Abstract
The plant growth-promoting rhizobacteria have developed many different (indirect and direct) mechanisms that have a positive effect on plant growth and development. Strains isolated from Astragaluscicer and Astragalusglycyphyllos root nodules were investigated for their plant growth-promoting properties such as production of indole-3-acetic acid (IAA) and siderophores, phosphate solubilization, ACC deaminase activity, and tolerance to heavy metals. IAA production and P-solubilization were frequent features in the analysed strains, while siderophores were not produced by any of them. In this work, we investigated the presence of the acdS genes and ACC deaminase activities in Astragalauscicer and A. glycyphyllos microsymbionts, classified within the genus Mesorhizobium. The results demonstrated that the acdS gene is widespread in the genome of Astragalus sp. microsymbionts; however, none of the tested strains showed ACC deaminase activity. The acdS gene sequence similarity of the analysed strains to each other was in the range from 84 to 99 %. On the phylogram of acdS gene sequences of milkvetch, the symbionts clustered tightly with the genus Mesorhizobium bacteria.
Electronic supplementary material
The online version of this article (doi:10.1007/s00203-016-1243-3) contains supplementary material, which is available to authorized users.
Keywords: Astragalus sp. microsymbionts, acdS gene, PGPR
Introduction
Ethylene affects plant growth and development. It is responsible for several processes in plants and, depending on the level, can e.g. promote root initiation, inhibit root elongation, activate plant hormone synthesis, and promote flower wilting. Ethylene is also involved in the response to both biotic and abiotic stresses. An increase in ethylene synthesis may accompany for example extreme temperatures, water flooding, drought, radiation, salinity, and presence of various pathogens. It has also been described that ethylene affects various stages of symbiosis (Vacheron et al. 2013; Glick 2014). Ethylene can inhibit nodule development in different fabacean plants, for example in Phaseolusvulgaris, Lotusjaponicas, and Trifoliumrepens (Tamimi and Timko 2003).
In the literature, there is a “stress ethylene” concept. The model of “stress ethylene” includes the synthesis of ethylene in two peaks. The first one is small and reflects ethylene that consumes the pool of ACC (1-aminocyclopropane-1-carboxylate) existing in stressed plant tissues. Probably, this ethylene is responsible for initiation of transcription of genes whose products are involved in defensive or protective mechanisms. The second, bigger ethylene peak reflects synthesis of additional ACC in plant response to stress. It is responsible for initiation of such processes as cell death, senescence, and chlorosis (Glick 2014; Singh et al. 2015).
Plant growth promotion by rhizosphere microorganisms (plant growth-promoting rhizobacteria, PGPR) is a result of various mechanisms such as production of indole-3-acetic acid (IAA), siderophores, 2,3-butanediol, lytic enzymes, and 1-aminocyclopropane-1-carboxylate deaminase (ACC deaminase) as well as induction of plant systemic resistance to pathogens. It has been demonstrated that inoculation of plants with ACC deaminase-producing rhizobia causes a decrease in the plant ethylene level, which in turn protects plant from the effects of biotic and abiotic stresses (Saleem et al. 2007; Bhattacharyya and Jha 2012; Beneduzi et al. 2012).
ACC deaminase (EC:4.1.99.4) synthesized by microorganisms converts ACC, the immediate precursor of ethylene in plants, into ammonia and ketobutyrate, which can be used as a source of nitrogen and carbon, respectively. This enzyme synthesized by soil bacteria decreases the level of ethylene in plants and, in consequence, stimulates plant growth. A low level of ethylene correlates with higher resistance of plants to various kinds of biotic and abiotic stresses such as high salt, extreme temperature, phytopathogenic infection. The presence of ACC deaminase-producing microorganisms in soil contributes to longer plant roots and shoots and to higher plant resistance to inhibition of growth by ethylene stress (Bhattacharyya and Jha 2012; Glick 2014).
It is worth noting that, under stress conditions, some microorganisms produce the phytohormone indole-3-acetic acid (IAA). IAA is a natural plant auxin that is the common product of l-tryptophan metabolism. The indole-3-acetic acid enhances development of longer roots with an increased number of root hairs and lateral roots. IAA inhibits or delays leaf abscission and affects plant flowering and fruiting. This auxine may stimulate such processes as tissue differentiation, xylem formation, nitrogen fixation, and plant stress resistance. It has been found that production of IAA by bacteria can promote plant growth and increase acdS gene transcription (Zaidi et al. 2010; Zhao 2010). The application of microorganisms producing both IAA and ACC deaminase as plants inoculates did not result in any increase of the ethylene level in contrast to plants inoculated only with IAA-producing bacteria (Glick 2014).
PGPR are able to synthesize different compounds that have a positive impact on the growth, development, and tolerance to different stresses in plants. These effects are related, inter alia, to nutrient enrichment of soil by phosphate solubilization, nitrogen fixation, or lipase and protease production. PGP microorganisms are often characterized by tolerance to abiotic stresses such as pH, drought, salinity, and heavy metal pollution (Zaidi et al. 2010; Zhao 2010; Beneduzi et al. 2012; Ahemad and Kibret 2014).
We have studied eight mesorhizobium strains isolated from nodules of Astragaluscicer and Astragallusglycyphyllos growing in Poland, Ukraine, and Canada (Wdowiak and Małek 2000; Gnat et al. 2014). In our earlier studies, A. glycyphyllos and A. cicer symbionts were classified into the genus Mesorhizobium, based on sequence analysis of 16S rRNA and housekeeping genes (Wdowiak-Wróbel and Malek 2010; Gnat et al. 2014, 2015a). Additionaly A. glycyphyllos nodule isolates were affiliated into the M. amorphae and M. ciceri species by DNA/DNA hybridization and to the new symbiovar, glycyphyllae” using nodA and nodC sequence analysis (Gnat et al. 2015a, b). The strains, representing different phenotypic and genomic groups of A. glycyphyllos and A. cicer isolates, were used to determine the ability to use ACC as a sole nitrogen source, the presence of the acdS gene in their genome, phylogeny of the acdS genes, tolerance of bacteria to heavy metals, and their capability of IAA synthesis and phosphate solubilization.
Materials and methods
Bacterial strains
Mesorhizobium strains ACMP18, USDA 3350, AW1/3, and CIAM0210 isolated from A. cicer and AG1, AG15, AG17, and AG27 isolated from root nodules of A. glycyphyllos, i.e. two fabacean plant species, were used in this study. Mannitol-yeast extract liquid medium YEM and mannitol-yeast extract agar YEMA were routinely used for culturing and maintenance of the rhizobia (Vincent 1970). The analysed rhizobia were maintained on the YEMA medium at 4 °C.
Phosphate solubilization
The phosphate solubilizing ability of the rhizobia was tested on Pikovskaya’s agar medium (Pikovskaya 1948). After 7 days of growth at 28 °C, bacteria that induced a clear zone around the colonies were considered to be positive for phosphate solubilization. The capability of the bacteria of phosphate solubilization was described by the solubilization index = the ratio of the total diameter (colony + halo zone) to the colony diameter (Edi-Premono et al. 1996).
IAA production
A. cicer and A. glycyphyllos microsymbionts were screened for their ability to produce IAA. The isolates were grown in Tris-TMRT medium (Manassila et al. 2007) and incubated at 28 °C for 5 days. The presence of IAA was estimated by adding 2 ml of Salkowski’s reagent (2 % 0.5 FeCl3 in 35 % HClO solution) into the bacterial culture and incubation of the mixture in the dark at 28 °C for 30 min. IAA concentrations ranging from 10 to 100 µg/ml were used as a positive control.
Detection of siderophores
Chrome-Azurol S (CAS) agar medium devoid of iron was used for detection of siderophores (Schwyn and Neilands 1987). The bacteria were grown in the synthetic medium described by Jadhav and Desai (1992) with and without 10 μM iron for 24 h on a rotary shaker at 28 ± 2 °C. Next, the cultures were centrifuged and the cell free supernatant was dropped onto CAS plates and incubated in the dark at 28 °C for 4–5 days. The blue colour of the CAS medium is due to the dye complexed with iron. In the presence of the siderophore, the ferric ions are bound, releasing a free dye, which is orange in colour (positive reaction).
Zn, Cd, Pb
The tolerance of A. cicer and A. glycyphyllos symbionts to heavy metals was investigated on yeast mannitol agar (YMA) medium supplemented with various soluble heavy metal salts, namely Cd, Pb, and Zn, at different concentrations. Pb was applied as Pb(CH3COO)2 (500 and 750 μg ml−1), Cd as CdSO4 × 8H2O (50 and 100 μg ml−1), and Zn as ZnSO4 × 7H2O (250, 500 and 750 μg ml−1). Resistance of the Astragalus sp. microsymbionts to heavy metals was determined by their growth on plates incubated at 28 °C for 4–5 days.
Ability to utilize ACC as a sole source of nitrogen
The analysed mesorhizobia were screened for their ability to utilize ACC as a sole nitrogen source in microtiter plates according to the method described by Shahzad et al. (2010).
ACC deaminase activity
To determine ACC deaminase activity, A. cicer and A. glycyphyllos symbionts were grown in 5 ml of TY medium (Beringer 1974) at 30 °C for 2–3 days until they reached the stationary phase. The bacterial cells were centrifuged and washed twice with 0.1 M Tris–HCl (pH 7.5). Next, the bacteria were suspended in 2 ml of M9 minimal medium supplemented with ACC (final concentration of 5 mM) and incubated at 30 °C with shaking for 36 h. ACC deaminase activity was determined according to the method described before (Ma et al. 2003; Penrose and Glick 2003). ACC deaminase activity was determined by measuring the production of α-ketobutyrate (Honma and Shimomura 1978).
The protein concentration in the cell extracts was determined by the method of Bradford (1976) using the Bio-Rad protein reagent (Bio-Rad; Protein Assay Dye Reagent Concentrate #500-0006).
PCR amplification and sequencing
The bacterial strains were grown in 5 ml of YEM liquid medium at 28 °C for 2–3 days until they reached the stationary phase. Next, the rhizobia were centrifuged at 20,000×g for 10 min. Genomic DNA was isolated from the rhizobial strains using the GES method (Pitcher et al. 1989).
To obtain the acdS gene sequences, the following set of degenerate primers was designed: primers acdSF (5′CAAGCTGCGCAAGCTCGAATA3′) and acdSR (5′CATCCCTTGCATCGATTTGC3′). The PCR assay was performed according to the manufacturer’s description using 25-μl of a reaction mixture (Sigma) under the following conditions: initial denaturation for 5 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 50 °C, and 1 min at 72 °C, and final 5 min elongation at 72 °C. The PCR products were purified using a Clean-up kit (A&A Biotechnology) and sequencing reactions were performed using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA). The products obtained were cleaned with an Ex-Terminator kit (A&A Biotechnology) and analysed in an automatic 3500 Genetic Analyzer sequencer (Applied Biosystems). The sequences of the acdS genes were compared with the sequences available in the GenBank and aligned using ClustalX2 multiple sequence alignment (Larkin et al. 2007). The phylogenetic tree of the acdS and AcdS sequences were constructed by MEGA 4.0 software (Tamura et al. 2007). The sequence similarity rate of the acdS sequence genes was determined according to the Kimura’s two-parameter model (Kimura 1980). The phylogenetic tree of the acdS gene sequences and deduced AcdS sequences were constructed using the neighbour-joining (NJ) method (Saitou and Nei 1987).
Labelling the probe with digoxigenin
The purified PCR product was tagged with digoxigenin (DIG) using a DIG Oligonucleotide 3′–end labelling kit (Roche Diagnostics GmbH, Mannheim, Germany). The reactions were carried out in a 50-μl reaction mixture containing 50 ng of template (DNA of Mesorhizobiumhuakuii MAFF303099), 5 μl reaction buffer (1×), 5 μl PCR DIG labelling Mix (200 μM dNTP), upstream and downstream primer (30 pmol each), and 0.75 μl enzyme mix (2.6 U). PCR was conducted in a DNA 2720 Thermal Cycler (Applied Biosystems) under the following conditions: 2 min initial denaturation at 94 °C, 10 cycles of 30 s denaturation at 95 °C, annealing at 60 °C for 30 s and 40 s of elongation at 72 °C, 20 cycles of 30 s denaturation at 95 °C, annealing at 60 °C for 30 s and 40 s plus 20 s for each successive cycle of elongation at 72 °C, followed by final extension at 72 °C for 7 min. The presence of the PCR labelling product (~710 bp) was checked by electrophoresis on 1 % agarose gel.
Preparation of the genomic DNA for Southern hybridization
Approximately 2 μg of genomic DNA isolated from each rhizobial strain was completely digested overnight with 10 U restriction enzyme HindIII (MBI Fermentas, Inc.) at 37 °C. The digested DNA was loaded and run on a 1 % agarose gel. 1-kb DNA ladder (MBI Fermentas, Inc.) was used as a molecular weight standard. HindIII digested genomic DNA of M. huakuii MAFF303099 was used as a positive control of the presence of the acdS gene. The DNAs were transferred onto a nylon membrane according to Sambrook and Russell (2001). The blot was hybridized with the acdS probe. Hybridization was performed according to the procedure described by the manufacturer (DIG Luminescent Detection Kit, Roche).
Accession numbers
The acdS sequences of the milkvetch microsymbionts studied in this work have been deposited in the GenBank database under accession numbers KU745724-KU745731.
Results
Growth of A. cicer and A. glycyphyllos microsymbionts on ACC medium as a sole nitrogen source
Four symbionts of A. cicer and four strains isolated from root nodules of A. glycyphyllos were tested for the capacity of utilization of ACC as a sole N source. The studied milkvetch microsymbionts showed good growth on the control medium-(NH4)2SO4 and much weaker growth on the ACC medium even after 96 h. The ability to use ACC as a sole nitrogen source suggests that the strains isolated from Astragalus sp. plants can possess ACC deaminase, i.e. an ACC-hydrolysing enzyme.
Southern analysis
The Southern hybridization was used to determine the number of acdS gene copies in each investigated mesorhizobium strain. The DNAs of the milkvetch microsymbionts were digested with the HindIII enzyme, which does not possess restriction sites within the amplified acdS genes. Then, restriction fragments were separated by agarose gel electrophoresis and blotted onto Hybond-N+. The blot was hybridized with a M. huakuii MAFF303099 strain acdS gene probe.
In the microsymbionts of the Astragalus sp., the copy of the acdS gene encoding ACC deaminase was detected using the Southern hybridization method. The results showed that strains AG1, AG15, AG17 and AG27 had a single band with the size of ca. 8000 bp. Strains ACMP18, USDA3350, CIAM0210, and AW1/3 also showed a single band, whose sizes were in the range of ca. 2500, 2200, 4000, and 4000, respectively.
Analysis of nucleotide and protein sequences of acdS genes of A. cicer and A. glycyphyllos symbionts
Phylogenetic analysis of the housekeeping genes of astragali isolates determined in the earlier studies indicated that the analysed isolates represent the genus Mesorhizobium (Wdowiak-Wróbel and Małek 2010; Gnat et al. 2015a, b). It was found that many bacteria of the genus Mesorhizobium have the ACC deaminase-coding acdS gene, which plays an important role in the growth and nodulation of fabacean legume plants (Ma et al. 2003, 2004; Glick et al. 2007; Conforte et al. 2010; Nascimento et al. 2012a, b).
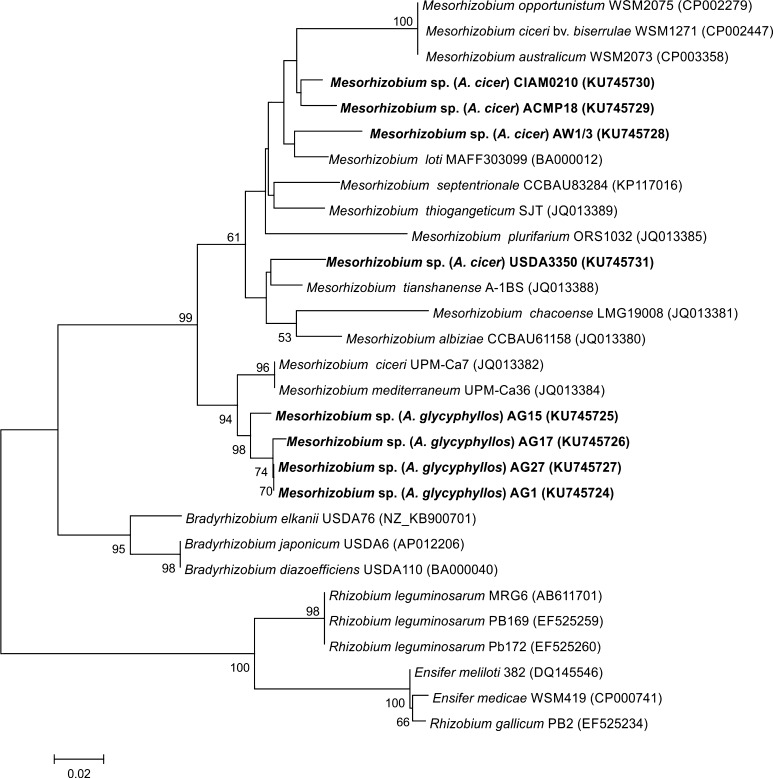
Although the astragali symbionts did not show ACC deaminase activity in vitro, the acdS gene sequences were obtained for all the analysed strains. The phylogenetic analysis of the aligned 529-bp acdS gene sequences resulted in the tree shown in Fig. 1. In the phylogenetic tree of partial acdS gene sequences, the A. cicer and A. glycyphyllos strains were split into 3 main well supported clusters and all of them grouped together with the genus Mesorhizobium strains within one monophyletic group (99 % bootstrap support). The phylogenetic analysis of the acdS genes revealed that A. glycyphyllos microsymbionts showed the greatest similarity to the sequences of the M. ciceri and M. mediterraneum strains (93–94 % sequence similarity), forming a strongly (99 % bootstrap) supported subgroup with these bacteria (Fig. 1). The sequences of the acdS genes of A. cicer microsymbionts AW1/3, ACMP18, CIAM0210 were related to the acdS of M. loti (with 89–90 % similarity). The phylogenetic analysis of the acdS gene of the USDA3350 strain used in this study showed that it belongs to a common group together with M. chacoense, M. albiziae, and M. tianshanense strains (86, 89, and 90 %, respectively).
Fig. 1.
Phylogenetic relationship between A. cicer and A. glycyphyllos microsymbionts and reference strains based on partial acdS sequences. Bootstrap values (1000 replicates) are shown when higher than 50 %. The scale bar represents the percentage of substitutions per site
The next cluster grouped bacteria of the genera Rhizobium and Ensifer. The last one comprised bacteria of the genus Bradyrhizobium. The high bootstrap values of both these groups showed robustness (100 and 95 %, respectively).
The phylogenetic tree derived from the AcdS protein sequences deduced from the acdS genes constructed by the neighbour-joining method is shown in Fig. S1. Only minor topological differences were found when the acdS nucleotide sequence tree and the tree based on AcdS amino acid sequences were compared. At the amino acid level, the AcdS protein sequences of A.cicer and A. glycyphyllos symbionts were 87–100 % identical to each other and 84–95 % to the AcdS proteins of reference Mesorhizobium strains.
Heavy metals
Heavy metal resistance patterns of Astragalus sp. microsymbionts were investigated using three heavy metals, i.e. zinc, cadmium, and lead at the concentration specified in the materials and methods. All isolates were found to be resistant to a 500 µg/ml concentration of Zn and Pb. Moreover, approximately 62 and 87 % of the analysed bacteria were found to be tolerant to 750 µg/ml of Zn and Pb, respectively (Table 1). Assessment of the tolerance of the A. cicer and A. glycyphyllos symbionts to cadmium revealed that they were much more sensitive to this heavy metal. They grew in the presence of 50 µg Cd in 1 ml medium but their growth was almost completely inhibited by 100 μg ml−1 of Cd in the medium. In the presence of 100 µg of Cd per 1 ml of medium, only two of the eight strains analysed showed weak growth. The inhibitory effect of heavy metals on the growth of the astragali symbionts was the following (in an increasing order): Pb(CH3COO)2, ZnSO4 × 7H2O, and CdSO4 × 8H2O.
Table 1.
Plant growth-promoting features of A. cicer and A. glycyphyllos microsymbionts
| Strain | P-solubilization | IAA production | Siderophore production | Tolerant to Zn | Tolerant to Cd | Tolerant to Pb | |||
|---|---|---|---|---|---|---|---|---|---|
| 500 μg ml−1 | 750 μg ml−1 | 50 μg ml−1 | 100 μg ml−1 | 500 μg ml−1 | 750 μg ml−1 | ||||
| ACMP18 | + | + | − | + | − | + | − | + | − |
| AW1/3 | + | − | − | + | + | + | − | + | + |
| CIAM0210 | + | + | − | + | − | + | − | + | + |
| USD3350 | + | + | − | + | − | + | − | + | + |
| AG1 | + | + | − | + | + | + | +/− | + | + |
| AG15 | + | + | − | + | + | + | − | + | + |
| AG17 | + | + | − | + | + | + | − | + | + |
| AG27 | + | + | − | + | + | + | +/− | + | + |
+ positive reaction; ± weak reaction; − negative reaction
Characterization of A. cicer and A. glycyphyllos rhizobia for phosphate solubilization ability, and production of IAA and siderophores
All the milkvetch nodulators analysed exhibited phosphate solubilizing activity, which was detected by the formation of a clear halo around bacterial colonies growing on Pitkovskay’s medium (Table 1). The isolates showed a varying P-solubilizing index, i.e. from 1.45 to 2.87. The highest P-solubilization index was shown by strains USDA3350 and ACMP18, i.e. the A.cicer symbionts, and the lowest index was noted for A. glycyphyllos isolate AG15.
The microsymbionts of A. cicer (3 strains) and A. glycyphyllos (4 strains) showed positive reaction in the test for IAA production. The strongest pink colour, i.e. the highest IAA concentration, was detected in the case of the CIAM0210 strain, while the AW1/3 strain did not synthesize IAA. No ability to produce siderophores was detected in any strains tested (Table 1).
Discussion
Some soil bacterial strains have a positive impact on plant growth and development. Such bacteria are called plant growth-promoting rhizobia (PGPR) (van Peer and Schippers 1989; Frommel et al. 1991; Kloepper et al. 1988). The positive effect of microorganisms on plant growth is related, inter alia, to protection of plants against phytopathogenic organisms or bacterial synthesis of compounds that improve plant growth and development (Glick 1995; Glick et al. 1999). PGPR can synthesize, e.g. phytohormones (auxin, cytokinins), siderophores, and enzymes that can positively influence plant growth and development or are able to solubilize minerals (i.e. phosphorus, potassium, zinc). One of the mechanisms is connected with production of the phytohormone indole-3-acetic acid (IAA). It was shown that IAA synthesized by bacteria participates in plant–microbe signalling and contributes in roots proliferation and elongation (Vessey 2003). It should be noted that bacterial IAA can also stimulate the activity of ACC synthetase, thereby increasing ACC synthesis (Glick 2012). The ability to synthesize IAA has been found in many rhizobia. Studies have shown that this auxin produced by rhizobia affects the nodulation process (Glick et al. 1998; Spaepen and Vanderleyden 2011). Our research showed that 87.5 % of A. cicer and A. glycyphyllos microsymbionts were able to produce indole-3-acetic acid. The ability to synthesize IAA has also been described, among others, for Rhizobiumleguminosarum, Mesorhizobiumcicer, and Mesorhizobiumloti strains (Chandra et al. 2007; Ahmad et al. 2008).
Phosphorus is one of the nutrients necessary for plant growth and development. Unfortunately, it occurs in soil mainly in the insoluble forms. Plants can absorb phosphorus only in two soluble forms, i.e. as the monobasic (H2PO4−) and the diabasic (HPO42−) ions (Glass 1989). Some PGPR are able to transform the insoluble form of phosphorus to a form available to the plant by acidification of the medium and by phosphorus chelation and transport to the cell (Hameeda et al. 2008; Richardson et al. 2009). It was noticed that inoculation of plants with phosphorus-solubilizing bacteria positively affects the development of the plants (Kucey et al. 1989; Chabot et al. 1996). The ability to solubilize inorganic phosphate has been described in many rhizobial species e.g. R. leguminosarum bv. viciae, Ensifermeliloti, Mesorhizobiummediterraneum, and M. loti (Peix et al. 2001; Chandra et al. 2007; Bianco and Defez 2010). Our studies showed that mesorhizobia isolated from the root nodules of A. cicer and A. glycyphyllos caused P-solubilization visible as a clear zone around the bacterial colony on Pikovskaya’s medium. It is worth noting that the P-solubilization zone for mesorhizobia isolated from the A. cicer root nodules was bigger than that for the A. glycyphyllos symbionts.
The development of rhizobium–fabacean plant symbiosis depends on various environmental factors, inter alia, salinity, pH, and the presence of heavy metals. It is worth noting that the presence of heavy metals at low concentrations is essential for many cellular processes in bacteria. For instance, molybdenum, zinc, and nickel are cofactors of many enzymes, cobalt is the central metal component of the vitamin B12 cofactor, and manganese can be an electron acceptor (Ahemad 2012). It has been shown, however, that heavy metals present in soil at higher concentrations interfere with the metabolism of soil bacteria and can decrease their activity and affect the rhizobium–fabacean plant interaction (Ahemad 2012). Bacteria have developed some mechanisms enabling them to survive in the presence of heavy metals, i.e. precipitation of metals as insoluble salts, efflux of metals from the cells, and chelation of metals (Wani et al. 2009; Zaidi et al. 2012). The mechanism of heavy metal toxicity in respect to rhizobia and rhizobium–fabacean symbiosis is poorly known. Wani et al. (2009) described tolerance of Mesorhizobium sp. RC1 and RC4 strains to Cr(VI). These strains exerted a beneficial effect on the development of chickpea growing in Cr-contaminated soil (Wani et al. 2009). Vidal et al. (2009) described highly metal-resistant Mesorhizobium metallidurans strains, symbionts of Anthyllisvulneraria, which tolerated even 16–32 mM Zn and 0.3–0.5 mM Cd in YEM liquid medium (Vidal et al. 2009). A majority of the strains isolated from A. cicer and A.glycyphyllos root nodules tolerated much lower concentrations of heavy metals, i.e. 0.65 mM Zn, 0.14 mM Cd, and 2.3 mM Pb. A similar resistance level to Zn (0.05–0.5 mM) and Cd (0.05 mM) was described by Vidal et al. (2009) in the case of M. tianshanense ORS 2640T and M. mediterraneum ORS 2739T.
The rhizobium–fabacean symbiosis is a complicated process regulated by both partners of this interaction. It is known that the plant hormone ethylene can inhibit rhizobial infection and nodule formation and limit the number of nodules. Some rhizobia are able to reduce “ethylene stress” by 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase synthesis. ACC deaminase breaks down ACC, an ethylene precursor, into ammonium and α-ketobutyrate. It was shown that bacteria producing ACC deaminase protect plants against various environmental stress factors and phytopathogens, delay plant senescence, and affect the nodulation process (Glick 2014; Nascimento et al. 2012a). The presence of the acdS gene encoding ACC deaminase has been described in many Gram-positive, i.e. Bacilluspumilus, Mycobacterium sp., and Rodococcus sp., and Gram-negative bacteria, e.g. rhizobial species such as R. leguminosarum bv. viciae, E. meliloti, Ensifer medicae, Bradyrhizobiumjaponicum, M. loti, and M. ciceri bv. biserrulae (Belimov et al. 2001, 2005; Sullivan et al. 2002; Madhaiyan et al. 2006; Murset et al. 2012; Nascimento et al. 2012a, b; Singh et al. 2015). Some investigations have also shown that many bacteria of the genus Rhizobium produce ACC deaminase under free-living conditions, whereas the free-living bacteria of the genus Mesorhizobium do not exhibit ACC deaminase activity (Nascimento et al. 2012a, b). In mesorhizobia, the activity of ACC deaminase can be observed during their symbiosis with the plant host, as shown in the case of M. huakuii bv. loti MAFF303099 (Uchiumi et al. 2004; Nukui et al. 2006). In this study, we identified acdS genes in the analysed A. cicer and A. glycyphyllos microsymbionts. The presence of a single copy of the acdS gene in the astragali strains was confirmed by Southern hybridization with the acdS gene of M. loti MAFF303099 as a hybridization probe. As in the case of the mesorhizobia studied by Nascimento et al. (2012a, b), no ACC deaminase activity was detected in the free-living A. cicer and A. glycyphyllos microsymbionts. It is also worth noting that the rhizobia display a low level of ACC deaminase activity in comparison with free-living and endophytic plant growth-promoting bacteria (Glick 2014). The induction of ACC deaminase activity in bacteria is a complex and relatively slow process. It was demonstrated that expression of this enzyme depends on the presence of ACC but some other amino acids such as l-alanine, DL-alanine, and d-serine are also capable to induce this process. It was also found that abiotic and biotic stress can also induce ACC deaminase activity (Glick 2014; Singh et al. 2015). This indicates that different factors may induce and affect the ACC deaminase activity.
The presence of the acdS genes in the genomes of A. cicer and A. glycyphyllos microsymbionts as well as the capability of these bacteria of P-solubilization, utilization of 1-aminocyclopropane-1-carboxylic acid (ACC) as a sole N source, and production of phytohormone IAA indicate that the analysed studied rhizobia can be treated as plant growth-promoting rhizobia. Further studies are needed to prove this assumption, especially these concerning the impact of these bacteria on plant growth and expression of astragali acdS genes during their symbiosis with fabaceans.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Phylogenetic relationship between A. cicer and A. glycyphyllos and reference strains based on deduced AcdS sequences. Bootstrap values (1000 replicates) are shown when higher than 50%. The scale bar represents the percentage of substitutions per site (JPEG 385 kb)
References
- Ahemad M. Implications of bacterial resistance against heavy metals in bioremediation: a review. J Inst Integr Omics Appl Biotechnol. 2012;3:39–46. [Google Scholar]
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci. 2014;26:1–20. doi: 10.1016/j.jksus.2013.05.001. [DOI] [Google Scholar]
- Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res. 2008;163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Belimov A, Safronova VI, Sergeyeva TA, Egorova TN, Matveyeva VA, Tsyganov VE, Borisov AY, Tikhonovich IA, Kluge C, Preisfeld A, Dietz KJ, Stepanok VV. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can J Microbiol. 2001;47:642–652. doi: 10.1139/w01-062. [DOI] [PubMed] [Google Scholar]
- Belimov A, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, Bullitta S, Glick BR. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.) Soil Biol Biochem. 2005;37:241–250. doi: 10.1016/j.soilbio.2004.07.033. [DOI] [Google Scholar]
- Beneduzi A, Ambrosini A, Passaglia LMP. Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Gen Mol Biol. 2012;35:1044–1051. doi: 10.1590/S1415-47572012000600020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya PN, Jha DK. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol. 2012;28:1327–1350. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- Bianco C, Defez R. Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl Environ Microbiol. 2010;76:4626–4632. doi: 10.1128/AEM.02756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chabot R, Antoun H, Cescas MP. Growth promotion of maize and lettuce by phosphate-solubilizing Rhizobium leguminosarum biovar. phaseoli. Plant Soil. 1996;184:311–321. doi: 10.1007/BF00010460. [DOI] [Google Scholar]
- Chandra S, Choure K, Dubey RC, Maheshwari DK. Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotinum and enhances growth of indian mustard (Brassica campestris) Braz J Microbiol. 2007;38:124–130. doi: 10.1590/S1517-83822007000100026. [DOI] [Google Scholar]
- Conforte VP, Echeverria M, Sanchez C, Ugalde RA, Menendez AB, Lepek VC. Engineered ACC deaminase-expressing free-living cells of Mesorhizobium loti show increased nodulation efficiency and competitiveness on Lotus spp. J Gen Appl Microbiol. 2010;56:331–338. doi: 10.2323/jgam.56.331. [DOI] [PubMed] [Google Scholar]
- Edi-Premono M, Moawad MA, Vleck PLG. Effect of phosphate solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones J Crop Sci. 1996;11:13–23. [Google Scholar]
- Frommel MI, Nowak J, Lazarovits G. Growth enhancement and development modifications of in vitro grown potato (Solanum tuberosum ssp. tuberosum) as affected by a nonfluorescent Pseudomonas sp. Plant Physiol. 1991;96:928–936. doi: 10.1104/pp.96.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM. Plant nutrition: an introduction to current concepts. Boston: Jones and Bartlett Publishers; 1989. [Google Scholar]
- Glick BR. The enhancement of plant growth by free living bacteria. Can J Microbiol. 1995;41:109–117. doi: 10.1139/m95-015. [DOI] [Google Scholar]
- Glick BR. Plant growth-promoting bacteria: mechanisms and applications. Waterloo: Hindawi Publishing Corporation Scientifica; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Glick BR, Penrose DM, Li J. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theor Biol. 1998;190:63–68. doi: 10.1006/jtbi.1997.0532. [DOI] [PubMed] [Google Scholar]
- Glick BR, Patten CL, Holguin G, Penrose DM. Biochemical and genetic mechanisms used by plant growth-promoting bacteria. London: Imperial College Press; 1999. [Google Scholar]
- Glick BR, Cheng Z, Czarny J, Duan J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur J Plant Pathol. 2007;119:329–339. doi: 10.1007/s10658-007-9162-4. [DOI] [Google Scholar]
- Gnat S, Wójcik M, Wdowiak-Wróbel S, Kalita M, Ptaszyńska A, Małek W. Phenotypic characterization of Astragalus glycyphyllos symbionts and their phylogeny based on the 16S rDNA sequences and RFLP of 16S rRNA gene. Ant Leeuwen. 2014;105:1033–1048. doi: 10.1007/s10482-014-0163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnat S, Malek W, Oleńska E, Wdowiak-Wróbel S, Kalita M, Rogalski J, Wójcik M. Multilocus sequence analysis supports taxonomic position of Astragalus glycyphyllos symbionts based on DNA–DNA hybridization. Int J Syst Evol Microbiol. 2015 doi: 10.1099/ijsem.0.000862. [DOI] [PubMed] [Google Scholar]
- Gnat S, Malek W, Oleńska E, Wdowiak-Wróbel S, Kalita M, Łotocka B, Wójcik M. Phylogeny of symbiotic genes and the symbiotic properties of rhizobia specific to Astragalus glycyphyllos L. Plos One. 2015 doi: 10.1371/journal.pone.0141504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameeda B, Harini G, Rupela OP, Wani SP, Reddy G. Growth promotion of maize by phosphate-solubilizing bacteria isolated from composts and macrofauna. Microbiol Res. 2008;163:234–242. doi: 10.1016/j.micres.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Honma M, Shimomura T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric Biol Chem. 1978;42:1825–1831. [Google Scholar]
- Jadhav RS, Desai AJ. Isolation and characterization of siderophore from cowpea Rhizobium (peanut isolate) Curr Microbiol. 1992;24:137–141. doi: 10.1007/BF01568978. [DOI] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Hume DJ, Schner FM, Singleton C, Tipping B, Laliberte M, Frauley K, Kutchaw T, Simonson C, Lifshitz R, Zaleska I, Lee L. Plant growth promoting rhizobacteria on canola. Plant Dis. 1988;72:42–46. doi: 10.1094/PD-72-0042. [DOI] [Google Scholar]
- Kucey RMN, Janzen HH, Leggett ME. Microbially mediated increases in plant available phosphorus. Adv Agron. 1989;42:199–225. doi: 10.1016/S0065-2113(08)60525-8. [DOI] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinform. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ma W, Sebestianova SB, Sebestian J, Burd GI, Guinel FC, Glick BR. Prevalence of 1-aminocyclopropane-1-carboxylate deaminase in Rhizobium spp. Ant Leeuwen. 2003;83:285–291. doi: 10.1023/A:1023360919140. [DOI] [PubMed] [Google Scholar]
- Ma W, Charles TC, Glick BR. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol. 2004;70:5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Ryu JH, Sa TM. Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase-containing Methylobacterium fujisawaense. Planta. 2006;224:268–278. doi: 10.1007/s00425-005-0211-y. [DOI] [PubMed] [Google Scholar]
- Manassila M, Nuntagij A, Kotepong S, Boonkerd N, Teaumroong N. Characterization and monitoring of selected rhizobial strains isolated from tree legumes in Thailand. Afr J Biotechnol. 2007;6:1393–1402. [Google Scholar]
- Murset V, Hennecke H, Pessi G. Disparate role of rhizobial ACC deaminase in root-nodule symbioses. Symbiosis. 2012;57:43–50. doi: 10.1007/s13199-012-0177-z. [DOI] [Google Scholar]
- Nascimento FX, Brígido C, Glick BR, Oliveira S. ACC deaminase genes are conserved between Mesorhizobium species able to nodulate the same host plant. FEMS Microbiol Lett. 2012;336:26–37. doi: 10.1111/j.1574-6968.2012.02648.x. [DOI] [PubMed] [Google Scholar]
- Nascimento FX, Brígido C, Glick BR, Oliveira S, Alho L. Mesorhizobium ciceri LMS-1 expressing an exogenous ACC deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett Appl Microbiol. 2012;55:15–21. doi: 10.1111/j.1472-765X.2012.03251.x. [DOI] [PubMed] [Google Scholar]
- Nukui N, Minamisawa K, Iyabe AS, Aoki T. Expression of the 1-aminocyclopropane-1-carboxylic acid deaminase gene requires symbiotic nitrogen fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl Environ Microbiol. 2006;72:4964–4969. doi: 10.1128/AEM.02745-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix A, Rivas-Boyero AA, Mateos PF, Rodrıguez-Barrueco C, Martınez-Molina E, Velázquez E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of Mesorhizobium mediterraneum under growth chamber conditions. Soil Biol Biochem. 2001;33:103–110. doi: 10.1016/S0038-0717(00)00120-6. [DOI] [Google Scholar]
- Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant. 2003;118:10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Pikovskaya RI. Mobilization of phosphorus in soil in connection with vital capacity of source microbial species. Microbiologiya. 1948;17:362–370. [Google Scholar]
- Pitcher DG, Saunders A, Owe RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009;321:305–339. doi: 10.1007/s11104-009-9895-2. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saleem M, Arshad M, Hussain S, Bhatti AS. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J Ind Microbiol Biotechnol. 2007;34:635–648. doi: 10.1007/s10295-007-0240-6. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Russell DW, editors. Molecular cloning: a laboratory manual. 3. Cold Spring: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shahzad SM, Khalid A, Arshad M, Tahir J, Mahmood T. Improving nodulation, growth and yield of Cicer arietinum L. through bacterial ACC-deaminase induced changes in root architecture. Eur J Soil Biol. 2010;46:342–347. doi: 10.1016/j.ejsobi.2010.05.007. [DOI] [Google Scholar]
- Singh RP, Shelke GM, Kumar A, Jha PN. Biochemistry and genetics of ACC deaminase: a weapon to “stress ethylene” produced in plants. Front Microbiol. 2015 doi: 10.3389/fmicb.2015.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harbor Perspect Biol. 2011;3:a001438. doi: 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, Weaver JE, Webby RJ, De Bruijn FJ, Ronson CW. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi SM, Timko MP. Effects of ethylene and inhibitors of ethylene synthesis and action on nodulation in common bean (Phaseolus vulgaris L.) Plant Soil. 2003;257:125–131. doi: 10.1023/A:1026280517660. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Ohwada T, Itakura M, Mitsui H, Nukui N, Dawadi P, Kaneko T, Tabata S, Yokoyama T, Tejima K, Saeki K, Omori H, Hayashi H, Maekawa T, Sriprang R, Murooka Y, Tajima S, Simomura K, Nomura M, Suzuki A, Shimoda Y, Sioya K, Abe M, Minamisawa K. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J Bacteriol. 2004;186:2439–2448. doi: 10.1128/JB.186.8.2439-2448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyé F, Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peer R, Schippers B. Plant growth responses to bacterization with selected Pseudomonas spp. strains and rhizosphere microbial development in hydroponic cultures. Can J Microbiol. 1989;35:456–463. doi: 10.1139/m89-070. [DOI] [Google Scholar]
- Vessey JK. Plant growth promoting rhizobacteria as biofertilizer. Plant Soil. 2003;255:571–586. doi: 10.1023/A:1026037216893. [DOI] [Google Scholar]
- Vidal C, Chantreuil C, Berge O, Maure L, Escarre J, Bena G, Brunel B, Cleyet-Marel JC. Mesorhizobium metallidurans sp. nov., a metal-resistant symbiont of Anthyllis vulneraria growing on metallicolous soil in Languedoc, France. Int J Syst Evol Microbiol. 2009;59:850–855. doi: 10.1099/ijs.0.003327-0. [DOI] [PubMed] [Google Scholar]
- Vincent JA. A manual for the practical study of root nodule bacteria. Oxford: Blackwell Scientific Publication Ltd; 1970. [Google Scholar]
- Wani PA, Zaidi A, Khan MS. Chromium reducing and plant growth promoting potential of Mesorhizobium species under chromium stress. Bioremed J. 2009;13:121–129. doi: 10.1080/10889860903124289. [DOI] [Google Scholar]
- Wdowiak S, Małek W. Numerical analysis of Astragalus cicer microsymbionts. Curr Microbiol. 2000;41:142–148. doi: 10.1007/s002840010108. [DOI] [PubMed] [Google Scholar]
- Wdowiak-Wróbel S, Małek W. Following phylogenetic tracks of Astragalus cicer microsymbionts. Ant Leeuwen. 2010;97:21–34. doi: 10.1007/s10482-009-9384-x. [DOI] [PubMed] [Google Scholar]
- Zaidi A, Ahemad M, Oves M, Ahmad E, Saghir Khan M. Role of phosphate-solubilizing bacteria in legume improvement, microbes for legume improvement. Vienna: Springer; 2010. [Google Scholar]
- Zaidi A, Wani PA, Khan MS, editors. Toxicity of heavy metals to legumes and bioremediation. Berlin: Springer; 2012. [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Ann Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationship between A. cicer and A. glycyphyllos and reference strains based on deduced AcdS sequences. Bootstrap values (1000 replicates) are shown when higher than 50%. The scale bar represents the percentage of substitutions per site (JPEG 385 kb)