Abstract
Background
At present, we do not have any biological tests which can contribute towards a diagnosis of depression. Neuroimaging measures have shown some potential as biomarkers for diagnosis. However, participants have generally been from the same ethnic background while the applicability of a biomarker would require replication in individuals of diverse ethnicities.
Aims
We sought to examine the diagnostic potential of the structural neuroanatomy of depression in a sample of a wide ethnic diversity.
Method
Structural magnetic resonance imaging (MRI) scans were obtained from 23 patients with major depressive disorder in an acute depressive episode (mean age: 39.8 years) and 20 matched healthy volunteers (mean age: 38.8 years). Participants were of Asian, African and Caucasian ethnicity recruited from the general community.
Results
Structural neuroanatomy combining white and grey matter distinguished patients from controls at the highest accuracy of 81% with the most stable pattern being at around 70%. A widespread network encompassing frontal, parietal, occipital and cerebellar regions contributed towards diagnostic classification.
Conclusions
These findings provide an important step in the development of potential neuroimaging-based tools for diagnosis as they demonstrate that the identification of depression is feasible within a multi-ethnic group from the community.
Declaration of interests
C.H.Y.F. has held recent research grants from Eli Lilly and Company and GlaxoSmithKline. L.M. is a former employee and stockholder of Eli Lilly and Company.
Copyright and usage
© The Royal College of Psychiatrists 2016. This is an open access article distributed under the terms of the Creative Commons Non-Commercial, No Derivatives (CC BY-NC-ND) licence.
Major depressive disorder (MDD) is characterised by a persistently low mood and an inability to experience pleasure that is accompanied by disturbances in neurovegetative symptoms, in particular in sleep and appetite, impairments in cognitive functioning, and often feelings of guilt and suicidal ideation of varying intensity.1 The diagnosis is made on the basis of clinical signs and symptoms from an assessment with the individual and additional information when available. However, at present, there are no biological measures which are diagnostic for depression or for any psychiatric disorder.
Findings from neuroimaging research though offer the potential to develop biomarkers which could aid in the diagnosis of depression.2 Structural neuroimaging studies of depression have revealed widespread cortico-limbic deficits in grey matter (GM)3 as well as in white matter (WM).4 Moreover, regional atrophy in the hippocampus may already be present in the first episode of depression5 and may worsen with recurrent episodes, in particular in patients with a more treatment-resistant form of depression.6 Similarly, alterations in WM integrity5 have been observed in the first episode7 and in adolescents at familial risk.8
A number of studies have examined how well structural magnetic resonance imaging (MRI) data are able to diagnose depression. Structural MRI scans are non-invasive, may be readily acquired, and are generally well tolerated as they are relatively short scans and do not require participants to perform a specific task while in the scanner, providing an accessibility which is important for a potential biomarker.9 Using structural MRI scans, the accuracy of diagnosis for depression has ranged from 48% to 91% (Table 1).10–16 We would not expect to achieve a perfect accuracy with current diagnostic criteria that are based on clinical signs and symptoms which are not necessarily linked to aetiology.17 Rather we would expect that there would be an iterative process by which the factors contributing to the neuroimaging classification would be delineated and the classification findings would in turn lead to new hypotheses.18
Table 1. Diagnosis of major depressive disorder from structural MRI studies.
| Study, year | MRI | Healthy controls | Patients | Diagnosis | Severity –HAMD: mean (s.d.) | Medication | Comparison | Classifier | Feature | Accuracy | Sensitivity | Specificity | P | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (males) | Age, years (s.d.) | N (males) | Age, years (s.d.) | ||||||||||||
| Costafreda10 2009 | 1.5 T | 37 (9) | 42.8 (6.7) | 37 (9) | 43.2 (8.8) | MDD | 20.6 (2.2) | Med free | MDD v. HV | SVM | GM | 67.6 | 64.9 | 70.3 | 0.027 |
| Gong11 | 3 T | 23 (12) | 38.2 | 23 (10) | 39.2 (12.9) | MDD | 24.2 (3.8) | Med naïve | MDD v. HV | SVM | GM | 76.1 | 69.6 | 82.6 | <0.001 |
| 2011 | WM | 84.7 | 73.9 | 95.7 | <0.001 | ||||||||||
| GM+WM | 76.1 | 73.9 | 78.3 | <0.001 | |||||||||||
| 23 (14) | 40.4 (12.6) | TRD | 23.5 (5.4) | Med naïve | TRD v. HV | GM | 67.4 | 65.2 | 69.6 | 0.01 | |||||
| WM | 58.7 | 60.9 | 56.5 | 0.13 | |||||||||||
| GM+WM | 65.2 | 65.2 | 65.2 | 0.02 | |||||||||||
| Liu12 2012 | 1.5 T | 17 (10) | 24.2 (4.4) | 17 (10) | 26.7 (7.7) | MDD | 25.6 (6.3) | Med naïve | MDD v. HV | Searchlight- | GM | 82.4 | – | – | – |
| PCA-SVM | WM | 91.2 | – | – | – | ||||||||||
| RFE-SVM | GM | 70.6 | – | – | – | ||||||||||
| WM | 76.5 | – | – | – | |||||||||||
| LLE- C Means | GM | 76.5 | – | – | – | ||||||||||
| WM | 88.2 | – | – | – | |||||||||||
| LLE-SVM | GM | 82.4 | – | – | – | ||||||||||
| WM | 88.2 | – | – | – | |||||||||||
| 18 (11) | 27.4 (7.7) | TRD | 23.9 (3.7) | On medsa | TRD v. HV | Searchlight- | GM | 85.7 | – | – | – | ||||
| PCA-SVM | WM | 85.7 | – | – | – | ||||||||||
| RFE-SVM | GM | 77.1 | – | – | – | ||||||||||
| WM | 85.7 | – | – | – | |||||||||||
| LLE-C Means | GM | 77.1 | – | – | – | ||||||||||
| WM | 85.7 | – | – | – | |||||||||||
| LLE-SVM | GM | 77.1 | – | – | – | ||||||||||
| WM | 85.7 | – | – | – | |||||||||||
| +Mwangi13 2012 | 1.5 T | 18 (7) | 40.6 (10.3) | 15 (6) | 46.1 (12.5) | TUD | 23.2 (4.3) | On meds | TUD v. HV | VBM-FBM-SVM | GM | 90.3 | 93.3 | 87.5 | 1×10–7b |
| 14 (7) | 43.0 (13.2) | 15 (5) | 44.7 (10.0) | TRD | 27.9 (5.8) | RVM | GM | 87.1 | 86.7 | 87.5 | 1×10–7b | ||||
| ++Kipli14 | – | – | – | – | – | – | – | – | – | SVM-EM | GM+WM+CSF | 85.3 | – | – | – |
| 2013 | Information gain-Rand Tree | 85.3 | – | – | – | ||||||||||
| SVM-K Means | 82.3 | – | – | – | |||||||||||
| Serpa15 2014 | 1.5 T | 38 (8) | 29.7 (7.9) | 19 (4) | 29.1 (8.3) | pMDD | 16.1c | On medsa | pMDD v. HV | SVM | GM+WM+ventricles | 59.6 | 31.6 | 73.7 | – |
| Qiu 201416 | 3 T | 32 (23) | 35.0 (11.2) | 32 (23) | 34.9 (11.1) | MDD | 24.3 (5.1) | Med naïve | MDD v. HV | SVM | Cortical thickness | 69 | 66 | 72 | 0.002 |
| Volume | 66 | 63 | 69 | 0.005 | |||||||||||
| Plial area | 69 | 69 | 69 | 0.001 | |||||||||||
| Curvature | 48 | 47 | 50 | 0.63 | |||||||||||
| Area | 59 | 66 | 53 | 0.10 | |||||||||||
| Sulcal depth | 58 | 56 | 59 | 0.12 | |||||||||||
| Jacobian Metric Distortion | 67 | 63 | 72 | 0.002 | |||||||||||
| +++Combination parametres | 69 | 69 | 69 | 0.002 | |||||||||||
Med, Medication; HAMD, 17-item Hamilton Depression Rating Scale; MDD, major depressive disorder; TRD, treatment-resistant depression; pMDD, psychotic MDD; HV, healthy volunteers; GM, grey matter; WM, white matter; CSF, cerebrospinal fluid; SVM, support vector machines; PCA, principle component analysis; RFE, recursive feature elimination; LLE, locally linear embedding; VBM, voxel based morphometry; RVM, relevance vector machine; FBM, feature based morphometry; EM, expectation-maximisation dustering algorithm; KMeans, simple K means classification via clustering; TUD, treatment unresponsive patients.
Depression status of MDD patients: first-episode – Liu (2012), Qiu (2014), Serpa (2014); first-episode and recurrent – Costafreda (2009); recurrent: Mwangi (2012); not stated: Gong (2011), Kipli (2013).
Some of the patients were medication free.
χ2P.
31-item HAMD.
Mwangi (2012): data were randomly divided into two sets (training set, testing set) of equal number of patients and controls (n=31). In patients, depression was considered to be treatment unresponsive. Minimum duration of illness was >3 months with antidepressant medication.
Kipli (2013), accuracy of 82.3% also obtained with other classifiers: information gain:-J48, information gain-RandomForest, SVM-K Means, SVM-RandomForest, ReliefF-RandomTree, all-naïve bayes.
Combined parameters: Qiu (2014) integrated all the morphometric parameters (i.e. cortical thickness, volume, plial area, curvature area, sulcal depth and Jacobian metric distortion) of the left and right hemispheres within a single model to investigate the discriminative power of the resulting combination.
Factors which contribute to the accuracy of the generated classification include the subtype of depression examined, features of the brain imaging measure which were included, and the method of analysis. Some of the highest accuracies have been achieved in patients with a well-recognised and more treatment-resistant form of depression11,13 who were taking a number of medications.13 The identification of depression seems to show an improved accuracy with the inclusion of WM rather than that from GM alone,11,12 and neuroimaging analysis methods which bring together measures from a number of regions are better able to detect widespread, subtle anatomical changes.2,19 However, studies to date had included individuals who were predominantly from the same ethnicity: Caucasian10,13 and Chinese.11,12,16 Ethnicity is moderating factor for functional brain responses in healthy participants,20 while an effect on structural neural features may be more mixed with evidence of differences in cortical thickness in frontal and parietal regions21 but not in hippocampal asymmetry.22 Ethnicity could thus impact on potential functional MRI-based markers while structural MRI-based markers may be more robust.
In the present study, we sought to examine how well structural MRI scans would identify patients with the form of depression that is most commonly observed in the community in an ethnically diverse sample of Asian, African and Caucasian descent, who were suffering from either their first or a recurrent form of depression and were all medication-free. We applied a multivariate imaging pattern analysis method which integrates regional cerebral data, namely optimally discriminative voxel-based analysis,23,24 and we examined the accuracy of diagnosis based on GM, WM and their combination using a support vector machine (SVM) method.25 We expected that GM would demonstrate a high diagnostic accuracy for depression and that an even higher accuracy may be achieved with WM in the multi-ethnic sample.
Method
Study design and participants
This is an open-label study which was approved by the Cambridgeshire 4 Research Ethics Committee. Patients were recruited from the local community by newspaper advertisements, and all participants provided informed written consent. The study was conducted in conformity with the Declaration of Helsinki and its amendments.
Participants were patients with MDD (n=23) and healthy controls (n=20), right-handed and matched for age, gender and IQ (all P>0.3) (Table 2). A diagnosis of MDD, single or repeated episode MDD without psychotic features, was made as defined by Diagnostic Statistical Manual of Mental Disorders, Fourth Edition, Text Revision26 and assessed with the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-IV)27 without any comorbid disorders. All patients had a minimum score of 18 on the 17-item Hamilton Depression Rating Scale (HAMD)28 at the time of study entry and were free of antidepressant medication for a minimum of 4 weeks before the start of the study (or 6 weeks for fluoxetine). Healthy controls had no history of psychiatric disorders, interviewed with SCID-IV,27 and had an HAMD score of ≤7. MRI scans were acquired at baseline (week 0) and at weeks 1, 8 and 12. MDD patients received treatment with an antidepressant medication following the baseline assessment.29
Table 2. Demographic and clinical characteristics.
| MDD patients | Healthy controls | P | |
|---|---|---|---|
| Participants, n | 23 | 20 | |
| Age, years: mean (s.d.) | 39.8 (11.2) | 38.8 (9.9) | 0.84 |
| Male, n (%) | 13 (56.5) | 12 (60.0) | 0.82 |
| Ethnicity, n (%) | |||
| Asian | 8 (34.8%) | 3 (15.0%) | |
| Black/African American | 2 (8.7%) | 5 (25.0%) | |
| Caucasian | 13 (56.5%) | 12 (60.0%) | |
| Baseline scores,a mean (s.d.) | |||
| HAMD | 22.0 (2.9) | 0.5 (1.1) | <0.001 |
| HAMA | 20.7 (5.4) | n/a | |
| Week 12 scores,a mean (s.d.) | |||
| HAMD | 6.9 (4.6) | 0.6 (1.3) | <0.001 |
| HAMA | 7.5 (4.4) | 0.6 (1.1) | <0.001 |
| Full IQ | 107.8 (10.7) | 109.2 (14.6) | 0.63 |
| Performance IQ | 103.2 (14.4) | 107.9 (15.2) | 0.3 |
| Verbal IQ | 110.0 (9.9) | 109.8 (12.4) | 0.95 |
HAMA, Hamilton Scale for Anxiety; HAMD, 17-item Hamilton Rating for Depression; MDD, major depressive disorder; n/a, not assessed.
Ratings were acquired at baseline, during an acute depressive episode and following 12 weeks of treatment for patients, and at baseline and at the 12-week follow-up for healthy participants.
The present study focuses on the baseline data while MDD patients were in an acute depressive episode. Baseline MRI scans were obtained from 29 MDD patients and 22 healthy controls. Data from two healthy volunteers had to be excluded due to excessive movement during the structural MRI scan. The present study consists of those participants who had completed all the scans in the study: 23 MDD and 20 healthy controls, as the SVM analysis requires approximately equal numbers of participants.
Data acquisition
Structural MRI scans were acquired on a 3-T GE SIGNA HDx (Milwaukee, USA) at King’s College London. A structural image was acquired at each session: Magnetisation Prepared Gradient Echo, resolution 1 mm3, acquisition parameters: echo time (TE): 2.85 ms, reaction time (TR): 6.98 ms, flip angle: 8°, slice thickness: 1.2 mm, interslice gap: 1.2 mm, field of view: 260 mm, matrix size: 64×64.
Image preprocessing
Preprocessing of the structural MRI T1 weighted images included bias correction, skull stripping and tissue segmentation. Bias correction was performed using N41TK, which is an improved MNI_N3 bias correction software package available at http://www.insight-journal.org/browse/publication/640package. Skull stripping was completed using Multi-Atlas Skull Stripping software (MASS, version 1.0), which is based on a multi-atlas registration framework and uses a set of templates from the study data set which best represents the anatomical variations.30 The images were then segmented into GM, WM and cerebrospinal fluid (CSF) using the automated algorithm Multiplicative Intrinsic Component Optimization (MICO).31 The quality of the processed images was ensured by automated quality control measures and manual review.
Spatial registration of all the skull-stripped images to the Jakob template was performed using the Deformable Registration via Attribute Matching and Mutual-saliencing Matching registration package (DRAMMS, version 1.1.0; http://www.cbica.upenn.edu/sbia/software/dramms/index.html).32 The deformation field from the resulting registration was used to obtain the Regional Analysis of Volumes Examined in Normalized Space (RAVENS) maps or regional volumetric maps for GM, WM and CSF. The maps were corrected for individual intracranial volume, down-sampled to 2×2×2 mm3.
Analysis of structural neuroanatomy: GM and WM
Regional differences in GM and WM between MDD patients and healthy controls were performed with the Optimally Discriminative Voxel Based Analysis (ODVBA, version 2.0) software package23 using the RAVENS maps. In ODVBA, the regional discriminative analysis which is restricted by the non-negativity constraints is firstly applied to a spatial neighbourhood around each voxel in order to identify the coefficients that best highlight the differences between two groups in that neighbourhood. Secondly, the statistical value of each voxel is then determined by a composition of contributions from all neighbourhoods to which it belongs. Finally, the statistical significances are obtained by using permutation tests. In the present study, 2000 permutations were used to derive the significances, and significance was assigned as P (uncorrected)<0.001 because of the relatively small sample size. ODVBA has shown greater sensitivity to detect subtle structural abnormities and improved delineation of the region of abnormality as compared with conventional GLM methods23,24 in various clinical studies.33,34
Classification using SVM
The GM and WM RAVENS maps were concatenated into a single feature vector for each participant. Classification was performed using SVM,35 a multivariate classification technique that can optimally use high dimensional information such as neuroimaging data.10,19 SVM identifies the optimal linear decision boundary, or hyperplane, that has the maximum margin separating the two training groups, namely MDD patients and healthy controls. SVM treats individual images as points located in high dimensional space. In SVM, both the hyperplane as well as the margin are important in classification accuracy, and usually the wider the margin the better the classification accuracy.35
SVM also extracts weight vectors as images, known as SVM discrimination maps, which represent the direction in which the two groups differ the most, and the significance levels were determined using analytical permutation testing.25 We used an advanced version which accounted for the SVM margins in addition to the SVM weights.36 The SVM analysis was performed using fivefold cross-validation strategy. The classification scores derived from the SVM analysis was evaluated using a receiver operating characteristic (ROC) curve to illustrate the diagnostic accuracy of the classifier. The area under the curve (AUC) was calculated from the ROC curve, which is a measure of the discriminative power of the classifier and is independent of the chosen P value or sample size.
The GM and WM RAVENS maps were also analysed using the SVM-based classification technique Classification of Morphological Patterns using Adaptive Regional Elements (COMPARE).37 The first step in this method is feature selection wherein regions that show high correlation between RAVENS maps and participant groups are extracted using a watershed algorithm. A further volume-increment algorithm is then applied to these regions to extract regional volumetric features, from which a feature selection method based on SVM classification criteria is used to identify the most relevant features for classification.37 The feature selection procedure produces a small number of volumetric measurements for more effective classification. The SVM classification technique with the most distinguishing features is then used to predict group differences using the leave-one-out cross-validation strategy.37
Results
Structural neuroanatomy
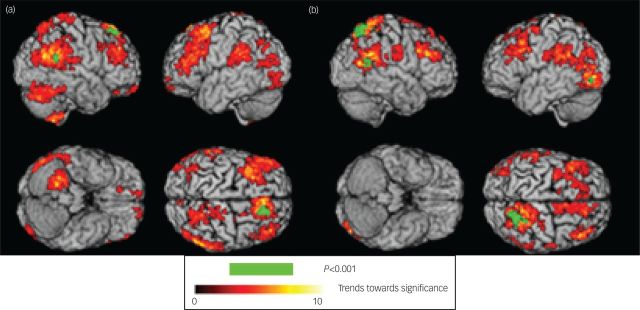
In GM density, reductions were revealed in the right superior frontal region (Talairach coordinates: x, y, z: 17.8, 31.5, 48.2; mass size: 74 voxels), right superior medial frontal region (x, y, z: 11.9, 31.7, 51.8; mass size: 14 voxels) and superior temporal regions (x, y, z: 53.5, −43.6, 20.6; mass size: 13 voxels) in MDD patients compared with healthy controls (P<0.001, uncorrected), and in WM density, reductions were evident in the right postcentral (x, y, z: 25.7, −41.8, 57.4; mass size: 14 voxels), superior parietal (x, y, z: 21.8, −57.3, 58.1; mass size: 212 voxels), middle temporal (x, y, z: 49.5, −51.6, 17.3; mass size: 46 voxels) and left inferior occipital regions (x, y, z: −37.6, −75.8, −1.3; mass size: 10 voxels) in MDD patients relative to healthy controls (P<0.001, uncorrected) (Fig. 1). There were no regions which showed greater volume in MDD patients relative to controls.
Fig. 1. Map of regions which showed a significant difference in grey and white matter in MDD patients and healthy controls: (a) grey matter regions demonstrating atrophy in MDD patients relative to healthy controls, (b) white matter regions demonstrating atrophy in MDD patients relative to healthy controls. Green indicates significant regions at P<0.001 uncorrected, and areas of red colour (threshold P<0.05) indicates the trend towards significance characterised by –log(p) values as shown in the colour bar.
Diagnostic classification
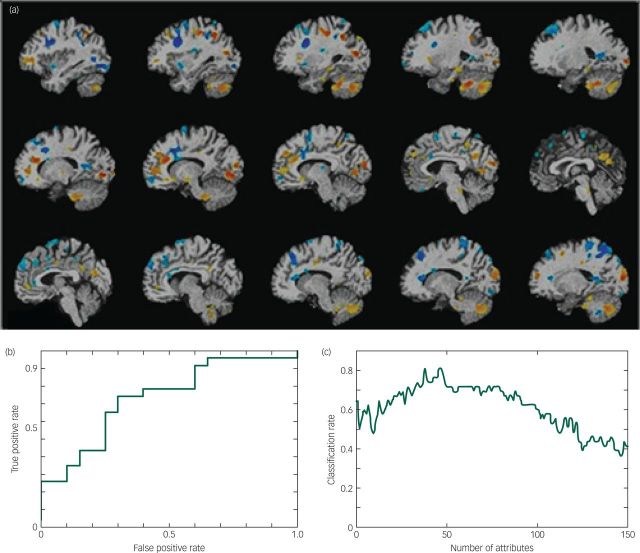
Using SVM analysis, 78.3% of MDD patients (sensitivity: 18 of 23 patients) and 55.0% of healthy controls (specificity: 11 of 20 healthy controls) for an overall accuracy of 67.4% (AUC=0.73, P=0.02, positive predictive value=66.67%, negative predictive value=68.75%) were correctly classified based on the combination of GM and WM density (Fig. 2). Based on GM only, the accuracy was reduced to 60.5%, which did not reach statistical significance (AUC=0.55, P=0.2), while the accuracy was higher at 65.1% (AUC=0.73, P=0.05) based on WM only. Regions which contributed to the classification of MDD patients included WM regions in the superior and medial frontal gyri, superior parietal and inferior occipital gyri, and in the cerebellum (Fig. 2).
Fig. 2. (a) Schematic map showing white matter regions which contributed towards diagnostic classification of MDD, regions are presented at P<0.05 uncorrected. Blue indicates regions showing atrophy in MDD patients relative to controls and yellow indicates regions of greater volume in MDD patients compared with healthy controls. (b) Receiver operating characterstic (ROC) curve for the comparison between MDD and healthy participants, area under curve (AUC)=0.73, P=0.02. The x-axis is the false positive rate (1-specificity) and the y-axis is the true positive rate (sensitivity). (c) Graph illustrating change in classification rate with number of attributes selected. The x-axis indicates the number of attributes and the y-axis the classification rate. The highest classification rate was 81.4% based on 47 features, while the most stable pattern was observed with an accuracy of around 70% based on 50–70 features.
From the COMPARE analysis which included feature selection, the highest classification rate was 81.4% based on 47 features, while the most stable pattern was observed with an accuracy of around 70% based on 50–70 features (Fig. 2).
Discussion
At present, the diagnosis of depression is based solely on clinical signs and symptoms, and there are no biological markers that can identify the disorder or predict clinical response. Using structural neuroimaging measures and machine-learning analysis, we have identified a potential non-invasive biomarker which can aid in the diagnosis of MDD in patients from a wide ethnic diversity from a community sample. Moreover, the features of depression which are commonly observed in the community were reflected in the present group as all the patients were in an acute depressive episode, were not taking any medications, and were suffering from some degree of psychosocial impairments. These findings provide an important step in the development of potential neuroimaging-based tools for clinical diagnosis as the findings demonstrate that the identification of depression is feasible within a multi-ethnic group from the general community.
It was notable that the highest accuracy was achieved from combining GM and WM features in the structural MRI scans. Regions which contributed to classification included a widespread network encompassing the superior and medial frontal, superior parietal and inferior occipital regions. MDD patients showed GM atrophy in the right superior frontal and temporal regions as well as loss of WM density in the parietal, inferior occipital and lateral temporal regions compared with the healthy controls. Deficits in GM have been consistently reported in the prefrontal regions extending into the precentral cortex and subcortically into the thalamus.3
Abnormalities in WM have been observed as reductions in volume as well as in fractional anisotropy, a measure of connectivity that is derived from diffusion tensor imaging, also extending from frontal to parietal and occipito-temporal regions,38 which may be associated with more severe symptoms.4 A higher accuracy of diagnosis has been reported in patients with a greater severity of illness and of the same ethnicity.11,12 Similar regions had contributed to the diagnosis, namely WM regions in the frontal, parietal, and occipital lobes.11,12 We also found that WM regions in the bilateral cerebellum contributed towards diagnostic classification, which has been previously observed12 but more frequently has not been examined.14–16 The cerebellum is involved in the regulation of emotional responses,39 and GM40 as well as WM41 deficits have been reported in depression. These findings underline the wide distribution of GM and WM abnormalities within fronto-limbic networks in depression, supporting the necessity of analyses which are capable of combining these features, rather than examining each region individually.
Ethnicity has a significant impact on the development and prognosis of psychiatric and neurological disorders, which may be reflected in their neuropathology.42 Perceived discrimination has been associated with greater activation and increased connectivity with the anterior cingulate cortex in healthy participants from an ethnic minority which was in part mediated by chronic stress.20 Effects on structural neuroanatomy may be more mixed as no significant effects were observed of ethnicity on hippocampal asymmetry in a community sample22 or on structural brain volumes when young Chinese Singaporeans were compared with White Americans,21 although differences in cortical thickness were reported as young White Americans had greater thickness in frontal and right superior parietal regions compared with Singaporeans, who in turn showed increased thickness in the left inferior temporal gyrus.21 Moreover, a comparison of hemispheric shape between Eastern and Western populations revealed shorter but wider hemispheres for Japanese relative to European participants.43 In multiple sclerosis, greater T1-weighted lesion volumes have been reported in patients of African compared with Caucasian ethnicity.44 Our present findings indicate potential reproducibility in the structural neuroanatomy in depression from diverse ethnicities.
Structural neuroimaging has also demonstrated a comparable accuracy for the diagnosis of schizophrenia, revealing an accuracy of 78% from a meta-analysis in comparison with healthy controls.45 In the present study, we achieved an accuracy of 81%, although the most stable findings were at an accuracy of around 70%. We would expect a higher accuracy for the diagnosis of schizophrenia, which is associated with greater global brain volume reductions, extensive regional atrophy as well as WM disruptions.46 Furthermore, we would not expect to fully replicate current diagnostic criteria systems. The process of developing potential neuroimaging-based biomarkers would involve an iterative process, in which the findings would lead to new hypotheses and would be incorporated as prior knowledge for subsequent studies.18
The majority of machine-learning analyses of neuroimaging measures in depression have examined the predictive potential of structural neuroanatomy and functional neural correlates for diagnosis and prognosis using SVMs.18 Major advantages of structural MRI measures is the ease with which the data can be acquired, its reliability and reproducibility, as well as its acceptability by patients. While we would not expect that neuroimaging-based diagnostic classification would wholly reproduce current symptom-based classifications, we would emphasise the need for biological markers to aid in the diagnosis of depression and other psychiatric disorders. This would be of particular importance for patients who may not be able to participate in the clinical assessment, who are unable to provide a sufficient clinical history, or when the specific diagnosis is unclear. Examining a single neuroimaging measure though may not fully encompass the multitudinous aspects of the disorder’s complex psychopathology. Moreover, the specificity of the diagnostic classification could have implications for healthy individuals. We would expect that the specificity of the classification would improve in an iterative process in the development of the potential biomarker.18 However, if the specificity were to remain low, this would have implications for the clinical applicability of the biomarker as well as for those healthy individuals who have structural neural features that are comparable with individuals with a diagnosis of depression, analogous to having genetic polymorphisms that are associated with increased risk for the development of a particular disorder or malignancy. Future longitudinal studies are needed to clarify the accuracy of diagnosis and could likely benefit from examining multiple neuroimaging measures as well as different machine-learning techniques. The challenge is to look for valid biomarkers of symptom conditions, perhaps incorporating neural and genetic measures. Such biomarkers could aid clinical interviews and decisions regarding diagnosis, prognosis and disease progression.
A limitation of the present study is the relatively small sample size, although comparable to previous studies.11–13 In order to develop a diagnostically meaningful tool, it is essential to ensure unbiased testing of the model. Ideally, the model should be tested in an independent sample of patients with MDD to evaluate and to validate the generalisability of the model. However, a significant challenge of the analysis of data sets from different scanners is the systematic difference in the scanner protocols and the acquired images, and an important step in the analysis would be to harmonise the data from the different scanners. In the present study, we applied a fivefold cross-validation strategy, by which the whole data set was randomly divided into five exclusive subsets of equal size, and for each fold one of these subsets was used as a test set, while the remaining four were used as training data sets. Furthermore, this process was repeated a thousand times with randomly permuted class labels in order to determine the significance of the classification accuracy. As we achieved converging results from both an SVM model on the imaging data and the COMPARE high-dimensional feature extraction and classification method, and we applied proper cross-validation in the analysis, we believe that our models provide unbiased and robust findings. Advantages of the current sample include that all patients were medication-free status while in an acute depressive episode and were recruited from the community, as one of the highest classification accuracy was observed in patients who were already on antidepressant medications;13 patients were in their first episode11,16 as well as having a history of recurrent episodes;10 and there was wide ethnicity which included Asian, African and Caucasian participants. Another concern is the access and cost of the MRI scan and the computational requirements of the analysis. We would envisage a database and analysis centre which the data could be sent to and the centre would provide a measure of the likelihood of the diagnosis, as well as the prognosis, and would form part of the clinical assessment and management.
In summary, we found that the diagnosis of MDD is feasible to a high level of accuracy in patients from a wide ethnic diversity. Diagnosis was based on a widespread network of WM as well as GM. The identification of individual patients with depression from an ethnically diverse, community sample indicates the generalisability of the diagnostic neural pattern and provides a valuable step in the development of diagnostic biomarkers. Multicentre validation studies would be important to test the robustness of the present findings.
Acknowledgements
We thank the participants, consultant psychiatrists, radiographers and study coordinators for their assistance in the study.
References
- 1.American Psychiatric Association. The Diagnostic and Statistical Manual of Mental Disorders. Fifth Edition (DSM-5) APA, 2013. [Google Scholar]
- 2.Fu CH, Steiner H, Costafreda SG. Predictive neural biomarkers of clinical response in depression: a meta-analysis of functional and structural neuroimaging studies of pharmacological and psychological therapies. Neurobiol Dis 2013; 52: 75–83. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson L, Sankar A, Adams TM, Fu CH. Recent advances in neuroimaging of mood disorders: structural and functional neural correlates of depression, changes with therapy, and potential for clinical biomarkers. Curr Treat Option Psychiatry 2014; 1: 278–93. [Google Scholar]
- 4.Cole J, Chaddock CA, Farmer AE, Aitchison KJ, Simmons A, McGuffin P, et al. White matter abnormalities and illness severity in major depressive disorder. Br J Psychiatry 2012; 201: 33–9. [DOI] [PubMed] [Google Scholar]
- 5.Cole J, Costafreda SG, McGuffin P, Fu CH. Hippocampal atrophy in first episode depression: a meta-analysis of magnetic resonance imaging studies. J Affect Disord 2011; 134: 483–7. [DOI] [PubMed] [Google Scholar]
- 6.MacQueen G, Frodl T. The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research? Mol Psychiatry 2011; 16: 252–64. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X, Wang X, Xiao J, Zhong M, Liao J, Yao S. Altered white matter integrity in first-episode, treatment-naive young adults with major depressive disorder: a tract-based spatial statistics study. Brain Res 2011; 1369: 223–9. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Fan X, Williamson DE, Rao U. White matter changes in healthy adolescents at familial risk for unipolar depression: a diffusion tensor imaging study. Neuropsychopharmacol 2011; 36: 684–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69: 89–95. [DOI] [PubMed] [Google Scholar]
- 10.Costafreda SG, Chu C, Ashburner J, Fu CH. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PLoS One 2009; 4: e6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Q, Wu Q, Scarpazza C, Lui S, Jia Z, Marquand A, et al. Prognostic prediction of therapeutic response in depression using high-field MR imaging. Neuroimage 2011; 55: 1497–503. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, Guo W, Yu D, Gao Q, Gao K, Xue Z, et al. Classification of different therapeutic responses of major depressive disorder with multivariate pattern analysis method based on structural MR scans. PLoS One 2012; 7: e40968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mwangi B, Ebmeier KP, Matthews K, Steele JD. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain 2012; 135: 1508–21. [DOI] [PubMed] [Google Scholar]
- 14.Kipli K, Kouzani AZ, Hamid IRA. Investigating machine learning techniques for detection of depression using structural MRI volumetric features. Int J Biosci Biochem Bioinform 2013; 3: 444–8. [Google Scholar]
- 15.Serpa MH, Ou Y, Schaufelberger MS. Neuroanatomical classification in a population-based sample of psychotic major depression and bipolar I disorder with 1 year of diagnostic stability. Bio Med Res Int 2014; 2014: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu L, Huang X, Zhang J, Wang Y, Kuang W, Li J, et al. Characterization of major depressive disorder using a multiparametric classification approach based on high resolution structural images. J Psychiatry Neurosci 2014; 39: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research Domain Criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry 2010; 167: 748–51. [DOI] [PubMed] [Google Scholar]
- 18.Fu C, Costafreda SG. In review neuroimaging-based biomarkers in psychiatry: clinical opportunities of a paradigm shift. Can J Psychiatry 2013; 58: 499–508. [DOI] [PubMed] [Google Scholar]
- 19.Fu CH, Mourao-Miranda J, Costafreda SG, Khanna A, Marquand AF, Williams SC, et al. Pattern classification of sad facial processing: toward the development of neurobiological markers in depression. Biol Psychiatry 2008; 63: 656–62. [DOI] [PubMed] [Google Scholar]
- 20.Akdeniz C, Tost H, Streit F, Haddad L, Wüst S, Schäfer A, et al. Neuroimaging evidence for a role of neural social stress processing in ethnic minority-associated environmental risk. JAMA Psychiatry 2014; 71: 672–80. [DOI] [PubMed] [Google Scholar]
- 21.Chee MWL, Zheng H, Goh JO, Park D, Sutton BP. Brain structure in young and old East Asians and Westerners: comparisons of structural volume and cortical thickness. J Cogn Neurosci 2011; 23: 1065–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucarelli RT, Peshock RM, McColl R, Hulsey K, Ayers C, Whittemore AR, et al. MR imaging of hippocampal asymmetry at 3T in a multiethnic, population-based sample: results from the Dallas Heart Study. Am J Neuroradiol 2013; 34: 752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Davatzikos C. ODVBA: optimally-discriminative voxel-based analysis. IEEE Trans Med Imaging 2011; 30: 1441–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, Davatzikos C. Optimally-discriminative voxel-based morphometry significantly increases the ability to detect group differences in schizophrenia, mild cognitive impairment, and Alzheimer’s disease. NeuroImage 2013; 79: 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaonkar B, Davatzikos C. Analytic estimation of statistical significance maps for support vector machine based multi-variate image analysis and classification. NeuroImage 2013; 78: 270–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) APA, 2000. [Google Scholar]
- 27.First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV Axis I (SCID-I), Clinician Version. APA, 1997. [Google Scholar]
- 28.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry 1960; 23: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu CH, Costafreda SG, Sankar A, Adams TM, Rasenick MM, Liu P, et al. Multimodal functional and structural neuroimaging effects in major depressive disorder before and after treatment with duloxetine. BMC Psychiatry 2015; 15: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-atlas skull-stripping. Acad Radiol 2013; 20: 1566–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Gore JC, Davatzikos C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn Reson Imaging 2014; 32: 913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 2011; 15: 622–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang T, Koutsouleris N, Meisenzahl E, Davatzikos C. Heterogeneity of structural brain changes in subtypes of schizophrenia revealed using magnetic resonance imaging pattern analysis. Schizophr Bull 2015; 41: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erus G, Battapady H, Zhang T, Lovato J, Miller ME, Williamson JD, et al. Spatial patterns of structural brain changes in type 2 diabetic patients and their longitudinal progression with intensive control of blood glucose. Diabetes Care 2015; 38: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vapnik V, Cortes C. Support-vector networks. Mach Learn 1995; 20: 273–97. [Google Scholar]
- 36.Goankar B, Shinohara MT, Davatzikos C. Interpreting support vector machine models for multivariate group wise analysis in neuroimaging. Med Image Anal 2015; 24: 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. COMPARE: Classification of Morphological Patterns Using Adaptive Regional Elements. IEEE Trans Med Imaging 2007; 26: 93–105. [DOI] [PubMed] [Google Scholar]
- 38.Liao Y, Huang X, Wu Q, Yang C, Kuang W, Du M, et al. Is depression a disconnection syndrome? Meta-analysis of diffusion tensor imaging studies in patients with MDD. J Psychiatry Neurosci 2013; 38: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blatt GJ, Oblak AL, Schmahmann JD. Cerebellar connections with limbic circuits: anatomy and functional implications In Handbook of the Cerebellum and Cerebellar Disorders (eds Manto M, Schmahmann JD, Rossi F, Gruol DL, Koibuchi N): 479–96. Springer, 2013. [Google Scholar]
- 40.Frodl TS, Koutsouleris N, Bottlender R, Born C, Jäger M, Scupin I, et al. Depression-related variation in brain morphology over 3 years: effects of stress? Arch Gen Psychiatry 2008; 65: 1156–65. [DOI] [PubMed] [Google Scholar]
- 41.Zeng LL, Liu L, Liu Y, Shen H, Li Y, Hu D. Antidepressant treatment normalizes white matter volume in patients with major depression. PLoS One 2012; 7: e44248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikram UZ, Snijder MB, Fassaert TJ, Schene AH, Kunst AE, Stronks K. The contribution of perceived ethnic discrimination to the prevalence of depression. Eur J Public Health 2014; 25: 243–8. [DOI] [PubMed] [Google Scholar]
- 43.Zilles K, Kawashima R, Dabringhaus A, Fukuda H, Schormann T. Hemispheric shape of European and Japanese brains: 3-D MRI analysis of intersubject variability, ethnical, and gender differences. Neuroimage 2001; 13: 262–71. [DOI] [PubMed] [Google Scholar]
- 44.Howard J, Battaglini M, Babb JS, Arienzo D, Holst B, Omari M, et al. MRI correlates of disability in African-Americans with multiple sclerosis. PLoS One 2012; 7: e43061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kambeitz J, Kambeitz-Ilankovic L, Leucht S, Wood S, Davatzikos C, Malchow B, et al. Detecting neuroimaging biomarkers for schizophrenia: a meta-analysis of multivariate pattern recognition studies. Neuropsychopharmacol 2015; 40: 1742–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bora E, Fornito A, Radua J, Walterfang M, Walterfang M, Seal M, et al. Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res 2011; 127: 46–57. [DOI] [PubMed] [Google Scholar]