Abstract
Purpose
Chemotherapy with platinum compounds and gemcitabine is frequently used in first-line treatment of advanced non-small cell lung cancer (NSCLC) patients in which tyrosine kinase inhibitors (EGFR or ALK) cannot be administered. Unfortunately, less than half of the patients achieve the benefit from chemotherapy. Gemcitabine is an analog of deoxycytidine (pyrimidine antimetabolite) with antitumor activity. The excess of deoxycytidine synthesized by RRM1 enzyme activity may be a cause of competitive displacement of gemcitabine, which reduces the efficacy of this cytostatic. The aim of this study was to determine the association between single nucleotide polymorphisms (SNPs) of the RRM1 promoter (−37C>A, −524C>T) and the effectiveness of first-line chemotherapy based on platinum compounds and gemcitabine in NSCLC patients.
Patients and methods
SNPs were determined by SNaPshot PCR® in DNA isolated from peripheral blood of 91 NSCLC patients.
Results
The median progression-free survival (PFS) was significantly longer in carriers of AA (−37C>A) as well as CC (−524C>T) genotype of RRM1 compared to patients with other genotypes (10.5 vs 3.5 months, p = 0.0437; HR = 2.17, 95 % CI 1.02–4.62 and 10.5 vs 3.5 months, p = 0.0343; HR = 2.12, 95 % CI 1.06–4.27). In addition, the CC genotype carriers (−37C>A) showed a significant increase in the risk of shortening overall survival (OS) in comparison to patients with AA or AC genotypes (9.5 vs 18 months, p = 0.0193; HR = 2.13, 95 % CI 1.13–4.03).
Conclusions
Presence of rare AA (−37C>A) and CC (−524C>T) genotypes of the RRM1 may be favorable predictive factors for chemotherapy with platinum compounds and gemcitabine in NSCLC patients.
Keywords: RRM1, Chemotherapy, Gemcitabine, Platinum compounds, Non-small cell lung cancer
Introduction
Non-small cell lung cancer (NSCLC, representing approx. 85 % of lung cancer cases) is still the most common cause of death (1.6 million in 2012) due to malignancies in developed countries [1]. Despite the dynamic development of medicine, the chemotherapy based on platinum compounds (cis- or carboplatin) and third generation drugs (e.g. gemcitabine) still remains standard regimen of the first-line treatment of advanced NSCLC. After administration of standard first-line chemotherapy, the objective response rates (ORR) varies between 20 and 30 %. Chemotherapy provides slight prolongation of patients’ survival time (1.5 months compared to the best supportive care, BSC); however, its use is associated with occurrence of relevant toxicity. Median overall survival (OS) of patients treated by systemic therapy range from 6 to 12 months [2]. Identification of driver mutations or other gene alterations (e.g. activating mutations in the EGFR and ALK rearrangements), which are potential molecular targets, seems to be an important advancement in optimization and individualization of NSCLC therapy. These drugs significantly improved treatment outcomes (over 60 % ORR, progression free survival (PFS) prolonged to 10 months) with acceptable toxicity [3–6]. Only 10–30 % (EGFR mutations) or 3–7 % (ALK rearrangements) Caucasian patients with advanced non-squamous NSCLC have molecular alterations and should be treated by target therapy. The lack of identified molecular targets for squamous cell carcinoma treatment is the reason why majority of patients still receive standard chemotherapy [7–9]. On the other hand, genetic predisposition (e.g. gene polymorphisms or mRNA expression) may be used to selection for potentially the most effective treatment regimens, which can prolong the life of patients and improve its quality [10–12]. Among the molecular changes, potentially the highest impact on the efficacy of chemotherapy have alterations of genes, which coding proteins involved in drug metabolism. Gemcitabine is frequently used in the treatment of NSCLC, ovarian and pancreatic cancer. Its mechanism of action is based on the incorporation to nucleic acids, which consequently induce apoptosis. The excess of deoxycytidine, biosynthesized with the participation of RRM1 causes competitive displacement of gemcitabine, reducing the efficacy of this chemotherapeutic agent [13, 14]. Some researchers have demonstrated that in patients with NSCLC the expression or SNPs of RRM1 may play prognostic and predictive role (e.g. for gemcitabine). The influence of single nucleotide polymorphisms (SNPs) on survival and the response to treatment with platinum compounds and gemcitabine in patients with advanced NSCLC are still not fully understood. Among the already known RRM1 SNPs, −37C>A (rs12806698) and −524C>T (rs11030918) seem to have the greatest importance as potential predictors of treatment regimens based on gemcitabine in NSCLC patients [15–17].
The aim of this study was to determine the association between SNPs of RRM1 promoter (−37C>A, −524C>T) and the effectiveness of chemotherapy based on platinum compounds and gemcitabine in patients with inoperable or advanced NSCLC.
Materials and methods
The study was performed on 91 Caucasian patients with inoperable, locally advanced or metastatic NSCLC (IIIB and IV), treated from 2010 to 2013 at the Department of Pneumonology, Oncology and Allergology, Medical University of Lublin. NSCLC diagnosis was based on histopathological or cytological examination. In the first-line treatment all patients received standard chemotherapy, based on platinum compounds and gemcitabine. The stage of disease was evaluated according to the TNM classification (VII edition by UICC). The median number of cycles of first-line chemotherapy was 4 (range 2–6). Subsequent lines of therapy: second or third were used in 56.1 and 16.5 % of patients, respectively (Table 1). Response to treatment was evaluated by RECIST V1.1 (Response Evaluation Criteria in Solid Tumors). Adverse events were estimated by Common Toxicity Criteria for Adverse Events (CTCAE) V4.0.
Table 1.
Patient characteristics
| Variable | Study group (n = 91) |
|---|---|
| Sex | |
| Male | 61 (67 %) |
| Female | 30 (33 %) |
| Age (years) | |
| Median | 62 |
| Mean ± SD | 62.5 ± 7.9 |
| Range | 38–78 |
| Smoking status (pack-years) | |
| Median | 30 |
| Mean ± SD | 31.4 ± 9.5 |
| Non-smokers | 5 (5.5 %) |
| Current smokers | 65 (71.4 %) |
| Former Smokers | 20 (22 %) |
| No data | 1 (1.1 %) |
| Histopathological diagnosis | |
| Adenocarcinoma | 46 (50.5 %) |
| Squamous cell carcinoma | 14 (15.4 %) |
| Large cell carcinoma | 16 (17.6 %) |
| NOS (not otherwise specified) | 15 (16.5 %) |
| Stage of disease | |
| IIIB | 28 (30.8 %) |
| IV | 63 (69.2 %) |
| Performance status | |
| PS = 0 | 12 (13.2 %) |
| PS ≥ 1 | 79 (86.8 %) |
| Weight loss before CTH | |
| Yes | 39 (42.9 %) |
| No | 43 (47.2 %) |
| No data | 9 (9.9 %) |
| Anemia before CTH | |
| Yes | 59 (64.8 %) |
| No | 32 (35.2 %) |
| Side effect after I line CTH | |
| Yes | 59 (64.8 %) |
| No | 24 (26.4 %) |
| No data | 8 (8.8 %) |
| Subsequent lines of treatment | |
| Yes | 51 (56.1 %) |
| No | 40 (43.9 %) |
| Second-line CTH (monotherapy) | 51 (56.1 %) |
| ERL | 12 (13.2 %) |
| PEM | 26 (28.6 %) |
| DCX | 13 (14.3 %) |
| Third-line CTH (monotherapy) | 15 (16.5 %) |
| ERL | 5 (5.5 %) |
| PEM | 6 (6.6 %) |
| DCX | 4 (4.4 %) |
ERL erlotinib, DCX docetaxel, PEM pemetrexed
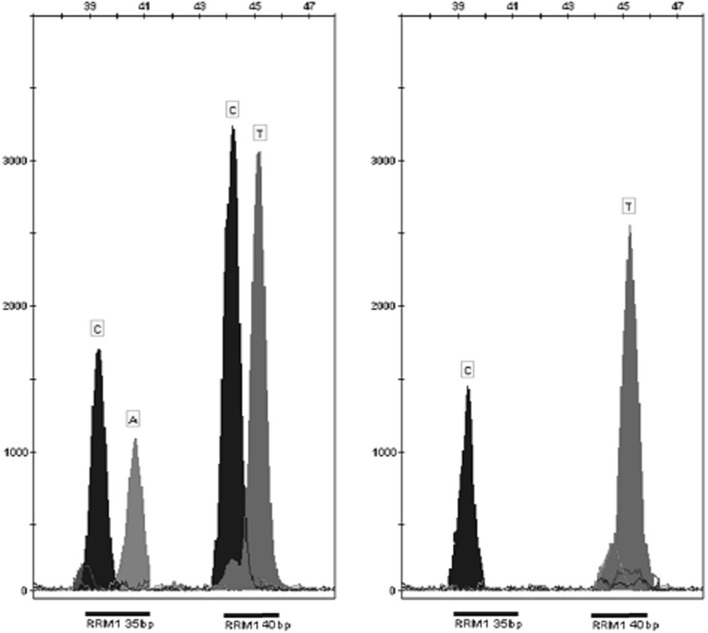
The isolation of DNA from peripheral blood leukocytes was performed using DNA Blood Mini Kit (Qiagen, Canada). Quality and quantity of extracted DNA were measured using a spectrophotometer BioPhotometer plus in cuvette equipped with UV/VIS filters (Eppendorf, Germany). Analysis of SNPs was conducted using the mini-sequencing technique (SNaPshot® PCR). For the reaction, a set of ABI PRISM SNaPshot® Multiplex (Life Technologies, USA) was used. An example of the results of genotyping is shown in Fig. 1.
Fig. 1.
Example of genotyping results of RRM1 gene obtained by capillary electrophoresis of the SNaPshot PCR products. From left (−37C>A and −524C>T respectively): AC and TT heterozygotes, CC and TT homozygotes
Statistical analysis
Statistical analysis was done using Statistica 10 (Statsoft, USA) and MedCalc 10 (MedCalc Software, Belgium). The results of p < 0.05 were considered to be statistically significant. Using the Chi-Square (χ2) balance of the Hardy–Weinberg (HW) equilibrium, associations between a series factors with the distribution of RRM1 polymorphisms were calculated. The Kaplan–Meier method was used to draw a comparison curve evaluating the survival probability (PFS and OS). Cox regression model with a stepwise selection with minimum AIC factor (Akaike Information Criterion) was used to determine the influence of clinical and genetic factors on survival. The median survival in the groups was compared by the use of the U-Mann–Whitney test.
Results
Distributions of genotypes in the RRM1 (−37C>A, −524C>T) did not depend on factors such as gender, age, histological type, stage of disease, performance status or smoking status (Table 2). The distribution of genotypes within the SNP −524C>T, unlike the −37C>A, was in Hardy–Weinberg equilibrium (p = 0.0966, χ2 = 2.7612 for −524C > T and p = 0.0279, χ2 = 4.8336 for −37C>A). Genotypes AA, AC and CC of RRM1 (−37C>A) occurred respectively in 5.7, 54.5 and 39.8 % of patients. Genotypes CC, CT and TT of RRM1 (−524C>T) occurred, respectively, in 7.7, 52.7 and 39.6 % of patients. Due to the fact that studied SNPs are located in the same gene (on chromosome 11) using the online tool SNAP (Broad Institute, USA) and data from the International HapMap project assesses coupling allele linkage disequilibrium (LD), LD was calculated based on data from population-based genetic studies (Utah state residents whose ancestors came from the north-western regions of Europe–CEU). Analyzed pair of SNPs has not reached the correlation coefficient R2 > 0.8 so, we concluded that they do not create haplotypes and are not subject to the common inheritance.
Table 2.
RRM1 gene genotypes distribution according to demographic and clinical factors
| Variable | RRM1 (−37C>A) | p, χ 2 | RRM1 (−524C>T) | p, χ 2 | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | CC | CT | TT | |||
| 5 (5.7 %) | 48 (54.5 %) | 35 (39.8 %) | 7 (7.7 %) | 48 (52.7 %) | 36 (39.6 %) | |||
| Sex | ||||||||
| Male | 2 (6.7 %) | 16 (53.3 %) | 12 (40 %) | 0.9557 | 3 (10 %) | 14 (46.7 %) | 13 (43.3 %) | 0.6755 |
| Female | 3 (5.2 %) | 32 (55.2 %) | 23 (39.6 %) | 0.091 | 4 (6.6 %) | 34 (55.7 %) | 23 (37.7 %) | 0.785 |
| Age (years) | ||||||||
| <70 | 3 (4.2 %) | 39 (54.9 %) | 29 (40.9 %) | 0.4751 | 6 (8.2 %) | 40 (54.8 %) | 27 (37 %) | 0.5932 |
| ≥70 | 2 (11.8 %) | 9 (52.9 %) | 6 (35.3 %) | 1.488 | 1 (5.6 %) | 8 (44.4 %) | 9 (50 %) | 1.045 |
| Smoking status | ||||||||
| Current smokers | 4 (6.2 %) | 35 (54.7 %) | 25 (39.1 %) | 0.5369 | 5 (7.7 %) | 35 (53.8 %) | 25 (38.5 %) | 0.0505 |
| Ex-smokers | – | 10 (55.6 %) | 8 (44.4 %) | 3.126 | – | 10 (50 %) | 10 (50 %) | 9.464 |
| Non-smokers | 1 (20 %) | 2 (40 %) | 2 (40 %) | 2 (40 %) | 2 (40 %) | 1 (20 %) | ||
| Histopathological diagnosis | ||||||||
| Adenocarcinoma | 3 (6.8 %) | 22 (50 %) | 19 (43.2 %) | 0.6648 | 4 (8.7 %) | 26 (56.5 %) | 16 (34.8 %) | 0.8393 |
| Squamous cell carcinoma | – | 8 (57.1 %) | 6 (42.9 %) | 4.088 | – | 7 (50 %) | 7 (50 %) | 2.752 |
| Large cell carcinoma | 1 (6.7 %) | 11 (73.3 %) | 3 (20 %) | 2 (12.5 %) | 7 (43.75 %) | 7 (43.75 %) | ||
| NOS NSCLC | 1 (6.6 %) | 7 (46.7 %) | 7 (46.7 %) | 1 (6.7 %) | 8 (53.3 %) | 6 (40 %) | ||
| Stage of disease | ||||||||
| IIIB | 3 (11.1 %) | 16 (59.3 %) | 8 (29.6 %) | 0.2032 | 2 (7.1 %) | 15 (53.6 %) | 11 (39.3 %) | 0.9891 |
| IV | 2 (3.3 %) | 32 (52.4 %) | 27 (44.3 %) | 3.187 | 5 (7.9 %) | 33 (52.4 %) | 25 (39.7 %) | 0.022 |
| Performance status | ||||||||
| PS = 0 | 1 (8.3 %) | 7 (58.3 %) | 4 (33.3 %) | 0.8381 | 1 (9.1 %) | 5 (45.45 %) | 5 (45.45 %) | 0.9889 |
| PS ≥ 1 | 4 (5.3 %) | 41 (53.9 %) | 31 (40.8 %) | 0.353 | 6 (9.2 %) | 28 (43.1 %) | 31 (47.7 %) | 0.022 |
Response to chemotherapy
There were no cases of complete remission (CR) as a result of first-line chemotherapy with platinum compounds and gemcitabine. Control of the disease was observed in 54.9 % of patients, of which partial remission (PR) and stable disease (SD) occurred, respectively, in 17.6 and 37.3 % of patients. Disease progression (PD) was observed in 45.1 % of patients. Patients with squamous cell carcinoma or those who develop anemia before chemotherapy had a significantly lower chance of disease control (p = 0.0392, OR = 0.27; p = 0.0189, OR = 0.33, respectively) when compared to other patients. Moreover, in patients with poor performance status (PS = 1) the risk of PD (p = 0.0495, OR = 4.87) was higher. There were no statistically significant differences in response according to other demographic and clinical factors. Both SNPs: −37C>A and −524C>T of RRM1 did not affect significantly the possibility of responses to treatment. In logistic regression analysis (including: sex, age, histopathological diagnosis, stage of disease, PS and SNPs of RRM1; overall fit of the model: χ2 = 22.45, p = 0.0021) only PS (p = 0.0002, OR = 0.0849 95 % CI 0.02–0.31) had an independent influence on ORR. Effect of SNPs of RRM1 was insignificant, however, the CC genotype (−37C>A) show a trend towards significance (p = 0.0655).
Progression-free survival
The median PFS of whole group of patients was 4 months. In patients who were diagnosed with adenocarcinoma or non-squamous cell carcinoma compared to squamous cell carcinoma patients a significantly lower risk of shortening PFS was noticed (respectively, 6 vs 3 months, p = 0.0456, HR = 0.61, 95 % CI 0.37–0.99 and 4.5 vs 2 months, p = 0.0140, HR = 0.34, 95 % CI 0.14–0.80). In patients with IIIB stage of NSCLC and patients without anemia compared to other patients, the risk of shortening of PFS was significantly lower (respectively, 7 vs 3, p = 0.0094, HR = 0.52, 95 % CI 0.32–0.85; 6.5 vs 3 months, p = 0.0154, HR = 0.54, 95 % CI 0.33–0.89). Other factors did not affect PFS significantly.
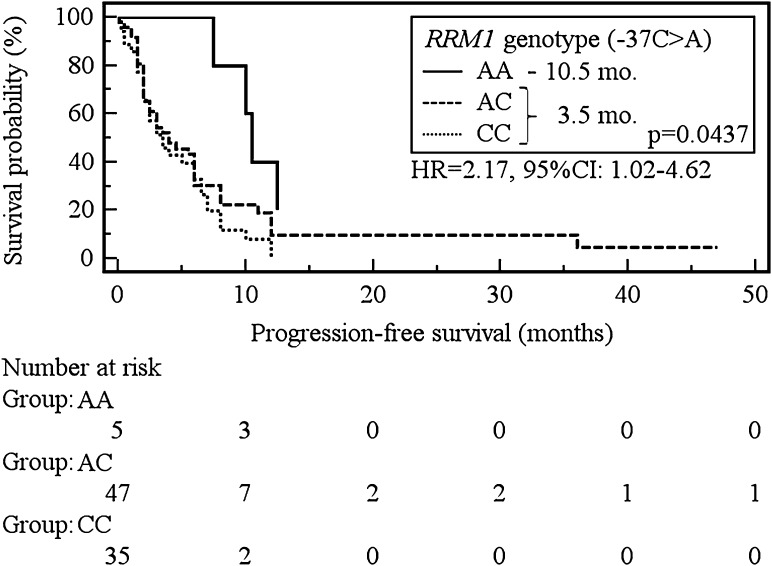
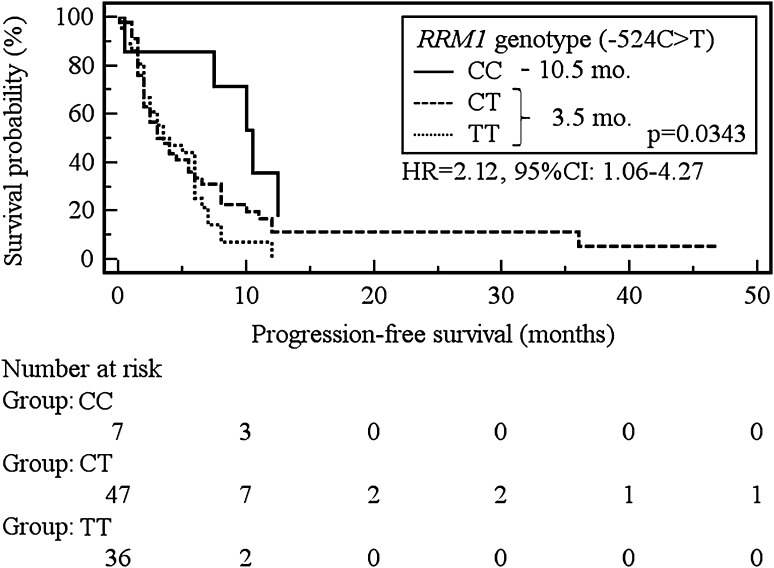
Carriers of the C allele of the RRM1 (−37C>A) showed a significant increase in the risk of PFS shortening in comparison to patients with the AA genotype (3.5 vs 10.5 months, p = 0.0437; HR = 2.17, 95 % CI 1.02–4.62, Fig. 2). Similarly, the presence of the T allele of RRM1 (−524C>T) was associated with a significant risk of shortening of PFS when confronted with the CC genotype carriers (3.5 vs 10.5 months, p = 0.0437; HR = 2.12, 95 % CI 1.06–4.27, Fig. 3).
Fig. 2.
The probability of progression-free survival change depending on RRM1 genotype (−37C>A)
Fig. 3.
The probability of progression-free survival change depending on RRM1 genotype (−524C>T)
In a Cox multivariate logistic regression analysis, poor (PS = 1) performance status (p = 0.0117, HR = 2.75, 95 % CI 1.26–6.02) and anemia prior to treatment (p = 0.0128, HR = 2.67, 95 % CI 1.24–5.74) were independently responsible for shortening of PFS in patients treated with chemotherapy using platinum compounds and gemcitabine (overall fit of the model: p = 0.0271, χ2 = 24.466) (Table 3).
Table 3.
Effect of demographic, clinical and genetic factors on overall response rates, progression-free survival and overall survival in the study group
| Variable | PD 41 (45.1 %) | SD, PR 50 (54.9 %) | p, OR 95 % CI | Median PFS (mo.) | p, χ 2 | HR 95 % CI | Median OS (mo.) | p, χ 2 | HR (95 % CI) |
|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||
| Male | 31 (50.8 %) | 30 (49.2 %) | 0.1179, 0.4839 | 4 | 0.8653 | 0.9560 | 13 | 0.7280 | 1.1177 |
| Female | 10 (33.3 %) | 20 (66.7 %) | 0.1948–1.2022 | 6 | 0.0288 | 0.5684–1.6079 | 11 | 0.1210 | 0.5971–2.0923 |
| Age (years) | |||||||||
| ≤70 | 34 (46.6 %) | 39 (53.4 %) | 0.5581, 0.7299 | 3.56 | 0.6344 | 0.8719 | 11.5 | 0.7812 | 0.9061 |
| >70 | 7 (38.9 %) | 11 (61.1 %) | 0.2546–2.0929 | 0.2262 | 0.4956–1.5339 | 16.5 | 0.0772 | 0.4519–1.8169 | |
| Smoking status | |||||||||
| Smokers | 39 (45.9 %) | 46 (54.1 %) | 0.7978, 0.7863 | 47.5 | 0.5347 | 0.7441 | 11.5 | 0.8851 | 0.9187 |
| Non-smokers | 2 (40 %) | 3 (60 %) | 0.125–4.9481 | 0.3854 | 0.2926–1.8921 | 18.5 | 0.0209 | 0.2909-2.9012 | |
| Histopathological diagnosis | |||||||||
| Adenocarcinoma | 18 (40 %) | 27 (60 %) | – | 6 | 0.1356 | – | 12 | 0.6073 | – |
| Squamous cell carcinoma | 10 (71.4 %) | 4 (28.6 %) | 2 | 5.5517 | 6.5 | 1.8352 | |||
| Large cell carcinoma | 8 (47.1 %) | 9 (52.9 %) | 4 | 13 | |||||
| NOS NSCLC | 5 (33.3 %) | 10 (66.7 %) | 3.5 | 9.5 | |||||
| Adenocarcinoma | 18 (39.1 %) | 28 (60.9 %) | 0.2520, 1.6263 | 6 | 0.0456 | 0.6074 | 12 | 0.4094 | 0.7802 |
| Other | 23 (51.1 %) | 22 (48.9 %) | 0.7077–3.7371 | 3 | 3.9962 | 0.3725–0.9903 | 11.5 | 0.6805 | 0.4326–1.4071 |
| Squamous cell carcinoma | 10 (71.4 %) | 4 (28.6 %) | 0.0392, 0.2696 | 2 | 0.0140 | 0.3399 | 6.5 | 0.1807 | 0.5301 |
| Non-squamous cell carcinoma | 31 (40.3 %) | 46 (59.7 %) | 0.0776–0.9369 | 4.5 | 6.0383 | 0.1437–0.8038 | 12 | 1.7916 | 0.2093–1.3427 |
| Large cell carcinoma | 8 (50 %) | 8 (50 %) | 0.6619, 0.7857 | 3.754 | 0.4972 | 0.8030 | 13 | 0.9921 | 1.0036 |
| Other | 33 (44 %) | 42 (56 %) | 0.2666–2,3157 | 0.4608 | 0.4263–1.5128 | 11.5 | 0.0001 | 0.4928–2.0439 | |
| Stage of disease | |||||||||
| IIIB | 10 (35.7 %) | 18 (64.3 %) | 0.2348, 1.7438 | 7 | 0.0094 | 0.5212 | 18 | 0.0701 | 0.5687 |
| IV | 31 (49.2 %) | 32 (50.8 %) | 0.6968–4.3641 | 3 | 6.7519 | 0.3188–0.8520 | 10 | 3.2800 | 0.3088–1.0475 |
| Performance status | |||||||||
| PS = 0 | 2 (16.7 %) | 10 (83.3 %) | 0.0495, 4.8750 | 7.5 | 0.2694 | 0.7024 | 21 | 0.0700 | 0.5124 |
| PS ≥1 | 39 (49.4 %) | 40 (50.6 %) | 1.0031–23.691 | 3.5 | 1.2197 | 0.3752–1.3148 | 11 | 3.2831 | 0.2485–1.0562 |
| Weight loss before CTH | |||||||||
| Yes | 17 (43.6 %) | 22 (56.4 %) | 0.9567, 1.0245 | 6 | 0.7040 | 0.9058 | 18 | 0.0376 | 0.4632 |
| No | 19 (44.2 %) | 24 (55.8 %) | 0.4278–2.4538 | 3 | 0.1443 | 0.5436–1.5092 | 7.5 | 4.3237 | 0.2242–0.9568 |
| Anemia before CTH | |||||||||
| Yes | 32 (54.2 %) | 27 (45.8%) | 0.0189, 0.3302 | 3 | 0.0154 | 0.5430 | 11 | 0.3844 | 0.7685 |
| No | 9 (28.1 %) | 23 (71.9 %) | 0.1309–0.8329 | 6.5 | 5.8656 | 0.3313–0.8901 | 13 | 0.7565 | 0.4246–1.3910 |
| Side effect after CTH | |||||||||
| Yes | – | – | – | 4.5 | 0.3008 | 0.7515 | 12 | 0.3435 | 0.7231 |
| No | 4 | 1.0706 | 0.4374–1.2911 | 21 | 0.8974 | 0.3697–1.4142 | |||
| Subsequent lines of treatment | |||||||||
| Yes | – | – | – | 5 | 0.7157 | 0.9131 | 16.5 | 0.0305 | 0.5011 |
| No | 3 | 0.1326 | 0.5596–1.4897 | 8 | 4.6802 | 0.2680–0.9371 | |||
| Family history of cancer (any malignant) | |||||||||
| Yes | 6 (33.3 %) | 12 (66.7 %) | 0.3367, 1.7500 | 3.5 | 0.8389 | 0.9349 | 7.5 | 0.4320 | 0.7412 |
| No | 21 (46.7 %) | 24 (53.3 %) | 0.5588–5.4810 | 3 | 0.0413 | 0.4887–1.7885) | 11.5 | 0.6175 | 0.3511–1.5645 |
| RRM1 (−37C>A) | |||||||||
| AA | – | 5 (100 %) | – | 10.5 | 0.0986 | – | 18.5 | 0.0677 | – |
| AC | 23 (47.9 %) | 25 (52.1 %) | 4 | 4.6330 | 18 | 5.3846 | |||
| CC | 15 (42.9 %) | 20 (57.1 %) | 3.5 | 9.5 | |||||
| AA | – | 5 (100 %) | 0.1352, 9.3077 | 10.5 | 0.0437 | 2.1736 | 18.5 | 0.8528 | 1.1004 |
| AC or CC | 38 (45.7 %) | 45 (54.3 %) | 0.4986–173.75 | 3.5 | 4.0687 | 1.0222–4.6220 | 11.5 | 0.0344 | 0.4006–3.0223 |
| AC | 23 (47.9 %) | 25 (52.1 %) | 0.3270, 0.6522 | 44 | 0.7840 | 1.0707 | 18 | 0.0351 | 1.9088 |
| AA or CC | 15 (37.5 %) | 25 (62.5 %) | 0.2775–1.533 | 0.0751 | 0.6569–1.7452 | 11 | 4.4381 | 1.0460–3.4833 | |
| CC | 15 (42.9 %) | 20 (57.1 %) | 0.9601, 1.0222 | 3.5 | 0.0928 | 1.5613 | 9.5 | 0.0193 | 2.1346 |
| AA or AC | 23 (43.3 %) | 30 (56.7 %) | 0.4316–2.4208 | 5.5 | 2.8249 | 0.9286–2.6251 | 18 | 5.4786 | 1.1312–4.0281 |
| RRM1 (−524C>T) | |||||||||
| CC | 1 (14.3 %) | 6 (85.7 %) | – | 10.5 | 0.0923 | – | 18.5 | 0.4129 | – |
| CT | 24 (50 %) | 24 (50 %) | 3.5 | 4.7664 | 11.5 | 1.7691 | |||
| TT | 16 (44.4 %) | 20 (55.6 %) | 3.5 | 10 | |||||
| CC | 1 (14.3 %) | 6 (85.7 %) | 0.1237, 5.4545 | 10.5 | 0.0343 | 2.1249 | 18.5 | 0.5983 | 1.2875 |
| CT or TT | 40 (47.6 %) | 44 (52.4 %) | 0.6291–47.293 | 3.5 | 4.4805 | 1.0574–4.2700 | 11 | 0.2776 | 0.5029–3.2959 |
| CT | 24 (50 %) | 24 (50 %) | 0.3174, 0.6538 | 3.5 | 0.9141 | 0.9740 | 11.5 | 0.3318 | 0.7505 |
| CC or TT | 17 (39.5 %) | 26 (60.5 %) | 0.2843–1.504 | 6 | 0.0116 | 0.6035–1.5720 | 11 | 0.9417 | 0.4204–1.3399 |
| TT | 25 (45.4 %) | 30 (54.6 %) | 0.9246, 0.9600 | 3.5 | 0.0983 | 1.5342 | 10 | 0.1807 | 1.5055 |
| CT or CC | 16 (44.4 %) | 20 (55.6 %) | 0.4124–2.2347 | 4 | 2.7336 | 0.9237–2.5484 | 13 | 1.7920 | 0.8270–2.7404 |
Overall survival
The median OS in the study population was 12 months. The median OS was significantly longer in patients without weight loss prior to chemotherapy compared to other patients (respectively, 18 vs 7.5 months, p = 0.0376; HR = 0.46, 95 % CI 0.22–0.96). Patients who were treated with subsequent lines of treatment showed significantly longer OS than patients who received only one line of treatment (16.5 vs 8 months, p = 0.0305; HR = 0.50, 95 % CI 0.27–0.94). However, when compared separately none of second-line scheme has not improve survival significantly (ERL vs other 11.5 vs 10 mo.; p = 0.4270, HR = 1.37, 95 % CI 0.63–2.99; PEM vs other 13 vs 11 mo.; p = 0.1813, HR = 1.51, 95 % CI 0.82–2.76; DCX vs other 16.5 vs 12 mo.; p = 0.3365, HR = 0.67, 95 % CI 0.29–1.52). There was no statistically significant effect of other factors on the OS.
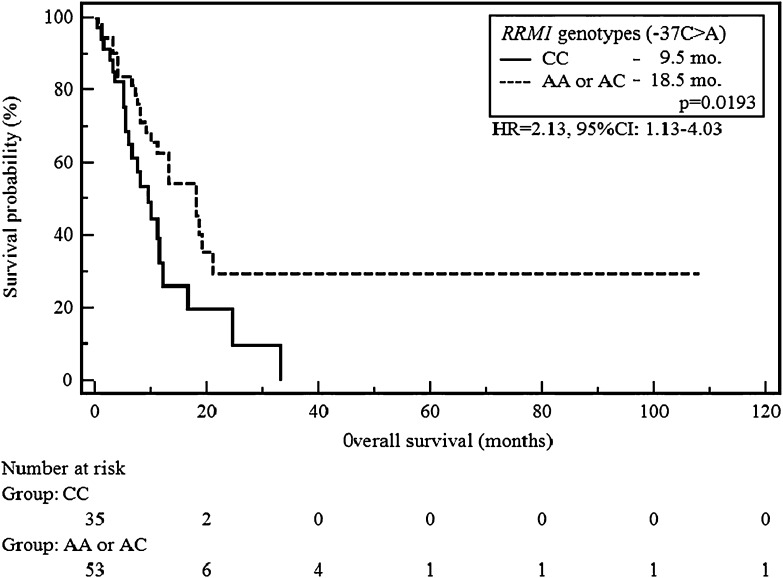
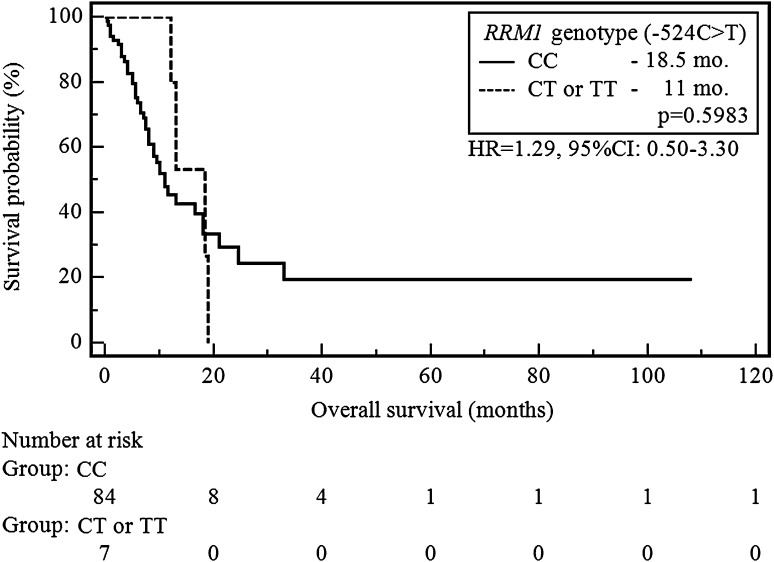
The CC genotype carriers of RRM1 (−37C>A) showed a significant increase in the risk of shortening of OS compared to patients with AA or AC genotypes (9.5 vs 18 months, p = 0.0193; HR = 2.13, 95 % CI 1.13–4.03, Fig. 4). None of the SNP −524C>T variants did significantly affect the length of OS in the study group (e.g. CC vs CT or TT, 18.5 vs 11 months, p = 0.5983, HR = 1.29, 95 % CI 0.50–3.30; Fig. 5).
Fig. 4.
The probability of overall survival change depending on RRM1 genotype (−37C>A)
Fig. 5.
The probability of overall survival change depending on RRM1 genotype (−524C>T)
Poor performance status (PS = 1, p = 0.0053, HR = 5.81, 95 % CI 1.70–19.93) and lack of subsequent lines of treatment (p = 0.0246, HR = 2.58, 95 % CI 1.15–7.53) were only factors that shorten OS in a Cox multivariate logistic regression analysis (overall fit of the model χ2 = 27.73, p = 0.0233).
Discussion
Despite progress in medicine, that has been made in recent years, there are still lack of predictive factors, which would allow qualify NSCLC patients to appropriate chemotherapy regimen. The current criteria of patients’ selection to cytostatic-based therapy are primarily performance status, treatment toxicity and clinician’s experience; however, it seems to be insufficient. In addition, most of the currently available chemotherapy regimens, regardless of the line of treatment, are characterized by significant differences in the effectiveness in various patients. This is another evidence that key to prediction of occurrence of resistance to cytostatics may be stored in genetic information [10–12, 18, 19].
Reliable evaluation of mRNA or protein expression requires access to the tumor tissue Tumor tissue in an advanced stages of NSCLC is difficult or, sometimes, impossible to obtain. Changes in the structure, function, stability, folding or expression of proteins may be caused by occurrence of specific SNPs in encoding or non-coding sequences (especially located in promoter region) of genes. Moreover, the analysis of SNPs may be carried out in materials that are easy to obtain (e.g. DNA from peripheral blood leukocytes) and thus easier introduced into routine clinical practice. Consequently, many studies (unfortunately mainly retrospective) assessed the effect of the individual polymorphic variants of different genes on the effectiveness of various treatment regimens.
There is lack of meta-analysis evaluating the impact of any RRM1 SNPs on response to treatment, PFS or OS in patients with NSCLC treated with gemcitabine. Among the eight original papers investigating relationship of SNPs (−37C>A and/or −524C>T) with response to treatment, four of them concerned Asian patients. Feng et al. analyzed both mentioned SNPs (genotypes) independently and in combination in 214 patients treated with different schemes chemotherapy based on platinum. Kim et al. evaluated allelotype of two mentioned SNPs in 97 patients received chemotherapy based on gemcitabine. Lin et al. estimated only relationship between −37C>A polymorphism and effectiveness of chemotherapy regimens containing gemcitabine in 40 NSCLC patients. Dong et al. assessed SNPs in 56 patients treated with chemotherapy based on gemcitabine [16, 17, 20, 21]. In studies concerning Caucasian population, analysis of −37C>A SNP was presented by Isla et al. (62 patients received platinum compounds in combination with docetaxel), Vinolas et al. (94 patients treated with platinum compounds in combination with vinorelbine) and Mlak et al. (62 patients treated with platinum compounds in combination with gemcitabine). While, Ludovini et al. assessed −524C>T SNPs in 168 patients received platinum-based chemotherapy [22–25].
Feng and co-workers did not demonstrate the association between the −37C>A SNP and the response to chemotherapy. However, the statistically significant relationship between −524C>T SNP of RRM1 and response to treatment (χ2 = 6.179, p = 0.046) was proved [16]. In contrast, Kim et al. reported a significantly higher response rate in patients with genotype AC/CT (−37C>A/−524C>T) in comparison to carriers of other combinations of RRM1 allelotypes (65.5 vs 42.6 %, p = 0.039) [18]. Lin et al. and Dong et al. have shown no significant differences in response to treatment depending on the RRM1 genotype [17, 19]. Isla et al., Vinolas et al., Ludovini et al. demonstrated no correlation between the genotype of RRM1 and response to the treatment [23–25].
The only evidence for association of −37A>C polymorphism and response to gemcitabine-based chemotherapy was reported in our previous study. We indicated that AC genotype was significantly related with less frequent response to chemotherapy (χ2 = 5.47, p = 0.0193). It was probably caused by the influence of A allele (importance of AA genotype was impossible to assess due to small study group). In addition, this study showed a significant association between the presence of genotype AA or AC and early progression of the disease (χ2 = 3.61; p = 0.0573) [22].
In the available literature six studies describe the relationship between SNPs of RRM1 and the duration of the PFS (Dong et al., Mlak et al., Vinolas et al., Isla et al., Kim et al., Ludovini et al.) and OS (Ryu et al., Mlak et al., Vinolas et al., Isla et al., Kim et al., Ludovini et al.) in patients with NSCLC treated in the first-line with platinum and third generation drug scheme (usually gemcitabine) [16, 20, 22–26]. Dong et al. presented the positive study regarding the impact of RRM1 SNPs on the length of PFS. Authors demonstrated significant differences in the PFS depending on −37C>A SNP (23.3, 30.7, 24.7 weeks for Asian patients with CC, CA, AA genotype, respectively, p = 0.043) [17]. Among the studies concerning Caucasian patients, only in our previous publication the presence of a significant effect of CC genotype (−37C>A) on reduction of the risk of shortening both PFS (6 months for patients with CC genotype vs 2 months for others patients, HR = 0.51, 95 % CI 0.29–0.89, p = 0.0087) and OS (16.5 months for patients with CC and 8 months for patients CA or AA genotypes, HR = 0.47; 95 % CI 0.22–0.99, p = 0.0448) was demonstrated [22]. Other studies revealed no significant association between polymorphic variants of RRM1 (−37C>A and/or −524C>T) and the length of PFS and/or OS in patients with NSCLC treated with first-line chemotherapy.
Therefore, statistically significant relationship between −524C>T SNP of RRM1 and the PFS length (CC genotype was significantly associated with prolongation of PFS) in patients with advanced NSCLC who were treated first-line chemotherapy based on platinum compounds and gemcitabine was described for the first time in present study. We also verified the results of the impact of −37C>A SNP on the PFS (AA genotype was significantly associated with PFS prolongation) and OS (CC genotype was significantly associated with OS shortening), described previously in smaller populations of NSCLC [17, 22]. The limitations of our research are retrospective character of analysis and heterogeneous study group.
Unfortunately, despite growing evidence suggesting that SNPs in genes encoding proteins involved in drug metabolism and DNA repair may help to explain the inter-individual variability of response or resistance to chemotherapy, most of available studies present conflicting results.
Differences between studies may be due to: (1) race differences (Asian vs Caucasian patients), which are reflected both in the incidence of SNPs and in phenotypic disparity; (2) NSCLC has many subtypes (characterized by, e.g.: differences in driver mutations occurrence and clinical course) thus in most studies different proportion of each subtype may occur. Differences in course of treatment: in some studies; (3) part of patients received chemoradiation. In first-line regimens different platinum compound (cis- or carboplatin) may be used; (4) In subsequent lines most patients are treated with multiple drugs, including: pemetrexed, docetaxel, and an TKI, as well as may undergo surgery, which significantly affects course of disease and survival.
Accordingly, it is crucial to conduct a large randomized prospective studies taking into account the respective proportions of race, subtypes of NSCLC, as well as based on suitable standards and uniform regimens of treatment.
Conclusions
The presence of rare genotypes: AA (−37C>A) and CC (−524C>T) of RRM1 promoter are favorable predictors associated with prolongation of PFS in NSCLC patients treated with first-line chemotherapy with platinum compounds and gemcitabine. Moreover, occurrence of CC genotype (−37C>A) is unfavorable predictor of OS shortening. Evaluation of selected SNPs of RRM1 may in the future, become a useful tool in the qualification of patients with NSCLC to the appropriate chemotherapy regimen. However, our results should be previously confirmed in sufficiently large and prospective studies.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Committee Ethics and Research at the Medical University of Lublin (no. consent: KE-0254/142/2010). This article does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Non-Small Cell Lung Cancer version 1.2015 (2015) http://www.nccn.org. Accessed 25 June 2015.
- 3.Gridelli C, de Marinis F, Cappuzzo F, Di Maio M, Hirsch FR, Mok T, et al. Treatment of advanced non-small-cell lung cancer with epidermal growth factor receptor (EGFR) mutation or ALK gene rearrangement: results of an international expert panel meeting of the Italian Association of Thoracic Oncology. Clin Lung Cancer. 2014;15:173–181. doi: 10.1016/j.cllc.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Simon GR, Somaiah N. A tabulated summary of targeted and biologic therapies for non-small-cell lung cancer. Clin Lung Cancer. 2014;15:21–51. doi: 10.1016/j.cllc.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 6.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 7.Krawczyk P, Ramlau R, Chorostowska-Wynimko J, Powrózek T, Lewandowska MA, Limon J, et al. The efficacy of EGFR gene mutation testing in various samples from non-small cell lung cancer patients: a multicenter retrospective study. J Cancer Res Clin Oncol. 2015;141(1):61–68. doi: 10.1007/s00432-014-1789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scagliotti G, Stahel RA, Rosell R, Thatcher N, Soria JC. ALK translocation and crizotinib in non-small cell lung cancer: an evolving paradigm in oncology drug development. Eur J Cancer. 2012;48:961–973. doi: 10.1016/j.ejca.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Simon GR, Schell MJ, Begum M, Kim J, Chiappori A, Haura E, et al. Preliminary indication of survival benefit from ERCC1 and RRM1-tailored chemotherapy in patients with advanced nonsmall cell lung cancer: evidence from an individual patient analysis. Cancer. 2012;118:2525–2531. doi: 10.1002/cncr.26522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Zhu X, Zhang L, Sun S, Huang J, Lin Y. A prospective study of biomarker-guided chemotherapy in patients with non-small cell lung cancer. Cancer Chemother Pharmacol. 2014;74:839–846. doi: 10.1007/s00280-014-2513-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzoni F, Cecere FL, Meoni G, Giuliani C, Boni L, Camerini A, et al. Phase II trial of customized first line chemotherapy according to ERCC1 and RRM1 SNPs in patients with advanced non-small-cell lung cancer. Lung Cancer. 2013;82(2):288–293. doi: 10.1016/j.lungcan.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine) Drug Resist Updat. 2002;5(1):19–33. doi: 10.1016/S1368-7646(02)00002-X. [DOI] [PubMed] [Google Scholar]
- 14.Rosell R, Crino L, Danenberg K, Scagliotti G, Bepler G, Taron M, et al. Targeted therapy in combination with gemcitabine in non-small cell lung cancer. Semin Oncol. 2003;30(Suppl 10):19–25. doi: 10.1016/S0093-7754(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 15.Bepler G, Zheng Z, Gautam A, Sharma S, Cantor A, Sharma A, et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer. 2005;47:183–192. doi: 10.1016/j.lungcan.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 16.Feng JF, Wu JZ, Hu SN, Gao CM, Shi MQ, Lu ZH, et al. Polymorphisms of the ribonucleotide reductase M1 gene and sensitivity to platin-based chemotherapy in non-small cell lung cancer. Lung Cancer. 2009;66:344–349. doi: 10.1016/j.lungcan.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Dong S, Guo AL, Chen ZH, Wang Z, Zhang XC, Huang Y, et al. RRM1 single nucleotide polymorphism—37C > A correlates with progression-free survival in NSCLC patients after gemcitabine-based chemotherapy. J Hematol Oncol. 2010;3:10. doi: 10.1186/1756-8722-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Sun H, Ren S, Kim Curran V, Zhang L, Zhou S, et al. Association of XRCC3 and XPD751 SNP with efficacy of platinum-based chemotherapy in advanced NSCLC patients. Clin Transl Oncol. 2012;14(3):207–213. doi: 10.1007/s12094-012-0785-3. [DOI] [PubMed] [Google Scholar]
- 19.Deng JH, Deng J, Shi DH, Ouyang XN, Niu PG. Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clin Transl Oncol. 2015 (Epub ahead of print). [DOI] [PubMed]
- 20.Kim SO, Jeong JY, Kim MR, Cho HJ, Ju JY, Kwon YS, et al. Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms of the ribonucleotide reductase M1 gene. Clin Cancer Res. 2008;14(10):3083–3088. doi: 10.1158/1078-0432.CCR-07-4591. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Liu X, Song S, Wang S. A study on the relationship between RRM1 single nucleotide polymorphisms and clinical characteristics in lung cancer patients. Zhongguo Fei Ai Za Zhi. 2008;11(6):784–788. doi: 10.3779/j.issn.1009-3419.2008.06.17. [DOI] [PubMed] [Google Scholar]
- 22.Mlak R, Krawczyk P, Ramlau R, Kalinka-Warzocha E, Wasylecka-Morawiec M, Wojas-Krawczyk K, et al. Predictive value of ERCC1 and RRM1 gene single-nucleotide polymorphisms for first-line platinum- and gemcitabine-based chemotherapy in non-small cell lung cancer patients. Oncol Rep. 2013;30(5):2385–2398. doi: 10.3892/or.2013.2696. [DOI] [PubMed] [Google Scholar]
- 23.Viñolas N, Provencio M, Reguart N, Cardenal F, Alberola V, Sánchez-Torres JM, et al. Single nucleotide polymorphisms in MDR1 gen correlates with outcome in advanced non-small-cell lung cancer patients treated with cisplatin plus vinorelbine. Lung Cancer. 2011;71(2):191–198. doi: 10.1016/j.lungcan.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Isla D, Sarries C, Rosell R, Alonso G, Domine M, Taron M, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15(8):1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 25.Ludovini V, Floriani I, Pistola L, Minotti V, Meacci M, Chiari R, et al. Association of cytidine deaminase and xeroderma pigmentosum group D polymorphisms with response, toxicity, and survival in cisplatin/gemcitabine-treated advanced non-small cell lung cancer patients. J Thorac Oncol. 2011;6(12):2018–2026. doi: 10.1097/JTO.0b013e3182307e1f. [DOI] [PubMed] [Google Scholar]
- 26.Ryu JS, Shin ES, Nam HS, Yi HG, Cho JH, Kim CS, et al. Differential effect of polymorphisms of CMPK1 and RRM1 on survival in advanced non-small cell lung cancer patients treated with gemcitabine or taxane/cisplatinum. J Thorac Oncol. 2011;6(8):1320–1329. doi: 10.1097/JTO.0b013e3182208e26. [DOI] [PubMed] [Google Scholar]