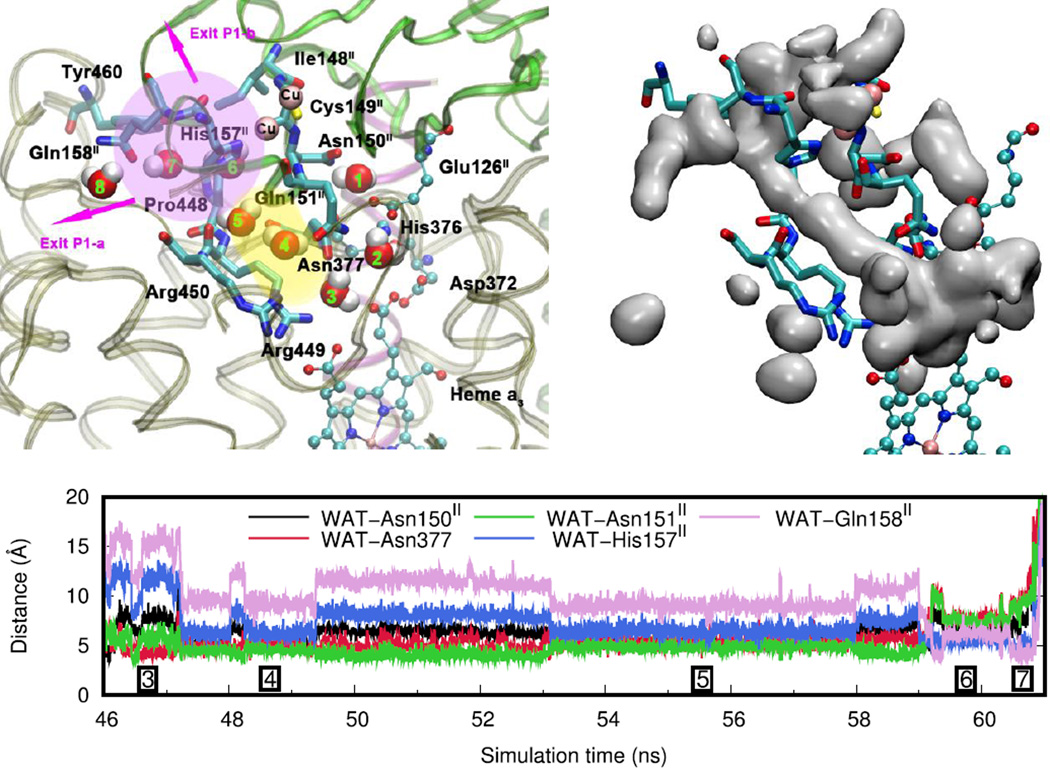
Pathway P1 is depicted in detail in Figure 7 along with an isosurface plot of the water occupancy averaged over the entire simulation trajectory and a distance analysis for a typical trajectory of a water molecule leaving the water pool. Water occupancy plots for simulations of all states A to D can be found in the Supporting Information. The entry to pathway P1 is located around the guanidinium groups of Arg449 and Arg450 and the carboxamide group of Asn377 (position 3). It extends as a pocket from the water pool along the space between the side chains of Arg449 and Arg450 and residues Gln151II and Asn150II (positions 4 and 5, yellow region). In the crystal structure there are two water molecules within this pocket. It connects to a highly solvent-accessible area of the protein (position 6, pink region), which also contains two water molecules in the crystal structure. To reach this region, water molecules have to pass through an opening between the side chains of both residues His157II and Pro448 and the backbone atoms of residue Asn150II. Residue His157II coordinates to CuA and it is thus possible that redox changes are able to influence this pathway and potentially mediate the exit of protons. In our simulations, we have found residue His157II to be very flexible. The solvent-accessible region is connected to the bulk water via two branches, indicated as Exit P1-a and Exit P1-b. The first exit passes at residue Gln158II on the left side of Tyr460, downwards and towards the left from the pink region in Figure 7. The second exit extends upwards and passes at the right side of residue Tyr460, which is also in vicinity of the crystal structure water molecules in this solvent accessible area.
Figure 7.
Water exit pathway P1. Top left: Detailed water pathway highlighting relevant residues. The pathway connects the water pool above the DNC to a solvent-accessible area (pink) via a pocket along the side-chains of residues Arg449 and Arg450 (above water position 3; yellow). Water position 1 is close to residue Glu126II that could be involved in proton transport. Top right: Water occupancy averaged over the entire trajectory (isosurface plot at 25% occupancy), clearly showing the path that connects the water pool to the protein exterior. Bottom: Distances between the center of mass of the water molecule leaving the water pool (WAT) and relevant residues along pathway P1 (Simulation E, see Table 1). The boxed numbers refer to the position of the water molecule as depicted in the top left figure.
Exit P1-b of this pathway is consistent with the alternative exit-R pathway that has been proposed by Sugitani et al. for the ba3 CcO from Thermus Thermophilus based on structural comparisons to water transport pathways that have been identified for aa3-type CcOs (see Figure S5 in the Supporting Information for a detailed comparison). [50] Key residues that surround the water pocket of exit pathway P1 above the water pool (His157II, Arg449, Arg450) are conserved in the exit-R pathway of aa3-type CcO from different species (bovine heart, PDB ID: 1V55; Paracoccus denitrificans, PDB ID: 1AR1; Rhodobacter sphaeroides, PDB ID: 2GSM). However, exit-R in aa3-type CcO follows a different path after entering the solvent-accessible region (position 6), which would be perpendicular to the plane of Figure 7. As a consequence, pathway P1 in ba3-type CcO is significantly shorter than exit-R in aa3-type CcOs.
The water molecules exiting through pathway P1 are in hydrogen bonding distance to at least one other water molecule throughout the entire passage from the water pool to the bulk. This suggests that protons could be conducted from the water pool to the P-side of the membrane via a Grotthuss mechanism along a continuous chain of water molecules that can transiently form within this pathway. The most probable point of interruption of such a water chain is at the transition between the water pocket above residues Arg449 and Arg450 and the solvent-accessible area (transition from position 5 to position 6 in Figure 7). Implications for proton transport will require a more detailed analysis.
Pathway P2 is depicted in Figure 8 together with a distance analysis for a typical trajectory of a water molecule leaving the water pool. The starting point of this pathway is the same water pool above the DNC (position 1), however, the exit is located on the opposite side of the water pool along the surface between protein subunits I and II. The entry to this pathway is composed of the tyrosyl group of Tyr136 and the side chain of Thr134 on one side, and the side chain indole of Trp229 and the guanidinium group of Arg225 on the other side (position 2). This entry connects to a small water pocket surrounded by Arg225, Tyr136, Pro137 and Pro221 (position 3 and yellow region in Figure 8 left). From this position onwards the water exit pathway bifurcates and two different branches can be followed, as indicated by pink arrows with corresponding labels. One of the branches is going along residues Pro221 and Pro129II upwards to a region (pink, labeled Gap 1) that connects to bulk water in vicinity of residues Arg52II and Asp220 (Exit P2-a). The other branch follows a path along the side chain pyrrolidine of Pro137 (position 4) to another solvent-exposed region (position 5, pink region labeled Gap 2), which is delimited by Ala224 and Glu203. This pink region subsequently connects to the protein exterior bulk water at residues Leu553 and Phe207 (Exit P2-b). The entrance region to pathway P2 exhibits similarities to the entrance region of the exit-P pathway in aa3-type CcO as described by Sugitani et al. [50] The exit-P pathway in aa3-type CcO, however, extends into a different direction beyond the proline residues and no spontaneous water transport along this pathway was observed in MD simulations. This is in contrast to our results for ba3-type CcO from Thermus thermophilus.
Figure 8.
Water exit pathway P2. Top left: Detailed water pathway highlighting relevant residues. The pathway connects the water pool above the DNC to the outer side of the membrane via a water pocket (yellow) and two regions (pink, Gap1 and Gap2) that open towards the bulk solvent. Gap 1 is surrounded by residues Arg52II, Pro129II, Pro221, Asp220 and Leu553; Gap 2 is surrounded by residues Leu553, Asp220, Ala224, Glu203 and Phe207. Top right: Water occupancy averaged over the entire trajectory (isosurface plot at 25% occupancy), showing the path that connects the water pool to the protein exterior. Bottom: Distances between the center of mass of the water molecule leaving the water pool (WAT) and relevant residues along pathway P2 (Simulation F, see Table 1). The numbers refer to the position of the water molecule as depicted on the left.
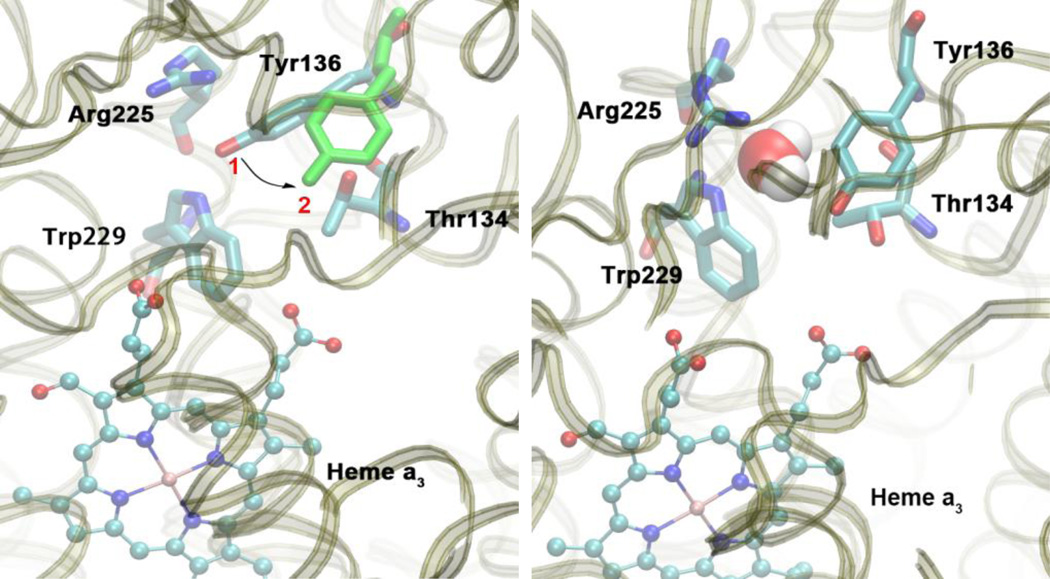
Very importantly, Tyr136 is a key residue at the entrance of pathway P2. It is able to block the entrance by changing its conformation as shown in Figure 9. When in position 1, the distance between Tyr136 and residues Arg225 and Trp229 is too small for water to enter the pathway. A conformational change into position 2 is required to vacate space that allows water to pass. Therefore, it is reasonable to assume that Tyr136 plays an important role, potentially acting as gate that prevents water and proton backflow in water pathway P2. MD simulations of aa3-type CcO from Paracoccus denitrificans have also shown a significant reorientation of residue Tyr167, [52] which is located at an equivalent position as Tyr136 in the ba3-type CcO studied in this work. It was speculated that Tyr167 plays an important role in the formation of a hydrogen-bonded connection to nearby Arg474 and proton exit pathways, although experimental evidence concerning the role of this residue is not available.
Figure 9.
Left: Residue Tyr136 is able to change its conformation from position 1 to position 2 (green). Right: In position 2, the entrance to water exit pathway P2 is open and water is able to pass through this gate. Snapshots are taken from simulation F.
As mentioned above, the protonation state seems to influence the rate of water exchange between the water pool above the DNC and the bulk solvent. Protonation state D seems to be special, since no water is found to leave or enter the water pool during the entire simulation time (Table 4). In this protonation state, there are strong hydrogen bond interactions between the doubly protonated His376 and both Glu126II and PRAa3, as described in the proton transport mechanism section above. This leads to structural changes in the enzyme that are sufficiently large to block water exit through the two water pathways. For pathway P1, the strong hydrogen bonding between Glu126II and doubly protonated His376 results in an increased distance between Glu126II and both Asn377 and Gln151II. This in turn leads to a stronger interaction between Gln151II and Arg449/Arg450, which narrows the entrance to pathway P1 as well as the “channel walls” of the water pocket, effectively blocking this pathway. This mechanism of blocking pathway P1 may work in conjunction with other mechanisms that could involve for instance redox changes of the CuA site as mentioned above and as suggested by others. [50] For pathway P2, it is notable that residue Tyr136 does not undergo any conformational change. It remains in position 1 (see Figure 9) during the entire simulation of protonation state D, thus preventing water molecules from passing through pathway P2. This clearly demonstrates that Tyr136 does play an important role in gating pathway P2. Analysis of the water occupancy within the exit pathways also shows that this results in an interruption or complete absence of water density within the channels (see Figures S6 and S7 in the Supporting Information). This indicates that blocking these pathways disrupts the hydrogen bond network and hence impairs proton transfer. Proton transport could, however, still be possible via ionizable residues in sufficient proximity.
During each CcO reaction cycle two water molecules are generated in a cavity near the active site of the DNC, which have to leave the enzyme. Two water molecules are present in the crystal structure in this region, which corresponds to the hydrophobic cavity that has been described by Sugitani et al. [50] for aa3-type enzymes. Through visualization of our simulation trajectories with a focus on water residing within this cavity, we have confirmed that water molecules can exchange between this region close to the DNC and the water pool. This confirms that water produced during the reaction cycle is able to be transported out of the enzyme via pathways P1 and P2 that we have identified above.
In agreement with what Fee et al. [40] proposed, our simulations also show that the carboxylate side chain group of Glu126II is able to rotate about its Cβ-Cγ and Cγ-Cδ bonds, thereby being unconstrained to swing into different positions. Protonated Glu126II (state E in Figure 1) forms a hydrogen bond with Asn150II, thus being in a position to transfer a proton to water in position 1 of Figure 7. From there, a proton could be transported through pathway P1. In other simulations, we have observed that Glu126II swings into the water pool at a location close to the entrance of pathway P2, which indicates that it could transfer a proton to a water molecule at position 1 in Figure 8. It is thus likely that proton transfer from Glu126II can proceed via either of the two pathways.
4 Conclusions
We have obtained new insight into the mechanism of proton pumping in ba3-type CcO from Thermus thermophilus and identified two water exit pathways that connect the water pool above the DNC to the outer P-side of the membrane. Our results are based on unbiased long-time scale, all-atom MD simulations of a variety of relevant protonation states of CcO with reduced heme b and CuA pair and DNC with a bridging hydroperoxide, Fe3+-(OOH−)-Cu2+. We present a potential mechanism for proton transport above the DNC that involves PRAa3, Asp372, His376 and Glu126II through analyzing changes in the protein structure and dynamics of the hydrogen bond network upon protonation of the different residues. The simulations support a mechanism, in which residue Asp372 and PRAa3 transfer the proton onto His376, which acts as the PLS. Key to this proton transfer mechanism is a rotation of residue His376 with alternate δ- and ε- protonation for every second proton transfer. It seems plausible that the proton is subsequently transferred to residue Glu126II, which is able to interact with the salt bridge Arg225–Asp287. Results from potential of mean force calculations with different protonation states of His376 and Glu126II demonstrate that the location of a proton can have a significant effect on the strength of this salt bridge. However, the salt bridge remains stable in all protonation states and thus is unlikely to act as a simple electrostatic gate as had been suggested before. The two water exit pathways that connect the water pool above the DNC to the outer P-side of the membrane can likely also act as proton exit transport pathways. Residue Glu126II is located nearby the salt bridge Arg225–Asp287 and interacts with the hydrogen bond network around Arg225, which is at the entrance of the first water exit pathway. Residue Glu126II is also able to swing out into the water pool close to the entrance of the second water exit pathway, and thus is likely to facilitate transfer of the proton into either of the two pathways. Importantly, both of these two water exit pathways can be blocked, which could prevent proton backflow. Protonation of the PLS His376 leads to a reduced distance between residues Gln151II and Arg449/Arg450 thus narrowing the entrance channel to the first pathway, while residue Tyr136 remains in a conformation that blocks the entrance to the second pathway. These observations from our simulations could be a valuable starting point for experimental verification of the closing mechanism of these water and proton transport pathways.
Supplementary Material
Highlights.
We have performed long-time scale, all-atom molecular dynamics simulations on ba3-type cytochrome c oxidase from Thermus thermophilus.
A mechanism for proton transport in the region above the dinuclear center (DNC) is proposed, with residue His376 as proton-loading site.
The salt bridge Arg225–Asp287 is stable in several protonation states and unlikely to act as a simple electrostatic gate.
Two water exit pathways that connect the water pool above the DNC to the outer P-side of the membrane are identified.
Protonation of His376 blocks both water pathways by narrowing the entrance channel or conformational changes of Tyr136.
Acknowledgments
We gratefully acknowledge financial support by NIH (Grant R01 GM100934) and the China Scholarship Council (CSC) (No, 201306820006) for funding a two-year research visit of LY in the USA. We also acknowledge the support and help for LY by Professor Sanguo Hong, Ning Zhang, and Hongming Wang of the Deparment of Chemisty, Institute for Advanced Study, Nanchang University, China. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575. We appreciate computer time from the San Diego Supercomputer Center through award TG-CHE130010 to AWG. We also thank the Scripps Research Institute for computational resources. This work was also supported in part through computer time provided by the National Biomedical Computation Resource (NBCR) NIH (P41 GM103426) and Exxact Corporation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoshikawa S, Shimada A. Reaction Mechanism of Cytochrome c Oxidase. Chem. Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 2.Popovic DM. Current advances in research of cytochrome c oxidase. Amino Acids. 2013;45:1073–1087. doi: 10.1007/s00726-013-1585-y. [DOI] [PubMed] [Google Scholar]
- 3.Wikström M, Sharma V, Kaila VRI, Hosler JP, Hummer G. New Perspectives on Proton Pumping in Cellular Respiration. Chem. Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 4.Wikström M. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim. Biophys. Acta, Bioenerg. 2004;1655:241–247. doi: 10.1016/j.bbabio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta, Bioenerg. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 6.Hemp J, Gennis R. Diversity of the Heme–Copper Superfamily in Archaea: Insights from Genomics and Structural Modeling, in. In: Schäfer G, Penefsky H, editors. Bioenergetics. Springer Berlin Heidelberg; 2008. pp. 1–31. [DOI] [PubMed] [Google Scholar]
- 7.Lee HJ, Reimann J, Huang YF, Adelroth P. Functional proton transfer pathways in the heme-copper oxidase superfamily. Biochim. Biophys. Acta, Bioenerg. 2012;1817:537–544. doi: 10.1016/j.bbabio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 8.von Ballmoos C, Adelroth P, Gennis RB, Brzezinski P. Proton transfer in ba(3) cytochrome c oxidase from Thermus thermophilus. Biochim. Biophys. Acta, Bioenerg. 2012;1817:650–657. doi: 10.1016/j.bbabio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Siletsky SA, Belevich I, Jasaitis A, Konstantinov AA, Wikström M, Soulimane T, Verkhovsky MI. Time-resolved single-turnover of ba3 oxidase from Thermus thermophilus. Biochim. Biophys. Acta, Bioenerg. 2007;1767:1383–1392. doi: 10.1016/j.bbabio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Rauhamäki V, Wikström M. The causes of reduced proton-pumping efficiency in type B and C respiratory heme-copper oxidases, and in some mutated variants of type A. Biochim. Biophys. Acta, Bioenerg. 2014;1837:999–1003. doi: 10.1016/j.bbabio.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 11.von Ballmoos C, Gonska N, Lachmann P, Gennis RB, Adelroth P, Brzezinski P. Mutation of a single residue in the ba(3) oxidase specifically impairs protonation of the pump site. Proc. Natl. Acad. Sci. USA. 2015;112:3397–3402. doi: 10.1073/pnas.1422434112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaila VRI, Verkhovsky M, Hummer G, Wikström M, Prevention of leak in the proton pump of cytochrome c oxidase. Biochim. Biophys. Acta. Bioenerg. 2008;1777:890–892. doi: 10.1016/j.bbabio.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Belevich I, Verkhovsky MI. Molecular mechanism of proton translocation by cytochrome c oxidase. Antioxid. Redox Signaling. 2008;10:1–29. doi: 10.1089/ars.2007.1705. [DOI] [PubMed] [Google Scholar]
- 14.Michel H. The mechanism of proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 1998;95:12819–12824. doi: 10.1073/pnas.95.22.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegbahn PEM, Blomberg MRA. Proton Pumping Mechanism in Cytochrome c Oxidase. J. Phys. Chem. A. 2008;112:12772–12780. doi: 10.1021/jp801635c. [DOI] [PubMed] [Google Scholar]
- 16.Siegbahn PEM, Blomberg MRA. Energy diagrams and mechanism for proton pumping in cytochrome c oxidase. Biochim. Biophys. Acta, Bioenerg. 2007;1767:1143–1156. doi: 10.1016/j.bbabio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Kim YC, Wikström M, Hummer G. Kinetic models of redox-coupled proton pumping. Proc. Natl. Acad. Sci. USA. 2007;104:2169–2174. doi: 10.1073/pnas.0611114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daskalakis V, Farantos SC, Guallar V, Varotsis C. Regulation of Electron and Proton Transfer by the Protein Matrix of Cytochrome c Oxidase. J. Phys. Chem. B. 2011;115:3648–3655. doi: 10.1021/jp1115993. [DOI] [PubMed] [Google Scholar]
- 19.Hemp J, Han H, Roh JH, Kaplan S, Martinez TJ, Gennis RB. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb(3) oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46:9963–9972. doi: 10.1021/bi700659y. [DOI] [PubMed] [Google Scholar]
- 20.Fadda E, Yu CH, Pomes R. Electrostatic control of proton pumping in cytochrome c oxidase. Biochim. Biophys. Acta. 2008;1777:277–284. doi: 10.1016/j.bbabio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8-Angstrom Resolution of Cytochrome-C-Oxidase from Paracoccus-Denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 22.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyoma H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 24.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 angstrom. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 25.Konstantinov AA, Siletsky S, Mitchell D, Kaulen A, Gennis RB. The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. Proc. Natl. Acad. Sci. USA. 1997;94:9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brzezinski P, Gennis RB. Cytochrome c oxidase: exciting progress and remaining mysteries. J. Bioenerg. Biomembr. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wikström M, Jasaitis A, Backgren C, Puustinen A, Verkhovsky MI. The role of the D- and K-pathways of proton transfer in the function of the haem-copper oxidases. Biochim. Biophys. Acta, Bioenerg. 2000;1459:514–520. doi: 10.1016/s0005-2728(00)00191-2. [DOI] [PubMed] [Google Scholar]
- 28.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba(3) oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc. Natl. Acad. Sci. USA. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woelke AL, Wagner A, Galstyan G, Meyer T, Knapp EW. Proton Transfer in the K-Channel Analog of B-Type Cytochrome c Oxidase from Thermus thermophilus. Biophys. J. 2014;107:2177–2184. doi: 10.1016/j.bpj.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popovic DM, Stuchebrukhov AA. Proton exit channels in bovine cytochrome c oxidase. J. Phys. Chem. B. 2005;109:1999–2006. doi: 10.1021/jp0464371. [DOI] [PubMed] [Google Scholar]
- 31.Belevich I, Bloch DA, Belevich N, Wikström M, Verkhovsky MI. Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc. Natl. Acad. Sci. USA. 2007;104:2685–2690. doi: 10.1073/pnas.0608794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaila VRI, Sharma V, Wikström M. The identity of the transient proton loading site of the proton-pumping mechanism of cytochrome c oxidase. Biochim. Biophys. Acta, Bioenerg. 2011;1807:80–84. doi: 10.1016/j.bbabio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Sugitani R, Medvedev ES, Stuchebrukhov AA. Theoretical and computational analysis of the membrane potential generated by cytochrome c oxidase upon single electron injection into the enzyme. Biochim. Biophys. Acta, Bioenerg. 2008;1777:1129–1139. doi: 10.1016/j.bbabio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita T, Voth GA. Insights into the Mechanism of Proton Transport in Cytochrome c Oxidase. J. Am. Chem. Soc. 2012;134:1147–1152. doi: 10.1021/ja209176e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goyal P, Lu JX, Yang S, Gunner MR, Cui Q. Changing hydration level in an internal cavity modulates the proton affinity of a key glutamate in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2013;110:18886–18891. doi: 10.1073/pnas.1313908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim. Biophys. Acta, Bioenerg. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Lu JX, Gunner MR. Characterizing the proton loading site in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2014;111:12414–12419. doi: 10.1073/pnas.1407187111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang HY, Choi SK, Vakkasoglu AS, Chen Y, Hemp J, Fee JA, Gennis RB. Exploring the proton pump and exit pathway for pumped protons in cytochrome ba(3) from Thermus thermophilus. Proc. Natl. Acad. Sci. USA. 2012;109:5259–5264. doi: 10.1073/pnas.1107345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fee JA, Case DA, Noodleman L. Toward a Chemical Mechanism of Proton Pumping by the B-Type Cytochrome c Oxidases: Application of Density Functional Theory to Cytochrome ba(3) of Thermus thermophilus. J. Am. Chem. Soc. 2008;130:15002–15021. doi: 10.1021/ja803112w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koutsoupakis C, Soulimane T, Varotsis C. Probing the Q-proton pathway of ba(3)-cytochrome c oxidase by time-resolved Fourier transform infrared spectroscopy. Biophys. J. 2004;86:2438–2444. doi: 10.1016/S0006-3495(04)74300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muramoto K, Hirata K, Shinzawa-Itoh K, Yoko-O S, Yamashita E, Aoyama H, Tsukihara T, Yoshikawa S. A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2007;104:7881–7886. doi: 10.1073/pnas.0610031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Qin L, Ferguson-Miller S. Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump. Proc. Natl. Acad. Sci. USA. 2011;108:1284–1289. doi: 10.1073/pnas.1012846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blomberg MR, Borowski T, Himo F, Liao RZ, Siegbahn PE. Quantum chemical studies of mechanisms for metalloenzymes. Chem. Rev. 2014;114:3601–3658. doi: 10.1021/cr400388t. [DOI] [PubMed] [Google Scholar]
- 45.Noodleman L, Du WGH, Fee JA, Götz AW, Walker RC. Linking Chemical Electron Proton Transfer to Proton Pumping in Cytochrome c Oxidase: Broken-Symmetry DFT Exploration of Intermediates along the Catalytic Reaction Pathway of the Iron-Copper Dinuclear Complex. Inorg. Chem. 2014;53:6458–6472. doi: 10.1021/ic500363h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Ballmoos C, Lachmann P, Gennis RB, Adelroth P, Brzezinski P. Timing of Electron and Proton Transfer in the ba(3) Cytochrome c Oxidase from Thermus thermophilus. Biochemistry. 2012;51:4507–4517. doi: 10.1021/bi300132t. [DOI] [PubMed] [Google Scholar]
- 47.Kaila VRI, Verkhovsky MI, Hummer G, Wikström M. Glutamic acid 242 is a valve in the proton pump of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2008;105:6255–6259. doi: 10.1073/pnas.0800770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt B, McCracken J, Ferguson-Miller S. A discrete water exit pathway in the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2003;100:15539–15542. doi: 10.1073/pnas.2633243100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma V, Enkavi G, Vattulainen I, Rog T, Wikström M. Proton-coupled electron transfer and the role of water molecules in proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2015;112:2040–2045. doi: 10.1073/pnas.1409543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugitani R, Stuchebrukhov AA. Molecular dynamics simulation of water in cytochrome c oxidase reveals two water exit pathways and the mechanism of transport. Biochim. Biophys. Acta, Bioenerg. 2009;1787:1140–1150. doi: 10.1016/j.bbabio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng XH, Medvedev DM, Swanson J, Stuchebrukhov AA. Computer simulation of water in cytochrome c oxidase. Biochim. Biophys. Acta, Bioenerg. 2003;1557:99–107. doi: 10.1016/s0005-2728(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 52.Olkhova E, Hutter MC, Lill MA, Helms V, Michel H. Dynamic water networks in cytochrome c oxidase from Paracoccus denitrificans investigated by molecular dynamics simulations. Biophys. J. 2004;86:1873–1889. doi: 10.1016/S0006-3495(04)74254-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wikström M, Verkhovsky MI, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta, Bioenerg. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 54.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High Resolution Structure of the ba3 Cytochrome c Oxidase from Thermus thermophilus in a Lipidic Environment. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Du W-G, Noodleman L. Density Functional Study for the Bridged Dinuclear Center Based on a High-Resolution X-ray Crystal Structure of ba(3) Cytochrome c Oxidase from Thermus thermophilus. Inorg. Chem. 2013;52:14072–14088. doi: 10.1021/ic401858s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anandakrishnan R, Aguilar B, Onufriev AV. H++3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40:W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics Modell. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 59.Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham ITE, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA. AMBER 14. San Francisco: University of California; 2014. [Google Scholar]
- 60.Salomon-Ferrer R, Case DA, Walker RC. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- 61.Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 63.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 65.Joung IS, Cheatham TE. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Comba P, Remenyi R. A new molecular mechanics force field for the oxidized form of blue copper proteins. J. Comput. Chem. 2002;23:697–705. doi: 10.1002/jcc.10084. [DOI] [PubMed] [Google Scholar]
- 67.Capece L, Boechi L, Perissinotti LL, Arroyo-Manez P, Bikiel DE, Smulevich G, Marti MA, Estrin DA. Small ligand-globin interactions: Reviewing lessons derived from computer simulation. Biochim. Biophys. Acta, Proteins Proteomics. 2013;1834:1722–1738. doi: 10.1016/j.bbapap.2013.02.038. [DOI] [PubMed] [Google Scholar]
- 68.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general Amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 69.Yang L, Skjevik AA, Du W-GH, Noodleman L, Walker RC, Götz AW. Parameters for the cofactors in B-type cytochrome c oxidase for molecular dynamics simulations with the Amber force field. Data in Brief, submitted. 2016 doi: 10.1016/j.dib.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baerends EJ, Ziegler T, Autschbach J, Bashford D, Bérces A, Bickelhaupt FM, Bo C, Boerrigter PM, Cavallo L, Chong DP, Deng L, Dickson RM, Ellis DE, van Faassen M, Fan L, Fischer TH, Fonseca Guerra C, Franchini M, Ghysels A, Giammona A, van Gisbergen SJA, Götz AW, Groeneveld JA, Gritsenko OV, Grüning M, Gusarov S, Harris FE, van den Hoek P, Jacob CR, Jacobsen H, Jensen L, Kaminski JW, van Kessel G, Kootstra F, Kovalenko A, Krykunov MV, van Lenthe E, McCormack DA, Michalak A, Mitoraj M, Morton SM, Neugebauer J, Nicu VP, Noodleman L, Osinga VP, Patchkovskii S, Pavanello M, Philipsen PHT, Post D, Pye CC, Ravenek W, Rodríguez JI, Ros P, Schipper PRT, van Schoot H, Schreckenbach G, Seldenthuis JS, Seth M, Snijders JG, Solà M, Swart M, Swerhone D, te Velde G, Vernooijs P, Versluis L, Visscher L, Visser O, Wang F, Wesolowski TA, van Wezenbeek EM, Wiesenekker G, Wolff SK, Woo TK, Yakovlev AL. ADF2014, SCM, Theoretical Chemistry. Amsterdam, The Netherlands: Vrije Universiteit; http://www.scm.com. [Google Scholar]
- 71.te Velde G, Bickelhaupt FM, Baerends EJ, Guerra CF, Van Gisbergen SJA, Snijders JG, Ziegler T. Chemistry with ADF. J. Comput. Chem. 2001;22:931–967. [Google Scholar]
- 72.Guerra CF, Snijders JG, te Velde G, Baerends EJ. Towards an order-N DFT method. Theor. Chem. Acc. 1998;99:391–403. [Google Scholar]
- 73.Handy NC, Cohen AJ. Left-right correlation energy. Mol. Phys. 2001;99:403–412. [Google Scholar]
- 74.Lee CT, Yang WT, Parr RG. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phys. Rev. B: Condens. Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 75.Van Lenthe E, Baerends EJ. Optimized Slater-type basis sets for the elements 1–118. J. Comput. Chem. 2003;24:1142–1156. doi: 10.1002/jcc.10255. [DOI] [PubMed] [Google Scholar]
- 76.Klamt A, Schuurmann G. Cosmo - a New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient. J. Chem. Soc., Perkin Trans. 1993;2:799–805. [Google Scholar]
- 77.Klamt A. Conductor-Like Screening Model for Real Solvents - a New Approach to the Quantitative Calculation of Solvation Phenomena. J. Phys. Chem. 1995;99:2224–2235. [Google Scholar]
- 78.Klamt A, Jonas V. Treatment of the outlying charge in continuum solvation models. J. Chem. Phys. 1996;105:9972–9981. [Google Scholar]
- 79.Chen JL, Noodleman L, Case DA, Bashford D. Incorporating Solvation Effects into Density-Functional Electronic-Structure Calculations. J. Phys. Chem. 1994;98:11059–11068. [Google Scholar]
- 80.Liu TQ, Han WG, Himo F, Ullmann GM, Bashford D, Toutchkine A, Hahn KM, Noodleman L. Density functional vertical self-consistent reaction field theory for solvatochromism - Studies of solvent-sensitive dyes. J. Phys. Chem. A. 2004;108:3545–3555. [Google Scholar]
- 81.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Grand S, Götz AW, Walker RC. SPFP: Speed without compromise-A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 83.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 84.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 85.Loncharich RJ, Brooks BR, Pastor RW. Langevin Dynamics of Peptides - the Frictional Dependence of Isomerization Rates of N-Acetylalanyl-N'-Methylamide. Biopolymers. 1992;32:523–535. doi: 10.1002/bip.360320508. [DOI] [PubMed] [Google Scholar]
- 86.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 87.Allen M, Tildesley D. Computer Simulation of Liquids. Oxford: Oxford University Press; 1988. [Google Scholar]
- 88.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 89.Miyamoto S, Kollman PA. Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 90.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 91.Perdew JP, Chevary JA, Vosko SH, Jackson KA, Pederson MR, Singh DJ, Fiolhais C. Atoms, Molecules, Solids, and Surfaces - Applications of the Generalized Gradient Approximation for Exchange and Correlation. Phys Rev B. 1992;46:6671–6687. doi: 10.1103/physrevb.46.6671. [DOI] [PubMed] [Google Scholar]
- 92.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 93.Du W-GH, Götz AW, Yang L, Walker RC, Noodleman L. A broken-symmetry density functional study of structures, energies, and protonation states along the catalytic O-O bond cleavage pathway in ba3 cytochrome c oxidase from Thermus thermophilus. Phys. Chem. Chem. Phys. 2016 doi: 10.1039/c6cp00349d. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Becke AD. Density-Functional Thermochemistry .3. The Role of Exact Exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 95.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab-Initio Calculation of Vibrational Absorption and Circular-Dichroism Spectra Using Density-Functional Force-Fields. J. Phys. Chem. 1994;98:11623–11627. [Google Scholar]
- 96.Tissandier MD, Cowen KA, Feng WY, Gundlach E, Cohen MH, Earhart AD, Coe JV, Tuttle TR. The proton's absolute aqueous enthalpy and Gibbs free energy of solvation from cluster-ion solvation data. J. Phys. Chem. A. 1998;102:7787–7794. [Google Scholar]
- 97.Marenich AV, Ho JM, Coote ML, Cramer CJ, Truhlar DG. Computational electrochemistry: prediction of liquid-phase reduction potentials. Phys. Chem. Chem. Phys. 2014;16:15068–15106. doi: 10.1039/c4cp01572j. [DOI] [PubMed] [Google Scholar]
- 98.Tawa GJ, Topol IA, Burt SK, Caldwell RA, Rashin AA. Calculation of the aqueous solvation free energy of the proton. J. Chem. Phys. 1998;109:4852–4863. [Google Scholar]
- 99.Torrie GM, Valleau JP. Non-Physical Sampling Distributions in Monte-Carlo Free-Energy Estimation - Umbrella Sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 100.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 101.Grossfield A. “WHAM: the weighted histogram analysis method”, version 2.0.2. http://membrane.urmc.rochester.edu/content/wham/
- 102.Woelke AL, Galstyan G, Galstyan A, Meyer T, Heberle J, Knapp EW. Exploring the Possible Role of Glu286 in CcO by Electrostatic Energy Computations Combined with Molecular Dynamics. J. Phys. Chem. B. 2013;117:12432–12441. doi: 10.1021/jp407250d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.