Abstract
The ventral nuclei of the lateral lemniscus (VNLL) are part of the central auditory system thought to participate in temporal sound processing. While the timing and location of VNLL neurogenesis have been determined, the genetic factors that regulate VNLL neuron development are unknown. Here, we use genetic fate-mapping techniques to demonstrate that all glycinergic and glycinergic/GABAergic VNLL neurons derive from a cellular lineage that expresses the homeobox transcription factor Engrailed 1 (En1). We also show that En1 deletion does not affect migration or adoption of a neuronal cell fate but does lead to VNLL neuron death during development. Furthermore, En1 deletion blocks expression of the transcription factor FoxP1 in a subset of VNLL neurons. Together, these data identify En1 as a gene important for VNLL neuron development and survival.
Keywords: auditory, brainstem, deafness, ear, hearing
INTRODUCTION
The ventral nucleus of the lateral lemniscus (VNLL) is a group of auditory neurons embedded in a large fiber tract that connects brainstem auditory nuclei with the auditory midbrain. VNLL neurons receive inputs mainly from the contralateral ventral cochlear nucleus (VCN), with smaller projections arising from the ipsilateral VCN and superior olivary complex (SOC) (Covey and Casseday, 1986; Huffman and Covey, 1995; Saint Marie et al., 1997; Schofield, 1995). The VNLL consists predominantly of glycinergic and GABAergic inhibitory neurons that project broadly to other NLL neurons and to the inferior colliculus (Riquelme et al., 2001; Roberts and Ribak, 1987; Saint Marie et al., 1997; Saint Marie and Baker, 1990). VNLL neurons are thought to participate in temporal sound processing and decoding of amplitude-modulated sounds, both of which are important for recognition of acoustic patterns present in speech (Batra, 2006; Covey and Casseday, 1991; 1986; Nayagam et al., 2005; Pollak et al., 2011; Recio-Spinoso and Joris, 2014; Yavuzoglu et al., 2011; Zhang and Kelly, 2006).
In the mouse, VNLL neurons are generated in rhombomere 4 (r4) between embryonic days 9.5 and 13.5 (E9.5 and E13.5) (Di Bonito et al., 2013; Pierce, 1973; Simon et al., 2001). VNLL neuron generation depends upon the brainstem segmentation gene Hoxb1, likely because Hoxb1 is required for r4 specification (Di Bonito et al., 2013; Domowicz et al., 2008). Beyond this, the genes and genetic pathways that control VNLL neuron development are unknown.
The mouse En1 gene encodes a transcription factor containing a highly conserved homeobox domain that acts as both a transcriptional activator and repressor (Davis et al., 1991; Joyner and Martin, 1987). Loss of En1 function can cause homeotic transformations such as ventral-to-dorsal transformations in the mouse limb (Garcia-Bellido and Santamaria, 1972; Loomis et al., 1996; Morata and Lawrence, 1975). In the CNS, En1 regulates neuronal specification in the developing cerebellum, midbrain, and spinal cord (Couchman et al., 2010; Matise and Joyner, 1997; Simon et al., 2005; Wurst et al., 1994). En1 is also crucial for maturation and survival, but not specification, of serotonergic and midbrain dopaminergic neurons (Fox and Deneris, 2012; Simon et al., 2001). We recently demonstrated that SOC expression of En1 is necessary for survival and proper positioning of glycinergic lateral superior olive (LSO) and medial nucleus of the trapezoid body (MNTB) neurons, GABAergic lateral nucleus of the trapezoid body (LNTB) neurons and cholinergic/glycinergic ventral nucleus of the trapezoid body (VNTB) neurons (Altieri et al., 2015; Jalabi et al., 2013). Since SOC neurons arise from r3–5 (Karis et al., 2001; Maricich et al., 2009), we speculated that En1 might also play a role in VNLL neuron development. To test this hypothesis, we generated En1 constitutive and conditional knockout (CKO) mice. We found that En1 plays a pivotal role in survival, but not generation or positioning, of VNLL neurons.
MATERIALS AND METHODS
Mice and mating paradigms
All experimental protocols were approved by Case Western Reserve University or Children’s Hospital of Pittsburgh of UPMC Animal Care Facilities. Mice were housed under pathogen-free conditions on a 12h light/dark cycle. The generation of Hoxb1Cre, En1Cre, En1flox, ROSALacZ and ROSAtdTomato mice were described previously (Arenkiel et al., 2003; Sgaier et al., 2007; Soriano, 1999). Hoxb1; En1CKO mice were produced by first mating Hoxb1Cre/+ mice with En1flox/flox mice to generate Hoxb1Cre/+; En1+/flox double-transgenic animals. These mice were bred with En1flox/flox mice to generate transgenic mice with four genotypes: Hoxb1+/+; En1+/flox, Hoxb1+/+; En1flox/flox, Hoxb1Cre/+; En1+/flox, and Hoxb1Cre/+; En1flox/flox. Only Hoxb1Cre/+; En1flox/flox (Hoxb1; En1CKO) mice have no En1 expression in Hoxb1 expressing cells. Mice of the other three genotypes are collectively referred to as “control” because they displayed no abnormal phenotypes, and their VNLL histology and immunostaining for all markers tested were indistinguishable at all ages examined.
For fate mapping experiments, En1Cre/+; ROSAtdTomato/+ mice were intercrossed to generate En1Cre/+; ROSAtdTomato mice and En1Cre/Cre; ROSAtdTomato mice. Alternatively, En1Cre/+ mice were mated to En1+/+; ROSALacZ/LacZ or En1flox/flox; ROSALac/LacZ mice to generate En1Cre/+; ROSALacZ/+ or En1Cre/flox; ROSALacZ/+ mice, respectively. This strategy allowed us to track En1-lineal cells in the presence or absence of En1 gene function using two different reporters.
Tissue harvesting and processing
For embryonic and postnatal day 0 (P0) ages, timed matings were used with the plug date designated as E0.5. Embryos were dissected into cold 1X PBS, brains isolated and fixed overnight at 4°C in fresh 4% paraformaldehyde (PFA)/0.1M phosphate buffer. For tissue harvested from P0 and adult mice, animals were transcardially perfused with 4% PFA or 4% PFA/0.2% glutaraldehyde and tissue was post-fixed for 2h-overnight at 4°C. Brains were cryoprotected in 30% sucrose/1X PBS for 48 h, embedded in Tissue-Tek O.C.T. (Sakura Finetek) and serially-sectioned at 10–25μm on a Leica CM1950 cryostat (Leica Microsystems, Wetzlar, Germany). Sections were mounted on Superfrost/Plus slides (Fisher scientific) and stored at −80°C. Two-three mice/genotype/age were analyzed.
Histology
β-galactosidase activity was determined using 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal) staining. Embryonic and adult tissue sections were prepared as described above and stained with Xgal for 4–24h at 37°C followed by 1X PBS washes and overnight fixation in 4% PFA at 4°C. Slides were then counterstained with Cresyl violet or nuclear fast red, dehydrated and mounted with Cytoseal 60 (Richard Allan Scientific). For all histological analyses, series of slides were processed to allow exact matching of VNLL levels between control and mutant brains.
Immunohistochemistry
Tissue cryosections were washed in 1X PBS then blocked for 1h at room temperature (RT) in 1X PBS/0.3% Triton X-100/3% normal donkey or goat serum (blocking solution). Slides were incubated overnight at 4°C with blocking solution containing dilutions of the following antibodies: rabbit anti-ALDH1L1 (Abcam) 1:500; rabbit anti-cleaved caspase-3 (Biocare Medicare) 1:250; rabbit anti-FoxP1 (Abcam Inc.) 1:400; mouse anti-GAD67 (EMD Millipore) 1:5000; rabbit anti-glycine (Millipore) 1:100; chicken anti-MAP2 (Abcam Inc.) 1:5000; rabbit anti-Olig2 (EMD Millipore) 1:250; mouse anti-TUJ1 (Abcam) 1:500. Sections were washed in 1X PBS and incubated for 1h with secondary antibodies conjugated to DyLight 488 or 549 (Jackson Immunoresearch) at a 1:500 dilution. All slides were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) or NeuroTrace fluorescent Nissl stain (Molecular Probes). After staining, sections were mounted with ProLong Gold to preserve the fluorescent signals and imaged using a Leica DM5500B epifluorescence microscope (Leica Microsystems, Exton, PA) or an inverted Zeiss Axio Observer on a PerkinElmer UltraVIEW VoX spinning disk confocal with a Hamamatsu C9100-13 camera and Volocity software.
In situ hybridization
The probe for GlyT2 (642bp) was amplified from pBluescript II plasmid using PCR primers flanked with T7/T3 sequences (forward T3, 5′-AATTAACCCTCACTAAAGGGAATGTGTGCATCTGTGTATGCA-3′; reverse T7, 5′-GTAATACGACTCACTATAGGGCCGGTATGGTAGTGGTGGCCACG-3′). The amplified PCR product was purified using a gel purification kit (Qiagen) and transcribed with the Ambion Maxiscript transcription kit (Life technologies). After transcription, the probe was precipitated with 4M LiCl in 100% ethanol for 1h at -80°C, centrifuged at 4°C for 20 minutes, the pellets rinsed with 70% ethanol, air dried, and resuspended in 30μl diethylpyrocarbonate-treated water. In situ hybridization was performed as previously described (Domowicz et al., 2008; Garcia-Bellido and Santamaria, 1972; Loomis et al., 1996; Morata and Lawrence, 1975). Briefly, frozen mouse brain sections were post-fixed with 4% PFA for 15 minutes and incubated with riboprobe in hybridization buffer (50% formamide, 5x Saline Sodium Citrate buffer-SSC, 1% SDS, 500μg/ml yeast tRNA and 200μg/ml heparin) overnight at 55°C. After hybridization, the slides were rinsed sequentially in solution X (50% formamide, 2xSSC, 1% SDS) for 1.5h at 65°C and Tris saline buffer containing 1%Tween 20 (TBST) for 45 min at RT. Sections were then blocked with 10% lamb serum for 1h and incubated with anti-digoxigenin antibody (Roche Applied Science) for 2h at RT. After staining, sections were rinsed with TBST for 45 min and NTMT buffer (100 mM Tris HCl, 100 mM NaCl, 50mM MgCl2, 1%Tween 20, and 2mM levamisol) for 30min at RT. Color development was processed with NBT/BCIP (Roche Applied Science) incubation at RT. Sense probe hybridization showed clean background and was used as a negative control. After staining, sections were counterstained with Cresyl violet, dehydrated with ethanol and mounted using Cytoseal.
Cell counts and statistics
Serially-sectioned, immunostained coronal tissue sections spaced every 40μm (E14.5, E17.5, P0, adult) and Xgal-stained coronal tissue sections spaced every 40μm (E11.5, E15.5) or 100μm (P0) through all anteroposterior levels of the VNLL were photographed at 25X on a Leica DM5500B epifluorescence microscope. Anteroposterior levels of mutant and control littermate brains were matched using the pontine nucleus as a reference point. The borders of the VNLL in mutant mice were delineated based on the location of neuronal cell bodies in the white matter of the lateral lemniscus identified by Nissl staining or FoxP1 immunostaining. Photographs used for counting imaged a 72,900μm2 area centered on the middle of the dorsoventral axis of the VNLL. Numbers of Xgal+ VNLL neurons (n =2 mice/genotype) at E11.5, E15.5 and P0 were counted. We analyzed cell death by calculating the percentage of tdTomato+ cells with pyknotic nuclei or that were also caspase-3+ at E17.5 (n =3 mice/genotype). We counted numbers of tdTomato+, FoxP1+ and tdTomato+/FoxP1+ VNLL neurons (n =3 mice/genotype) at E14.5 and P0; tdTomato+, MAP2+ and tdTomato+/MAP2+ cells (n =3 mice/genotype) in adult mice; and FoxP1+, GAD67+ and FoxP1+/GAD67+ neurons in adult mice (n =3 mice/genotype). No correction factors were applied. Cell counts are reported as mean ±SEM, and statistical significance determined by independent sample one- or two-tailed t-tests or one-way ANOVA followed by Tukey’s multiple comparisons (GraphPad Prism, La Jolla, CA).
RESULTS
En1-lineal VNLL cells are glycinergic and glycinergic/GABAergic neurons
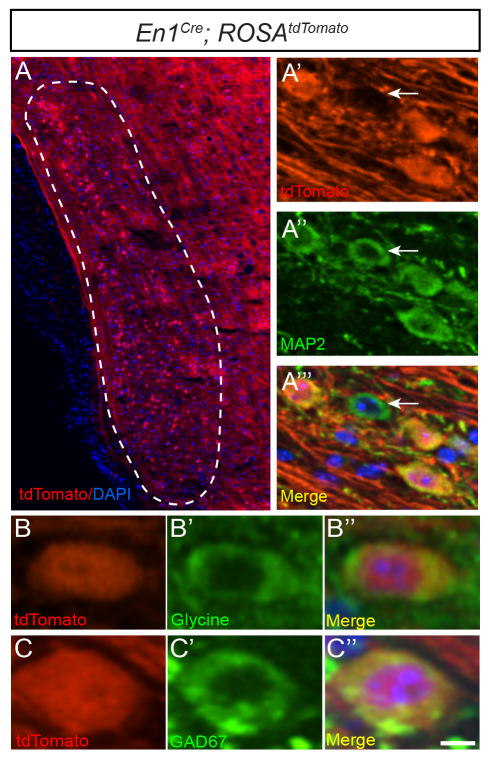
To determine the distribution of En1-lineal cells in the VNLL, we generated En1Cre/+; ROSAtdTomato/+ and En1Cre/+; ROSALacZ/+ mice, where En1-lineal cells are irreversibly labeled and easily visualized by tdTomato fluorescence or Xgal staining, respectively (Figs. 1A, 3G). Immunostaining with the neuronal marker MAP2 showed that all En1-lineal VNLL cells (n =342 tdTomato+ cells from n =3 mice) in adult En1Cre/+; ROSAtdTomato/+ mice were neurons, and that 91 ±2.7% (n =374 MAP2+ cells from n =3 mice) of VNLL neurons derive from the En1 lineage (Fig. 1A′–A‴). Immunostaining for glycine or GAD67 revealed that all (n =155 tdTomato+ neurons from n =3 mice) En1-lineal VNLL neurons were glycinergic while 26.4 ±5.8% (n =124 GAD67+/tdTomato+ cells of n =461 total tdTomato+ cells from n =3 mice) were also GABAergic in neurotransmitter phenotype (Fig. 1B–B″, C–C″). These data indicate that the vast majority of VNLL neurons are derived from the En1 lineage.
Figure 1. En1-lineal cells in the VNLL are glycinergic and glycinergic/GABAergic neurons.
Dotted lines in this and subsequent figures delineate the VNLL or the region in which VNLL neurons are typically found. Coronal brainstem section of adult En1Cre; ROSAtdTomato mouse showing endogenous tdTomato expression (A′, B, C) overlaid with DAPI (A) and immunostaining for MAP2 (A″), glycine (B′), GAD67 (C′) or merged images with DAPI (A‴, B″, C″). Arrows in (A′–A‴) denote tdTomato- VNLL neuron. Scale bar: 70μm (A), 16μm (A′–A‴), 10μm (B–C″).
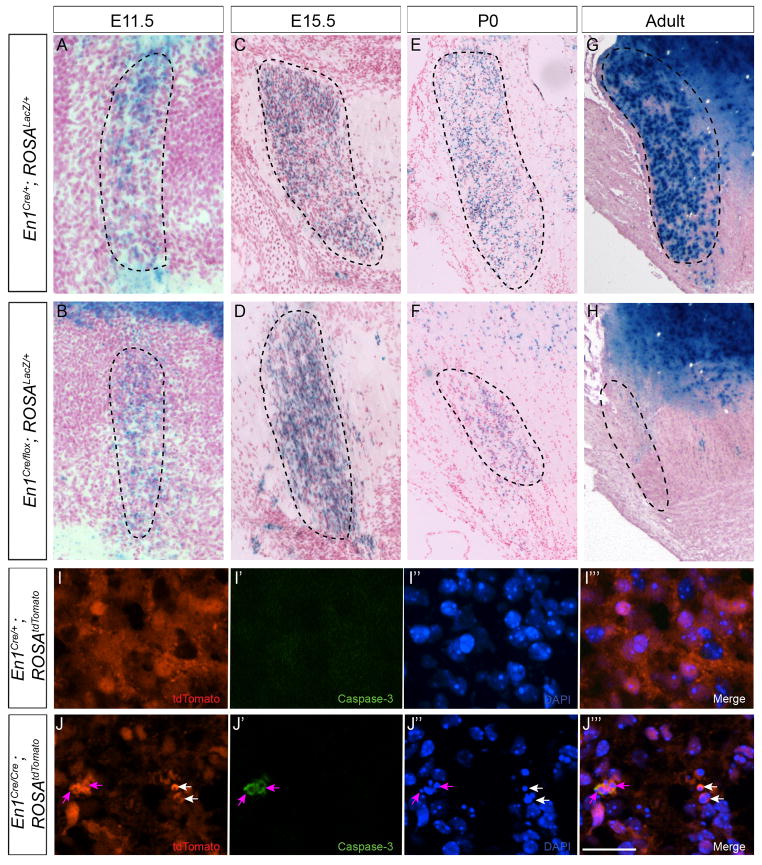
Figure 3. En1 is required for VNLL neuron survival.
Xgal staining of brainstem sections from E11.5 (A, B), E15.5 (C, D), P0 (E, F) and adult (G, H) En1Cre/+; ROSALacZ (A, C, E, G) and En1Cre/flox; ROSALacZ (B, D, F, H) mice. Endogenous tdTomato (I, J), immunostaining for activated caspase-3 (I′, J′), DAPI staining (I″, J″) and merged images (I″′, J‴) show cell death in the VNLL of E17.5 En1Cre/Cre; ROSAtdTomato (J–J‴) but not En1Cre/+; ROSAtdTomato (I–I‴) mice. Purple and white arrowheads (J–J‴) denote caspase-3+/tdTomato+/pyknotic cells and tdTomato+ cells with pyknotic nuclei, respectively. Scale bar: 150μm (A, B), 250μm (C–H), 20μm (I–J‴).
En1 deletion causes glycinergic VNLL neuron death
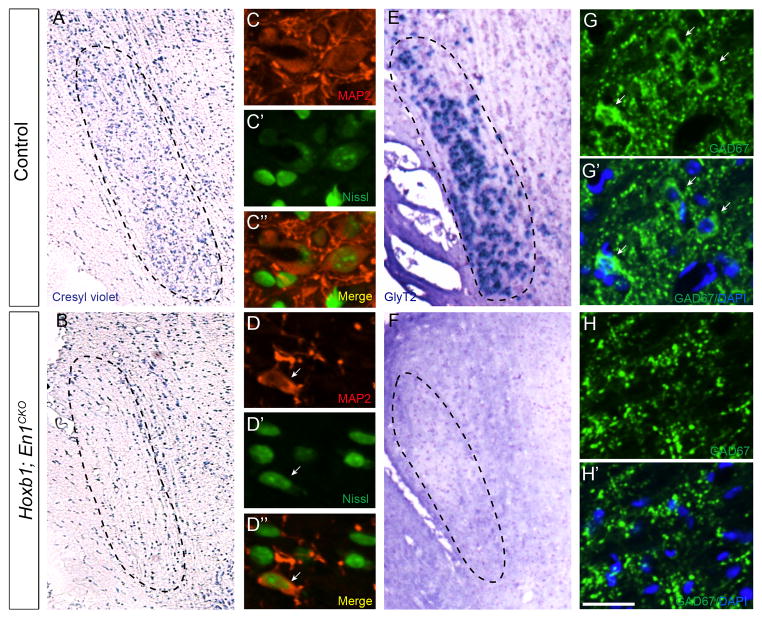
VNLL neurons originate from rhombomere 4 (r4) starting at E9.5 (Di Bonito et al., 2013; Pierce, 1973). Immunostaining for En1 in E12.5 Egr2; ROSAtdTomato mice, where all cells in r3 and r5 are tdTomato+, showed the presence of En1+ cells in r4 (Supplemental Fig. 1). To determine the effects of En1 deletion on VNLL neurons, we conditionally deleted the gene in r4 by generating Hoxb1Cre; En1flox/flox (Hoxb1; En1CKO) mice. Adult Hoxb1; En1CKO mice had very few VNLL neurons based on Cresyl violet and MAP2 immunostaining (Fig. 2A–D″, Supplemental Fig. 2), and none of the remaining neurons were glycinergic based on in situ hybridization for GlyT2 (Fig. 2E, F). Furthermore, GAD67 immunostaining revealed a 99% reduction in GABAergic VNLL neuron numbers in adult Hoxb1; En1CKO mice compared to littermate controls (0.33 ±.33 vs. 38.5 ±3.2; p<0.01; n =3 mice/genotype) (Fig. 2G–H′). These data indicate that En1 is essential for generation, cell fate acquisition or survival of glycinergic and glycinergic/GABAergic VNLL neurons.
Figure 2. En1 deletion leads to loss of glycinergic and GABAergic neurons.
Cresyl violet staining (A, B), MAP2 immunostaining (C, D), Neurotrace Nissl staining (C′, D′), merged images (C″, D″), in situ hybridization for the glycinergic neuron marker GlyT2 (E, F), GAD67 immunostaining (G–H′) and DAPI (G′, H′) of coronal brainstem sections of adult littermate control (A, C–C″, E, G, G′) and Hoxb1; En1CKO (B, D–D″, F, H, H′) mice. GAD67+ staining in (H, H′) marks only axon terminals but not neuronal cell bodies (white arrowheads in G, G′). White arrowhead (D–D″) indicates rare MAP2+ neuron retained in VNLL of Hoxb1; En1CKO mice (D–D″). Scale bar: 200μm (A, B), 25μm (C–C″, D–D″, G–H′), 250μm (E, F).
En1 is expressed by cells in the brainstem extending from r1 to the spinal cord (Davis and Joyner, 1988). To verify that En1-lineal VNLL neurons derived from r4 and not other brainstem levels, we compared VNLL region morphology in adult Hoxb1; En1CKO and En1-null mice. Cresyl violet staining on sections throughout the anteroposterior dimension of the VNLL region revealed similar acellular appearances of the VNLL region in both cases (Supplemental Figure 2). These data show that constitutive En1-deletion produced the same phenotype as conditional deletion using the Hoxb1Cre driver, and further support the conclusion that VNLL neurons are derived from r4.
To determine whether failure of neuronal generation, aberrant cell fate acquisition and/or cell death explained the decrease in VNLL neurons seen in adult Hoxb1; En1CKO mice, we fate-mapped the En1 lineage in En1Cre/+; ROSALacZ and En1Cre/flox; ROSALacZ mice at different embryonic and postnatal ages. In En1Cre/flox; ROSALacZ mice, En1 “self-deletes” within 24 hours following initiation of its expression (Sgaier et al., 2007), enabling us to study how En1 deletion affected the distribution of En1-lineal cells. Xgal+ cells were first detected in the developing VNLL at E11.5 (Fig. 3A). The distribution and number of X-gal+ cells were similar in En1Cre/+; ROSALacZ and En1Cre/flox; ROSALacZ mice at E11.5 (417 ±57 vs. 514 ±33; p =0.48) and E15.5 (2477 ±50 vs. 2224 ±37; p =0.21) (n =2 mice/genotype/age) (Fig. 3A–D). However, numbers of X-gal+ cells in P0 En1Cre/flox; ROSALacZ mice were reduced by 64% compared to En1Cre/+; ROSALacZ littermates (855 ±15 vs. 2379 ±37; p =0.02) (n =2 mice/genotype) (Fig. 2E, F). No Xgal+ cells were present in the region of the VNLL in adult En1Cre/flox; ROSALacZ mice (Fig. 3G, H).
The decrease in Xgal+ cell numbers between E15.5 and adulthood suggested that En1-null VNLL cells died. To examine this possibility, we immunostained brainstem tissue sections from E17.5 En1Cre/+; ROSAtdTomato and En1Cre/Cre; ROSAtdTomato embryos for the apoptotic cell death marker caspase-3 and counterstained with the nuclear marker DAPI (Fig. 3I–J‴). We found that the VNLL of En1Cre/Cre; ROSAtdTomato mice contained ~24x more tdTomato+/caspase-3+ cells (1.68 ±0.54% vs. 0.071 ±0.071%; p =0.021; n =3 mice/genotype), and there was a trend toward an 8-fold increase in the percentage of tdTomato+ cells with pyknotic nuclei (7.2 ±3.1% vs. 0.89 ±0.47%; p =0.059; n =3 mice/genotype) compared to En1Cre/+; ROSAtdTomato littermate controls. Taken together with the Xgal+ cell count data, these data suggest that En1 is important for survival but not specification of VNLL neurons.
En1 deletion does not alter neuronal cell fate
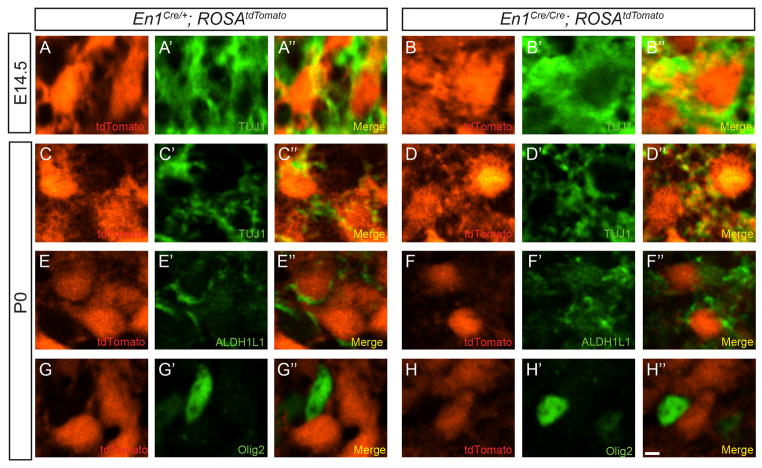
En1 deletion can cause cell fate specification errors (Loomis et al., 1996), so we investigated the possibility that VNLL neuron death occurred secondary to a cell fate switch in En1-null cells. Immunostaining with the neuron-specific marker β-III tubulin (TUJ1) (Menezes and Luskin, 1994) in E14.5 and P0 En1Cre/+; ROSAtdTomato and En1Cre/Cre; ROSAtdTomato mice revealed that all tdTomato+ VNLL cells co-expressed TUJ1 (Fig. 4A–D″). Furthermore, no tdTomato+ VNLL cells in P0 En1Cre/+; ROSAtdTomato or En1Cre/Cre; ROSAtdTomato mice were immunoreactive for the astrocyte marker ALDH1L1 or the oligodendrocyte marker Olig2 (Fig. 4E–H″). These data demonstrate that En1 deletion does not affect adoption or maintenance of VNLL neuronal cell fate.
Figure 4. En1-null VNLL cells maintain neuronal identity.
Single-plane confocal images of brainstem sections from E14.5 (A–B″) and P0 (C–H″) En1Cre/+; ROSAtdTomato (A–A″, C–C″, E–E″, G–G″) and En1Cre/Cre; ROSAtdTomato (B–B″, D–D″, F–F″, H–H″) mice immunostained for the neural marker TUJ1 (A′, B′, C′, D′), the astrocyte marker ALDH1L1 (E′, F′) or the oligodendrocyte marker Olig2 (G′, H′). Scale bar: 12μm.
En1 deletion results in reduced expression of FoxP1
The forkhead transcription factor FoxP1 is widely expressed in the central nervous system and is important for neuron development, positioning and survival (Bacon et al., 2014; Palmesino et al., 2010; Rousso et al., 2008). FoxP1 is also expressed by En1-lineal SOC cells during embryonic and early postnatal development (Marrs et al., 2013), and this expression requires En1(Altieri et al., 2015). We found that FoxP1+ cells were present in the VNLL of E14.5, P0 and adult control mice, that all FoxP1+ cells were neurons, and that all adult VNLL neurons were FoxP1+ (Fig. 5A–A″, C′C″, E–E″). Notably, a small number of FoxP1+/tdTomato- cells were present at E14.5 and P0 (Fig. 5A–A″, C–C″). These data suggest that a minority of FoxP1+ VNLL neurons are not derived from the En1 lineage.
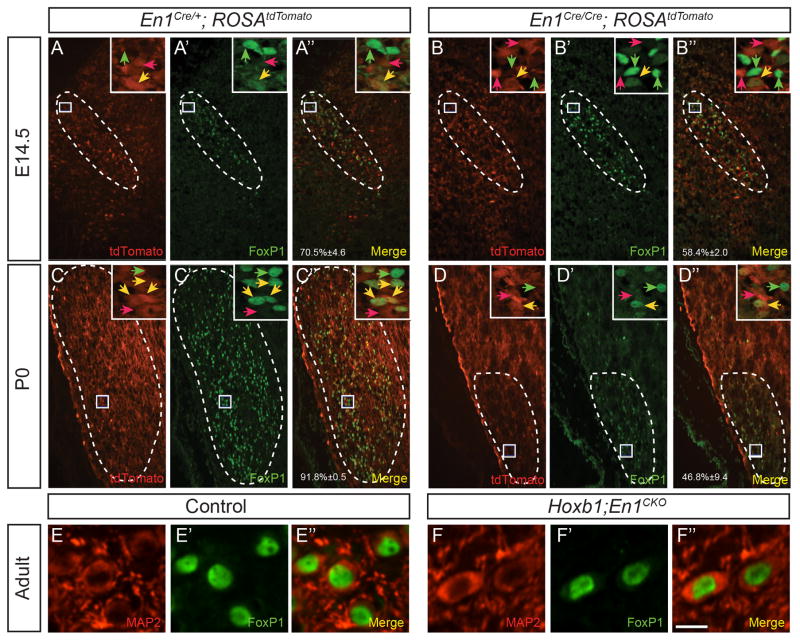
Figure 5. A subset of VNLL neurons require En1 function to initiate FoxP1 expression.
Endogenous tdTomato expression (A, B, C, D), immunostaining for FoxP1 (A′, B′, C′, D′) and merged images (A″, B″, C″, D″) in coronal brainstem sections through the VNLL of E14.5 (A–B″) and P0 (C–D″) En1Cre/+; ROSAtdTomato (A–A″, C–C″) and En1Cre/Cre; ROSAtdTomato (B–B″, D–D″) mice. The percentage of tdTomato+ cells that are also FoxP1+ (A″-D″) is shown as mean ±SEM (n =3 mice/genotype/age). Boxed areas are shown in insets. Red, green and yellow arrowheads indicate tdTomato+, FoxP1+ and tdTomato+/FoxP1+ cells, respectively. Double immunostaining for MAP2 (E, F), FoxP1 (E′, F′) and merged images (E″, F″) in coronal brainstem sections in the VNLL of adult control (E–E″) and Hoxb1; En1CKO (F–F″) mice. Scale bar: 120μm (AD″), 12μm (insets), 8 μm (E–F″).
We next sought to determine whether En1 deletion might affect FoxP1 expression in the VNLL. The percentage of tdTomato+ cells that were also FoxP1+ was similar in E14.5 En1Cre/+; ROSAtdTomato and En1Cre/Cre; ROSAtdTomato mice (Fig. 5A–B″; p =0.43, Tukey’s pairwise test; n =3 mice/genotype). However, this percentage was significantly lower in P0 En1Cre/Cre; ROSAtdTomato vs. En1Cre/+; ROSAtdTomato mice (Fig. 5C–D″; p<0.01; n =3 mice/genotype). In adult Hoxb1; En1CKO mice that lack all En1-lineal VNLL cells, double immunostaining for FoxP1 and MAP2 revealed that the small number of remaining FoxP1+ VNLL cells were neurons (Fig. 5E–F″). We questioned whether remaining neurons in the VNLL of Hoxb1; En1CKO mice were GABAergic. Whereas 18.0 ±2.5% of FoxP1+ VNLL neurons in control mice co-expressed GAD67, none of the few remaining FoxP1+ VNLL neurons in Hoxb1; En1CKO mice were GAD67+ (p<0.05; n =3 mice/genotype). These data suggest that En1 is necessary for initiation and/or maintenance of FoxP1 expression in a subset of VNLL neurons, and that En1-lineal/FoxP1+ neurons comprise the glycinergic and glycinergic/GABAergic VNLL neuron population.
DISCUSSION
We show that En1 is necessary for glycinergic and glycinergic/GABAergic VNLL neuron survival during late embryonic and early postnatal development. This role is consistent with En1 actions in other CNS regions, as the gene also acts as a survival factor in midbrain dopaminergic, brainstem serotonergic and SOC neurons (Altieri et al., 2015; Fox and Deneris, 2012; Simon et al., 2001). Why En1 deletion triggers neuronal death is not clear. Given En1’s role in regulating expression of axon guidance molecules such as Eph family members (Friedman and O’Leary, 1996; Logan et al., 1996; Rétaux and Harris, 1996), it is possible that aberrant axon targeting and subsequent loss of trophic support could cause death of VNLL neurons. Further studies are needed to identify the mechanism by which En1 regulates VNLL neuron survival.
One interesting difference between En1 deletion phenotypes in the SOC and VNLL concerns neuron positioning. In the SOC, En1 deletion causes the appearance of an ectopic medial cell group composed of En1-lineal neurons (Altieri et al., 2015). In contrast, VNLL neuron positioning appears to be unaffected by En1 deletion. This suggests that En1 participates in nuclear formation in the SOC but not in the VNLL. Different mechanisms may control SOC and VNLL neuron positioning because non-glutamatergic SOC neurons migrate radially from the ventricular neuroepithelium while VNLL neurons follow a tangential migratory route to move from r4 to the rostral hindbrain (Di Bonito et al., 2013). The molecules that control brainstem nuclear formation are for the most part unknown; future studies will aim to determine En1 downstream target genes involved in this process.
Another interesting difference between the SOC and VNLL concerns the relationship of En1 and FoxP1. We found that the forkhead transcription factor FoxP1 is expressed by developing VNLL neurons and, similar to the SOC, that most FoxP1+ VNLL neurons are derived from the En1-lineage (Marrs et al., 2013). In both regions, a small subset of FoxP1+ neurons survive following En1 deletion, suggesting that En1 deletion acts in a cell-autonomous manner. The difference lies in the fact that FoxP1 expression is lost from En1-lineal neurons in the SOC following En1 deletion (Altieri et al., 2015), while many En1-null VNLL neurons continue to express FoxP1 until their death (Fig. 5). That the percentage of tdTomato+ VNLL cells that express FoxP1 does not increase between E14.5 and P0 in En1Cre/Cre; ROSAtdTomato mice as it does in En1Cre/+; ROSAtdTomato mice indicates that En1 may be required for initiation or maintenance of FoxP1 expression in only a subset of En1-lineal VNLL neurons. These data imply that there are three populations of VNLL neurons: En1-lineal neurons whose FoxP1 expression requires En1, En1-lineal neurons whose FoxP1 expression does not require En1, and non-En1-lineal neurons that express FoxP1. Whether these three classes correlate with functional differences in these neuronal populations is not clear. Further experiments aimed at determining the roles of FoxP1 in the development of VNLL neurons might provide insight into these differences.
Previous studies reported that the majority of VNLL neurons contain glycine or are dual glycinergic/GABAergic in neurotransmitter phenotype (Riquelme et al., 2001; Saint Marie et al., 1997). Consistent with this, we found that all VNLL neurons derived from the En1 lineage were glycinergic while 26% of these were also GABAergic. This is again different from the situation in the SOC, where En1-lineal LSO and MNTB neurons are glycinergic, LNTB neurons are GABAergic, and VNTB neurons are cholinergic/glycinergic (Altieri et al., 2015). Therefore, it is unlikely that En1 plays a direct role in neurotransmitter phenotype acquisition. Rather, they support the interpretation that En1 controls other aspects of neuronal phenotypic maturation.
The VNLL participates predominantly in monaural sound pathways that link the brainstem and inferior colliculus (Covey and Casseday, 1991; Riquelme et al., 2001; Schofield and Cant, 1997). However, comprehensive analysis of VNLL function has been difficult secondary to the relative inaccessibility of this nucleus and a lack of genetic tools with which to probe its function in living animals. Our data indicate that En1Cre transgenic mice could be used to drive available optogenetic or other genetically-regulated functional tools in VNLL neurons to provide more information about their function.
Supplementary Material
Horizontal section of E12.5 Egr2Cre; ROSAtdTomato mouse brainstem visualized for tdTomato (red) and En1 immunostaining (green). Brainstem midline is indicated by dotted white line. Solid white lines delineate boundaries of rhombomeres r3, r4 and r5. An En1+ cell group (dotted circle) is present in r4. Scale bar: 100μm.
Cresyl violet-stained coronal sections through the region of the VNLL in control adult (A), En1-null (B) and Hoxb1Cre; En1CKO mice. Sections were aligned at similar brainstem levels using the pontine nucleus (outlined in blue) and median raphe nucleus (outlined in red) as landmarks. Sections throughout the VNLL region are shown for each brain, with the most anterior sections on the left. Dotted black lines indicate the VNLL in control mice; solid black lines identify the midline. Scale bar: 400μm.
Acknowledgments
We thank members of the Maricich lab and Dr. Sharyl Fyffe-Maricich for critical discussions concerning the data and the manuscript. We thank Dr. Gary Landreth at Case Western Reserve University for supplying laboratory space to WJ. Confocal imaging was done at Children’s Hospital of Pittsburgh with the generous assistance of Dr. Tim Sanders and his laboratory. This work was supported by the Richard King Mellon Institute for Pediatric Research at the University of Pittsburgh (SMM), the Child Neurology Society (SMM), the American Hearing Research Foundation (SMM), the Research Advisory Committee of Children’s Hospital of Pittsburgh of UPMC Postdoctoral Grant (TZ), the National Institute on Deafness and other Communication Disorders (NIDCD) of the National Institutes of Health (NIH) T32DC011499 (SCA), NIDCD F32DC014896 (SCA), and NIDCD F32DC011982 (WJ).
Footnotes
AUTHOR CONTRIBUTIONS
SCA, TZ and SMM designed the study; SCA, TZ, WJ and RRR-G performed the experiments; SCA, TZ and SMM analyzed the data; SCA, TZ and SMM wrote the manuscript with input from the other authors.
The authors declare no conflicts of interest.
References
- Altieri SC, Jalabi W, Zhao T, Romito-DiGiacomo RR, Maricich SM. En1 directs superior olivary complex neuron positioning, survival, and expression of FoxP1. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–386. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Batra R. Responses of neurons in the ventral nucleus of the lateral lemniscus to sinusoidally amplitude modulated tones. Journal of Neurophysiology. 2006;96:2388–2398. doi: 10.1152/jn.00442.2006. [DOI] [PubMed] [Google Scholar]
- Couchman K, Grothe B, Felmy F. Medial superior olivary neurons receive surprisingly few excitatory and inhibitory inputs with balanced strength and short-term dynamics. Journal of Neuroscience. 2010;30:17111–17121. doi: 10.1523/JNEUROSCI.1760-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey E, Casseday JH. The monaural nuclei of the lateral lemniscus in an echolocating bat: parallel pathways for analyzing temporal features of sound. J Neurosci. 1991;11:3456–3470. doi: 10.1523/JNEUROSCI.11-11-03456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey E, Casseday JH. Connectional basis for frequency representation in the nuclei of the lateral lemniscus of the bat Eptesicus fuscus. J Neurosci. 1986;6:2926–2940. doi: 10.1523/JNEUROSCI.06-10-02926.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Holmyard DP, Millen KJ, Joyner AL. Examining pattern formation in mouse, chicken and frog embryos with an En-specific antiserum. Development. 1991;111:287–298. doi: 10.1242/dev.111.2.287. [DOI] [PubMed] [Google Scholar]
- Davis CA, Joyner AL. Expression patterns of the homeo box-containing genes En-1 and En-2 and the proto-oncogene int-1 diverge during mouse development. Genes & Development. 1988;2:1736–1744. doi: 10.1101/gad.2.12b.1736. [DOI] [PubMed] [Google Scholar]
- Di Bonito M, Narita Y, Avallone B, Sequino L, Mancuso M, Andolfi G, Franzè AM, Puelles L, Rijli FM, Studer M. Assembly of the auditory circuitry by a Hox genetic network in the mouse brainstem. PLoS Genet. 2013;9:e1003249. doi: 10.1371/journal.pgen.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domowicz MS, Sanders TA, Ragsdale CW, Schwartz NB. Aggrecan is expressed by embryonic brain glia and regulates astrocyte development. Dev Biol. 2008;315:114–124. doi: 10.1016/j.ydbio.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox SR, Deneris ES. Engrailed is required in maturing serotonin neurons to regulate the cytoarchitecture and survival of the dorsal raphe nucleus. J Neurosci. 2012;32:7832–7842. doi: 10.1523/JNEUROSCI.5829-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman GC, O’Leary DD. Retroviral misexpression of engrailed genes in the chick optic tectum perturbs the topographic targeting of retinal axons. J Neurosci. 1996;16:5498–5509. doi: 10.1523/JNEUROSCI.16-17-05498.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A, Santamaria P. Developmental analysis of the wing disc in the mutant engrailed of Drosophila melanogaster. Genetics. 1972;72:87–104. doi: 10.1093/genetics/72.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman RF, Covey E. Origin of ascending projections to the nuclei of the lateral lemniscus in the big brown bat, Eptesicus fuscus. J Comp Neurol. 1995;357:532–545. doi: 10.1002/cne.903570405. [DOI] [PubMed] [Google Scholar]
- Jalabi W, Kopp-Scheinpflug C, Allen PD, Schiavon E, Digiacomo RR, Forsythe ID, Maricich SM. Sound localization ability and glycinergic innervation of the superior olivary complex persist after genetic deletion of the medial nucleus of the trapezoid body. Journal of Neuroscience. 2013;33:15044–15049. doi: 10.1523/JNEUROSCI.2604-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Martin GR. En-1 and En-2, two mouse genes with sequence homology to the Drosophila engrailed gene: expression during embryogenesis. Genes & Development. 1987;1:29–38. doi: 10.1101/gad.1.1.29. [DOI] [PubMed] [Google Scholar]
- Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, Fritzsch B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::AID-CNE8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Logan C, Wizenmann A, Drescher U, Monschau B, Bonhoeffer F, Lumsden A. Rostral optic tectum acquires caudal characteristics following ectopic engrailed expression. Curr Biol. 1996;6:1006–1014. doi: 10.1016/s0960-9822(02)00645-0. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Xia A, Mathes EL, Wang VY, Oghalai JS, Fritzsch B, Zoghbi HY. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brainstem accessory auditory nuclei. J Neurosci. 2009;29:11123–11133. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs GS, Morgan WJ, Howell DM, Spirou GA, Mathers PH. Embryonic origins of the mouse superior olivary complex. Dev Neurobiol. 2013;73:384–398. doi: 10.1002/dneu.22069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Expression patterns of developmental control genes in normal and Engrailed-1 mutant mouse spinal cord reveal early diversity in developing interneurons. J Neurosci. 1997;17:7805–7816. doi: 10.1523/JNEUROSCI.17-20-07805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes JR, Luskin MB. Expression of neuron-specific tubulin defines a novel population in the proliferative layers of the developing telencephalon. J Neurosci. 1994;14:5399–5416. doi: 10.1523/JNEUROSCI.14-09-05399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Lawrence PA. Control of compartment development by the engrailed gene in Drosophila. Nature. 1975;255:614–617. doi: 10.1038/255614a0. [DOI] [PubMed] [Google Scholar]
- Nayagam DAX, Clarey JC, Paolini AG. Powerful, onset inhibition in the ventral nucleus of the lateral lemniscus. Journal of Neurophysiology. 2005;94:1651–1654. doi: 10.1152/jn.00167.2005. [DOI] [PubMed] [Google Scholar]
- Pierce ET. Time of origin of neurons in the brain stem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- Pollak GD, Gittelman JX, Li N, Xie R. Inhibitory projections from the ventral nucleus of the lateral lemniscus and superior paraolivary nucleus create directional selectivity of frequency modulations in the inferior colliculus: a comparison of bats with other mammals. Hearing Research. 2011;273:134–144. doi: 10.1016/j.heares.2010.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio-Spinoso A, Joris PX. Temporal properties of responses to sound in the ventral nucleus of the lateral lemniscus. Journal of Neurophysiology. 2014;111:817–835. doi: 10.1152/jn.00971.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rétaux S, Harris WA. Engrailed and retinotectal topography. Trends in Neurosciences. 1996;19:542–546. doi: 10.1016/s0166-2236(96)10062-x. [DOI] [PubMed] [Google Scholar]
- Riquelme R, Saldaña E, Osen KK, Ottersen OP, Merchán MA. Colocalization of GABA and glycine in the ventral nucleus of the lateral lemniscus in rat: an in situ hybridization and semiquantitative immunocytochemical study. J Comp Neurol. 2001;432:409–424. doi: 10.1002/cne.1111. [DOI] [PubMed] [Google Scholar]
- Roberts RC, Ribak CE. GABAergic neurons and axon terminals in the brainstem auditory nuclei of the gerbil. J Comp Neurol. 1987;258:267–280. doi: 10.1002/cne.902580207. [DOI] [PubMed] [Google Scholar]
- Saint Marie RL, Baker RA. Neurotransmitter-specific uptake and retrograde transport of [3H]glycine from the inferior colliculus by ipsilateral projections of the superior olivary complex and nuclei of the lateral lemniscus. Brain Research. 1990;524:244–253. doi: 10.1016/0006-8993(90)90698-b. [DOI] [PubMed] [Google Scholar]
- Saint Marie RL, Shneiderman A, Stanforth DA. Patterns of gamma-aminobutyric acid and glycine immunoreactivities reflect structural and functional differences of the cat lateral lemniscal nuclei. J Comp Neurol. 1997;389:264–276. doi: 10.1002/(sici)1096-9861(19971215)389:2<264::aid-cne6>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Schofield BR. Projections from the cochlear nucleus to the superior paraolivary nucleus in guinea pigs. J Comp Neurol. 1995;360:135–149. doi: 10.1002/cne.903600110. [DOI] [PubMed] [Google Scholar]
- Schofield BR, Cant NB. Ventral nucleus of the lateral lemniscus in guinea pigs: cytoarchitecture and inputs from the cochlear nucleus. J Comp Neurol. 1997;379:363–385. doi: 10.1002/(sici)1096-9861(19970317)379:3<363::aid-cne4>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, Joyner AL. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–2335. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HH, Saueressig H, Wurst W, Goulding MD, O’Leary DD. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. Journal of Neuroscience. 2001;21:3126–3134. doi: 10.1523/JNEUROSCI.21-09-03126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HH, Scholz C, O’Leary DDM. Engrailed genes control developmental fate of serotonergic and noradrenergic neurons in mid- and hindbrain in a gene dose-dependent manner. Mol Cell Neurosci. 2005;28:96–105. doi: 10.1016/j.mcn.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- Yavuzoglu A, Schofield BR, Wenstrup JJ. Circuitry underlying spectrotemporal integration in the auditory midbrain. Journal of Neuroscience. 2011;31:14424–14435. doi: 10.1523/JNEUROSCI.3529-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kelly JB. Responses of neurons in the rat’s ventral nucleus of the lateral lemniscus to amplitude-modulated tones. Journal of Neurophysiology. 2006;96:2905–2914. doi: 10.1152/jn.00481.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Horizontal section of E12.5 Egr2Cre; ROSAtdTomato mouse brainstem visualized for tdTomato (red) and En1 immunostaining (green). Brainstem midline is indicated by dotted white line. Solid white lines delineate boundaries of rhombomeres r3, r4 and r5. An En1+ cell group (dotted circle) is present in r4. Scale bar: 100μm.
Cresyl violet-stained coronal sections through the region of the VNLL in control adult (A), En1-null (B) and Hoxb1Cre; En1CKO mice. Sections were aligned at similar brainstem levels using the pontine nucleus (outlined in blue) and median raphe nucleus (outlined in red) as landmarks. Sections throughout the VNLL region are shown for each brain, with the most anterior sections on the left. Dotted black lines indicate the VNLL in control mice; solid black lines identify the midline. Scale bar: 400μm.