Summary
Williams syndrome (WS) is a genetic neurodevelopmental disorder characterized by an uncommon hypersociability and a mosaic of retained and compromised linguistic and cognitive abilities. Nearly all clinically diagnosed individuals with WS lack precisely the same set of genes, with breakpoints in chromosome band 7q11.231–5. The contribution of specific genes to the neuroanatomical and functional alterations, leading to behavioral pathologies in humans, remains largely unexplored. Here, we investigate neural progenitor cells (NPCs) and cortical neurons derived from WS and typically developing (TD) induced pluripotent stem cells (iPSCs). WS NPCs have an increased doubling time and apoptosis compared to TD NPCs. Using an atypical WS subject6, 7, we narrowed this cellular phenotype to a single gene candidate, FZD9. At the neuronal stage, WS-derived layers V/VI cortical neurons were characterized by longer total dendrites, increased numbers of spines and synapses, aberrant calcium oscillation and altered network connectivity. Morphometric alterations observed in WS neurons were validated after Golgi staining of postmortem layers V/VI cortical neurons. This human iPSC model8 fills in the current knowledge gap in WS cellular biology and could lead to further insights into the molecular mechanism underlying the disorder and the human social brain.
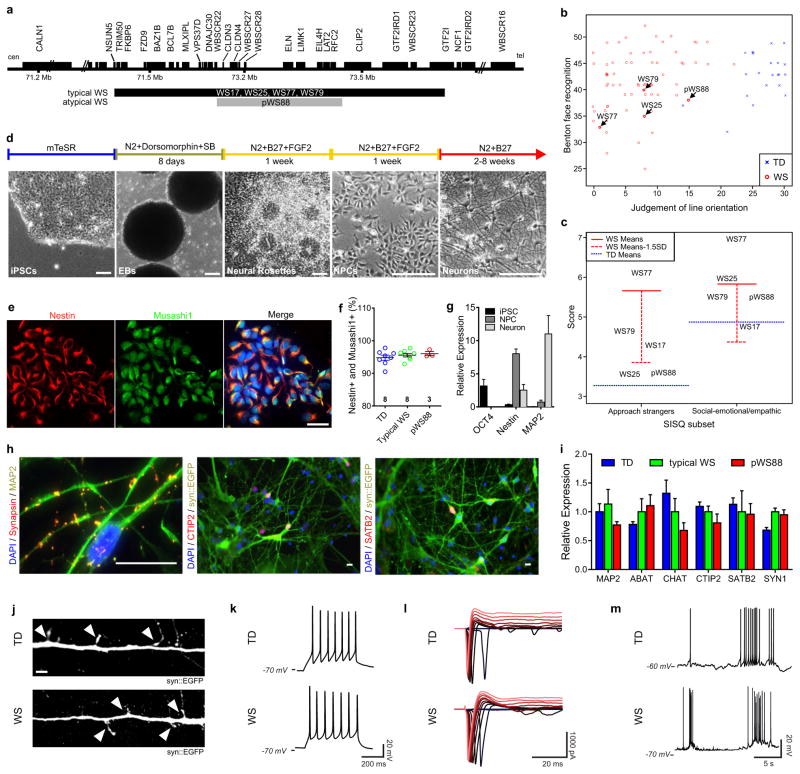
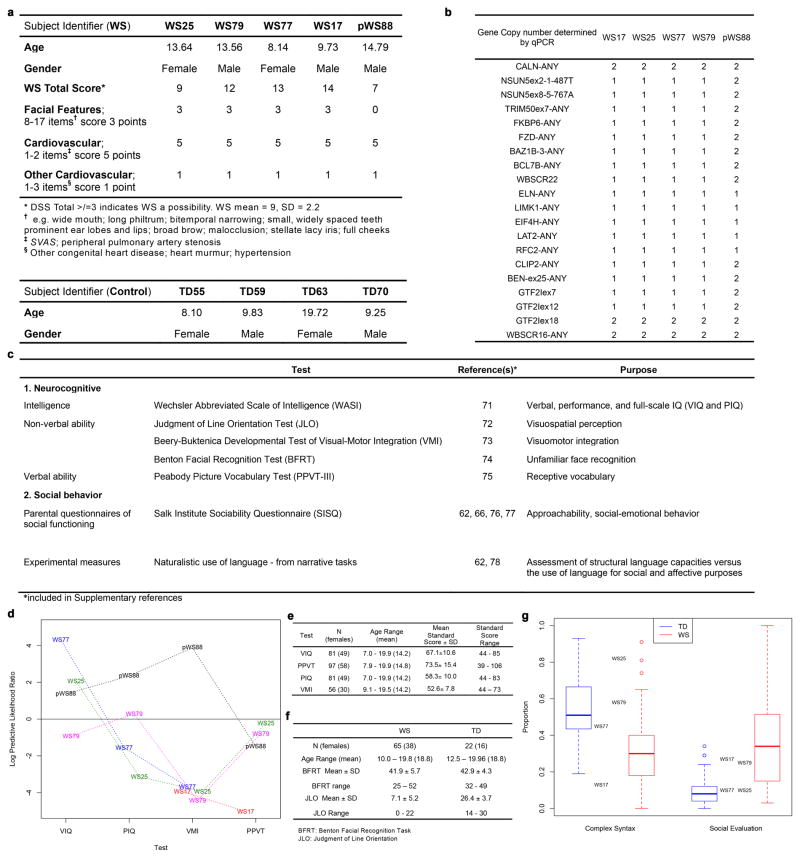
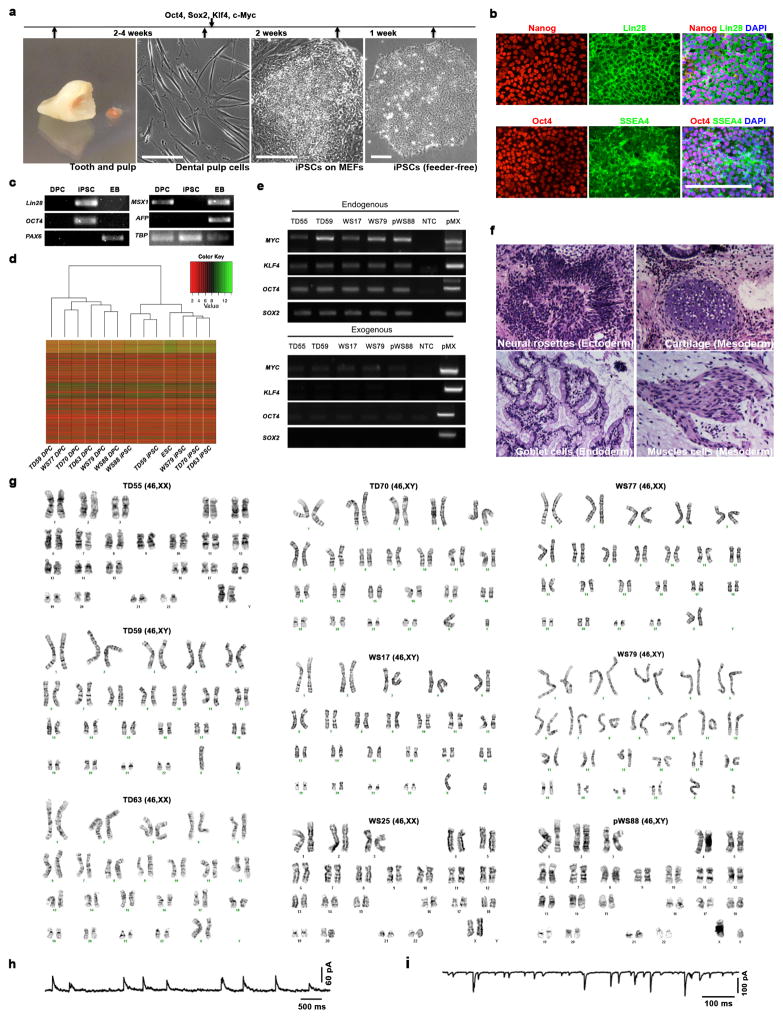
This study included participants with clinical diagnosis of WS: individuals harboring typical gene deletions in the Williams-Beuren Syndrome Critical Region1, 9 (WS17, 25, 77 and 79) and atypical WS subject with a partial deletion (pWS88) as well as TD subjects (TD55, 59, 63 and 70) (Fig. 1a and Extended Data Fig. 1a). After a series of cognitive and social profiles2, 5, 10, we confirmed that the typically-deleted WS participants were a representative cohort of the disorder (Fig. 1b–c; Extended Data Fig. 1b–g and Supplementary Note 1). To generate a human cellular model of WS11, 12, dental pulp cells obtained from subjects’ deciduous teeth were reprogrammed into iPSCs (Extended Data Fig. 2a). We selected two to three clones from each individual for further investigation (Extended Data Fig. 2b–g and Supplementary Table 1, 2). To obtain the relevant cells, iPSC clones underwent neural induction (Fig. 1d) and were further characterized (Fig. 1e–g). Cortical neurons were obtained using a modified protocol from our previous publication11 (Fig. 1h–j). Finally, iPSC-derived neurons exhibited a complete set of electrophysiological properties (Fig. 1k–m and Extended Data Fig. 2h, i).
Figure 1. Characterization of participating individuals and iPSC differentiation.
a, Diagram showing genes and deletion region of WS subjects. b, Scatter plot of Benton Face Recognition and Judgment of Line Orientation scores (jitter added) for n = 69 WS subjects and n = 22 TD subjects. c, Solid red lines depict mean test scores for WS (n = 101 for Approach Strangers; n = 100 for Social-emotional/empathic), and dotted blue lines depict mean test scores for TD (n = 80 for Approach Strangers; n = 79 for Social-emotional/empathic). d, Neural induction and neuronal differentiation protocol. Scale bar, 50 μm. e, Stage-specific protein expression in iPSC-derived NPCs. Scale bar, 50 μm. f, High percentage of Nestin and Musashi1-positive population was comparably observed in TD, typical WS and pWS88 NPCs by FACS. Data are shown as mean ± s.e.m. n = number of clones. g, Stage-specific markers for iPSC (OCT4), NPC (Nestin) and neuron (MAP2) by qPCR. h, Stage-specific protein expression in 6-week-old neurons. Scale bar, 25 μm. i, Expression of different neuronal markers in neurons indicating multiple neuronal subtypes in 6-week-old culture by qPCR. Data are shown as mean ± s.e.m. j, A representative image of neuronal protrusions (spine-like; arrowheads) from iPSC-derived neurons. Scale bar, 2 μm. k–m, 4-week-old TD and WS iPSC-derived neurons show evoked action potentials (k), evoked voltage-dependent sodium and potassium currents (l), and spontaneous bursts of action potentials (m).
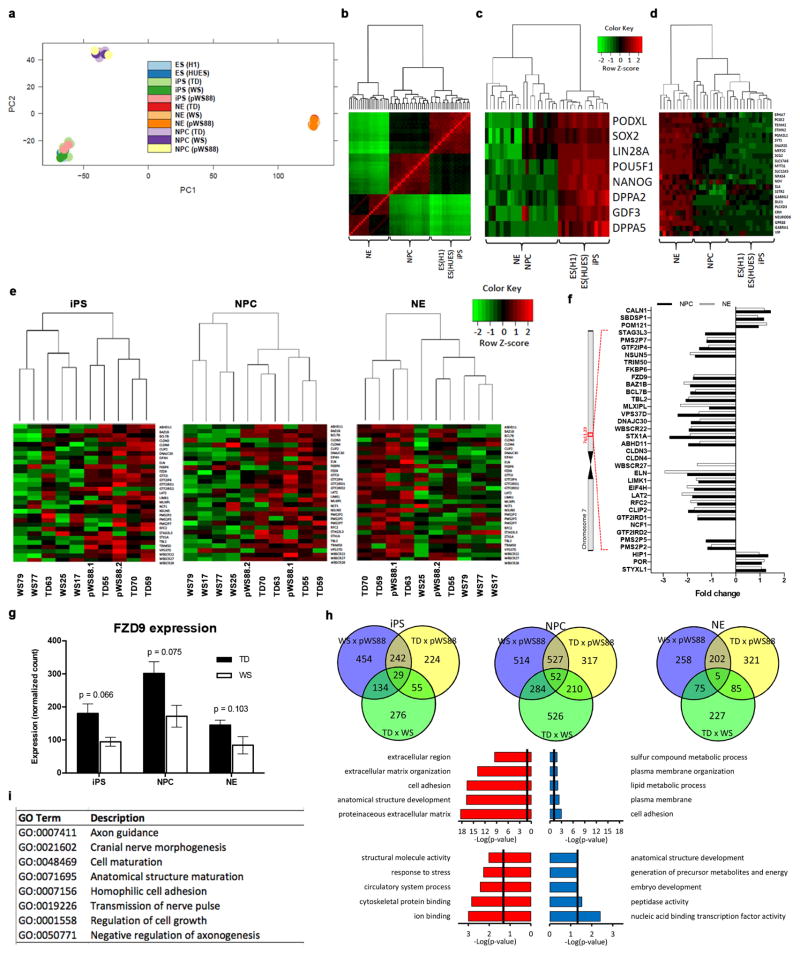
The impact of the genome-wide WBSCR deletion was determined by unbiased RNA-seq (Extended Data Fig. 3a–d). Differential expression analyses revealed misregulated genes among the three genotypes (Extended Data Fig. 3e–h; Extended Data Table 1 and Supplementary Table 3–9). Gene ontology analyses (GO) in NPCs and neurons revealed biological processes that are relevant to the condition (Extended Data Fig. 3h, i; Extended Data Table 2–3 and Supplementary Table 10). Remarkably, “cell adhesion,” “axon guidance” and “cell maturation” were also among the top-ranking categories detected in an independent publication using WS NPCs gene expression analysis13.
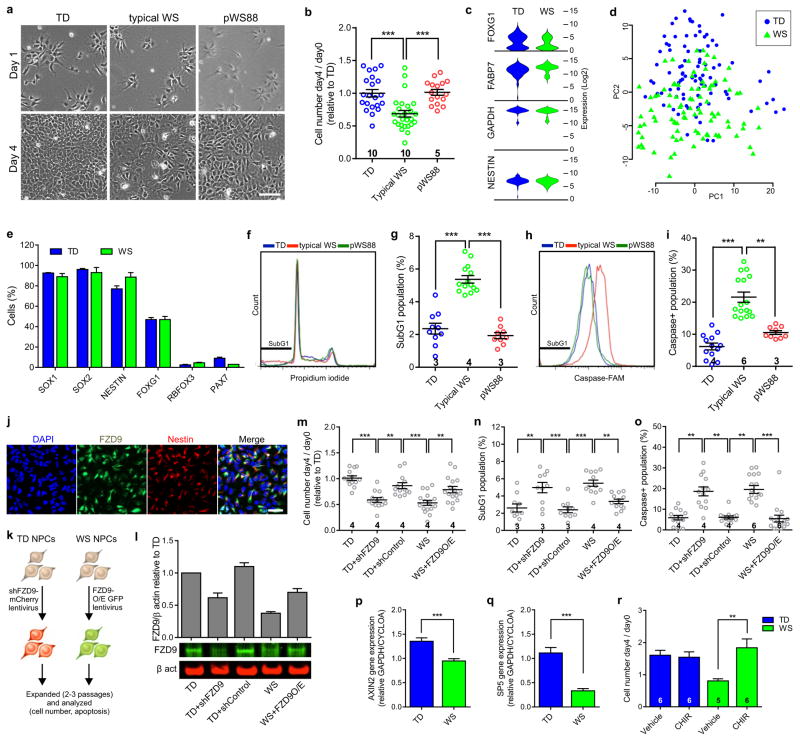
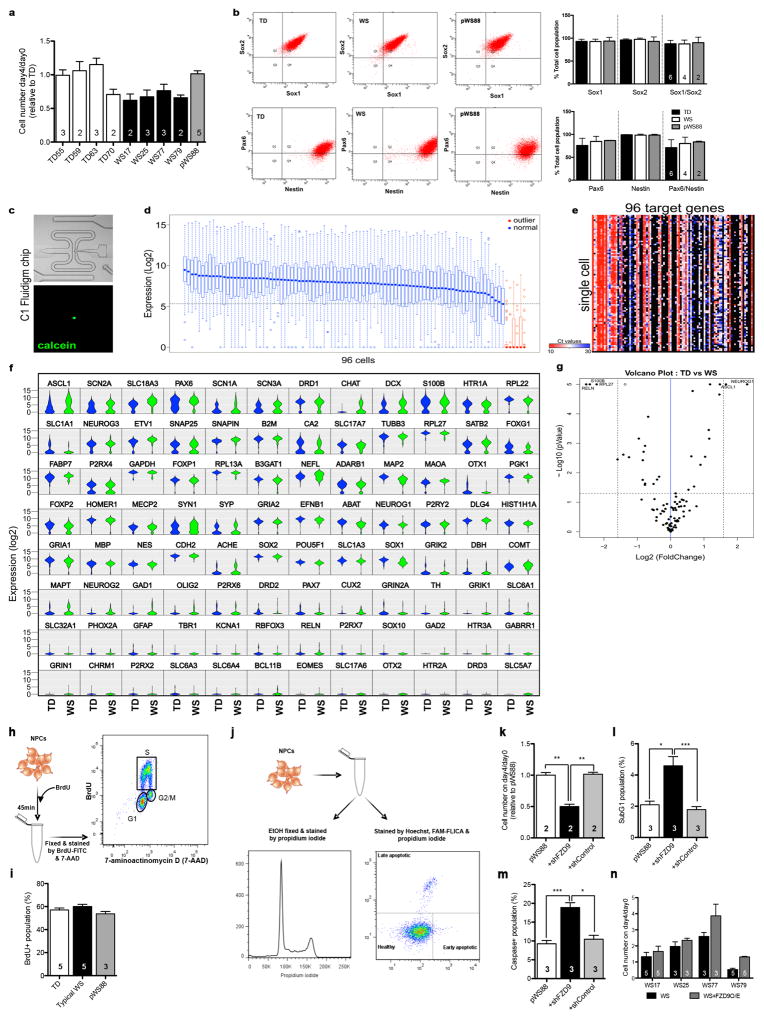
As suggested by the NPC global gene expression analyses, during the culture maintenance, typical WS NPCs became confluent more slowly than TD NPCs (Fig. 2a). After plating the same number of NPCs, we verified that the number of typical WS NPCs on day 4 was less than the TD NPCs (Fig. 2b and Extended Data Fig. 4a). To rule out the possibility that the difference in heterogeneity of iPSC-derived NPCs could result in this observation, the NPC population was fully characterized and no difference between WS and TDs were found (Fig. 1e–g and Extended Data Fig. 4b). We also used single-cell gene expression profiling to access the homogeneity of the NPCs (Fig. 2c–e; Extended Data Fig. 4c–g). We further investigated the proliferation of WS NPCs by performing BrdU labeling, immunostaining and FACS (Extended Data Fig. 4h, i). Since no difference was found, we assessed apoptosis in WS NPCs using DNA fragmentation (propidium iodide) and caspase assay (Extended Data Fig. 4j). We found a significant increase in subG1 (Fig. 2f, g) and caspase-positive populations (Fig. 2h, i) in WS NPCs, indicating increased apoptosis.
Figure 2. Defect in apoptosis of WS-derived NPCs due to haploinsufficiency of FZD9.
a, Representative images showing the difference in confluency between TD, typical WS and pWS88 iPSC-derived NPCs on day 4. Scale bar, 100 μm. b, Ratio of NPC number on day 4 over day 0 relative to TD. c, Violin plots of representative genes expressed in NPCs from single cell analyses. d, Principal component analysis (PCA) was used to compare the expression levels in individual cells based on the first two principal components. e, Percentage of cells expressing NPC, neuronal (RBFOX3) and neural crest (PAX7, contaminant population) related genes. WS and TD iPSC-derived NPCs show similar percentages of cells expressing target genes over defined Ct control value. f, Representative propidium iodide histogram showing an increase in subG1 population in typical WS NPCs. g, Percentage of subG1 population. h, Representative histogram showing an increase in caspase activity (caspase-FAM intensity) in typical WS NPCs. i, Percentage of population with high caspase activity. j, FZD9 protein expression in TD iPSC-derived NPCs. k, Schematic of FZD9 gain/loss of function experiments in NPCs. l, Expression level of FZD9 protein after treatment with shFZD9, shControl and FZD9 overexpression vectors, assessed by Western blot analysis. m–o, Ratio of NPC number on day 4 over day 0 relative to TD (m), percentage of subG1 population (n) and percentage of population with high caspase activity (o) when TD NPCs were treated with shFZD9 and shControl, and WS NPCs were overexpressed with FZD9. p–q, Significant decrease in expression of Axin2 (p) and SP5 (q) of WS NPCs compared to TDs. r, Rescue of WS NPC viability after CHIR98014 treatment. All data are shown as mean ± s.e.m. and n = number of clones. **P<0.01, ***P<0.001, Kruskal-Wallis test and Dunn’s multiple comparison test (b, g, i), one-way ANOVA and Tukey’s post hoc test (g, m–o), two-sided unpaired Student’s t test (p, q), two-sided unpaired Mann Whitney test (r).
Frizzled 9 (FZD9) is expressed in NPCs14 (Fig. 2j) and has been shown to regulate cell division and programmed cell death in different cell types15, 16. In our study, FZD9 was hemizygously deleted in the typical WS participants, but retained in atypical pWS88 (Fig. 1a and Extended Data Fig. 1b). Thus, we hypothesized that FZD9 regulates human NPC apoptosis. We transduced TD NPCs with a lentivirus carrying either shRNA against FZD9 (shFZD9) or non-specific shRNA (shControl) and WS NPCs with lentiviruses carrying a FZD9 cDNA construct (Fig. 2k, l). TD NPCs transduced with shFZD9 showed a reduction in the number of cells on day 4 (Fig. 2m) and an increase in the subG1 population (Fig. 2n) and caspase activity (Fig. 2o) compared to TD NPCs expressing the shControl. Similar results were observed in atypical pWS88 (Extended Data Fig. 4k–n). Restoring FZD9 expression in typical WS NPCs brought the number of NPCs on day 4/day 0 to a similar level to TD NPCs. It also significantly reduced the apoptotic population to the TD level.
Since several Wnt genes were downregulated in WS NPCs (Extended Data Tables 2, 3) and FZD9 can be activated by Wnt ligands, we tested if we could rescue the NPC viability by treating cells with the GSK3 inhibitor CHIR9801417. First, we confirmed that the canonical Wnt pathway was affected in WS by measuring the Axin2 and SP5 expression levels, two universal Wnt target genes18,19. Both genes were significantly downregulated in WS cells compared to TDs (Fig. 2p, q). By treating WS NPCs with CHIR98014, we were able to rescue cell viability (Fig. 2r). Together, our results indicate a role for FZD9 in NPC viability.
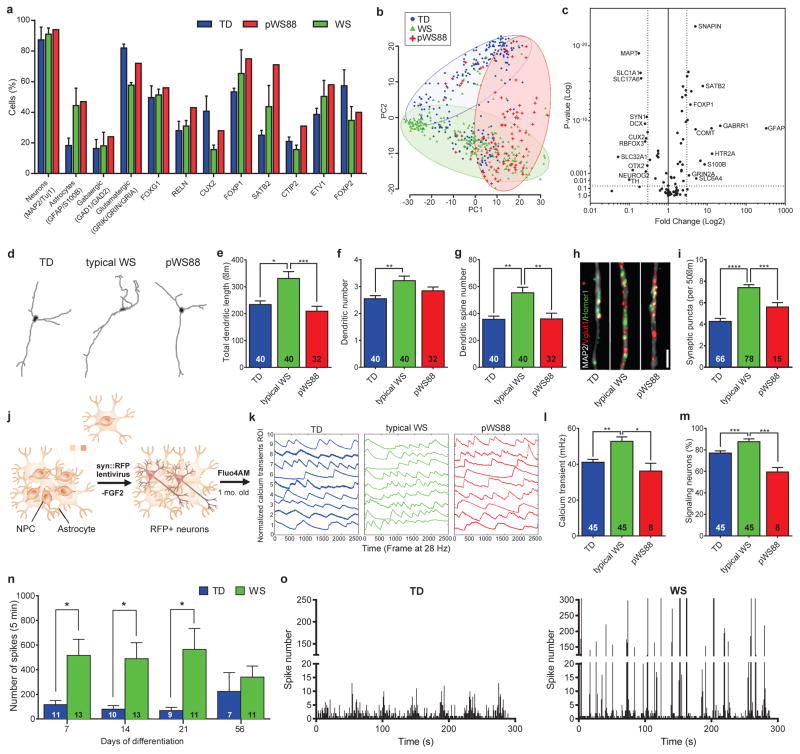
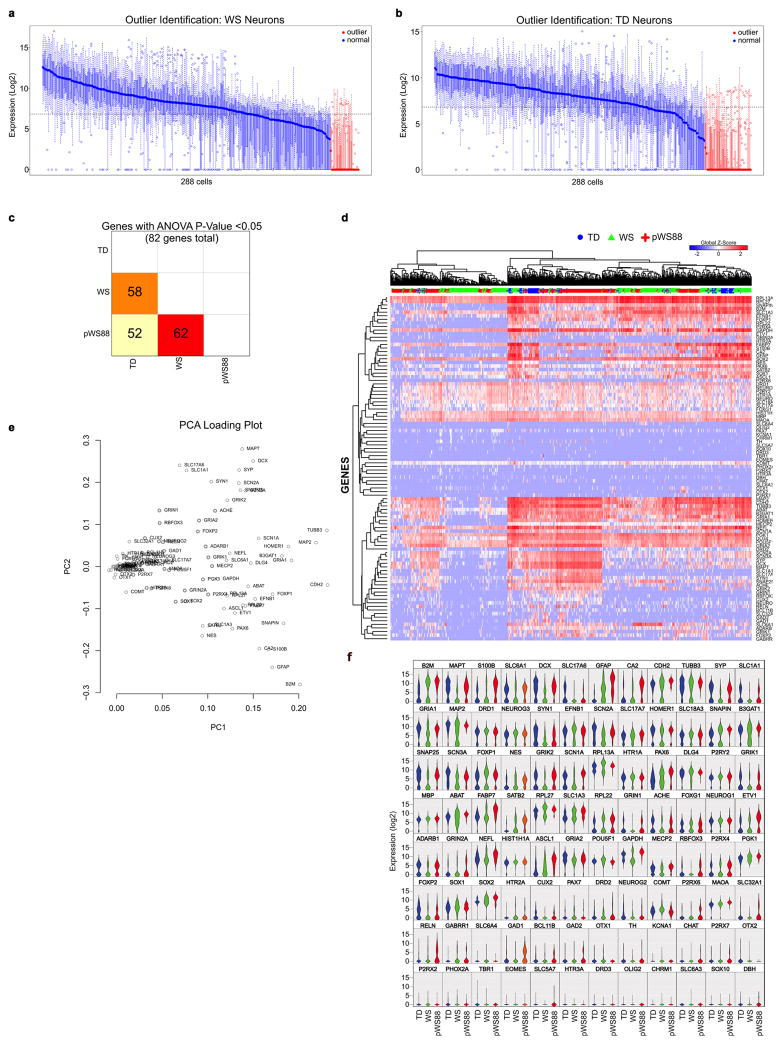
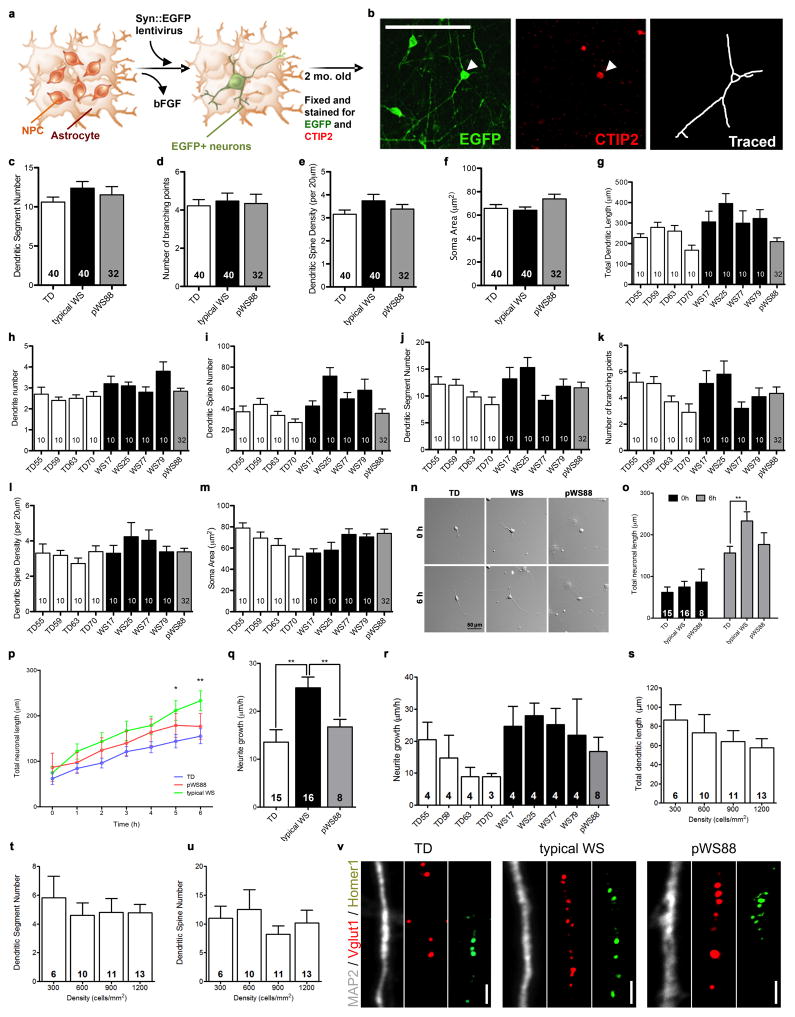
Our protocol generated a consistent population of forebrain neurons, confirmed by the pan-neuronal and subtype-specific cortical markers such as CTIP2 (Layers V/VI)20–22 and SATB2 (Layers III) (Fig. 1h, i). The neuronal population was also characterized by single-cell gene expression profiling (Fig. 3a–c and Extended Data Fig. 5a–f), revealing mostly glutamatergic neurons, with a small population of GABAergic neurons and glia (Fig. 3a). We did not detect significant variability in these subtypes of neurons expressing target genes or in the expression levels of several markers for cortical layers and neurotransmitters among the genotypes. However, we did detect differences in the expression of specific genes in these populations (Fig. 3b, c) that could lead to specific alterations in mature neurons. We focused specifically on markers for cortical layers V/VI, since pathologies affecting these layers have been reported in disorders with compromised social functioning, such as autism23. We found that typical WS iPSC-derived CTIP2-positive neurons had significantly higher total dendritic length, dendrite number, and number of dendritic spines than TDs (Fig. 3d–g and Extended Data Fig. 6a–m). Interestingly, atypical pWS88 neurons were morphologically similar to TD neurons except for dendrite number (Fig. 3f). To determine if the WS neuronal phenotype was cell autonomous or dependent on other cells or culture conditions, we recorded the dendritic growth over time. The result showed a faster dendritic growth rate in WS neurons compared to TD or pWS88 (Extended Data Fig. 6n–r). Also, no differences were observed in the total dendritic length, segment number or spine density using NPCs plated at different cellular densities (Extended Data Fig. 6s–u).
Figure 3. Altered morphology of WS-derived cortical neurons and network activity.
a, Percentage of cells expressing neural markers, neurotransmitter and cortical layer-related genes. WS, pWS88 and TD iPSC-derived neurons show non-significant percentage of cells expressing target genes over defined control Ct value. b, PCA of 672 cells projected onto the first two components. Overlaid populations of TD, pWS88 and WS neurons are shown. c, Volcano plot illustrates differences in expression patterns of target genes of iPSC-derived neurons from the single cell analyses. The dotted lines represent more than or equal to 3.0-fold differentially expressed genes between the groups at P<0.05 (unpaired Student’s t test). d, Representative images of tracings from TD, typical WS and atypical pWS88 iPSC-derived neurons (Syn::eGFP- and CTIP2-positive neurons). e–g, Morphometric analyses showing significant differences between TD, typical WS and pWS88 in total dendritic length (e), between TD and typical WS in dendrite number (f) and between TD, typical WS and pWS88 in dendritic spine number (g). h and i, Puncta quantification of post- and pre-synaptic markers. Scale bar, 2 μm. For e–g and i, data are shown as mean ± s.e.m. n = number of traced neurons. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, Kruskal-Wallis test and Dunn’s multiple comparison test (e–g), one-way ANOVA and Tukey’s post hoc test (i). j, Schematic diagram summarizing preparation of neurons for calcium transient analysis. k, Representative images of the calcium tracing from iPSC-derived neurons. Fluorescence intensity changes reflecting intracellular calcium fluctuations in neurons in different Regions of Interest (ROI). l and m, Typical WS-derived neurons exhibited significant increase in calcium transient frequency (l) and percentage of signaling neuron in the culture (m) when compared to TD or pWS88 neurons. Data are shown as mean ± s.e.m. n = number of fields analyzed; 3198 neurons for TD, 4446 neurons for WS and 48 neurons for pWS88. *P<0.05, **P<0.01, ***P<0.001, Kruskal-Wallis test and Dunn’s multiple comparison test. n, MEA analyses revealed an increase in spontaneous neuronal spikes in WS during differentiation compared to TD. o, Although the number of total network bursts do not differ, WS shows a higher number of spikes in each burst compared to TD. Data are shown as mean ± s.e.m. n = number of MEA wells analyzed *P<0.05, **P<0.01, two-sided unpaired Student’s t test.
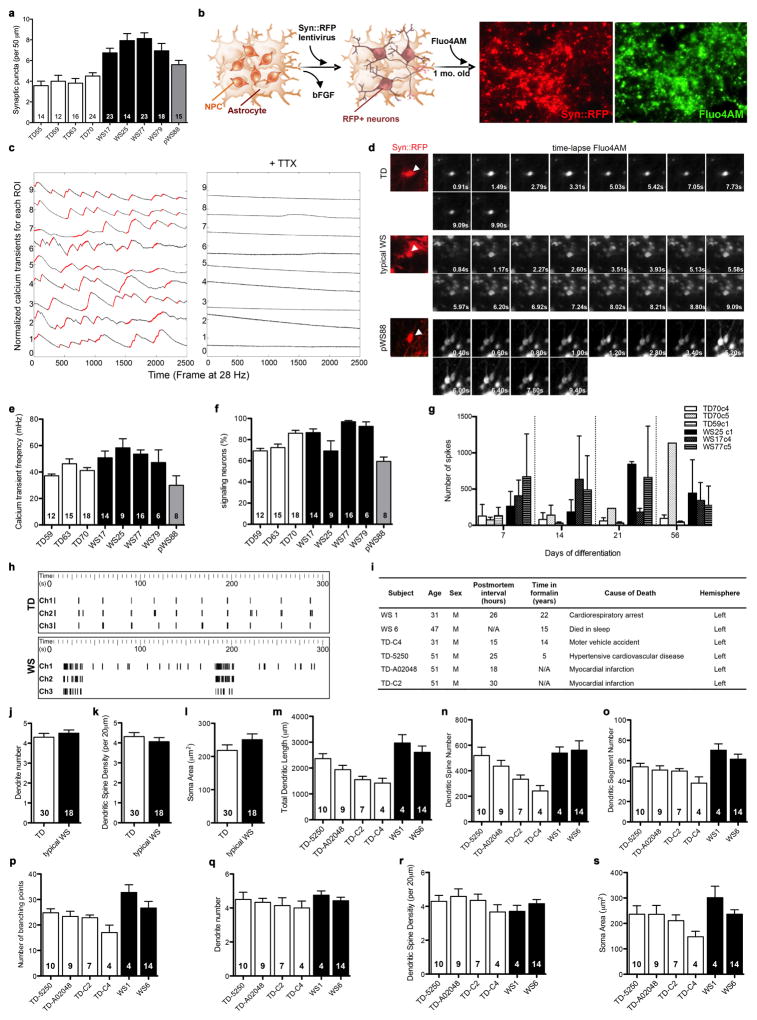
An increase in the number of dendritic spines per neuron could lead to an increase in synaptic contacts and, therefore, synaptic activity24, which could result in functional alterations. WS neurons had significantly more glutamatergic excitatory synapses compared to TD and pWS88 (Fig. 3h, i and Extended Data Fig. 6v and 7a) and an increased frequency of calcium transients with a higher percentage of signaling neurons in WS cultures (Fig. 3j–m and Extended Data Fig. 7b–f). Using multi-electrode array (MEA) electrophysiology, our data showed that WS neuronal cultures had a significant increase in spike frequency compared to TD-derived neurons (Fig. 3n, o and Extended Data Fig. 7g, h).
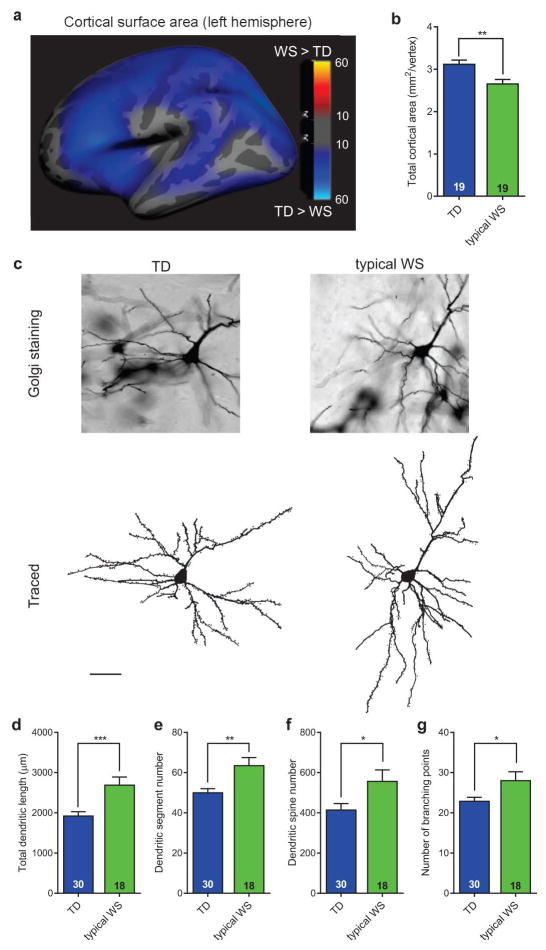
In an attempt to place our iPSC findings in a larger context of the cortical morphology of human subjects at the gross anatomical and cellular levels, we conducted two sets of additional experiments to test predictions based on NPC and neuronal differences found in vitro. In addition to the total volume reduction in WS brains previously reported25, multivariate analyses of variance (MANOVA) from our structural brain imaging of living subjects, revealed a significant decrease in overall cortical surface area in WS compared to TD individuals (Fig. 4a, b), but not in cortical thickness. Additionally, we conducted a separate set of experiments on postmortem brains from WS and TD tissue donors to investigate possible alterations in the morphology of cortical neurons, as predicted by our iPSC findings. Similar to WS iPSC-derived CTIP2-positive neurons, postmortem layers V/VI pyramidal neurons displayed larger total dendritic length and higher number of dendritic spines (Fig. 4c–e), similar spine density and soma area than TDs (Extended Data Fig. 7i–s). Postmortem layer V/VI neurons also showed significantly increased numbers of dendritic segments and branching points (Fig. 4f, g), and similar number of dendritic trees.
Figure 4. Neuroanatomical and morphological alterations in WS human brains.
a, Statistical parametric map of the vertex-wise group differences between TD and WS in cortical surface area (left hemisphere shown) assessed by structural MRI scans. Color scales indicate the p-value for statistical test: blue indicates decrease; gray indicates no difference. The statistics are displayed on a template group-averaged cortical surface rendering of healthy adult subjects. b, Reduction in overall cerebral cortical surface area in WS. Data are shown as mean ± s.e.m. n = number of brains analyzed. **P<0.01, one-sided unpaired Student’s t test. c, Representative images of postmortem cortical layer V/VI pyramidal neurons using Golgi staining (top) and their corresponding tracing (bottom) from TD and WS. d–g, Morphometric analysis showing significant increases in total dendritic length (d), dendritic spine numbers (e), dendritic segment number (f) and number of branching points (g) in WS compared to TD postmortem cortical layer V/VI pyramidal neurons. Data are shown as mean ± s.e.m. n = number of traced neurons. *P<0.05, **P<0.01, ***P<0.001, two-sided unpaired Student’s t test (d), two-sided unpaired Mann Whitney test (e–g).
The morphometric data in combination with the increased glutamatergic gene expression and number of co-localized synaptic puncta observed in WS neurons suggest that an increased number of synapses may result in the altered network activity, which could contribute to the characteristic behavior of WS individuals. Our study reveals that the WS phenotypes described here are the foundation for the understanding of the complex human social behavior. This approach provides an additional strategy to study the cellular and molecular underpinnings of complex human attributes, such as language in a social environment.
Methods
Subjects for behavioral study and source of cells for reprogramming
The study protocols were approved by USCD and Salk Institute IRB/ESCRO committees. Four TD individuals (ages 8 – 19) and five individuals with WS (ages 8 – 14; Extended Data Fig. 1a) were included in the analysis: four of the latter had typical WS gene deletions and one (pWS88) had a partial deletion in the WS region. Informed consents were obtained from all subjects or their parents as appropriate. Genetic diagnosis of WS was established using fluorescent in situ hybridization (FISH) probes for elastin (ELN), a gene consistently associated with the deletion in the typical WS region1, 9. All of the WS participants with confirmed genetic deletion exhibited the medical and clinical characteristics of the WS phenotype, including previously established cognitive, behavioral, and physical features associated with the syndrome4. WS diagnosis was confirmed based on the Diagnostic Score Sheet for WS (DSS; American Academy of Pediatrics Committee on Genetics, 2001), with a particular focus on the cardiovascular abnormalities and the characteristic facial features associated with the ELN deletion. The scores for the participants were at the mean for WS (9) or higher, with the individual with partial deletion in the WS chromosomal region (pWS88) scoring lower than the individuals with typical WS deletion. Similarly, pWS88 reported fewer symptoms with connective tissue and growth, his cognitive scores were slightly higher than the typical WS subjects, and he did not demonstrate the disparity between verbal and visual-spatial abilities typical of WS. However, pWS88 did display behavioral and developmental features consistent with WS, including developmental delay, over-friendliness, and anxiousness.
Behavioral and neurocognitive tests
The participants were administered standard tests to quantify their non-verbal and verbal abilities, as well as versions of the WS cognitive (WSCP) and social (WSSP) profiles to capture the distinct pattern of strengths and weaknesses both within and across domains associated with the WS cognitive and social phenotype. Details of the tests and the measures tapping into the two profiles are presented in Extended Data Figure 1. The WSCPs for the five WS participants were constructed by calculating log predictive likelihood ratios under assumed normality for age-appropriate TD vs. WS classifications based on VIQ, PIQ, VMI and PPVT standard scores, subject to availability. Predictive distributions were based on published normative means and SDs for each of the tests employed, whereas for the WS classification the predictive distributions26 were determined using data from n = 81 (VIQ and PIQ), n = 56 (VMI), and n = 97 (PPVT) subjects in a broader WS sample (described in Extended Data Fig. 1d). A Tobit model was used to estimate parameters for WS subjects on the VMI due to the presence of floor effects. The WSSPs for the five WS participants were constructed using measures of social approach behavior, emotionality/empathy, and language use.
WS deletion confirmation
Quantitative PCR was used to define the breakpoints of deleted regions in DNA isolated from iPSCs, or lymphoblast cell lines for WS participants, with probes spanning from CALN1 to WBSCR16 and template DNA. Taqman expression assay probes detecting the WS region genes were designed and synthesized with sequences shown in Supplementary Table 11. RNase P (VIC) was used as control. Quantitative PCR was performed on the ABI PRISM 7900HT system and the results were analyzed using SDS 3.2.
Cell collection, reprogramming and characterization
We avoided invasive sample collection methods such as skin biopsy or blood withdrawal by taking advantage of the natural loss of deciduous teeth as a source of somatic cells. We chose to reprogram dental pulp cells (DPCs) because these cells develop from the same set of early progenitors that generate neurons. Furthermore, the neurons derived from iPSCs generated from DPCs express higher levels of forebrain genes when compared to the ones generated from skin fibroblast-derived iPSCs27, serving the purpose of this study. Deciduous teeth were collected when they fell out and were shipped to our laboratory in DMEM 1X (Mediatech) with 4% Pen/Strep (Mediatech). Dental pulp was pulled out, washed in PBS with 4% Pen/Strep and incubated in 5% TrypLE (Gibco) for 15 min. Pulp was partially dissociated using needles and plated in culture medium (DMEM/F12 50:50, 15% FBS, 1%NEAA, 1% fungizone and 2% Pen/Strep). In one to four weeks, DPCs migrated out of the pulp and could be passaged and frozen as stock. DPCs in early passage (two to three) were reprogrammed using pMXs retroviruses expressing Yamanaka transcription factors (obtained from Addgene, Cambridge, MA, USA)28. After 4 days, transduced DPCs were trypsinized, plated on mouse embryonic fibroblasts and cultured using human embryonic stem cell (hESC) medium. After manually picked and clonally expanded, feeder-free iPSCs were grown on matrigel-coated dishes (BD Bioscience, San Jose, CA, USA) with mTeSR1 (StemCell Technologies) or iDEAL29.
Karyotyping
All G-banding karyotyping analyses were performed by Molecular Diagnostics Service (San Diego, CA) and Children’s Hospital Los Angeles (Los Angeles, CA).
Genotyping
Two hundred nanograms of DNA were processed and hybridized to the Illumina Infinium Human Core Exome BeadChip following manufacturer’s instructions. Illumina GenomeStudio V2011.1 with the Genotyping Module Ver. 1.9.4 was used to normalize data and call genotypes using reference data provided by Illumina. Illumina’s cnv Partition and gada R package were used to automatically detect aberrant copy number region. In addition, the B Allele Frequency (BAF) and Log R Ratio (LRR) distributions were manually checked to determine additional CNVs not detected by the software. Sample identification/relatedness was assessed by comparing called genotypes for each sample. The absolute number of different genotypes was counted and the Euclidean distances were calculated to identify relatedness of the samples.
Teratoma assay
Dissociated iPSC colonies were centrifuged and resuspended in 1:1 matrigel and phosphate buffer saline solution. The cells were injected subcutaneously in nude mice. After 1–2 months, teratomas were dissected, fixed and sliced. Sections were stained with hematoxylin and eosin for further analysis. Protocols were previously approved by the University of California San Diego Institutional Animal Care and Use Committee.
Neural induction and neuronal differentiation
iPSCs were cultured on matrigel-coated dishes and fed daily with mTeSR for 7 days. On the next day, mTeSR was substituted by N2 medium (DMEM/F12 supplemented with 0.5X N2 supplement (Life Technologies), 1 μM dorsomorphin (Tocris) and 1 μM SB431542 (Stemgent)) for 1–2 days. iPSC colonies were lifted off, cultured in suspension on the shaker (95 rpm at 37°C) for 8 days to form EBs and fed with N2 media. EBs then were mechanically dissociated, plated on a matrigel-coated dish and fed with N2B27 medium (DMEM/F12 supplemented with 0.5X N2 supplement, 0.5X B27 supplement (Life Technologies), 1% penicillin/streptomycin, and 20 ηM FGF). The emerging rosettes were picked manually, dissociated completely using accutase and plated on a poly-ornithine/laminin-coated plate. NPCs were expanded in N2B27 medium and fed every other day. To differentiate NPCs into neurons, FGF was withdrawn from the N2B27 medium. NPCs and neurons were characterized for stage-specific markers by immunostaining and flow cytometry (NPCs only), expression profile by single cell RT-PCR and RNA sequencing and electrophysiological property (neurons).
Total RNA extraction
Total RNA of DPCs, iPSCs, NPCs and neurons was extracted using TRIzol reagent (Life Technologies) according to the manufacturer’s protocols. Contaminating DNA in RNA samples was removed using TURBO DNase (Life Technologies) according to the manufacturer’s protocols. Quality and quantity of DNase-treated RNA were assessed using NanoDrop 1000 (Thermo Scientific).
PCR for exogenous retrovirus DNA silencing
RNA was extracted from iPSCs as previously described using Trizol reagent (Life Technologies). cDNA was generated from the RNA using super script III protocol according to manufacturer’s instructions. PCR was performed using primers listed below at the following cycles: 94°C for 10 minutes; 35 repeats of 94°C for 30 seconds, 62°C for 30 seconds and 72°C for one minute; and finally, 72°C for seven minutes. As a positive control, the pMX plasmid of the four vectors used on the reprogramming of the cells was placed along the samples as well as water as a negative template control for amplification. As an additional positive control for the endogenous genes, two hESC lines were used along with our iPSCs: H1 and HUES6 cells. Primers used were:
| Endo-cMyc | F TTGAGGGGCATCGTCGCGGGA |
| R GCGTCCTGGGAAGGGAGATCC | |
| Endo-Klf4 | F GAA ATT CGC CCG CTC CGA TGA |
| R CTG TGT GTT TGC GGT AGT GCC | |
| Endo-OCT3/4 | F TCT TTC CAC CAG GCC CCC GGC TC |
| R TGC GGG CGG ACA TGG GGA GAT CC | |
| Endo-SOX2 | F GCCGAGTGGAAACTTTTGTCG |
| R GGCAGCGTGTACTTATCCTTCT | |
| Exo transgenes F pMXs-TgUS | F GTG GTG GTA CGG GAA ATC AC |
| Exo-Oct4 R pMXs-Oct3/4-TgDS | R TAG CCA GGT TCG AGA ATC CA |
| Exo-Sox2 R pMXs-Sox2-TgDS | R GGT TCT CCT GGG CCA TCT TA |
| Exo-Klf4 R pMXs-Klf4-TgDS | R GGG AAG TCG CTT CAT GTG AG |
| Exo-c-Myc R pMXs-c-Myc-TgDS | R AGC AGC TCG AAT TTC TTC CA |
Embryoid body (EB) formation for pluripotency characterization
Partially dissociated iPSCs were re-suspended in EB medium (DMEM/F12 medium, 1X N2 supplement and 1% FBS) and cultured on shaker (95 rpm) at 37°C. Medium was changed every 3–4 days. After 20 days, total RNA of EBs was extracted for further gene expression analyses by qPCR.
Mycoplasma testing
All tissue culture samples were routinely tested for mycoplasma by PCR. One mL of media supernatants (with no antibiotics or fungizone) was collected for all cell lines, spun down, and resuspended in TE buffer. Ten μL of each sample were used in PCR reaction with the following primers: Forward: GGCGAATGGGTGAGTAAC; Reverse: CGGATAACGCTTGCGACCT. Any positive sample was immediately discarded.
Microarray
Three hundred nanograms of total extracted RNA from each sample were subjected to microarray by using the Affymatrix GeneChip one-cycle target labeling kit (Affymatrix, Santa Clara, CA) according to the manufacturer’s recommended protocols. The resultant biotinylated cRNA was fragmented and then hybridized to the GeneChip Human 1.0 ST Array [764885 probes, 28869 genes, 19734 gene-level probe sets with putative full-length transcript support (GenBank and RefSeq)] based on human genome, Hg18. Arrays were prepared at the University of California DNA Core Facility. Arrays were analyzed by the Affy (Affymetrix pre-processing)30 Bioconductor software package for microarray data. Data were then normalized by the RMA (robust multichip averaging) method to background-corrected and normalized probe levels to obtain a summary expression of normalized values for each probe set. Normalized microarray samples were then clustered by a hierarchical approach based on a matrix of distances. Normalized expression data were used to create a distance matrix that is calculated based on Euclidean distance between the transcripts over a pair of samples representing a variation between two samples. Having the distances for all pairs of samples, a linkage method is used to cluster samples in a dendrogram by using calculated distances (sample expression similarities). This method also creates a heatmap to graphically show the expression correlation between the samples.
Gene expression analyses by qPCR
RNA samples were reverse transcribed into cDNA using the Super Script III First Strand Synthesis System (Invitrogen, CA) according to manufacturer’s instructions. Reactions were run on the Bio-Rad detection system using Sybr-green master mix (Bio-Rad). Primers were selected from Primerbank; validated database (http://pga.mgh.harvard.edu/primerbank/) and specificity were confirmed by melting curve analysis through Bio-Rad detection system (Bio-Rad). Sequences of the primers are described in Supplementary Table 12. Quantitative analysis was performed using the comparative threshold cycle method31. GAPDH was used as housekeeping gene. Each sample was run in triplicate.
RNA-seq and global gene expression analyses
The RNA-seq analyses were previously described by our group32. Briefly, RNAs were isolated using the RNeasy Mini kit (QIAGEN). A total of 1,000 ng of RNA was used for library preparation using the Illumina TruSeq RNA Sample Preparation Kit. The RNAs were sequenced on Illumina HiSeq2000 with 50bp paired-end reads, generating 50 million high quality sequencing fragments per sample on average. For validation purposes of biological samples subjected to RNA-seq, hESC and iPSC data available from the literature were downloaded and used to compare with our sequenced cell lines. The two hESC lines used are available through the accession codes SRR873630 (HUES-6, referred as ES(HUES)) and SRR873631 [H1, referred to here as ES(H1)]. The two human iPSC lines used are available through the accession codes SRR873619 [referred to here as iPS(TD,1)] and SRR873620 [referred to here as iPS(TD,2)].
Gene ontology (GO) enrichment analysis
RNA-seq enrichment analysis was performed using WebGestalt33 and Cytoscape34 software plugins, considering only categories having statistical significance (p-value < 0.05). Genes tested for differential expression were used as the background for GO annotation and enrichment analysis.
NPC counting
NPCs were seeded onto poly-ornithine/laminin-coated six-well plates at a total number of 105 cells/well on day 0. Medium change was done on day 2. Cells were collected and counted on day 4.
NPC flow characterization
NPCs were resuspended, dissociated with accutase and fixed using fixation buffer (BioLegend) for 15 minutes followed by three PBS washes. The cell pellet was incubated and kept in Perm III buffer (BD Biosciences) in the −20C until needed for the experiment. A total of 106 cells were incubated with antibodies Sox1 (PE), Sox2 (APC) or Nestin (PE) and Pax6 (APC) (Bd Biosciences) for 30 minutes and then washed three times before being resuspended for cell analyses. Cells were analyzed in a plate reader mode using FACS Canto II machine (BD Biosciences).
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde for 10–20 min, washed with PBS three times (5 min each), permeabilized with 0.1% triton X-100 for 15 min, incubated in blocking solution (2% BSA) for 1 hour at RT and then in primary antibodies (goat anti-Nanog, Abcam ab77095, 1:500; rabbit anti-Lin28, Abcam ab46020, 1:500; rabbit anti-Oct4, Abcam ab19857, 1:500; mouse anti-SSEA4, Abcam ab16287, 1:200; mouse anti-Nestin, Abcam ab22035, 1:200; rabbit anti-Musashi1, Abcam ab52865, 1:250; rat anti-CTIP2, Abcam ab18465, 1:250; rabbit anti-SATB2, Abcam ab34735, 1:200; chicken anti-MAP2, Abcam ab5392, 1:1000; rabbit anti-FZD9, Origene TA314730, 1:150; chicken anti-EGFP, Abcam ab13970, 1:1000; rabbit anti-Synapsin1, EMD-Millipore AB1543P, 1:500; mouse anti-Vglut1, Synaptic Systems 135311, 1:500; rabbit anti-Homer1, Synaptic Systems 160003, 1:500) overnight at 4°C. The next day, cells were washed with PBS three times (5 min each), incubated with secondary antibodies (Alexa Fluor 488, 555 and 647, Life Technologies, 1:1000) for 1 hour at RT, and washed with PBS three times (5 min each). Nuclei were stained using DAPI (1:10,000). Slides or coverslips were mounted using ProLong Gold antifade mountant (Life Technologies).
DNA fragmentation analysis
One million NPCs were harvested to single cell suspension in 1mL PBS, then fixed by addition of 3 mL of 100% ethanol and stored at 4°C for at least two hours. NPC pellets were washed once with 5 mL PBS. After removal of PBS, cells were resuspended in 1 mL of propidium iodide (PI) staining solution (0.1% (v/v) Triton X-100, 10 μg/mL PI, and 100 μg/mL RNase A in 1X PBS). WS and TD NPC samples were analyzed by FACS on a Becton Dickinson LSRI, and gating of subG1 population (cells with fragmented DNA) was examined using FLOWJO-Flow Cytometry Analysis Software.
Caspase assay
Caspase activity was measured using Green FLICA Caspases 3 & 7 Assay kit (ImmunoChemistry Technologies, LLC.). Briefly, NPCs were harvested, washed and stained with 1X carboxyfluorescein Fluorochrome Inhibitor of Caspase Assay (FAM-FLICA) reagent, 10 μg/mL Hoechst and 10 μg/mL propidium iodide (PI). Samples were analyzed on the NC-3000 using the pre-optimized Caspase Assay. The population with caspase activity was used to analyze for apoptosis.
Proliferation assay
NPC proliferation was assessed using BD Pharmingen BrdU Flow Kits (BD Biosciences) according to the manufacturer’s protocol. Briefly, NPCs were incubated with 1 μM BrdU for 45 min at 37°C and harvested to single cell suspension. NPCs were then fixed and permeabilized using BD Cytofix/Cytoperm Buffer and stained using FITC-conjugated anti-BrdU antibody and 7-aminoactinomycin D (7-AAD), a fluorescent dye for labeling DNA. Fluorescence-activated cell sorting (FACS) was done on LSRFortessa (BD Biosciences) and, to obtain the percentage of the BrdU-positive population, the cell cycle profiles were analyzed using Flowjo-Flow Cytometry Analysis Software.
Construction and characterization of lentiviruses
Commercially available lentiviral vectors (pLKO.1) expressing short-hairpin RNAs (shRNAs) against FZD9 under the control of the U6 promoter (Thermo Scientific) were engineered to express the Discosoma sp. red fluorescent protein mCherry under the control of the hPGK (human phosphoglycerate kinase) promoter. The following shRNAs against FZD9 and a non-silencing scrambled control shRNA were selected (Thermo Scientific):
shRNA-control, 5′-TTCTCCGAACGTGTCACGT-3′
shRNA-FZD9, 5′-ATCTTGCGGATGTGGAAGAGG-3′
For rescue experiments, FZD9 cDNA was amplified from TD NPC cDNA as template by the following primer pair:
5′-CCGAGATCTTCGAGGTGTGTGGGGTTCTCCAAAG-3′;
5′-TCTAGAGCCACCATGGCCGTAGCGCCTCTG-3′
The reaction was performed using Phusion High-Fidelity DNA polymerase (New England Biolabs) according to the manufacturer’s protocol. The FZD9 cDNA was cloned into a lentiviral vector driven by the ubiquitin promoter followed by a self-clevage peptide and GFP sequence. The specificity and efficiency of shRNA-control, shRNA-FZD9, and the FZD9-WT constructs were verified by co-transfection into HEK-293 cells. Cell lysates were collected and analyzed by Western blot analysis with anti-FZD9 antibodies (Aviva OAEC02415, 1:1000).
CHIR-98014 treatment
CHIR-98014 (Selleckchem) was resuspended according to manufacturer’s instructions into 10 mM stock using DMSO and then diluted to 100 μM. Final concentration used in cells was 100 ηM of CHIR-98014, whereas the vehicle cells received only DMSO. For qPCR experiments, NPCs were propagated in 6-well plates until 70% confluency and then treated with CHIR-98014 for six hours to have their RNA collected using Trizol as previously described. For the NPC counting experiment, cells were seeded in 6-well plates as described in the presence of CHIR-98014 or DMSO, in triplicates (TD and WS). After 48 hours, the culture medium was changed and treatment was repeated. Cells were collected and counted after 96 hours of incubation.
Astrocyte differentiation
The TD NPCs were lifted into suspension and maintained on a shaker (95 rpm) to form neurospheres for three weeks. For the first week, the spheres were grown with N2B27 medium. The neurospheres were overlaid with the astrocyte medium (Lonza) for the remaining two weeks. The neurospheres were plated onto poly-ornithine- and laminin-coated plates and expanded for two to three passages before experimentation. Co-cultures of neurons and astrocytes were prepared for morphometric and functional analyses.
Western blotting
NPCs were lysed in RIPA buffer with protease inhibitor. Rabbit anti-FZD9 antibody (Aviva OAEC02415, 1:1000) and mouse anti-β-actin (Abcam ab8226, 1:3000) were used as primary antibodies. IRDye 800CW goat anti-rabbit and IRDye 680RD goat anti-mouse (1:10000) were used as secondary antibodies. The Odyssey system was used for signal detection. Signal intensities were measured using the Odyssey Image Studio and semi- quantitative analysis of FZD9 signal intensity was corrected with respect to β-actin relative quantification. A paired t-test analysis with a p-value < 0.05 was used in the comparison of TD and WS FZD9 signal intensity normalized data.
Synaptic puncta quantification
Co-localized Vglut (pre-synaptic) and Homer1 (post-synaptic) puncta were quantified after three-dimensional reconstruction of z-stack random images for all individuals and from two different experiments. Slides were analyzed under a fluorescence microscope (Z1 Axio Observer Apotome, Zeiss). Only puncta in proximity of MAP2-positive processes were scored.
Single cell qRT-PCR and analysis
Specific target amplification was performed in individual dissociated NPCs or 6-week-old neurons using C1 Single-Cell and BioMark HD Systems (Fluidigm), according to the manufacturer’s protocol and as described previously35–37. Briefly, single cells were captured on a C1 chip (10–17 μm cells) and cell viability was checked using a LIVE/DEAD Cell Viability/Cytotoxicity kit (Life Technologies). After lysis, RNA was reverse transcribed into cDNA with validated amplicon-specific DELTAgene Assays (Supplementary Table 13) using SuperScript III RT Platinum Taq Mix. Specific target amplification was performed by 18 cycles of 95°C denaturation for 15 seconds and 60°C annealing and amplification for 4 min. Each preamplified cDNA was mixed with 2X SsoFast EvaGreen Supermix with Low ROX (Bio-Rad) and then pipetted into an individual sample inlet in a 96.96 Dynamic Array IFC chip (Fluidigm). DELTAgene primer pairs (Supplementary Table 13) were diluted and pipetted into individual assay inlets in the same 96.96 Dynamic Array IFC chip. Quantitative PCR results were analyzed using Fluidigm’s Real-time PCR Analysis software using Linear (Derivative) Baseline Correction Method and the Automatic (Gene) Ct Threshold Method with 0.65 curve quality threshold. Hierarchical clustering heatmap, PCA analyses, violin plots of Log2-expression Ct values (LoD = 24) and ANOVA statistical analysis were carried out using Singular Analysis Toolset 3.0 (Fluidigm).
Calcium imaging
Neuronal networks derived from human iPSCs were transduced with lentivirus carrying the Syn::RFP reporter construct. Cell cultures were washed with Krebs HEPES Buffer (KHB) (10 mM HEPES, 4.2 mM NaHCO3, 10 mM dextrose, 1.18 mM MGSO4, 1.18 mM KH2PO4, 4.69 mM KCl, 118 mM NaCl, 1.29 mM NaCl2; pH 7.3) and incubated with 2–5 μM Fluo-4AM (Molecular Probes/Invitrogen, Carlsbad, CA) in KHB for 40 min. 5,000 frames were acquired at 28 Hz with a region of 256 × 256 pixels (100x magnification), using a Hamamatsu ORCA-ER digital camera (Hamamatsu Photonics K.K., Japan) with a 488 nm (FITC) filter on an Olympus IX81 inverted fluorescence confocal microscope (Olympus Optical, Japan). Images were acquired with MetaMorph 7.7 (MDS Analytical Technologies, Sunnyvale, CA), processed and analyzed using individual circular regions of interest (ROI) on ImageJ and Matlab 7.2 (Mathworks, Natick, MA). Syn::RFP+ neurons were selected after confirmation that calcium transients were blocked with 1 mM of tetrodotoxin (TTX). The amplitude of signals was presented as relative fluorescence changes (ΔF/F) after background subtraction. The threshold for calcium spikes was set at the 95th percentile of the amplitude of all detected events.
Electrophysiology
For whole-cell patch-clamp recordings, individual coverslips containing live 1-month-old neurons were transferred into a heated recording chamber and continuously perfused (1 mL/min) with artificial cerebrospinal fluid (ACSF) bubbled with a mixture of CO2 (5%) and O2 (95%) and maintained at 25 °C. ACSF contained (in mM) 121 NaCl, 4.2 KCl, 1.1 CaCl2, 1 MgSO4, 29 NaHCO3, 0.45 NaH2PO4-H2O, 0.5 Na2HPO4 and 20 glucose (all chemicals from Sigma). Whole-cell recordings were performed using a digidata 1440A/ Multiclamp 700B and Clampex 10.3 (Molecular devices). Patch electrodes were filled with internal solutions containing 130 mM K-gluconate, 6 mM KCl, 4 mM NaCl, 10mM Na-HEPES, 0.2 mM K-EGTA; 0.3 mM GTP, 2 mM Mg-ATP, 0.2 mM cAMP, 10mM D-glucose, 0.15% biocytin and 0.06% rhodamine. The pH and osmolarity were adjusted for physiological conditions. Data were all corrected for liquid junction potentials, electrode capacitances were compensated on-line in cell-attached mode and a low-pass filtered at 2 kHz were used. The access resistance of the cells in our sample was around 37 MOhm with resistance of the patch pipettes 3–5 MOhm. Spontaneous synaptic AMPA events were recorded at the reversal potential of Cl− and could be reversibly blocked by AMPA receptor antagonist (10 μM NBQX, Sigma). Spontaneous synaptic GABA events were recorded at the reversal potential of Na+ and could be reversibly blocked with GABAa receptor antagonist (10 μM SR95531, Sigma).
Multi-electrode array (MEA)
Using 12-well MEA plates from Axion Biosystems, we plated the same density of NPCs from TD and WS individuals in triplicate. Each well was seeded with 10,000 NPCs that were induced into neuronal differentiation as previously described. Each well was coated with poly-L-ornithine and laminin prior to cell seeding. Cells were fed once a week and measurements were taken before the medium was changed. Recordings were performed using a Maestro MEA system and AxIS software (Axion Biosystems), using a band-pass filter with 10 Hz and 2.5 kHz cutoff frequencies. Spike detection was performed using an adaptive threshold set to 5.5 times the standard deviation of the estimated noise on each electrode. Each plate first rested for 5 minutes in the Maestro, and then 5 to 10 minutes of data were recorded to calculate the spike rate per well. MEA analysis was performed using the Axion Biosystems Neural Metrics Tool, wherein electrodes that detected at least 5 spikes/minute were classified as active electrodes. Bursts were identified in the data recorded from each individual electrode using an adaptive Poisson surprise algorithm. Network bursts were identified for each well, using a nonadaptive algorithm requiring a minimum of ten spikes with a maximum inter-spike interval of 100 ms. Only channels that exhibited bursting activity (more than 10 spikes in 5-min interval) were included in this analysis. After measurement, neurons were immunostained to check morphology and density.
Postmortem brain specimens and cortical sampling
We used six postmortem brains (2 WS and 4 TD) that were gender, age and hemisphere matched. All brain specimens were harvested within a postmortem interval of 18 to 30 hours and had been immersed and fixed in 10% formalin for up to 20 years. For the purpose of the present experiments, samples were obtained from anatomically well-identified cortical areas in a consistent manner across specimens. Tissue blocks approximately 5 mm3 were removed from primary somatosensory cortex (Brodmann area 3) and primary motor cortex (BA 4) in the arm/hand knob region of the pre- and postcentral gyri, respectively, and from the secondary visual area (BA 18) from approximately 1.4 cm dorsally to the occipital pole and 2 cm from the midline38, 39. We focused specifically on these parts of the cortex because pathologies in dendritic morphology in these areas have been reported in other neurodevelopmental disorders40–42. In addition, pyramidal neurons in the selected areas reach their mature-like morphology early in development and start displaying dendritic pathologies sooner than high integration areas, such as the prefrontal cortex, allowing for a comparison of post-mortem findings with iPSC-derived neurons in early stages of development43, 44.
Postmortem brain tissue processing
Sampled tissue blocks were processed using an adaptation of the Golgi-Kopsch method45, which has been shown to give good results with tissue that has been fixed for long periods of time46. Briefly, blocks were immersed in a solution of 3% potassium dichromate, 0.5% formalin for eight days, followed by immersion into 0.75% silver nitrate for 2 days. Blocks were then sectioned on a vibratome, perpendicular to the pial surface, at a thickness of 120 μm. Golgi sections were cut into 100% ethyl alcohol and transferred briefly into methyl salicylate followed by toluene, mounted onto glass slides and cover-slipped. Adjacent blocks from each region were sectioned at 60 μm and stained with thionin for visualization of cell bodies and laminar organization, which enabled identifying the position of each individual neuron within a specific cortical layer. Cytoarchitectonic analysis of histological sections from each block confirmed that tissue was sampled from the ROI and that the Golgi-impregnated pyramidal neurons were located in cortical layers V/VI.
Cell selection and quantification
Golgi-impregnated neurons
Cortical neurons from all six postmortem brains were used in the study. Neurons included in the morphological analysis did not display degenerative changes47. Only neurons with fully impregnated soma, apical dendrites with present oblique branches, and at least two basal dendrites with third order segments were chosen for the analysis48. To minimize the effects of cutting on dendritic measurements, we included neurons with cell bodies located near the center of 120-μm thick histological sections, with natural terminations of higher-order dendritic branches present where possible38, 48. Inclusion of the neurons completely contained within 120-μm sections biases the sample toward smaller neurons, leading to the underestimation of dendritic length49; therefore, we applied the same criteria blinded across all WS and TD specimens, and we thus included the neurons with incomplete endings if they were judged to otherwise fulfill the criteria for successful Golgi impregnation. All neurons were oriented with apical dendrite perpendicular to the pial surface; inverted pyramidal cells as well as magnopyramidal neurons were excluded from the analysis. Neuronal morphology was quantified along x-, y-, and z-coordinates using Neurolucida v.10 software (MBF Bioscience, Williston, VT) connected to a Nikon Eclipse 80i microscope, with 40x (0.75) Plan Fluor dry objective. Tracings were conducted on both apical and basal dendrites, and the results reflect summed values for both types of dendrites per neuron. Following the recommendation that the applications of Sholl’s concentric spheres or Eayrs’ concentric circles for the analysis of neuronal morphology are not adequate when neuronal morphology is analyzed in three dimensions49, we conducted dendritic tree analysis with the following measurements 38, 48: (1) soma area – cross sectional surface area of the cell body; (2) dendritic length – summed total length of all dendrites per neuron; (3) dendrite number – number of dendritic trees emerging directly from the soma per neuron; (4) dendritic segment number – total number of segments per neuron; (5) dendritic spine/protrusion number – total number of dendritic spines per neuron; (6) dendritic spine/protrusion density – average number of spines/20 μm of dendritic length; and (7) branching point number – number of nodes (points at the dendrite where a dendrite branches into two or more) per neuron. Dendritic segments were defined as parts of the dendrites between two branching points - between the soma and the first branching point in the case of first order dendritic segments, and between the last branching point and the termination of the dendrite in the case of terminal dendritic segments. Since the long formalin-fixation time may result in degradation of dendritic spines, spine values may be underestimated and are thus reported here with caution. All of the tracings were accomplished blind to brain region and diagnostic status.
iPSC-derived neurons
The iPSC-derived sample consisted of EGFP-positive eight-week-old neurons with pyramidal- or ovoid-shaped soma and at least two branched neurites (dendrites) with visible spines/protrusions. Protrusions from dendritic shaft, which morphologically resembled dendritic spines in postmortem specimens, were considered and quantified as dendritic spines in iPSC-derived neurons. The neurites were considered dendrites based on the criteria applied in postmortem studies: (1) thickness that decreased with the distance from the cell body; (2) branches emerging under acute angle; and (3) presence of dendritic spines. In addition, only EGFP-positive neurons with nuclei co-stained with CTIP2, indicative of layer V/VI neurons, and with the dendrites displaying evenly distributed fluorescent stain along their entire length were included in the analysis. The morphology of the neurons was quantified along x-, y-, and z-coordinates using Neurolucida v.9 software (MBF Bioscience, Williston, VT) connected to a Nikon Eclipse E600 microscope with 40x oil objective. No distinction was made between apical and basal dendrites, and the results reflect summed length values of all neurites/dendrites per neuron, consistent with what was done for the postmortem neurons. The same set of measurements used in the analysis of Golgi-impregnated neurons was applied to the analysis of iPSC-derived neurons, and all of the tracings were accomplished blind to the diagnostic status and were conducted by the same rater (B.H-M.). Intra-rater reliability was assessed by having the rater traced the same neuron after a period of time. The average coefficient of variation between the results of retraced neurons was 2% for SA, TDL, DSN, and BPN, and 3% for DPN; there was no variation in TN in different tracings of the same neuron. The accuracy was further checked by having three individuals (B.H-M., B.J. and L.S.) trace the same neuron.
Brain imaging data acquisition and quality control
MRI scanning was completed in 19 participants with WS (aged 19 to 43 years; mean = 29.0, SD = 8.8; 11 males, 8 females) and 19 TD control subjects (aged 16 to 43 years; mean = 26.2, SD = 7.3; 8 males, 11 females). There was no significant difference between the groups in age (t = 1.0, p < 0.30) or in gender ratio (Pearson χ2 = 0.95, p < 0.33). A standardized multiple modality high-resolution structural MRI protocol was implemented, involving 3D T1- and T2-weighted volumes and a set of diffusion-weighted scans. Imaging data were obtained at the UCSD Radiology Imaging Laboratory on a 1.5 Tesla GE Signa HDx 14.0M5 TwinSpeed system (GE Healthcare, Waukesha, WI) using an eight-channel phased array head coil. A 3D inversion recovery spoiled gradient echo (IR-SPGR) T1-weighted volume was acquired with pulse sequence parameters optimized for maximum gray/white matter contrast (TE = 3.9 ms, TR = 8.7 ms, TI = 270 ms, flip angle = 8°, TD = 750 ms, bandwidth = ± 15.63 kHz, FOV=24 cm, matrix=192×192, voxel size=1.25×1.25×1.2 mm). All MRI data were collected using prospective motion (PROMO) correction for non-diffusion imaging50. This method has been shown to improve image quality, reduce motion-related artifacts, increase the reliability of quantitative measures, and improve the clinical diagnostic utility of MRI data obtained in children and clinical groups 51, 52. Standardized quality control procedures were followed for both raw and processed data, including visual inspection ratings by a trained imaging technician and computer algorithms testing general image characteristics as well as aspects specific to each imaging modality, such as contrast properties, registrations, and artifacts from motion and other sources. Subjects included in the current analyses were only those who passed all raw and processed quality control measures.
MRI data post-processing
Image post-processing and analysis were performed using FreeSurfer software suite (http://surfer.nmr.mgh.harvard.edu/). Surface-based cortical reconstruction and subcortical volumetric segmentation procedures have been shown elsewhere53–59. Briefly, three-dimensional model of the cortical surface was generated using MRI scans with 4 attributes: white matter segmentation; tessellation of the gray/white matter boundary; inflation of the folded, tessellated surface; and correction of topological defects54, 55. Cortical thickness was measured using the distances from each point on the white matter surface to the pial surface58. Cortical surface area was measured at the pial surface for the entire cerebrum and for each parcel of the Desikan and Destrieux atlases 54, 55, 59, 60.
Statistical analysis
Means ± s.e.m. for each parameter were obtained from samples described in Supplementary Table 1. There were no statistical methods used to predetermine sample size and no adjustments for multiple comparisons. All statistical analyses were done using Prism (Graphpad). Prior to statistical analysis comparing means between three to five unmatched groups of data, normal distribution was tested using D’Agostino & Pearson omnibus normality test and variance similarity was tested using Bartlett’s test for equal variances. Means of 3–5 unmatched groups, where normal distribution and equal variances between groups were confirmed, were statistically compared using one-way ANOVA and Tukey’s post hoc test. Otherwise, Kruskal-Wallis test and Dunn’s multiple comparison test were used. Prior to statistical analysis comparing means between two unmatched groups of data, normal distribution was tested using D’Agostino & Pearson omnibus normality test and variance similarity was tested using F test to compare variances. To compare the means of two groups where normal distribution and similar variance between groups were confirmed, Student’s t test was used. Otherwise, Mann Whitney test was used. Significance was defined as P<0.05(*), P<0.01(**), P<0.001(***) or P<0.0001(****).
Extended Data
Extended Data Figure 1. WS participants in iPSC study and their neurocognitive and social profiles.
a, Summary of scores on the Diagnostic Score Sheet (DSS) for WS subjects. b, Table showing allele number of genes in WS-deleted region in each subject obtained from qPCR. c, Summary of all neurocognitive and social behavioral tests used on this study. d–e, WS neurocognitive profiles. Log predictive likelihood ratio for iPSC subjects (identified by subject number) calculated as the log of the ratio of the likelihoods for each individual test score based on the predictive distributions for TD and WS subjects (d). Values < 0 indicate depressed scores consistent with expectations for WS. Predictive distributions for TD subjects used published norms (means and standard deviations with assumed normality). Predictive distributions for WS subjects were calculated using available WS data (VIQ/PIQ n = 81, VMI n = 56, PPVT n = 97) (e), assuming normality and least squares estimation, and according to the procedures described elsewhere26. WS parameter estimates for the VMI were calculated using censored regression due to a number of WS subjects scoring at the instrument floor. f, Description of population included in Benton Face Recognition and Judgment of Line Orientation in Figure 1b (TD n = 22 vs. WS n = 65). g, Boxplots for WS (red) and TD (blue) subjects on Complex Syntax (WS n = 45; TD n = 47) and Social Evaluation (WS n = 44; TD n = 49). Red and blue circles depict scores that are more than 1.5 times the inter-quartile range away from the median.
Extended Data Figure 2. Generation and characterization of iPSCs.
a, Diagram summarizing reprogramming protocol using retrovirus carrying Yamanaka transcription factors (see Supplementary Information for details). Scale bar, 200 μm. b, Representative images of iPSCs expressing pluripotent markers including Nanog, Lin28, Oct4 and SSEA4 assessed by immunofluorescence staining. Scale bar, 200 μm. c, Expression of three germ layer markers in iPSC-derived embryoid bodies (EBs); PAX6 (ectoderm), MSX1 (mesoderm) and AFP (endoderm) assessed by semi quantitative RT-PCR. TBP, housekeeping control. d, Cluster analysis showing correlation coefficients of microarray profiles of 3 WS dental pulp cells (DPCs), 3 TD DPCs, 3 WS iPSCs, 3 TD iPSCs and one ESCs. e, Representative PCR showing silencing of the four transgenes (exogenous) in iPSCs. f, Representative images of teratoma from iPSCs showing tissues of three germ layers; neural rosettes (ectoderm), cartilage (mesoderm), muscle cells (mesoderm) and goblet cells (endoderm). g, Representative image of iPSC chromosomes showing its genetic stability assessed by G-banding karyotype analysis. h–i, Spontaneous synaptic GABA events (h) and spontaneous synaptic AMPA events (i) in one-month-old iPSC-derived neurons.
Extended Data Figure 3. Global gene expression analysis during neuronal differentiation.
a, Principal component (PC) analysis plot of embryonic stem cells (ES), induced pluripotent stem cells (iPS), neuronal progenitor cells (NPC) and neurons (NE) for TD, WS and pWS88. c, Euclidian matrix distance-based heatmap and hierarchical clustering-based dendrogram of ES, NPC and NE cells for WD, WS and pWS88 samples. Expression variability between samples is indicated by Z-score, varying from green (negative variation) to red (positive variation). c, Euclidian matrix distance-based heatmap and hierarchical clustering-based dendrogram of pluripotency gene markers for ES, NPC and NE cells for TD, WS and pWS88 samples. d, Euclidian matrix distance-based heatmap and hierarchical clustering-based dendrogram of neuronal gene markers for iPS, NPC and NE cells for TD, WS and pWS88 samples. Expression variability between samples is indicated by Z-score, varying from green (negative variation) to red (positive variation). e, Specific cell type-based clustering analysis of biological replicates subjected to RNA-seq for the WS-related genes in three stages during differentiation (iPS, NPC and NE). f, Fold change variation of WS-related genes in different cell lines. Ideogram of chromosome 7 (band 7q11.23) corresponding to the commonly deleted region with the WS-related genes. Fold change variation of normalized WS-related gene expression in NPCs and neurons (NE) compared to TDs. Non-represented fold change corresponds to those genes having high expression variability between biological replicates, or having very low expression values. g, Expression of FZD9 gene in iPSC, NPCs and neurons from TD and WS. Error bars are represented by standard error. h, Venn diagram showing correlation of significant differentially expressed genes between TD, pWS88 and WS during neuronal differentiation. Significantly enriched GO terms found for down-regulated (red histogram) and up-regulated (blue histogram) differentially expressed genes between TD and WS in NPC. Significantly enriched GO terms found for down-regulated (red histogram) and up-regulated (blue histogram) differentially expressed genes between TD and WS in neurons (NE). Vertical line (black) corresponds to significant p-value (0.05). i, Enriched GO metabolic process terms found in NPC of WS samples correlated with the GO found by a similar comparison performed by Adamo et al. (2014)13.
Extended Data Figure 4. Defect in WS NPC apoptosis and role of FZD9.
a, Ratio of NPC number on day 4 over day 0 relative to TD. Data are shown as mean ± s.e.m. n = number of clones. b, High percentage (>95%) of Sox1/Sox2-positive and Pax6/Nestin-positive cell population was comparably observed in TD, typical WS and pWS88 NPCs assessed by FACS. Data are shown as mean ± s.e.m. n = number of clones. c, Microfluidics of C1 chip used to capture live single cells (calcein+ cell). d, Outliers exclusion based on the recommended/default LoD value 24, analyzed by Fluidigm Singular 3.0. Outliers were removed manually based on the sample median Log2Ex values. e, Representative example of non-normalized Ct Plot, indicated with the rectangle in the heat map. Cells are shown in rows and genes in columns. The range of cycle threshold (Ct) values is color coded from low (blue) to high (red) and absent (black). f, Violin plots of all 96 genes showing the comparison between TD and WS NPCs from the single cell analyses (Log2ex values). The majority of genes show unimodal expression distribution. g, Volcano plot of single-cell expression data. Plot illustrates differences in expression patterns of target genes of iPSC-derived NPCs. The dotted lines represent more than or equal to 3.0-fold differentially expressed genes between the groups at P<0.05 (unpaired two-sample t-test). h, Schematic diagram summarizing NPC preparation for proliferation assay and representative scatter plot showing cells in each cycle phase (G1, S and G2/M). i, No significant differences in percentage of the BrdU-positive population between TD, typical WS and pWS88 NPCs. j, Schematic diagram summarizing NPC preparation for apoptosis analysis and representative analyzed data for DNA fragmentation (left) and caspase assay (right). k–m, Changes in ratio of NPC number on day 4 over day 0 relative to TD (k), percentage of subG1 population (l) and percentage of population with high caspase activity (m) of pWS88 NPCs when treated with shFZD9 and shControl. n, Increase in cell number day 4/day 0 upon overexpression of FZD9 in WS iPSC-derived NPCs. Data are shown as mean ± s.e.m. for each individual. n = technical replicates. For i and k–m, data are shown as mean ± s.e.m. n = number of clones, *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA and Tukey’s post hoc test (i), Kruskal-Wallis test and Dunn’s multiple comparison test (k–m).
Extended Data Figure 5. Single-cell analysis of WS and TD iPSC-derived neurons.
a–b, Outliers exclusion based on LoD = 24, analyzed by Fluidigm Singular 3.0. Outliers were removed manually based on the sample median Log2Ex values. c, Heatmap of number of genes with ANOVA P-value < 0.05 (82 genes in total). d, Unsupervised hierarchical clustering of 672 single-cell of WS and TD iPSC-derived neurons identified cell sub-populations not linked with the genotype. Cells are shown in rows and genes in columns. Log2- gene expression levels were converted to a global Z-score (blue is the lowest value and red is highest). Genes were clustered using the Pearson correlation method and cells were clustered using Euclidean method. e, PCA projections of the 96 genes, showing the contribution of each gene to the first two PCs. f, Violin plots of all 96 genes showing the comparison between TD, WS and pWS88 neurons from the single cell analyses (Log2ex values).
Extended Data Figure 6. Morphometric analysis of WS-derived CTIP2-positive cortical neurons.
a, Schematic diagram summarizing preparation of neurons for evaluation through morphometric analysis. b, Representative images of EGFP- and CTIP2-positive neuron (arrowhead) and tracing. Scale bar, 200 μm. c–f, No significant differences in dendritic segment numbers (c), number of branching points (d), dendritic spine density (e) and soma area (f) between TD, typical WS and pWS88 were observed. g–m, Morphometric analysis shown as individual subject for total dendritic length (g), dendritic tree number (h), dendritic spine number (i), dendritic segment number (j), number of branching points (k), dendritic spine density (l) and soma area (m). n, Four-weeks-old neurons were dissociated and plated to trace total neurite length every hour, in a total of 6 h. Representative images of traced neurons plated after 0 and 6 h from TD, typical WS and atypical pWS88 iPSC-derived neurons. o–r, Morphometric analysis showing significant differences among TD, typical WS and pWS88 in the initial neurite growth velocity (6h period). r, Morphometric analysis shown for individual subjects for neurite growth velocity for 6h interval. n = number of traced neurons. s–u, No significant changes were observed in the total dendritic length (s), dendritic segment number (t) and dendritic spine number (u) of TD neurons plated in different densities (300–1200 cells/mm2). v, Individual channels of puncta quantification of post- and pre-synaptic markers (Homer1 / Vglut1). Scale bar, 2 μm. For c–m and o–u, data are shown as mean ± s.e.m. n = number of traced neurons, *P<0.05, **P<0.01, Kruskal-Wallis test (c–f), one-way ANOVA and Tukey’s post hoc test (o–q, r–u).
Extended Data Figure 7. Alteration in calcium transient in WS iPSC-derived neurons and morphometric analysis of cortical layer V/VI pyramidal neurons in postmortem tissue.
a, Puncta quantification of post- and pre-synaptic markers. The synaptic proteins Vglut (pre-synaptic) and Homer1 (post-synaptic) were used as markers and only co-localized puncta on MAP2+ cells were quantified and graphed. Data are shown as the mean ± s.e.m. n = number of neurons. b, Schematic diagram summarizing preparation of neurons for calcium transient analysis. Representative images of live neuronal culture expressing RFP driven by synapsin promoter and the uptake of Fluo-4AM calcium dye. c, Blockade of calcium transient by TTX inhibition of synaptic activity. d, Representative images of calcium transient in single neurons (RFP-positive, arrowhead) from TD (top), typical WS (middle) and pWS88 (bottom). Number in the lower right of each figure represents each time point (second) when change in Fluo-4AM occurs. e–f, Calcium transient analysis shown as individual for frequency (e) and percentage of signaling neurons (f). Data are shown as mean ± s.e.m. n = number of fields analyzed. g, MEA analyses revealed an increase in spontaneous neuronal spikes. Data shows individual clones. i, Table showing subjects used for the analysis. h, Raster plot of TD and WS iPSC-derived neurons analyzed by multi-electrode array. i, Table showing subjects used for the analysis. j–l, No significant differences in dendrite number (j), dendritic spine density (k) and soma area (l) between TD and typical WS were observed. Data are shown as mean ± s.e.m. n = number of traced neurons, two-sided unpaired Student’s t test. m–s, Morphometric analysis shown for each individual for total dendritic length (m), dendritic spine number (n), segment number (o), branching point number (p), dendrite number (q), dendritic spine density (r) and soma area (s). Data are shown as mean ± s.e.m. n = number of traced neurons.
Extended Data Table 1.
List of top ten most significant differentially expressed genes in WS compared to TD for NPC and neurons.
| NPC: TD x WS
| |||
|---|---|---|---|
| Gene name | Description | Fold-change | p-value |
| SCN4A | sodium channel, voltage gated, type IV alpha subunit | −11.92 | 9.72E-10 |
| SLC7A14 | solute carrier family 7, member 14 | −15.85 | 1.85E-12 |
| SLC38A5 | solute carrier family 38, member 5 | −8.51 | 5.67E-09 |
| ADGRA2 | adhesion G protein-coupled receptor A2 | −30.50 | 3.26E-08 |
| SLC1A6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | 8.03 | 1.56E-08 |
| CXCL12 | chemokine (C-X-C motif) ligand 12 | −7.51 | 3.33E-08 |
| SLC30A3 | solute carrier family 30 (zinc transporter), member 3 | 10.28 | 8.32E-09 |
| SLC8A2 | solute carrier family 8 (sodium/calcium exchanger), member 2 | −16.84 | 5.36E-13 |
| HTR1B | 5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled | −10.96 | 8.20E-09 |
| SLC24A2 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | −13.25 | 4.43E-10 |
| NPC: TD x pWS88
| |||
|---|---|---|---|
| Gene name | Description | Fold-change | p-value |
| GABRA3 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 14.71 | 6.30E-10 |
| SYT13 | synaptotagmin XIII | 6.55 | 1.87E-08 |
| PPP2R2C | protein phosphatase 2, regulatory subunit B, gamma | 15.74 | 7.69E-09 |
| CELF4 | CUGBP, Elav-like family member 4 | 7.27 | 1.22E-07 |
| TRIM67 | tripartite motif containing 67 | 15.08 | 6.52E-10 |
| ADRA2A | adrenoceptor alpha 2A | 8.44 | 1.42E-07 |
| JAKMIP1 | janus kinase and microtubule interacting protein 1 | 19.56 | 7.21E-09 |
| CA10 | carbonic anhydrase X | 74.54 | 8.50E-10 |
| LHFPL4 | lipoma HMGIC fusion partner-like 4 | 8.31 | 5.71E-08 |
| ACSL6 | acyl-CoA synthetase long-chain family member 6 | 7.07 | 7.63E-08 |
| Neuron: TD x pWS88
| |||
|---|---|---|---|
| Gene name | Description | Fold-change | p-value |
| CRYM | crystallin, um | 7.68 | 3.21E-05 |
| RASL12 | RAS-like, family 12 | 13.41 | 6.04E-08 |
| PDLIM1 | PDZ and LIM domain 1 | 5.44 | 1.00E-05 |
| ZSCAN10 | zinc finger and SCAN domain containing 10 | 58.80 | 6.22E-06 |
| ANO1 | anoctamin 1, calcium activated chloride channel | 9.61 | 1.78E-06 |
| DUSP23 | dual specificity phosphatase 23 | 5.09 | 0.000114507 |
| SLC16A5 | solute carrier family 16 (monocarboxylate transporter), member 5 | 33.27 | 8.90E-05 |
| KRT19 | keratin 19, type I | 9.94 | 0.000130985 |
| TMEM30B | transmembrane protein 30B | 17.09 | 2.11E-07 |
| KIF18B | kinesin family member 18B | 4.50 | 0.000217123 |
| Neuron: TD x WS
| |||
|---|---|---|---|
| Gene name | Description | Fold-change | p-value |
| FAM19A5 | family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 | 1076.39 | 2.23E-10 |
| TPM2 | tropomyosin 2 (beta) | −5.88 | 2.99E-08 |
| SCN4A | sodium channel, voltage gated, type IV alpha subunit | −45.81 | 1.63E-07 |
| IGSF21 | immunoglobin superfamily, member 21 | 8.20 | 2.33E-07 |
| TNNT2 | troponin T type 2 (cardiac) | −29.87 | 3.58E-07 |
| PXMP4 | peroxisomal membrane protein 4, 24kDa | −7.39 | 3.83E-05 |
| ZSCAN10 | zinc finger and SCAN domain containing 10 | 35.64 | 9.74E-05 |
| MYOZ1 | filamin-, Actinin- And Telethonin-Binding Protein | −6.64 | 0.000119159 |
| LAD1 | ladinin 1 | −24.81 | 0.000213205 |
| PHOSPHO1 | phosphatase, Orphan 1 | 33.95 | 0.000266061 |
Extended Data Table 2.
Most significant (p-value<0.05) enriched GO terms in NPC of WS compared to TD samples.
| Down-regulated genes in WS compared to TD in NPC cells
| ||||
|---|---|---|---|---|
| GO TERM | GO Description | −Log (p-value) | P-value | Genes |
| GO:0005578 | proteinaceous extracellular matrix | 18.3452855 | 4.52E-19 | WNT16,MMP25,DCN,LAMC3,COL16A1,WNT11,ADAMTS2,LAMB1,PAPLN,MMP9,WISP1,ECM2,COL1A1,CPZ,EFEMP1,LTBP2,COL10A1,COL21A1,OMD,ADAMTS8,WNT10A,EMILIN1,ADAMTS14,KERA, FBLN5,ADAMTS17,COL6A1,COL6A2,ADAMTS10,HMCN1,ADAMTS16,HAPLN1,ADAMTS4,COL6A3,COL1A2,MFAP4,SOST,NPNT,COL6A5,HPSE2,VWC2,PRELP,SPOCK3,LAMA2,ADAMTSL2,COL28A1 |
| GO:0048856 | anatomical structure development | 16.80415844 | 1.57E-17 | WNT16,MYLIP,TFAP2B,BAZ1B,CD4,DCN,SEMA3B,ADGRA2,RUNX3,IBSP,TG,ALX4,CAMK2B,TLE2,MAOB,RORA,EPHA8,TP63,PLXNA2,RARB,MEF2C,WNT11,TFAP2C,LAMB1,DSP,NEFH,MMP9,HCK, SGCG,FLT1,CRISPLD2,CPQ,MET,COBL,ENG,PITX3,COL1A1,CPT1A,CYP27B1,VDR,EYA4,TFEB,PCDHB2,PCDHB5,PCDHB6,PDGFRB,FN1,TFAP2E,LPPR4,RCAN3,TNNT2,BCL11A,PCDHB14, PCDHB12,INHBA,COL10A1,BMP2,TCF15,SIX1,HSPA2,PTPRB,CDH15,SERPINF1,LYVE1,PDGFRA,TBX3,HEY2,AGT,WNT10A,RAPGEF5,MYO7A,MSTN,ARHGAP24,CDH11,COL6A1,COL6A2,ITGA10, CASQ1,LEFTY2,EPHA5,HAPLN1,ARHGAP26,DCDC2,CACNA1C,EDNRA,UNC5D,ZIC3,MMP14,CACNA1D,NBL1,FGF17,ADAMTS4,STC1,ITGB2,PTH1R,PLXDC1,ALOX15,BRINP3,MSX1,FZD5,COL6A3, ELF3,SERPINI1,HEYL,PITX2,NPY1R,NPY5R,ITGA2,DACT2,COL1A2,SYK,DACT1,ANPEP,BATF2,NPNT,EFNA1,GPR183,BNC1,ALCAM,SIX2,SGCD,HAS2,PTGER4,NINJ2,SCG2,RARG,SSH3,ABLIM3, CSPG4,CMKLR1,LY6H,PCDHB9,TH,MAB21L1,GREM2,ADGRB1,FES,CSF1R,NTM,CAMK1D,GAS6,OPCML,SCN5A,TBX1,ALDH1A3,SLITRK6,SGCZ,TNFAIP2,NTF3,RTN4RL1,PDE2A,DNER,VWC2, PRELP,TDRD7,AKR1C3,RYR1,MME,LAMA2,PCDHB11,PAPSS2,MAFB,POU5F1,PCDHA7,PCDHA5,DPF3,ITGA1,GSTA1,PCDHA11,PCDHA10 |
| GO:0007155 | cell adhesion | 16.70678997 | 1.96E-17 | CD4,CLDN11,IBSP,TNC,LAMC3,RORA,EPHA8,COL16A1,LAMB1,FERMT1,HCK,SRPX,WISP1,ECM2,ENG,CXCL12,PRPH2,PCDHB2,PCDHB5,PCDHB6,FN1,PCDH17,PCDHB14,PCDHB12,ADGRE5, TNFAIP6,OMD,ADGRE2,ISLR,CDH15,MYBPH,LYVE1,PDGFRA,EMILIN1,PCDH10,FBLN5,CDH11,COL6A1,COL6A2,ITGA10,HAPLN1,CNTNAP5,ITGB2,AZGP1,SNED1,COL6A3,IGFBP7,ITGA2,EDIL3, SYK,MFAP4,NPNT,EFNA1,PCDH7,ALCAM,LRRN2,PTGER4,NINJ2,COL6A5,PCDHB9,ADGRB1,NTM,GAS6,OPCML,PKP3,THBS2,LAMA2,PCDHB11,ITGBL1,PCDHA7,PCDHA5,ITGA1,COL28A1, PCDHA13,PCDHGC3,ACTN3,PCDHA11,PCDHA10,PCDHGA7 |
| GO:0030198 | extracellular matrix organization | 13.94978657 | 1.12E-14 | DCN,IBSP,TNC,LAMC3,COL16A1,ADAMTS2,LAMB1,MMP9,ENG,COL1A1,EFEMP1,FN1,COL10A1,COL21A1,BMP2,EMILIN1,ADAMTS14,FBLN5,COL6A1,COL6A2,ITGA10,CTSK,HAPLN1,MMP14, ADAMTS4,ITGB2,CTSS,COL6A3,ITGA2,KLKB1,COL1A2,MFAP4,LTBP3,NPNT,HAS2,COL6A5,GAS6,LAMA2,ITGA1,COL28A1 |
| GO:0005576 | extracellular region | 9.533946889 | 2.92E-10 | WNT16,SCIN,MMP25,SYT7,DCN,SEMA3B,CLDN11,GPRC5A,ACPP,EHD2,IBSP,TNC,FAM65C,TG,LAMC3,ENTPD2,LY75,TLE2,BCL3,MAOB,FRMPD1,COL16A1,WNT11,ADAMTS2,LAMB1,DSP,SUSD2, APOL4,PAPLN,MMP9,BPI,MCF2,PLP2,ACP5,FLT1,WFDC1,CRISPLD2,CRYM,PDGFRL,CPQ,WISP1,NDRG1,MET,STX1A,PCOLCE,ECM2,ENG,PTGDS,CXCL12,PNPO,COL1A1,CPZ,OAS3,PDGFRB, POMC,EFEMP1,FN1,IL1R1,QPCT,ANGPTL1,VAMP8,LTBP2,ENOX1,CAT,CPXM2,XPNPEP2,INHBA,ADGRE5,COL10A1,EDN3,COL21A1,CPNE5,BMP2,PCSK2,CFP,HSPA2,OMD,TUBA4A,FGL2,ISLR, CDH15,DPP6,SERPINF1,GSTT2B,LYVE1,IAH1,ADAMTS8,DYSF,AGT,WNT10A,EMILIN1,ADAMTS14,MSTN,RBP5,KERA,SLC46A3,FBLN5,ADAMTS17,CDH11,COL6A1,COL6A2,NTN5,ADAMTS10,FCN3, HMCN1,CTSK,LEFTY2,TMEFF2,ADAMTS16,HAPLN1,GABRB2,PLA2G7,IGSF1,LCN9,SLC5A12,CCDC3,IGSF10,PLA2R1,TMPRSS11D,MMP21,GNA14,WIF1,NBL1,FGF17,ADAMTS4,STC1,C1R,ITGB2, PTH1R,AZGP1,FGFR4,PLXDC1,CXCL16,ITIH3,BRINP3,SNED1,HAAO,CTSS,S100A11,COL6A3,IGFBP7,SERPINI1,ALB,CDCP1,PRSS12,EDIL3,KLKB1,COL1A2,CHMP4C,AKR1E2,VWA2,MFAP4,ACSM1, ANPEP,GPT,SOST,LTBP3,NPNT,FSTL5,EFNA1,ALCAM,CYTL1,SOSTDC1,SCG2,COL6A5,HPSE2,CSPG4,ADGRG2,C1QTNF1,VPS37D,CLEC14A,EPS8L2,GREM2,PENK,C1S,GRID1,GAS6,OPCML, OLFML1,ALDH1A3,FAM19A3,MUC6,TNFAIP2,FAM212A,NTF3,RTN4RL1,THBS2,THSD4,VWC2,PRELP,PLAC9,SPOCK3,HLA-DRB1,AKR1C3,RYR1,MME,LAMA2,ADAMTSL2,SULT1C2,HLA- DRB5,ITGBL1,RASSF9,TGM2,ITGA1,COL28A1,APOL6,PCDHGC3,GSTA1,C4A,ACTN3,PCDHA10 |
| GO:0005615 | extracellular space | 8.287607079 | 5.16E-09 | WNT16,DCN,ACPP,IBSP,TNC,TG,TLE2,WNT11,LAMB1,APOL4,MMP9,FLT1,WFDC1,CPQ,WISP1,PCOLCE,ENG,PTGDS,COL1A1,OAS3,POMC,EFEMP1,FN1,ANGPTL1,LTBP2,ENOX1,CPXM2,INHBA, ADGRE5,EDN3,BMP2,PCSK2,CFP,HSPA2,FGL2,SERPINF1,AGT,WNT10A,MSTN,FBLN5,COL6A2,FCN3,CTSK,LEFTY2,PLA2G7,NBL1,FGF17,ADAMTS4,C1R,AZGP1,PLXDC1,CXCL16,CTSS,S100A11, COL6A3,IGFBP7,SERPINI1,ALB,KLKB1,COL1A2,VWA2,ACSM1,ANPEP,SOST,CYTL1,SOSTDC1,SCG2,C1QTNF1,C1S,GAS6,TNFAIP2,VWC2,SPOCK3,C4A |
| GO:0005886 | plasma membrane | 8.166494631 | 6.82E-09 | CALCR,ABCC8,TBXA2R,SCN4A,CACNA2D2,MYLIP,MMP25,CD4,HFE,CLDN11,GPRC5A,ACPP,SLC38A5,ATP1A2,ADGRA2,EHD2,PTGER3,CYBA,ENTPD2,LY75,MCOLN3,CAMK2B,FRMPD1,CNGB1, EPHA8,MGLL,PLXNA2,PAG1,ATP8B1,IL12RB2,KCNK2,GABRP,DSP,KCNK6,SUSD2,SLC10A1,SLC8A3,FERMT1,HCK,BPI,PLP2,NALCN,SGCG,FLT1,NDRG1,MET,VIPR2,COBL,STX1A,ENG,RNF43, TCIRG1,TNS2,KCNA1,OAS3,ADGRD1,PCDHB2,PCDHB5,PCDHB6,SLC27A6,PDGFRB,IL1R1,LPPR4,SLC8A2,SGK1,VAMP8,PCDH17,GPR68,GRIA2,PCDHB14,PCDHB12,ENOX1,XPNPEP2,ADGRE5, PMEPA1,HSPA2,PTPRB,ADGRE2,ADCY4,CDH15,ATP1A4,LYVE1,TRPM1,PDGFRA,HTR1B,DYSF,SLC19A3,PCDH10,FBLN5,DISP2,CDH11,BEST4,ITGA10,RGS16,UNC80,SLC22A14,EPHA5,GABRB2, SLC22A3,HTR2C,SLC26A7,SLC5A12,PLCH2,LYPD6B,CACNA2D4,CACNA1C,ANO4,EDNRA,GABRA2,PLA2R1,TMPRSS11D,MCOLN2,FGD5,SLC24A2,GNA14,MMP14,CACNA1D,LYPD5,ABCG1,ITGB2, PTH1R,AZGP1,FGFR4,LY6E,PLXDC1,ALOX15,CXCL16,DHRS3,FZD5,CDCP1,MST1R,PRSS12,NPY1R,NPY5R,ITGA2,KLKB1,ACSL6,GRIK2,HCN1,SYK,ANPEP,PARM1,AFAP1L2,GSG1L,EFNA1,PTAFR, NPR1,GPR183,PCDH7,TM4SF1,SLC16A5,SIX2,SGCD,PRKCDBP,HAS2,KCND3,PTGER4,NINJ2,SYNPO,GPR22,HPSE2,EVC2,BNC2,CSPG4,ADGRG2,C1QTNF1,CMKLR1,SLCO3A1,LY6H,EPS8L2, PCDHB9,TH,GPR139,ADGRB1,FES,CSF1R,NTM,GRID1,RGS7,OPCML,SCN5A,PKP3,SLITRK6,TMEM173,ANKS1B,SGCZ,IFNLR1,PRKG1,EVI2B,RTN4RL1,PDE2A,TPCN1,QRFPR,NPSR1,GJA4,DNER, OR7D2,CLEC2A,NKAIN2,HLA-DRB1,RYR1,MME,CHRNG,PCDHB11,HLA-DRB5,PPAPDC1A,PCDHA7,PCDHA5,ITGA1,PCDHA13,PCDHGC3,C4A,FMN1,PCDHA11,PCDHA10,PCDHGA7 |
| GO:0055085 | transmembrane transport | 7.872963268 | 1.34E-08 | ABCC8,SCN4A,CACNA2D2,SLC7A14,SLC38A5,ATP1A2,MCOLN3,ATP2C2,CNGB1,ATP2A3,RARB,SLC25A43,ATP8B1,KCNK2,FXYD5,SLC17A6,GABRP,KCNK6,SLC10A1,SLC8A3,PLP2,NALCN,CPT1A, TCIRG1,KCNA1,SLC27A6,PEX5L,STEAP3,SLC8A2,SGK1,GRIA2,ADCY4,ATP1A4,TRPM1,ADAMTS8,SLC19A3,SLC46A3,BEST4,CASQ1,UNC80,SLC22A14,SLC25A48,GABRB2,SLC22A3,SLC26A7, SLC5A12,CACNA2D4,CACNA1C,ANO4,GABRA2,MCOLN2,SLC24A2,CACNA1D,ABCG1,AZGP1,COX7A1,ALB,GRIK2,HCN1,SLC16A5,KCND3,RARG,SLCO3A1,SLC17A8,SLC9A9,GRID1,SLC25A18, GAS6,SCN5A,PDE2A,TPCN1,RYR1,CHRNG |
| Up-regulated genes in WS compared to TD in NPC cells
| ||||
|---|---|---|---|---|
| GO TERM | GO Description | −Log (p-value) | P-value | Genes |
| GO:0007155 | cell adhesion | 2.964750421 | 0.00108455 | ITGB5,TNR,TEK,PARVG,CDHR1,MPZL2,GRID2,ACAN,PIK3CD,EGFL7,CLDN6,GCNT1,CLDN4,PTPRT,DPP4 |
| GO:0005886 | plasma membrane | 2.391582019 | 0.00405899 | GABRA3,ERBB3,ITGB5,TNFRSF10A,SLC1A6,SLC6A12,GLP1R,SLC30A3,ABCG2,TEK,CSMD2,SDC4,BTN1A1,F12,PDE6B,MAP7,PARVG,RHCG,PPAP2C,ZNF185,PLIN2,CDHR1,KIRREL3,DLG2,GPR158, GRID2,ADCY8,XKR8,F11R,CLIC6,WNT4,OXGR1,OSCAR,PIK3CD,MRGPRF,GRM8,KCNB2,EPHA10,ADRA2C,CLDN6,CLDN4,PTPRT,DPP4,OCLN,CD247,HLA-DOA,SHISA9,HLA-DMB |
| GO:0006629 | lipid metabolic process | 2.063032094 | 0.00864904 | ST3GAL6,ST8SIA5,SDC4,APOC1,CYP2J2,CYP11A1,PPAP2C,PLIN2,WNT4,HACD1,PIK3CD,ACOT12,FADS6,UGT8,SPTSSB |
| GO:0007009 | plasma membrane organization | 1.872296029 | 0.0134185 | MAP7,XKR8,F11R,WNT4 |
| GO:0006790 | sulfur compound metabolic process | 1.823703224 | 0.0150071 | MGST1,ST3GAL6,SDC4,ACOT12,OPLAH,CHST6 |
| GO:0007267 | cell-cell signaling | 1.725551179 | 0.0188126 | GABRA3,PTPRN,SLC1A6,SLC6A12,TEK,DLG2,GRID2,ADCY8,KCNB2,MAFA,ADRA2C |
| GO:0048856 | anatomical structure development | 1.689882589 | 0.0204229 | ERBB3,MYLK,ITGB5,HTATIP2,TNR,HPCAL4,PADI2,TEK,LIN28A,CHI3L1,TMOD1,RHCG,RP11-35N6.1,KIRREL3,MPZL2,DLG2,GRID2,ACAN,F11R,WNT4,HACD1,KRT19,PIK3CD,EGFL7,UGT8,ALX1,EPHA10, ADRA2C,CLDN4 |
| GO:0005576 | extracellular region | 1.604486647 | 0.0248607 | ST3GAL6,ERBB3,MYLK,ITGB5,ZDHHC15,CPVL,ENPP5,TNR,PADI2,TEK,NPPB,SDC4,BTN1A1,SPINK2,PRRG3,APOC1,F12,CHI3L1,CYP2J2,RHCG,CBLC,PLIN2,KIRREL3,ACAN,ITLN2,F11R,VWA5B1,CLIC6, WNT4,MUC3A,OSCAR,KRT19,EGFL7,MRGPRF,CREG2,EPHA10,BEX5,ADAMTSL5,PABPC1L2A,DPP4,GPX3,FAM19A5 |
| GO:0007010 | cytoskeleton organization | 1.404009141 | 0.0394449 | MYLK,ITGB5,CORO2A,MAP7,TMOD1,PARVG,F11R,CDC42BPG,KRT19 |
| GO:0016757 | transferase activity, transferring glycosyl groups | 1.395223326 | 0.040251 | ST3GAL6,ST8SIA5,GALNT14,UGT8,GCNT1 |
Extended Data Table 3.
Most significant (p-value<0.05) enriched GO terms in neurons of WS compared to TD samples.
| Down-regulated genes in WS compared to TD in neurons
| ||||
|---|---|---|---|---|
| GO TERM | GO Description | −Log(p-value) | P-value | Genes |
| GO:0043167 | ion binding | 2.995842055 | 0.00100962 | BAZ1B,FMO1,VRK2,DSG2,RFC2,ME1,ATP2A3,ADAMTS2,MYL9,TNNC2,ENO3,FOLR1,ACSS3,TRIM38,PCDHB5,TNNC1,ABCG2,CAT, ADGRE5,MYH2,ADGRE2,ZNF835,ATP8B3,MTL5,GSTM1,ACTA1,SH3RF2,ACTC1,ALOX15,CAPN13,ZNF283,ZNF558,ACOX2,GSTM4, SLFN12,RHOD,RNF212,ZNF626,SERPINA5,S100A13,S100A4,ZNF502,ZFP28,ZNF560,ZNF667,PEG3,ZNF726,ZNF737,ZNF578 |
| GO:0008092 | cytoskeletal protein binding | 2.841323964 | 0.00144104 | USH1C,MYBPC2,TNNC2,STX1A,TNNC1,TNNT2,MYH2,ACTA1,ACTC1,MYOZ1,TPM2 |
| GO:0003013 | circulatory system process | 2.42626763 | 0.00374742 | ELN,TNNC1,AGT,ACTC1,CD34 |
| GO:0006950 | response to stress | 2.270514469 | 0.00536396 | TBXA2R,BAZ1B,VRK2,RFC2,ATP2A3,IL4R,LAT2,MYL9,TRIM38,CAT,ADGRE5,MYH2,ADGRE2,MTL5,GSTM1,PARP9,ALOX15,ACOT11, BATF2,CD34,BHLHA15,SIGIRR,IFITM2,SERPINA5,HLA-DRB1,HLA-DRB5,POU5F1 |
| GO:0005198 | structural molecule activity | 2.015682009 | 0.00964535 | MYOM2,ELN,MYBPC2,MYL9,MYH2,TINAGL1,ACTA1,LAD1,TPM2 |
| GO:0005829 | cytosol | 1.675077895 | 0.0211311 | USH1C,ME1,MYBPC2,MYL9,TNNC2,STX1A,EIF4H,ENO3,TRIM38,TNNC1,TNNT2,MYH2,GSTM1,PARP9,ACTA1,ACTC1,ALOX15, GSTM4,RHOD,ALDH1A3,S100A13,TPM2,POU5F1 |
| GO:0005777 | peroxisome | 1.574933909 | 0.0266113 | PXMP4,CAT,ACOX2 |
| GO:0003674 | Molecular function | 1.54856974 | 0.0282768 | USH1C,TBXA2R,SCN4A,BAZ1B,FMO1,ACPP,VRK2,MYOM2,DSG2,PREX2,ELN,RFC2,ME1,ATP2A3,IL4R,LAT2,MYBPC2,ADAMTS2, PROKR2,MYL9,PXMP4,TNNC2,STX1A,TBL2,EIF4H,ENO3,FOLR1,ACSS3,TRIM38,SLC22A2,PCDHB5,SLC27A6,TNNC1,TNNT2, ABCG2,CAT,ADGRE5,MYH2,ADGRE2,ZNF835,ATP8B3,NSUN5,MTL5,DMGDH,GSTM1,AGT,THUMPD2,PARP9,TINAGL1,ACTA1, RASSF3,SH3RF2,LAD1,ACTC1,ALOX15,ACOT11,UBXN10,CAPN13,RBM47,TC2N,ZNF283,ZNF558,BATF2,ACOX2,GSTM4,CYTL1, PRKCDBP,SLFN12,RHOD,CD34,CHRNA9,GCNT4,MYOZ1,RNF212,BHLHA15,MAATS1,ALDH1A3,RNLS,RBM43,SIGIRR,CCK, ZNF626,LRRIQ4,SERPINA5,S100A13,HLA-DRB1,S100A4,ZNF502,ZFP28,SLC2A10,ZNF560,ZNF667,PEG3,TPM2,HLA-DRB5,LAYN, POU5F1,ZNF726,ANKRD65,ZNF737,ZNF578,SLC22A31,TCF24 |
| GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups | 1.475366347 | 0.0334683 | GSTM1,GSTM4 |
| GO:0016887 | ATPase activity | 1.459248951 | 0.0347337 | RFC2,ATP2A3,TNNT2,ABCG2,ATP8B3,ACTC1 |
| GO:0016874 | ligase activity | 1.406285389 | 0.0392387 | RFC2,ACSS3,TRIM38,SLC27A6,SH3RF2,RNF212 |
| GO:0015979 | photosynthesis | 1.387345392 | 0.0409878 | RFC2 |
| GO:0005615 | extracellular space | 1.320715681 | 0.0477842 | ACPP,ENO3,ADGRE5,AGT,TINAGL1,ACTA1,ACTC1,CYTL1,SERPINA5,S100A13,S100A4 |
| Up-regulated genes in WS compared to TD in neurons
| ||||
|---|---|---|---|---|
| GO TERM | GO Description | −Log(p-value) | P-value | Genes |
| GO:0001071 | nucleic acid binding transcription factor activity | 2.409059017 | 0.00389889 | PITX1,TFAP2C,HAND1,NR5A2,IRF6,ZSCAN10,KLF4,ALX1,DMBX1,FOXB2 |
| GO:0008233 | peptidase activity | 1.531733923 | 0.0293945 | HPN,PRSS16,RHBDF2,ADAMTS16,USP41,TMPRSS2 |
| GO:0009790 | embryo development | 1.354676981 | 0.0441899 | HPN,HAND1,NR5A2,KLF4 |
| GO:0006091 | generation of precursor metabolites and energy | 1.330439536 | 0.0467262 | ALDOC,MT-ND2,MT-ND4,MT-ND1 |
| GO:0048856 | anatomical structure development | 1.312454095 | 0.0487019 | PITX1,TFAP2C,HPN,ALDOC,HAND1,NR5A2,IRF6,ZSCAN10,KLF4,PRDM14,BMP6,ISL2,PROK2,PHOSPHO1,ALX1,DMBX1 |
Supplementary Material
Acknowledgments
This work was supported by grants from the California Institute for Regenerative Medicine (CIRM) TR2-01814 and TR4-06747, the National Institutes of Health through the P01 NICHD033113, NIH Director’s New Innovator Award Program 1-DP2-OD006495-01, R01MH094753, R01MH103134, U19MH107367 and a NARSAD Independent Investigator Grant to A.R.M., grants from the Engmann Foundation, the JPB Foundation and Helmsley Foundation to F.H.G., the Royal Thai Government Scholarship to T.C., CIRM postdoctoral fellowship to C.A.T., the Rita L. Atkinson Graduate fellowship to B.H-M. Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. We would like to acknowledge Dr. Kristen Jepsen at Institute of Genomic Medicine at UCSD for the DNA bead arrays and members of the Willert laboratory (UCSD) for the assistance with the Wnt pathway experiments. We would like to thank all TD participants and WS individuals and their families.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
A.R.M., T.C. and C.A.T. designed the experiments and wrote the manuscript with input from K.S. and all authors. T.C. processed DPCs, generated and characterized iPSCs, NPCs and neurons, and performed cell number, proliferation, and apoptosis experiments as well as FZD9 knockdown and overexpression and statistical analysis. C.A.T. performed C1 single cell analyses, synaptic quantification, calcium imaging, cell density experiments, live neuronal morphology analysis and statistical analysis. B.C.F. performed MEA recording, PCR for retrovirus silencing and Wnt pathway gene expression analysis. B.C.F. and S.E.R prepared astrocytes for co-culture experiments, NPC characterization by flow cytometry and CHIR 98014 experiments. K.S. designed all morphometry experiments with B.H-M. and B.J., and co-wrote the manuscript to link the various levels of investigation from the whole brain imaging findings to the cellular level. L.S. prepared Golgi staining for postmortem neurons with help from K.L.H. and B.J. B.H-M. obtained morphometric data on iPSC-derived neurons and postmortem neurons. D.X.Y., M.C.N.M., C.A.T. and L.M. performed calcium transient experiments and statistical analysis. T.T.B. performed brain scan and statistical analysis with help from A.M.D. C.B. performed electrophysiological test. M.D., W.W., P.L. and Y.M.S performed and neurocognitive and social tests. A.J., Y.M.S, and M.C.A., performed analyses and interpretation of social/neurocognitive tests. R.H.H. performed bioinformatics analysis. L.D. and J.R.K. confirmed deletion of all WS participants who donated cells for reprogramming. E.H., U.B., F.H.G, K.S. and A.R.M. edited the manuscript for publication.
The authors declare no competing financial interests.
References
- 1.Korenberg JR, et al. VI. Genome structure and cognitive map of Williams syndrome. Journal of cognitive neuroscience. 2000;12(Suppl 1):89–107. doi: 10.1162/089892900562002. [DOI] [PubMed] [Google Scholar]
- 2.Meyer-Lindenberg A, et al. Neural basis of genetically determined visuospatial construction deficit in Williams syndrome. Neuron. 2004;43:623–631. doi: 10.1016/j.neuron.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Bellugi U, Lichtenberger L, Mills D, Galaburda A, Korenberg JR. Bridging cognition, the brain and molecular genetics: evidence from Williams syndrome. Trends in Neurosciences. 1999;22:197–207. doi: 10.1016/s0166-2236(99)01397-1. [DOI] [PubMed] [Google Scholar]
- 4.Bellugi U, Lichtenberger L, Jones W, Lai Z, St George MI. The neurocognitive profile of Williams Syndrome: a complex pattern of strengths and weaknesses. J Cogn Neurosci. 2000;12(Suppl 1):7–29. doi: 10.1162/089892900561959. [DOI] [PubMed] [Google Scholar]
- 5.Doyle TF, Bellugi U, Korenberg JR, Graham J. “Everybody in the world is my friend” hypersociability in young children with Williams syndrome. American Journal of Medical Genetics Part A. 2004;124A:263–273. doi: 10.1002/ajmg.a.20416. [DOI] [PubMed] [Google Scholar]
- 6.Dai L, et al. Is it Williams syndrome? GTF2IRD1 implicated in visual-spatial construction and GTF2I in sociability revealed by high resolution arrays. Am J Med Genet A. 2009;149A:302–314. doi: 10.1002/ajmg.a.32652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelmann L, et al. An atypical deletion of the Williams-Beuren syndrome interval implicates genes associated with defective visuospatial processing and autism. Journal of Medical Genetics. 2007;44:136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chailangkarn T, Acab A, Muotri AR. Modeling neurodevelopmental disorders using human neurons. Curr Opin Neurobiol. 2012;22:785–790. doi: 10.1016/j.conb.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewart AK, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 10.Jarvinen-Pasley A, et al. Defining the social phenotype in Williams syndrome: a model for linking gene, the brain, and behavior. Dev Psychopathol. 2008;20:1–35. doi: 10.1017/S0954579408000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltrao-Braga PCB, et al. Feeder-Free Derivation of Induced Pluripotent Stem Cells From Human Immature Dental Pulp Stem Cells. Cell Transplantation. 2011;20:1707–1719. doi: 10.3727/096368911X566235. [DOI] [PubMed] [Google Scholar]
- 13.Adamo A, et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 14.Van Raay TJ, et al. frizzled 9 is expressed in neural precursor cells in the developing neural tube. Dev Genes Evol. 2001;211:453–457. doi: 10.1007/s004270100174. [DOI] [PubMed] [Google Scholar]
- 15.Zhao C, et al. Hippocampal and visuospatial learning defects in mice with a deletion of frizzled 9, a gene in the Williams syndrome deletion interval. Development. 2005;132:2917–2927. doi: 10.1242/dev.01871. [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto T, Tomizawa M, Yokosuka O. SiRNA of frizzled-9 suppresses proliferation and motility of hepatoma cells. Int J Oncol. 2009;35:861–866. doi: 10.3892/ijo_00000400. [DOI] [PubMed] [Google Scholar]
- 17.Lian X, et al. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem cell reports. 2014;3:804–816. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jho EH, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimura N, et al. Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem. 2007;282:1225–1237. doi: 10.1074/jbc.M605851200. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. P Natl Acad Sci USA. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. P Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Current Opinion in Neurobiology. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 24.Spitzer NC, Root CM, Borodinsky LN. Orchestrating neuronal differentiation: patterns of Ca2+ spikes specify transmitter choice. Trends in Neurosciences. 2004;27:415–421. doi: 10.1016/j.tins.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Chiang MC, et al. 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. Neuroimage. 2007;36:1096–1109. doi: 10.1016/j.neuroimage.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawless JF, Fredette M. Frequentist prediction intervals and predictive distributions. Biometrika. 2005;92:529–542. [Google Scholar]
- 27.Chen J, et al. Transcriptome Comparison of Human Neurons Generated Using Induced Pluripotent Stem Cells Derived from Dental Pulp and Skin Fibroblasts. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltrao-Braga PCB, et al. Feeder-Free Derivation of Induced Pluripotent Stem Cells From Human Immature Dental Pulp Stem Cells. Cell Transplantation. 2011;20:1707–1719. doi: 10.3727/096368911X566235. [DOI] [PubMed] [Google Scholar]
- 29.Marinho PA, Chailangkarn T, Muotri AR. Systematic optimization of human pluripotent stem cells media using Design of Experiments. Sci Rep. 2015;5:9834. doi: 10.1038/srep09834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Marchetto MC, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llorens-Bobadilla E, et al. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, et al. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59:71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermann BP, et al. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biology of reproduction. 2015;92:54. doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs B, et al. Regional dendritic and spine variation in human cerebral cortex: a quantitative golgi study. Cereb Cortex. 2001;11:558–571. doi: 10.1093/cercor/11.6.558. [DOI] [PubMed] [Google Scholar]
- 39.Semendeferi K, et al. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex. 2011;21:1485–1497. doi: 10.1093/cercor/bhq191. [DOI] [PubMed] [Google Scholar]
- 40.Marin-Padilla M. Structural abnormalities of the cerebral cortex in human chromosomal aberrations: a Golgi study. Brain Res. 1972;44:625–629. doi: 10.1016/0006-8993(72)90324-1. [DOI] [PubMed] [Google Scholar]
- 41.Takashima S, Becker LE, Armstrong DL, Chan F. Abnormal neuronal development in the visual cortex of the human fetus and infant with down’s syndrome. A quantitative and qualitative Golgi study. Brain Res. 1981;225:1–21. doi: 10.1016/0006-8993(81)90314-0. [DOI] [PubMed] [Google Scholar]
- 42.Jay V, Chan FW, Becker LE. Dendritic arborization in the human fetus and infant with the trisomy 18 syndrome. Brain research Developmental brain research. 1990;54:291–294. doi: 10.1016/0165-3806(90)90153-p. [DOI] [PubMed] [Google Scholar]
- 43.Marin-Padilla M. Prenatal and early postnatal ontogenesis of the human motor cortex: a golgi study. II. The basket-pyramidal system. Brain Res. 1970;23:185–191. doi: 10.1016/0006-8993(70)90038-7. [DOI] [PubMed] [Google Scholar]
- 44.Vuksic M, Petanjek Z, Rasin MR, Kostovic I. Perinatal growth of prefrontal layer III pyramids in Down syndrome. Pediatr Neurol. 2002;27:36–38. doi: 10.1016/s0887-8994(02)00380-6. [DOI] [PubMed] [Google Scholar]
- 45.Jacobs B, et al. Quantitative analysis of cortical pyramidal neurons after corpus callosotomy. Ann Neurol. 2003;54:126–130. doi: 10.1002/ana.10620. [DOI] [PubMed] [Google Scholar]
- 46.Riley JN. A reliable Golgi-Kopsch modification. Brain Res Bull. 1979;4:127–129. doi: 10.1016/0361-9230(79)90067-4. [DOI] [PubMed] [Google Scholar]
- 47.Williams RS, Ferrante RJ, Caviness VS., Jr The Golgi rapid method in clinical neuropathology: the morphologic consequences of suboptimal fixation. J Neuropathol Exp Neurol. 1978;37:13–33. doi: 10.1097/00005072-197801000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Jacobs B, Scheibel AB. A Quantitative Dendritic Analysis of Wernicke Area in Humans 1. Life-Span Changes. J Comp Neurol. 1993;327:83–96. doi: 10.1002/cne.903270107. [DOI] [PubMed] [Google Scholar]
- 49.Uylings HB, Ruiz-Marcos A, van Pelt J. The metric analysis of three-dimensional dendritic tree patterns: a methodological review. J Neurosci Methods. 1986;18:127–151. doi: 10.1016/0165-0270(86)90116-0. [DOI] [PubMed] [Google Scholar]
- 50.White N, et al. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2010;63:91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown TT, et al. Prospective motion correction of high-resolution magnetic resonance imaging data in children. Neuroimage. 2010;53:139–145. doi: 10.1016/j.neuroimage.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuperman JM, et al. Prospective motion correction improves diagnostic utility of pediatric MRI scans. Pediatr Radiol. 2011;41:1578–1582. doi: 10.1007/s00247-011-2205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jovicich J, et al. Reliability in multi-site structural MRI studies: effects of gradient nonlinearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 54.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 55.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 56.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Fischl B, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 58.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 60.Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.