Abstract
Melatonin receptors are seven transmembrane‐spanning proteins belonging to the GPCR superfamily. In mammals, two melatonin receptor subtypes exist ‐ MT1 and MT2 ‐ encoded by the MTNR1A and MTNR1B genes respectively. The current review provides an update on melatonin receptors by the corresponding subcommittee of the International Union of Basic and Clinical Pharmacology. We will highlight recent developments of melatonin receptor ligands, including radioligands, and give an update on the latest phenotyping results of melatonin receptor knockout mice. The current status and perspectives of the structure of melatonin receptor will be summarized. The physiological importance of melatonin receptor dimers and biologically important and type 2 diabetes‐associated genetic variants of melatonin receptors will be discussed. The role of melatonin receptors in physiology and disease will be further exemplified by their functions in the immune system and the CNS. Finally, antioxidant and free radical scavenger properties of melatonin and its relation to melatonin receptors will be critically addressed.
Abbreviations
- AD
Alzheimer's disease
- ADHD
attention‐deficit/hyperactivity disorder
- ASDs
autism spectrum disorders
- fMLP
N‐formyl‐l‐methionyl‐l‐leucyl‐l‐phenylalanine
- FPG
fasting plasma glucose
- HD
Huntington's disease
- IOP
intraocular pressure
- MS
multiple sclerosis
- MTR
melatonin receptor
- NREM
non‐rapid eye movement
- PD
Parkinson's disease
- QR2
quinone reductase 2
- REM
rapid eye movement
- SCN
suprachiasmatic nucleus
- T2D
type 2 diabetes
Tables of Links
| TARGETS |
|---|
| GPCRs a |
| 5‐HT2C receptor |
| β2‐adrenoceptor |
| A2A adenosine receptor |
| GPR50 |
| MT1 receptor |
| MT2 receptor |
| Voltage‐gated ion channels b |
| Cav2.2 channels |
| Transporters c |
| GLUT1 |
| Enzymes d |
| CaMKII |
| ERK1 |
These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016) and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (a,b,c,dAlexander et al., 2015a, 2015b, 2015c, 2015d).
Introduction
The hormone melatonin is mainly produced by the pineal gland following a circadian rhythm, with high levels during the subjective night. Melatonin can also be produced by extra‐pineal sites like the retina, the gastrointestinal tract and the innate immune system. This hormone regulates a variety of physiological and neuroendocrine functions in mammals through activation of two GPCRs, the MT1 and MT2 receptors. Both receptors are typically coupled to Gi/o‐type proteins and the MT1 receptor is coupled, in addition, to Gq‐type proteins. In humans, the MTNR1A gene encoding MT1 is located on chromosome 4q35.1 and the MTNR1B gene encoding MT2 on chromosome 11q21‐q22.
This article will review and discuss recent updates by the melatonin receptor subcommittee of the International Union of Basic and Clinical Pharmacology (IUPHAR) database (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=39), which include the development of new MT receptor ligands, radioligands and structural perspectives of the MT receptors. The discovery of MT receptor dimers with physiological function in vivo as well as genetic variants and mutants of MT receptors will be discussed as they provide a new dimension to understand MT receptor pharmacology and function. An update on the latest results obtained with MT receptor knockout (KO) mouse models will be provided. Among the many physiological effects of MT receptors, we chose to focus on those of the immune system and the CNS. At the end of the review, MT receptor‐independent effects, including antioxidant and free radical scavenger properties of melatonin, will be critically addressed.
For more complete or other specific aspects of MT receptors, the reader is referred to other recent expert reviews (Jockers et al., 2008; Dubocovich et al., 2010; Markus et al., 2013; Tosini et al., 2014; Zlotos et al., 2014; Liu et al., 2016).
MT receptor ligands
MT1 and MT2 receptors share a high degree of sequence homology and bind both the natural ligand, melatonin, with high affinity. Important progress has been made in the development of synthetic MT receptor antagonists and agonists and subtype‐selective ligands by diversifying the chemical scaffolds. Indeed, MT receptor ligands from different structural classes show distinct structure–activity relationships on native and recombinant MT receptors (Dubocovich et al., 1997; Dubocovich et al., 2010; Zlotos, 2012; Zlotos et al., 2014). The methoxy group and the acetamido side chain of melatonin determine the intrinsic activity and the binding affinity, respectively, at both hMT1 and hMT2 receptors (Dubocovich et al., 1997; Browning et al., 2000; Audinot et al., 2003). Replacement of the amide methyl group by ethyl and propyl substituents enhances affinity (Sugden et al., 1995). Exchange of the indole ring by various aromatic scaffolds maintains high binding and agonist potency.
Substitutions at the two‐position with a halogen or a phenyl group generate agonists with ~10‐fold increased binding affinity. The majority of non‐selective MT1–MT2 receptor ligands, including drugs used in humans, for example, ramelteon [Rozerem®, (Kato et al., 2005; Mini et al., 2007; Rawashdeh et al., 2011)], agomelatine [Valdoxan®, (de Bodinat et al., 2010)] and tasimelteon [Hetlioz®, (Rajaratnam et al., 2009; Lavedan et al., 2015)], are agonists (Figure 1). Ramelteon and tasimelteon are MT receptor selective, while agomelatine is also an antagonist at the 5‐HT2C receptors, a pharmacological property believed to contribute to its antidepressant action. The therapeutic effects of approved drugs acting on hMT1 and/or hMT2 receptors as agonists were recently reviewed (Liu et al., 2016). Other non‐selective MT receptor agonists include 6‐chloromelatonin, 6‐hydroxymelatonin, 2‐iodomelatonin, GR 196429 (Dubocovich et al., 1997; Browning et al., 2000; Audinot et al., 2003), UCM 793 (Rivara et al., 2007) and 2‐methoxy‐α,β‐didehydro‐agomelatine (Morellato et al., 2013). This latter ligand shows the highest affinity for hMT1 (K i = 0.03 nM) and hMT2 (K i = 0.07 nM) receptors and ~3500‐fold greater potency than melatonin in the melanophore aggregation assay. TIK 301 (Mulchahey et al., 2004) acts also as an antagonist at the 5‐HT2C and 5‐HT2B receptors (Landolt and Wehrle, 2009). 5‐HEAT has a unique pharmacological profile acting as a full agonist at the hMT1 receptor and antagonist at the hMT2 receptor (Nonno et al., 2000). EFPPEA, a high‐affinity hMT1 (K i = 0.062 nM) and hMT2 receptor (K i = 0.420 nM) agonist, decreases the percentage of wakefulness and increases the percentage of slow wave sleep in cats (Koike et al., 2011). The competitive MT receptor antagonist luzindole lacks the methoxy group, which led to the suggestion that this group is necessary for intrinsic activity (Dubocovich, 1988). Similarly, S22153 acts as a partial agonist (Audinot et al., 2003). Luzindole, with a 15‐ to 25‐fold higher affinity for hMT2 than for hMT1 receptors, is widely used for pharmacological characterization of functional MT receptors (Dubocovich et al., 1997; Dubocovich et al., 1998; Browning et al., 2000; Dubocovich et al., 2010).
Figure 1.
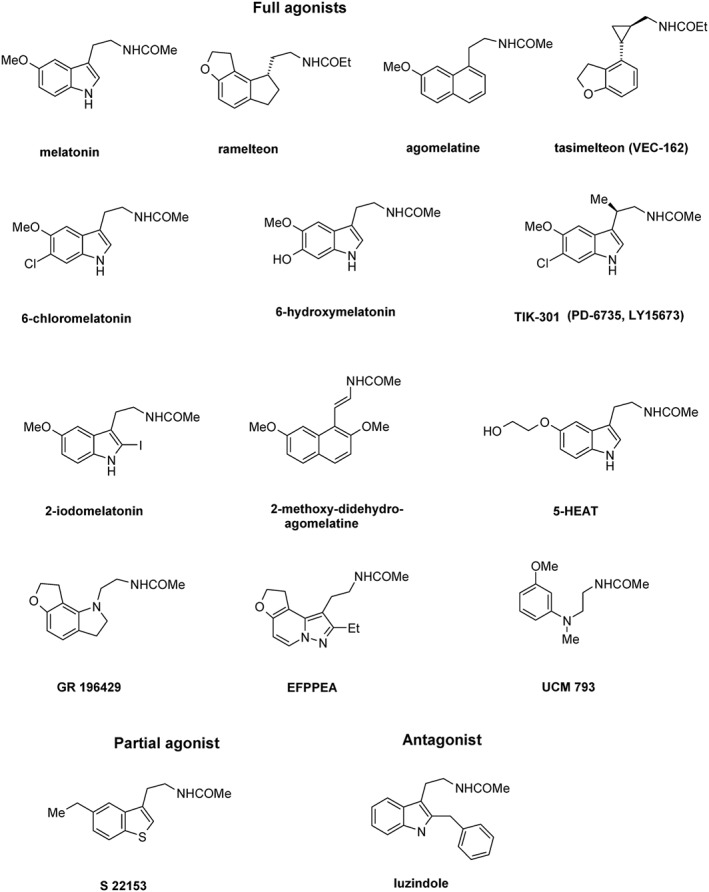
Structures of non‐selective MT1/MT2 receptor ligands. 5‐HEAT, 5‐hydroxyethoxy‐N‐acetyltryptamine; EFPPEA, ethyl‐furo‐pyrazolo‐pyridine‐ethyl‐acetamide.
A ligand is considered selective for a specific receptor type when its affinity or potency is at least 100‐times higher than that for the other(s) receptor types in the family (Dubocovich et al., 2010). This concept holds true for in vitro studies where ligand concentrations can be easily adjusted. However, ligand selectivity might be more difficult to reach in vivo, in the body fluids reaching the receptors. Depending on ligand dose and pharmacokinetics, concentrations could easily raise to levels activating both receptors (e.g. MT1 and MT2 receptors). This is particularly of concern for melatonin and synthetic MT receptor ligands activating receptors at picomolar concentrations (Dubocovich et al., 1997; Browning et al., 2000; Audinot et al., 2003). Therefore, caution should be taken when interpreting selective MT receptor activation in vivo using MT receptor‐selective ligands, unless pharmacological selectivity or lack of it, is confirmed by KO models with deletion of each receptor type.
Numerous ligands with high selectivity for hMT2 over hMT1 receptors have been identified (Zlotos et al., 2014). MT2 receptors possess a lipophilic pocket close to the N1–C2 binding region of melatonin, which is absent in MT1 receptors (Rivara et al., 2005). Accordingly, most MT2 receptor‐selective ligands bear a flexible bulky hydrophobic substituent in a position equivalent to C2 or N1 of melatonin (Figure 2). The tetrahydroquinoline analogue UCM1014 is the most potent MT2 receptor‐selective ligand reported to date. It shows picomolar binding affinity (K i = 0.001 nM) at hMT2 receptors, >10 000‐fold selectivity over hMT1 receptors and full agonist profile in the GTPγS test (Spadoni et al., 2015). Other agonists with approximately 800‐fold hMT2 receptor selectivity are BOMPPA (Hu et al., 2010; Heckman et al., 2011; Chan et al., 2013; Hu et al., 2013) and CIFEA (Koike et al., 2011). In imprinting control region mice, CIFEA reentrainment effects to a new light/dark cycle indicate the involvement of MT receptors in the regulation of chronobiotic activity (Koike et al., 2011). The dose of CIFEAA used in this study most likely reached micromolar concentrations, which would activate both MT1 and MT2 receptors, precluding any conclusion about the specific receptor type involved in the regulation of chronobiotic processes. Similarly, doses of the MT2 receptor‐selective antagonist 4P‐PDOT (90 μg/mouse s.c.) used to block the melatonin‐mediated phase advance of circadian activity rhythms in mice (Dubocovich et al., 1998) may have resulted in micromolar circulating 4P‐PDOT concentrations hence blocking both MT1 and MT2 receptors.
Figure 2.
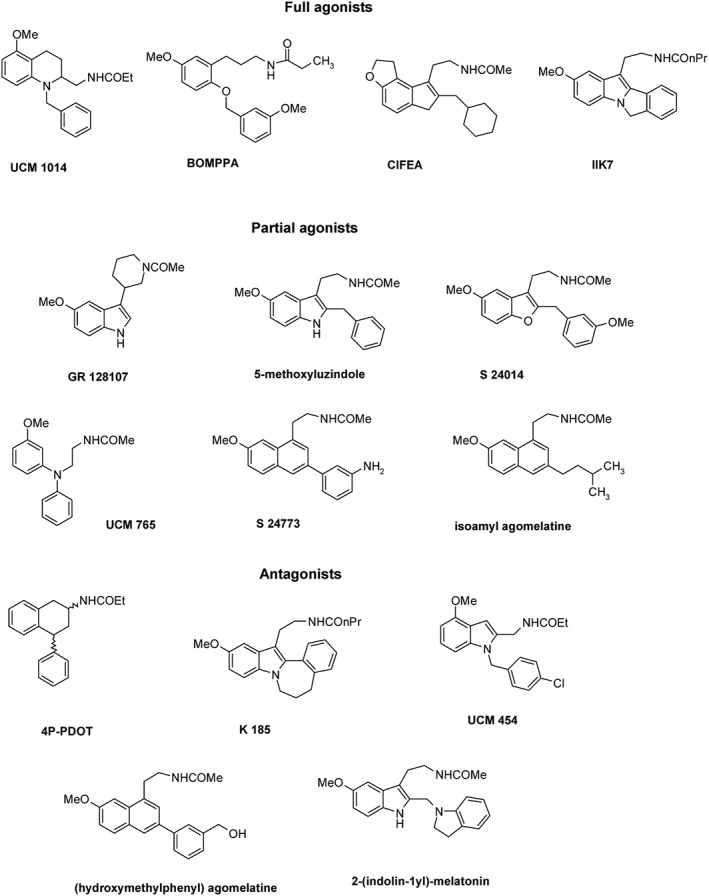
Structures of MT2 receptor‐selective ligands. BOMPPA, benzyloxy‐methoxyphenyl‐propylamide; CIFEA, cyclohexylmethyl‐indenofurane‐ethylacetamide; 4P‐PDOT, 4‐phenyl‐2‐propionamidotetralin.
Two moderately selective MT2 receptor ligands, the agonist IIK7 (Faust et al., 2000) and the partial agonist UCM 765 (Rivara et al., 2007), have been used to examine the role of each MT receptor type in the modulation of sleep architecture. UCM 765 promoted non‐rapid eye movement (NREM) sleep in rodents, and this effect was blocked by the MT2 receptor antagonist 4P‐PDOT (Ochoa‐Sanchez et al., 2011). In contrast, the non‐selective MT1–MT2 receptor agonist UCM793 decreased sleep onset without having an effect on NREM sleep maintenance suggesting that dual MT1 and MT2 receptor agonistic activity accounts for the effect on sleep onset, whereas selectivity for MT2 receptors has an additional effect on NREM sleep maintenance. IIK7 was also reported to reduce NREM sleep onset latency and transiently increase the time spent in NREM sleep in rats without altering rapid eye movement (REM) sleep latency or the amount of REM sleep (Fisher and Sugden, 2009).
Among the hMT2 receptor‐selective partial agonists GR 128107, 5‐methoxyluzindole, S 24014, S 24773 (Dubocovich et al., 1997; Audinot et al., 2003) and isoamyl agomelatine, the latter shows the highest affinity (K i = 0.01 nM) and selectivity (7200‐fold) (Ettaoussi et al., 2012). 4P‐PDOT, an hMT2 receptor‐selective antagonist with 300‐ to 1500‐fold higher affinity for hMT2 receptors, is still considered the gold standard for pharmacological characterization of MT receptors (Dubocovich et al., 1997). Other MT2 receptor‐selective antagonists, such as K185 (Sugden et al., 1999; Faust et al., 2000), UCM 454 (Rivara et al., 2005) and 2‐(indolin‐1yl) melatonin (Zlotos et al., 2009), display ~100‐fold higher affinity for hMT2 receptors. For (hydroxymethyl)phenyl agomelatine, the affinity for hMT2 receptors is 750‐times higher than for hMT1 receptors (Poissonnier‐Durieux et al., 2008).
Discovery of MT1 receptor‐selective ligands remains a challenge, and only few compounds with preference for hMT1 receptors have been reported (Zlotos et al., 2014). Ligands preferentially binding to these receptors reach maximally 100‐fold selectivity, and, when investigated, this selectivity is significantly reduced in functional in vitro studies (Figure 3). A common structural feature conferring MT1 receptor selectivity is a bulky, hydrophobic ether replacing the methoxy group. The first hMT1 receptor‐selective agents were obtained by connecting two agomelatine units via their ether oxygen by (CH2)3‐ and (CH2)4‐linker to give S 26131 (antagonist) and S 26284 (partial agonist), both displaying ~100‐fold selectivity (Audinot et al., 2003; Descamps‐Francois et al., 2003). A similar approach led to the UCM 793 dimer with 100‐fold hMT1 receptor selectivity and partial agonist activity (Spadoni et al., 2011). Monomeric ligands, such as CBOBNEA (Mesangeau et al., 2010), and AAE M PBP amine (Rivara et al., 2012) are partial agonists showing similar ~100‐fold selectivity for hMT1 receptors. N‐acetyl‐O‐phenoxypropyl serotonin is a full agonist obtained by exchange of the methoxy group of melatonin with an O(CH2)3OPh moiety. Although it shows only 10‐fold binding preference toward hMT1 receptors, its MT1–MT2 receptor binding ratio and hMT1 receptor affinity were higher than that for the MT1 receptor‐selective reference compound S 26131 that was retested under the same experimental conditions (Markl et al., 2011). A 140‐fold MT1 receptor selectivity could be attained by introduction of two fluorine atoms into the N‐acetyl group of agomelatine. The resulting difluoroagomelatine shows high hMT1 receptor binding (K i = 0.03 nM) and is a non‐selective MT1–MT2 receptor full agonist (Ettaoussi et al., 2012). Very recently, tetrafluoro S26131, the difluoroacetamide analogue of S 26131, has been reported to show higher affinity and selectivity toward hMT1 receptors than the parent ligand (Zlotos et al., 2015).
Figure 3.
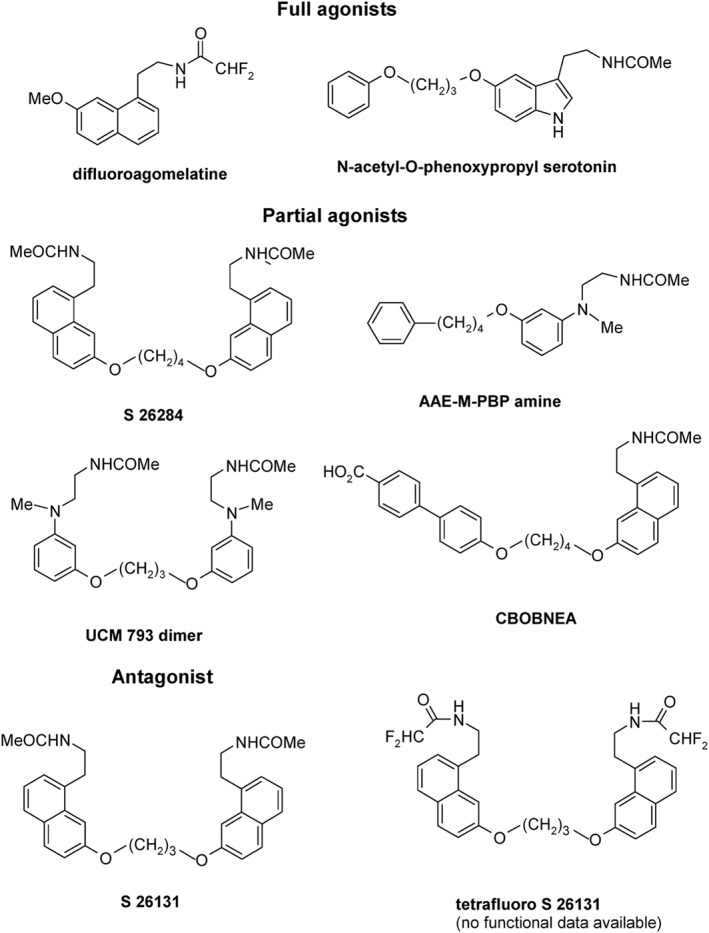
Structures of MT1 receptor‐selective ligands. CBOBNEA, carboxybiphenyloxy‐butoxy‐naphthalene‐ethylacetamide; AAE‐M‐PBP amine, acetylaminoethyl‐methyl‐phenylbutoxyphenyl‐amine.
In summary, while numerous ligands selective for the MT2 receptor subtype are available, discovery of ligands with at least >100‐fold selectivity for MT1 receptors remains a challenging task. None of the MT1 receptor‐selective ligands has been tested in vivo. Future progress on the elucidation of the structure of MT receptors will hopefully foster the discovery of such ligands. The MT1–MT2 non‐selective receptor antagonist luzindole and the MT2 receptor‐selective antagonists 4P‐PDOT are still considered the gold standards for pharmacological characterization of MT receptors.
Radioligands – update
Radioactive‐ and fluorescent‐labelled ligands are indispensable tools for the pharmacological characterization of GPCRs. A major breakthrough in the field of MT receptor research was the labelling of 2‐iodomelatonin with 125I at carbon 2 resulting in a high‐affinity radioligand with high specific activity (Vakkuri et al., 1984) for use in the localization (Vanecek, 1988) and pharmacological characterization of MT receptors in tissues (Dubocovich and Takahashi, 1987). The radioligand, 2‐[125I]iodomelatonin (2‐[125I]‐IMLT), has been extensively used as a high‐affinity radioligand for both MT1 and MT2 receptors, and was until recently the only available radioligand for the characterization and localization of melatonin binding sites in native tissues (Figure 4). Studies performed with [3H]‐melatonin ([3H]‐MLT) established the pharmacological profile of the human recombinant MT1 and MT2 receptors, as being identical to that established using 2‐[125I]‐IMLT as a radioligand. However, due to the rather low specific activity of this [3H]‐MLT, its use to characterize and/or localize melatonin sites in tissues with low MT receptor density is limited (Browning et al., 2000).
Figure 4.
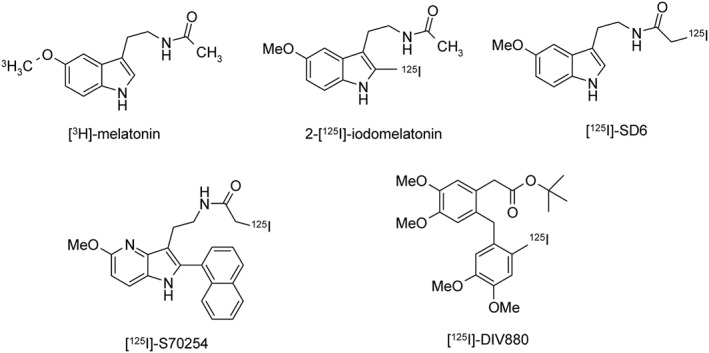
Structures of radioligands used to determine binding affinity for MT1 and MT2 receptors.
Three new iodinated radioligands have been recently characterized for use in the pharmacological characterization and localization of MT receptors (Figure 4). These radioligands are as follows: SD6 (N‐[2‐(5‐methoxy‐1Hindol‐3‐yl)ethyl]iodoacetamide), S70254 (2‐iodo‐N‐2‐[5‐methoxy‐2‐(naphthalen‐1‐yl)‐1H‐pyrrolo[3,2‐b]pyridine‐3‐yl])acetamide and DIV880 2‐(2‐[(2‐iodo‐4,5‐dimethoxyphenyl)methyl]‐4,5‐dimethoxy phenyl) (Legros et al., 2013; Legros et al., 2016). [125I]‐SD6 has a similar pharmacological profile to that of 2‐[125I]‐IMLT with the same affinity for MT1 and MT2 receptors. On the contrary, the two other radioligands [125I]‐S70254 and [125I]‐DIV880 show selectivity for MT2 receptors with pKd values of 9.6 and 9.7, respectively, in the absence of any specific binding to MT1 receptors. All radioligands are agonists, either partial agonists ([125I]‐S70254, [125I]‐DIV880) or full agonists (2‐[125I]‐IMLT, [125I]‐SD6, [3H]‐MLT), which means that their Kd values depend not only on the affinity of the ligand for the receptor but also on the activation of the G protein in the ternary Ligand‐Receptor‐G protein complex.
The extensive pharmacological characterization of these three new radioligands in comparison with 2‐[125I]‐IMLT and [3H]‐MLT on membrane preparations from CHO‐K1 cell lines stably expressing hMT1 or hMT2 receptors showed that [125I]‐S70254 and [125I]‐DIV880 mainly differ from 2‐[125I]‐IMLT in their dissociation kinetics, which are faster for [125I]‐S70254 and [125I]‐DIV880 than for 2‐[125I]‐IMLT (Legros et al., 2016). Interestingly, [125I]‐SD6 labelled only approximately half of the binding sites detected with 2‐[125I]‐IMLT in cells expressing hMT1 receptors while comparable amounts were detected in cells expressing hMT2 receptors (Legros et al., 2013). This suggests the existence of different receptor subpopulations for hMT1 receptors of which [125I]‐SD6 labels a more restricted number than 2‐[125I]‐IMLT. In contrast, for hMT2 receptors, similar subpopulations would be detectable by both radioligands. The nature of these receptor subpopulations is currently unknown but could be related to the differential engagement of hMT1 receptors into complexes with different G proteins or ß‐arrestins following the binding of these agonistic radioligands. [125I]‐SD6 detected as 2‐[125I]‐IMLT binding sites in sheep retinal membranes, while the MT2 receptor‐specific ligands [125I]‐S70254 and [125I]‐DIV880 failed to do so.
The MT2 receptor‐specific [125I]‐S70254 was successfully used for autoradiography studies in rat and sheep brain and retina slices (Legros et al., 2016). A similar labelling pattern to 2‐[125I]‐IMLT (detecting MT1 and MT2 receptors) was observed in several areas but also distinct labelling in others. Absence of labelling by [125I]‐S70254 in regions that are labelled by 2‐[125I]‐IMLT can be explained by low (undetectable) MT2 receptor levels. Absence of 2‐[125I]‐IMLT labelling in regions labelled by [125I]‐S70254 could be due to the detection of different receptor complexes (see above).
Altogether, the new radioligands considerably expand the repertoire of pharmacological tools for MT receptors with the development of MT2 receptor‐specific radioligands and radioligands detecting distinct receptor populations revealing a previously unrecognized diversity. The availability of a radiolabelled antagonist would largely contribute in a better characterization of these different populations. Further advances can be expected from the development of fluorescent‐labelled ligands.
Structural perspectives for MT receptors
Currently, crystal structures of MT1 and MT2 receptors are not available. Despite a sequence identity lower than 30% between MT receptors and the closest crystallized GPCRs, several three‐dimensional (3D) models have produced some structural hypotheses for binding of (non)selective MT1 and/or MT2 receptor agonists (Table 1). According to site‐directed mutagenesis data, most of these models corroborate the importance of both serine residues 3.35 and 3.39 in MT1 as well as His5.46 in both MT1 and MT2 receptors. Although His5.46 seems to be an anchoring residue for polar interactions with the methoxy or amide group of melatonin, only a few models of MT1 receptors display a direct participation of serine residues in melatonin binding (Chugunov et al., 2006; Farce et al., 2008), which could be otherwise involved in an essential bending of helix 3 for binding site plasticity. Models take also into account several receptor–ligand interactions with amino acids conserved within GPCRs and known to play a role in aromatic switch activation (F5.47, W6.48).
Table 1.
Summary of reported MT receptor homology models
| Homology model | Crystal template | Binding amino acid | Reference |
|---|---|---|---|
| MT1‐ramelteon (non‐selective agonist) | Inactive rhodopsin (Palczewski et al., 2000) | Y175(E2), S182(E2), V5.43, H5.46 | Uchikawa et al., 2002 |
| MT1‐agomelatine (non‐selective agonist) | Inactive rhodopsin (Okada et al., 2002) | L2.46, M3.32, S3.35, S3.39, H5.46, F5.47, P5.50 | Voronkov et al., 2005 |
| MT1‐melatonin (non‐selective agonist) | Inactive rhodopsin (Okada et al., 2002) | L2.46, M3.32, S3.35, V3.36, S3.39, H5.46, F5.47, F6.44, W6.48, L6.51, N6.52, N7.49 | Farce et al., 2008 |
| MT1‐2phenylmelatonin (selective agonist) | Active β2‐adrenergic (Rasmussen et al., 2011a, 2011b) | M3.32, G3.33, Y5.38, H5.46, W6.48, L6.51, N6.52, Y7.39, A7.42, Y7.43 | Rivara et al., 2012 |
| MT2‐2iodomelatonin (non‐selective agonist) | Inactive rhodopsin (Okada et al., 2002) | V5.42, H5.46, N6.52, L6.56, Y7.43 | Mazna et al., 2004 |
| MT2‐UCM454 (selective antagonist) | Inactive rhodopsin (Okada et al., 2004) | V3.36, I3.37, V3.40, Y183(E2), H5.46, F5.47, P5.50, I5.51, F6.44, W6.48 | Rivara et al., 2005 |
| MT2‐melatonin | Inactive rhodopsin (Okada et al., 2004) | L2.46, A2.49, S3.35, I3.37, S3.39, V5.42, V5.43, H5.46, F5.47 | Voronkov et al., 2005 |
| MT2‐melatonin | Inactive rhodopsin (Okada et al., 2002) | S3.35, V3.36, S3.39, V5.42, H5.46, W6.48, Y7.43 | Chugunov et al., 2006 |
| MT2‐melatonin | Inactive rhodopsin (Okada et al., 2002) | G3.33, V3.36, I3.37, N4.60, L4.57, T191(E2), Y5.38, H5.46 | Farce et al., 2008 |
| MT2‐melatonin | Active rhodopsin (Scheerer et al., 2008) | A3.29, V3.36, N4.60, H5.46, W6.48, L6.51 | Zefirova et al., 2011 |
| MT2‐acylaminoethyl tetralin (selective partial agonist) | Active β2‐adrenoceptor (Rasmussen et al., 2011a, 2011b) | M3.32, V3.36, N4.60, H5.46, W6.48, N6.52, Y7.43 | Pala et al., 2013 |
Such homology modelling methods make predictions of flexible receptor regions difficult. Although not directly proven for MT receptors, E2 and I3 loops are known to be key features for ligand accessibility and G protein binding of GPCRs. Moreover, amino acid sequences of MT receptors show several singularities like the presence of a 3.49NRY3.51 motif instead of the classical 3.49DRY3.51 motif of other rhodopsin‐like GPCRs. Another specificity is the replacement of the proline by an alanine residue in the conserved 7.49NPXXY7.53 motif. Buried in the vicinity of the cytoplasmic surface, these marked differences are likely to affect receptor activation and/or signalling specificity of MT receptors rather than the ligand binding process.
Whereas 3D models of MT receptors were up to now dedicated to the discovery and optimization of new efficient drugs, the next generation of 3D models should be expanded toward larger, multimeric systems and not be restricted to receptor monomers. Computation of the energy landscape of GPCRs by enhanced molecular dynamics simulations, together with NMR and X‐ray studies, provided valuable molecular insights on the dynamics of ligand recognition, receptor activation and oligomerization (Johnston and Filizola, 2014). Depicting free energy landscapes of MT receptors should address biasing molecular dynamics simulations from the inactive apoform transiting toward the active trimeric L‐R‐Gi or L‐R‐arrestin forms of receptors (Figure 5). As ligands modulate these free energy landscapes (Provasi et al., 2011; Dror et al., 2013), in silico optimization of new efficient ligand structures could be explored by predicting its functional selectivity through arrestin or Gi‐mediated pathways. These approaches also open the way for the exploration of homodimers and heterodimers, particularly MT1/MT2 and MT1/GPR50 receptor complexes, as discussed in the following section.
Figure 5.
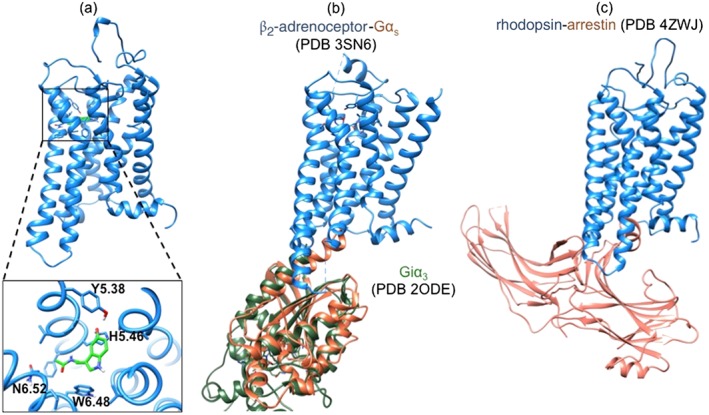
Structures useful for the study of MT1 receptor structure–function relationships. MT1*‐MLT (A) was derived from active forms of rhodopsin, β2‐adrenoceptor and A2A adenosine receptors (unpublished data, N.R.). Docking of melatonin (MLT) in the solvent‐accessible cavity was achieved by energy relaxation by 300‐ns molecular dynamics simulations. The structure of the MT1*‐MLT‐Giα3 complex could be modelled on the basis of the sequence homology with the β2‐adrenoceptor‐Gαs structure (Rasmussen et al., 2011b) and the homology between Gαs and Giα3 (Soundararajan et al., 2008) (B) whereas the structure of the MT1*‐MLT‐arrestin complex could be modelled based on the crystallized rhodopsin‐arrestin complex (Kang et al., 2015) (C).
MT receptor dimers
MT receptors are part of dynamic signalling complexes that contain proteins involved in receptor biosynthesis, export, signalling, desensitization, internalization and cytoskeleton modulation (Daulat et al., 2007; Maurice et al., 2008) (IntAct database, http://www.ebi.ac.uk/intact/search/do/search?searchString=pubid:26514267). The core of these complexes is often composed of receptor dimers, either homodimers of the same receptor or heterodimers composed of two different receptors (see Maurice et al., 2011 and Ferre et al., 2014). Initial observations have been made in 2002 in transfected HEK293 cells demonstrating the capacity of MT1 and MT2 receptors to form homodimers and heterodimers (Ayoub et al., 2002) with MT1/MT2 heterodimers showing a pharmacological profile distinct from MT2 homodimers (Ayoub et al., 2004). Shortly after, MT1 and MT2 receptors were reported to form heterodimers with the orphan GPR50, which completely abolished the function of MT1 receptors in MT1/GPR50 heterodimers (Levoye et al., 2006a). Sporadic reports on Western blots of endogenously expressed MT receptors in chicken astrocyte cultures (Adachi et al., 2002) and Xenopus tectal cells (Prada et al., 2005) further indicated the possible existence of MT receptor homodimers. However, the physiological relevance of these dimers remained largely unclear (Levoye et al., 2006b) until 2013 when compelling in vivo evidence for the functional significance of MT1/MT2 heterodimers was obtained. In retinal photoreceptor cells, melatonin enhances the light sensitivity during the night. The phenotype of MT1 receptor KO (MT1 −/−) and MT2 receptor KOs (MT2 −/−) mice, the use of type‐selective ligands and overexpression of a dominant negative form of MT2 receptors in photoreceptor cells of transgenic mice indicated the exclusive involvement of MT1/MT2 heterodimers in this physiological effect of melatonin (Baba et al., 2013). Interestingly, this effect was dependent on the activation of the Gq/PLC pathway by melatonin, an observation that could be confirmed in vitro in cells co‐expressing MT1 and MT2 receptors.
Whether MT receptor heterodimers could become novel drug targets remains an open question. Recent evidence on the antidepressant agomelatine suggests this possibility (Kamal et al., 2015). Previous studies showed that agomelatine is a high‐affinity agonist for MT1 and MT2 receptors and an antagonist with moderate affinity for 5‐HT2C receptors (Audinot et al., 2003; Millan et al., 2003). Of note, the antidepressant effect of agomelatine involves both pathways in a synergistic manner (Racagni et al., 2011). Formation of MT2/5‐HT2C heterodimers was demonstrated in transfected HEK293 cells, and these heterodimers are targeted by agomelatine (Kamal et al., 2015). Agomelatine behaved as a biased ligand, activating the Gi/cAMP pathway and antagonizing the Gq/PLC pathway. Whether the MT2/5‐HT2C heterodimer participates in the antidepressant effect of agomelatine remains to be shown.
Formation of receptor dimers offers the possibility to design dimeric ligands targeting receptor dimers. Several dimeric ligands with two identical pharmacophores have been synthesized for MT receptors and their binding properties have been determined (Audinot et al., 2003; Descamps‐Francois et al., 2003; Mesangeau et al., 2010; Spadoni et al., 2011; Journe et al., 2014). Binding of the two pharmacophores of these dimeric ligands to the two protomers of the same receptor dimer has been only shown in one study using a BRET approach (Journe et al., 2014). Compounds linked through 22–24 atom spacers were able to bind to MT1 and MT2 receptor protomers in pre‐existing homodimers and heterodimers and to induce conformational changes detected by BRET. Induction of receptor dimerization was not observed. The functional properties of these compounds remain to be studied. Taken together, the existence and physiological relevance of MT receptor dimers are increasingly recognized, but its functional role and pharmacological exploitation are still ongoing.
Genetic variants and mutants of MT receptors
The existence of many rare variants in the human population was discovered in recent genome sequencing programmes. The 1000 human genome project detected 38 million variants (Abecasis et al., 2012) and 172 variants, including 46 non‐synonymous variants, has been identified on average per GPCR in a population of 14 002 individuals (Nelson et al., 2012; Karamitri and Jockers, 2014). Numerous variants have been identified in the MTNR1A and MTNR1B genes, encoding MT1 and MT2 receptors respectively. Here, only non‐synonymous variants, modifying the amino acid sequence of the receptors, will be considered (Figure 6). Variants with altered receptor function can potentially participate in disease development. Ebisawa et al. (1999) were searching for variants in MTNR1A and MTNR1B genes in patients with circadian disorders. Two non‐synonymous variants were identified in the MTNR1A gene (R54W, A157V) that were threefold and twofold more frequent in people with non‐24 h sleep–wake syndrome (Table 2) (Ebisawa et al., 1999). Due to the small sample size (N = 22), statistical significance was not reached.
Figure 6.

Distribution of non‐synonymous MT1 (A) and MT2 (B) receptor variants identified in various human populations. Positions of variants are highlighted in light brown. Typical signatures of MT receptors such as the 3.49NRY3.51 motif and the 7.49NAXXY7.53 motif are highlighted in red. Residues suspected to be directly involved in melatonin binding (S3.35 and S3.39 in MT1 and H5.46 in both MT1 and MT2 receptors) are highlighted with a blue circle. The putative palmitoylation site at C314 is indicated in MT1 receptors.
Table 2.
Biologically important MT1 receptor variants
| Amino acid change | Type of variant | Description | Reference |
|---|---|---|---|
| I49N | Missense mutation | Rare variant identified in autism spectrum disorder patients, impaired cell surface expression, melatonin binding, cAMP inhibition and ERK1/1 activation | Chaste et al., 2010 |
| R54W | Missense mutation | Common variant identified in control population without obvious functional defect | Ebisawa et al., 1999 |
| A157V | Missense mutation | Common variant identified in control population without obvious functional defect | Chaste et al., 2010; Ebisawa et al., 1999 |
| G166E | Missense mutation | Common variant identified in control population, impaired cell surface expression, reduced cAMP inhibition and ERK1/2 activation | Chaste et al., 2010 |
| Y170X | Nonsense mutation | Rare variant identified in attention‐deficit hyperactivity disorder (ADHD) patient, premature STOP codon with impaired cell surface expression and cAMP inhibition | Chaste et al., 2011 |
| I212T | Missense mutation | Common variant identified in control population, impaired cell surface expression, cAMP inhibition and reduced ERK1/2 activation | Chaste et al., 2010 |
| A266V | Missense mutation | Common variant identified in control population with reduced ERK1/2 activation | Chaste et al., 2010 |
| K334N | Missense mutation | Rare variant identified in control population with reduced cAMP inhibition | Chaste et al., 2010 |
Common [minor allelic frequency (MAF) >1%], rare (MAF 0.1–1%) variants.
Alteration of melatonin synthesis has been reported in autism spectrum disorders (ASDs) triggering the search for variants in MTNR1A and MTNR1B genes in 295 patients with ASD, 362 controls and 284 individuals from the human genome diversity panel (Chaste et al., 2010). Six non‐synonymous mutations were identified for MTNR1A and 10 for MTNR1B (Tables 2 and 3). The majority of these mutants showed altered receptor function. Particularly deleterious mutants were MT1‐I49N, which is devoid of any melatonin binding and cell surface expression, and MT1‐G166E and MT1‐I212T, which showed severely impaired cell surface expression and biased behaviour toward the ERK1/2 pathway. No significant difference in the prevalence of these mutations was found indicating that they do not represent major risk factors for ASD.
Table 3.
Biologically important MT2 receptor variants
| Amino acid change | Type of variant | Description | Reference |
|---|---|---|---|
| A8S | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| A13V | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| G21S | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| W22L | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| G24E | Missense mutation | Common variant, not associated with type 2 diabetes risk but associated with prevalence of obesity and increased BMI shown in one study but not in another | Andersson et al., 2010; Bonnefond et al., 2012; Chaste et al., 2010; Ebisawa et al., 1999 |
| A25T | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| P36S | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| A52T | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| L66F | Missense mutation | Very rare variant identified in control population without obvious functional defect | Ebisawa et al., 1999 |
| A74T | Missense mutation | Very rare variant identified in control population and type 2 diabetes patients, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| G109A | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| M120V | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| M120I | Missense mutation | Very rare variant identified in population with impaired fasting glucose and control population without obvious functional defect | Bonnefond et al., 2012 |
| S123R | Missense mutation | Very rare variant identified in population with impaired fasting glucose and control population without obvious functional defect | Bonnefond et al., 2012 |
| V124I | Missense mutation | Very rare variant identified in several populations including type 2 diabetes and ADSD without obvious functional defect in one study and impaired ERK1/2 activation in another | Andersson et al., 2010; Bonnefond et al., 2012; Chaste et al., 2010 |
| R138C | Missense mutation | Rare variant, not associated with type 2 diabetes risk, no Gi and ERK1/2 activation | Andersson et al., 2010; Bonnefond et al., 2012; Chaste et al., 2010 |
| R138L | Missense mutation | Very rare variant identified in control population, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| R138H | Missense mutation | Very rare variant identified in control population, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| Y141F | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| M146V | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| R154H | Missense mutation | Very rare variant identified in control population and type 2 diabetes patients without obvious functional defect | Bonnefond et al., 2012 |
| L166I | Missense mutation | Very rare variant identified in control population, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| T201M | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| R222H | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| I223T | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| R231H | Missense mutation | Rare variant, not associated with type 2 diabetes risk | Andersson et al., 2010; Bonnefond et al., 2012; Chaste et al., 2010 |
| A234T | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| E237K | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| S238G | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| K243R | Missense mutation | Common variant, not associated with type 2 diabetes risk | Bonnefond et al., 2012 |
| D246N | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| F250V | Missense mutation | Very rare variant identified in type 2 diabetes patients with impaired ERK1/2 activation | Bonnefond et al., 2012 |
| R316H | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
| R330W | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| R330Q | Missense mutation | Very rare variant identified in control population without obvious functional defect | Chaste et al., 2010 |
| A342V | Missense mutation | Very rare variant identified in type 2 diabetes patients, without obvious functional defect | Bonnefond et al., 2012 |
| I353T | Missense mutation | Very rare variant identified in type 2 diabetes patients and control population, associated with type 2 diabetes risk, impaired Gi protein activation | Bonnefond et al., 2012 |
| A359E | Missense mutation | Very rare variant identified in control population without obvious functional defect | Bonnefond et al., 2012 |
Common [minor allelic frequency (MAF) >1%], rare (MAF 0.1–1%) and very rare (MAF < 0.1%) variants.
Four non‐synonymous mutations were identified for MTNR1A and four for MTNR1B in a cohort of 101 individuals with attention‐deficit/hyperactivity disorder (ADHD) (Tables 2 and 3); however, none of them was enriched in ADHD individuals as compared with the general population (Chaste et al., 2011). The MT1‐Y170X nonsense mutation was only detected in one ADHD patient and introduced a premature STOP codon resulting in complete loss of receptor function.
MT receptor variants have been most extensively sought in studies focused on type 2 diabetes (T2D) based on the discovery of several frequent polymorphisms associated with increased fasting plasma glucose (FPG) and T2D risk close to the MTNR1B gene in genome‐wide association studies (Bouatia‐Naji et al., 2009; Prokopenko et al., 2009). Sequencing of the coding region of the MTNR1B gene revealed six non‐synonymous variants (G24E, L60R, V124I, R138C, R231H and K243R) of which none was associated with T2D risk. The common 24E variant was associated with increased body mass and decreased FPG (Andersson et al., 2010), an observation that was not replicated in a later study (Bonnefond et al., 2012). Whereas only subtle changes in the capacity of G24E and V124I to activate a GαΔ6qi4myr chimeric G protein, the L60R variant was completely inactive in transfected COS cells. A more extensive sequencing study discovered 40 non‐synonymous variants in the coding region of MTNR1B (Tables 2, 3, 4) (Bonnefond et al., 2012) of which 36 very rare mutants associated with T2D risk. Functional analysis of the 40 variants revealed intact cell surface expression for all variants. There was complete loss of melatonin binding in four very rare cases (A42P, L60R, P95L and Y308S) and partially and severely blunted signalling (Gαqi9 chimera and ERK1/2 activation) in one rare case (R138C) and nine very rare cases (W22L, A52T, A74T, R138H, R138L, L166I, R222H, R330W and I353T). Carriers of the 13 very rare loss‐of‐function variants showed increased T2D risk establishing a functional link between MTNR1B and T2D (see Karamitri et al., 2013).
Table 4.
Mutations in the MTNR1B gene associated with susceptibility to type 2 diabetes
| Amino acid change | Type of variant | Description | Reference |
|---|---|---|---|
| A42P | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, no melatonin binding and signalling | Bonnefond et al., 2012 |
| L60R | Missense mutation | Very rare variant identified in control population and type 2 diabetes patients, associated with type 2 diabetes risk, no melatonin binding and signalling | Andersson et al., 2010; Bonnefond et al., 2012 |
| P95L | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, no melatonin binding and signalling | Bonnefond et al., 2012 |
| Y308S | Missense mutation | Very rare variant identified in type 2 diabetes patients, associated with type 2 diabetes risk, no melatonin binding and signalling | Bonnefond et al., 2012 |
Very rare (minor allelic frequency <0.1%) variants.
In conclusion, the genetic variability of the MTNR1B gene in terms of non‐synonymous variants has now been well defined and an association of very rare variants with T2D risk established. Less is known about the variability of the MTNR1A gene in terms of non‐synonymous variants.
MT receptor mouse models – update
MT1 −/− mice were created in the late 1990s followed by the generation of MT2 −/− mice in 2003 (Liu et al., 1997; Jin et al., 2003). Studies using these mice have provided important insights on the role that MT receptors play in the modulation of many different biological functions. In MT1 −/− mice, but not in MT2 −/− mice, the inhibitory effect of melatonin on neuronal activity in the suprachiasmatic nucleus (SCN) is impaired suggesting the involvement of MT1 receptors. In contrast, in SCN slices from MT1 −/− mice melatonin (1–10 pM) phase shifts the peak of circadian rhythms of neuronal firing by approximately 3 h suggesting the involvement of MT2 receptors (Liu et al., 1997; Dubocovich et al., 2005). Blockade of this effect using the MT2 receptor‐selective 4P‐PDOT antagonist confirmed the latter conclusion shaping a pathway where MT2 phase shifts the peak of neuronal firing through PLC‐PKC signalling pathway (Mc Arthur et al., 1997; Hunt et al., 2001; Dubocovich et al., 2005). Liu et al. (1997) reported that the phase shift of neuronal firing rhythms induced by 2‐iodomelatonin (10 pM) was of smaller magnitude in the SCN slice from MT1 −/− than in wild type (WT) mice suggesting a role for the MT1 receptors in this response (see detailed discussion in Dubocovich, 2007). Together, these findings suggest a potential role for both MT1 and MT2 receptors in the phase shift of circadian rhythms of neuronal firing in the SCN slice in vitro. The use of MT1 −/− mice demonstrated that the MT1 receptor is required for the melatonin‐meditated phase shift of the onset of overt circadian rhythm of locomotor activity (Dubocovich et al., 2005). An independent study demonstrated that C3H/HeN mice (melatonin‐proficient) entrained faster to a phase advance of dark onset than the C57BL/6J mice (melatonin‐deficient), suggesting a facilitating role of endogenous melatonin on circadian reentrainment (Pfeffer et al., 2012). However, we should note that faster entrainment could also result from genetic differences between the two mouse strains rather than different endogenous melatonin levels (Adamah‐Biassi et al., 2013). In a mouse strain producing endogenous melatonin, the faster entrainment to an abrupt advance of dark onset persisted in MT1 −/− C3H/HeN mice but was lost in MT2 −/− and double KOs (MT1 −/−/MT2 −/−) suggesting again the involvement of MT2 receptors. This apparent contradiction could be explained by the activation of MT2 and MT1 receptors by endogenous and exogenous melatonin, respectively, at different periods of sensitivity (subjective night vs. subjective day, respectively). Changes in efficacy could also result from desensitization and/or internalization of MT receptors in response to exposure to physiological and supraphysiological melatonin concentrations as demonstrated by the phase shift of the peak of neuronal firing in the SCN by physiological levels of melatonin, which involved the desensitization of MT2 receptors (Gerdin et al., 2004).
MT receptor KO mice have been also used to elucidate the role played by these receptors in the regulation of the sleep/wake cycle. In MT2 −/− NREM, sleep is decreased during the light phase (i.e. during the time that mice normally sleep), whereas MT1 −/− mice showed an increase in the amount of NREM sleep during the dark phase (i.e. during active phase) (Ochoa‐Sanchez et al., 2011). Further analysis of the data indicated that MT1 receptor signalling is implicated in the modulation of the daily rhythm of REM sleep (Ochoa‐Sanchez et al., 2011). An additional study in which double KOs (MT1 −/−/MT2 −/−) were used indicated that removal of both receptors induced an increase in wakefulness and a reduction in REM sleep (Comai et al., 2013). Hence, these data seem to indicate that removal of MT receptors may affect wakefulness rather than sleep.
The effect of MT receptor removal has been also investigated in the mouse retina, where these receptors are widely distributed (Baba et al., 2009; Baba et al., 2013). Removal of either receptor has profound effects on photoreceptors function as it abolishes the daily rhythms in the scotopic and photopic electroretinogram (Baba et al., 2009; Alcantara‐Contreras et al., 2011; Sengupta et al., 2011). Such a result also indicates that MT1 and MT2 receptors form heterodimers in mouse photoreceptors (Baba et al., 2013). Further studies have also demonstrated that removal of MT receptors in addition affects the viability of the photoreceptors and retinal ganglion cells during aging (Baba et al., 2009; Alcantara‐Contreras et al., 2011; Gianesini et al., 2016) as well as corneal biology (Baba et al., 2015).
As mentioned before, recent studies have also implicated MT receptors in the pathogenesis of T2D in humans (Bouatia‐Naji et al., 2009; Lyssenko et al., 2009; Bonnefond et al., 2012). Thus, a few studies used MT receptor KO mice to determine the mechanisms by which these receptors contribute to regulation of glucose homeostasis and insulin sensitivity (Stumpf et al., 2008; Muhlbauer et al., 2009; Contreras‐Alcantara et al., 2010). Mice lacking MT1 receptors exhibit higher mean blood glucose levels than controls (Muhlbauer et al., 2009) and tend to be more glucose intolerant and insulin resistant than WT and MT2 −/− mice (Contreras‐Alcantara et al., 2010). Furthermore, removal of MT1 or MT2 receptors abolishes the daily rhythm in blood glucose levels (Owino et al., 2016).
Finally, it is important to mention that although the reproductive system of mice is not sensitive to photoperiod, the development of MT receptor KO mice provided an important tool for dissecting the mechanisms by which melatonin regulates reproduction in photoperiodic species. For example, MT1 receptor signalling controls the rhythmic expression of the clock gene Period 1 in the pituitary gland (von Gall et al., 2002), and further studies have shown that the rhythmic expression of several other clock genes (Per1, Per 2, Bmal1 and Cry 1) in the mouse Pars tuberalis depends on MT1 receptor signalling as well (Jilg et al., 2005). MT1 receptor signalling has been also reported to be crucial for the photoperiodic response of gene expression in the ependymal cell layer and thus for the photoperiodic regulation of gonadal activity (Sheynzon and Korf, 2006; Yasuo et al., 2009). Finally, we should mention that a recent study reported that MT1 receptor signalling plays a key role in photoperiodic programming of serotonergic neurons as well as depression‐ and anxiety‐related behaviours in mice (Green et al., 2015).
In conclusion, studies in the last 20 years using MT receptor KO mice have greatly helped to understand the role(s) played by these receptors in the regulation of many physiological functions, and they have provided important insights on the mechanisms by which melatonin signalling affects these functions.
Functional role of MT receptors in physiology and pathophysiology
MT receptors are involved in many physiological processes that will however not all be covered by this review but can be consulted in other reviews (Dubocovich et al., 2010; Tosini et al., 2012; Karamitri et al., 2013; Tosini et al., 2014; Johnston and Skene, 2015). Here, we have focused our attention on two major processes, the immune system and the CNS. Important progress has been made recently in both fields, and links to diseases have been established justifying a review of our current knowledge on these aspects. Finally, we will make a critical assessment of reports of receptor‐independent effects of melatonin, such as its binding to additional binding sites and the intrinsic antioxidant and free radical scavenger properties of this hormone.
MT receptors in the immune system
The role of melatonin as a player in immunity, first proposed by Berman in 1926, is now well accepted (Carrillo‐Vico et al., 2013). Several reports have demonstrated that melatonin produced by the either pineal gland or immune cells can regulate the activation of an immune response. Melatonin derived from activated human lymphocytes induces the synthesis of IL‐2 and IL‐2 receptors (Carrillo et al., 2004; Carrillo‐Vico et al., 2013). Luzindole and targeted deletion of the MNTR1A gene (Lardone et al., 2006; Lardone et al., 2010) block the effect of lymphocyte‐derived melatonin. Interestingly, daily rhythms of plasma melatonin and IL‐2 are transiently lost in non‐infectious human inflammatory conditions, and the recovery of the IL‐2 rhythm follows the restoration of the daily melatonin rhythm (Pontes et al., 2007). In addition, the daily and seasonal variation of melatonin production contributes to the seasonality of some diseases. In multiple sclerosis (MS), melatonin blocks the differentiation of Th17 cells and boosts the generation of protective type 1 regulatory T‐cells by an MT1 receptor‐dependent mechanism, resulting in the seasonal variation of MS symptoms (Farez et al., 2015). Seasonality of regular immunity is also related to changes in the melatonin system (Weil et al., 2015). In the spleen of several species, extended light exposure decreases MT1 receptor expression (Maestroni, 1993; Lahiri and Haldar, 2009; Yadav and Haldar, 2013). In healthy conditions, rolling and adhesion of neutrophils to the endothelial cell layer are inhibited by activation of MT2 receptors and ligands binding to the putative MT3 binding site, respectively (Lotufo et al., 2001). In contrast, other effects of melatonin such as the inhibition of transcription factors that mediate acute inflammation induced by LPS (Tamura et al., 2010) or N‐formyl‐l‐methionyl‐l‐leucyl‐l‐phenylalanine (fMLP) (Cernysiov et al., 2015) were not blocked by luzindole suggesting a mode of action independent of MT receptors.
MT receptors also play an important role in promoting engulfing of bacteria, fungi and parasites. Melatonin facilitates the invasion of erythrocytes by Plasmodium falciparum (Hotta et al., 2000), the invasion of macrophages by Leishmania amazonensis (Laranjeira‐Silva et al., 2015) and the phagocytosis of zymosan by colostrum polymorphonuclear and mononuclear cells (Pires‐Lapa et al., 2013) and the RAW 264.7 macrophage cell line (Muxel et al., 2012). The entrance of different microorganisms in polymorphonuclear and mononuclear cells, including colostral and lineage‐established cell lineages, is blocked by luzindole. Indeed, parasites, bacteria and fungi activate the NF‐κB pathway in these two cell models resulting in the expression of arylalkyl‐N‐acetyltransferase and the synthesis of melatonin. Luzindole and 4P‐PDOT blocked the expression of dectin‐1, a protein that is important for phagocytosis, suggesting the participation of MT2 receptors in this effect (Muxel et al., 2012; Pires‐Lapa et al., 2013; Muxel et al., 2016). Thus, the evaluation of binding parameters and functional states of MT receptors in immune‐competent cells needs to consider the masking effect of on‐demand synthesized melatonin.
Although complex, the role of melatonin on the immune system is now beginning to be understood. MT1 and MT2 receptor types appear to play different roles, with MT1 receptors as the main target in acquired immune response and MT2 receptors as the target for innate immune responses.
MT receptors in the CNS
MT receptors are widely expressed throughout the CNS and are particularly well characterized in the SCN of the hypothalamus, where they are known to inhibit neuronal firing and mediate the phase shifting effect of melatonin on circadian rhythms (see above). In addition to its chronobiotic effect, melatonin participates in the modulation of neuronal functions, neurodevelopment at early and late stages (Kong et al., 2008; Chen et al., 2014) and affects brain structures underlying sleep regulation (Ochoa‐Sanchez et al., 2011), drug‐related learning (Wang et al., 2005; Savaskan et al., 2006) and reward (Hutchinson et al., 2012; Clough et al., 2014). MT receptors mediate the melatonin‐induced increase in dendrite length, thickness and complexity of hippocampal neurons, as these effects were partially blocked by luzindole (Dominguez‐Alonso et al., 2015). Similarly, melatonin‐induced differentiation and maturation of adult neural stem cells were almost abolished in the presence of luzindole (de la Fuente Revenga et al., 2015). A recent study using MT2 −/− mice showed that MT2 receptors were essential for axogenesis and for the formation of functional synapses (Liu et al., 2015). MT2 receptors were also involved in melatonin‐induced protection against oxidative stress and memory impairment in a mouse model of ageing (Shin et al., 2015). Recent advances in the understanding of presynaptic MT receptors and their role in neurodegenerative diseases are discussed in the following sections.
Presynaptic MT receptors
The role of melatonin on the regulation of calcium‐dependent dopamine release from axon terminals in brain and amacrine cells in the retina was shown in the early 1980s (Zisapel and Laudon, 1982; Dubocovich, 1983). However, more direct and global proof for the presence of presynaptic melatonin heteroreceptors (i.e. receptor for a transmitter or hormone other than the neuron's own neurotransmitter) capable of regulating neurotransmitter release was still insufficient. A recent protein interaction network analysis has established that MT1, but not MT2 receptors, are expressed on presynaptic axon terminal membranes in the hypothalamus, striatum, cortex and hippocampus, where they are part of the presynaptic protein network (Benleulmi‐Chaachoua et al., 2016). Notably, this study shows a strong physical association between MT1 receptors and presynaptic proteins such as synapsin, SNAP25, Munc‐18 and voltage‐gated Cav2.2 channels. Interaction with the latter was responsible for constitutive inhibition of calcium entry by MT1 receptors in a Gβγ‐dependent manner (Benleulmi‐Chaachoua et al., 2016).
These recent findings provide strong support for the involvement of MT receptors in synaptic functions, particularly in neurotransmitter release as indicated by previous studies. Indeed, activation of MT receptors has been implicated in the inhibition of 3H‐dopamine release from the ventral hippocampus, medulla pons, preoptic area and hypothalamus (median and posterior) (Zisapel and Laudon, 1982; Dubocovich, 1983). This effect followed a diurnal rhythm in the hypothalamus with a maximum and a minimum observed at ZT 5 and ZT 13–15, respectively (Zisapel et al., 1985). 6‐Chloromelatonin‐mediated modulation of noradrenaline turnover via activation of presynaptic melatonin heteroreceptors was demonstrated in hypothalamus (Fang and Dubocovich, 1990). In this model, luzindole, applied during the night when melatonin levels are high, accelerated noradrenaline turnover suggesting the involvement of MT receptors stimulated by endogenous melatonin (Fang and Dubocovich, 1990). The presence of presynaptic MT heteroreceptors on retino‐hypothalamic fibres innervating superficial retinorecipient layers of the avian optic tectum has been inferred by the presence of 2‐[125I]‐MLT binding sites and its decrease following transsection of the retinotectal pathway (Krause et al., 1992; Krause et al., 1994). The function of these presynaptic MT receptors is currently unknown, but a modulatory role of these receptors on the light input pathway to visual and circadian target responses is likely. Recent electrophysiological evidence suggests that melatonin acting through presynaptic MT receptors increases glutamatergic neurotransmission in the habenula, an effect blocked by luzindole (Evely et al., 2016). Finally, it is worth mentioning in this context that MT receptors were first shown to be involved in the inhibition of depolarization‐evoked calcium‐dependent neurotransmitter (dopamine) release from amacrine cells in the chick and rabbit retina (Dubocovich, 1983; Dubocovich, 1985). These mammalian functional presynaptic heteroreceptors were used to establish the first structure–activity relationship for MT receptor ligands, which correlated with the pharmacological profile of MT2 receptors (Dubocovich et al., 1997), and to identify and pharmacologically characterize the first competitive MT receptor ligands, luzindole and 4P‐PDOT (Dubocovich, 1988).
In summary, proteomic studies of the MT1 receptor interactome has revived interest in the function of presynaptic MT receptors and reinforced previous functional studies indicating the role of presynaptic MT receptors in neurotransmitter release. Use of MT receptor KO mouse models will be particularly instrumental in this context, as they will clarify the respective roles of MT1 and MT2 receptors. Based on current data, a predominant role of MT1 receptors in presynaptic functions, such as neurotransmitter release, and a potential role of MT2 receptors in axogenesis and synapse formation can be postulated.
MT receptors in neurodegenerative diseases
Altered expression of MT receptors has been frequently reported in neurodegenerative diseases and psychiatric disorders, including Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD) and ASD. In AD patients, MT1 receptor expression in the SCN and MT2 receptor expression in the hippocampus are reduced compared to control subjects in post‐mortem brains (Wu et al., 2007). Intriguingly, higher expression of MT1 receptors was detected in hippocampal arteries of AD brains (Savaskan et al., 2002), which might be due to a compensatory response to the low levels of circulating melatonin in these patients (Zhou et al., 2003). These observations suggest that the expression of MT receptors under pathological conditions can be differentially regulated depending on the brain area. In PD patients, down‐regulation of MT1 and MT2 receptor expression was observed in the substantia nigra and amygdala, the two most relevant areas in PD pathogenesis (Adi et al., 2010). Small case–control studies accessing MT1 and MT2 receptor expression in HD patients showed no changes in the SCN (van Wamelen et al., 2013), while decreased expression of MT1, but not of MT2 receptors, was detected in the striatum (Wang et al., 2011). Interestingly, the progressive loss of MT1 receptors correlates with HD severity, also confirmed in a mouse model of HD (Wang et al., 2011). In ASD patients, no information on MT1 and MT2 receptor expression is available, but several MT1 and MT2 receptor mutants with strongly reduced function have been identified (Chaste et al., 2010).
Additional evidence supports the emerging concept of MT receptor dysfunction as a permissive condition favouring the development and/or progression of neurodegenerative diseases. The neuroprotective effect of endogenous and exogenous melatonin has been demonstrated in different systems (see Escribano et al., 2014). In a neuroinflammatory model induced by LPS administration, cerebellar neuronal death was observed only in animals pretreated with luzindole (Pinato et al., 2015). Similarly, depletion of endogenous melatonin by pinealectomy caused spontaneous neuronal loss in the hippocampal CA1 area, which was prevented by treatment with agomelatine (Tchekalarova et al., 2016). The requirement of MT receptors for the neuroprotective action of melatonin has also been elegantly demonstrated in a series of in vitro studies in which luzindole treatment or siRNA‐mediated knockdown of MT1 receptors enhanced neuronal vulnerability to cell death (Wang et al., 2011). Different cell stressor conditions such as temperature shift or treatments with hydrogen peroxide, TNF or with the HD‐related protein huntingtin, resulted in reduced levels of MT1 receptors. Accordingly, the AD‐related neurotoxic amyloid β peptide (Aβ) impairs the function of MT receptors (Cecon et al., 2015), implying that these receptors and melatonin signalling are among the primary molecular targets affected in the course of AD.
Insights in the effects of MT receptors on cognitive functions are also obtained from MT receptor KO mice. MT2 −/− mice show impaired long‐term potentiation and performance in memory tests (Larson et al., 2006). However, the double KO MT1 −/−/MT2 −/− mice show no clear differences from WT mice in memory test performances and show increased long‐term potentiation responses, even though the deletion of MT receptors negatively affected the expression of important proteins for synaptic activity, such as phospho‐synapsin and spinophilin (O'Neal‐Moffitt et al., 2014). The relevance of MT receptors for cognitive performance was clearly shown with an AD mouse model lacking MT1 and MT2 receptors, in which melatonin treatment failed to improve performance on hippocampal‐dependent spatial learning tasks, as observed in the AD mouse model in the presence of MT1 and MT2 receptors. Impressively, the lack of MT receptors per se markedly increased the mortality in young AD mice (O'Neal‐Moffitt et al., 2015). Finally, the therapeutic use of melatonin has been proposed and tested in a number of murine models and clinical trials in several neurodegenerative conditions, including AD (Olcese et al., 2009; Cardinali et al., 2010; Peng et al., 2013; Wade et al., 2014; Zhang et al., 2016), amyotrophic lateral sclerosis (Weishaupt et al., 2006; Zhang et al., 2013), PD (Medeiros et al., 2007; Naskar et al., 2015; Zhang et al., 2016) and HD (van Wamelen et al., 2015). The therapeutic use of melatonin is usually associated with sleep improvement and better alignment of circadian parameters, and its beneficial effect on neuroprotection and cognitive performance is starting to be recognized (Joshi et al., 2015; Wade et al., 2014). Dysfunction or down‐regulation of MT receptors is likely to be part of the primary pathophysiological mechanisms rather than a consequence of advanced neurodegeneration and, thus, prophylactic hormonal replacement and/or early stage intervention strategies to restore MT receptor expression and function might provide the most efficient result.
Taken together, the subcellular localization and role of MT receptors in neuronal functions and their participation in neurodegererative diseases are now starting to be understood and suggest a broad modulatory role of melatonin in neuronal function, development and plasticity.
Melatonin as antioxidant and free radical scavenger
The IUPHAR classifies only clearly identified pharmacological targets in mammals. However, some effects of melatonin persist even in the absence of MT1 and MT2 receptors or upon complete pharmacological blockade of MT receptors, indicating the existence of MT receptor‐independent mechanisms, which are still not fully understood. In addition, MT receptor‐dependent and ‐independent mechanisms can participate simultaneously, as demonstrated by O'Neal‐Moffitt et al. (2015) regarding the antioxidant and pro‐cognitive effects of melatonin on AD mice models, for example. Two main mechanisms have been put forward to explain the antioxidant and free radical properties of melatonin: these are melatonin binding to the MT3 binding site (Nosjean et al., 2000; Dubocovich et al., 2003) and to the cytosolic enzyme quinone reductase 2 (QR2) (Nosjean et al., 2000; Dubocovich et al., 2003), and melatonin scavenging of free radicals, as this hormone has been suggested to be an electron donor (see Tan et al., 2015). Binding of melatonin to intracellular targets is readily achieved, due to the hydrophilic nature of this indolamine. Melatonin binds with nanomolar affinity to MT3/QR2 binding sites but shows a pharmacological profile distinct from MT1 and MT2 receptors. The order of affinities for the MT3 binding site is 2‐iodomelatonin > N‐acetyl‐serotonin > melatonin (Dubocovich, 1995; Nosjean et al., 2000), the order for MT1 and MT2 receptors is 2‐iodomelatonin > melatonin >>>>N‐acetyl‐serotonin. MCA‐NAT (5‐methoxycarbonylamino‐N‐acetyltryptamine), prazosin and N‐acetyltryptamine are selective ligands for the membrane MT 3 binding site (Dubocovich, 1995; Molinari et al., 1996; Nosjean et al., 2000). Nosjean et al. (2000) showed that a cytosolic binding site identified as QR2 has the pharmacological characteristics of the membrane MT3 binding site. QR2 is a cytosolic flavin adenine dinucleotide (FAD)‐dependent flavoprotein that reduces menadione and other quinones by using N‐ribosyl‐ and N‐alkyldihydronicotinamides as the co‐substrates (Liao and Williams‐Ashman, 1961), thus acting as a detoxifying enzyme to increase the antioxidant defence (Jockers et al., 2008). There are still open questions as to whether the melatonin binding site on QR2 corresponds to the MT3 binding site, in particular regarding those sites that are membrane‐associated.
Several physiological effects of melatonin such as inhibition of leukocyte adhesion to rat endothelial cell layers were mimicked by MT3 agonists (Lotufo et al., 2001). Similar observations were made for the expression of adhesion molecules by granulocytes (Cernysiov et al., 2015), the increase in dopamine levels in chick retina (Sampaio Lde et al., 2014) and the reduction of intraocular pressure (IOP) in rabbits (Alarma‐Estrany et al., 2009). However, it has been questioned whether the functional effects of MCA‐NAT are indeed mediated by QR2, as the lack of QR2 did not prevent the MCA‐NAT‐induced reduction on IOP, and overexpression of QR2 did not promote receptor‐like responses (Vincent et al., 2010). In addition, MCA‐NAT turned out to be a partial agonist for MT1 and MT2 receptors at submicromolar concentrations suggesting the possibility that some of the effects of MCA‐NAT might be mediated by MT1 and/or MT2 receptors (Vincent et al., 2010).
Melatonin and its metabolites, with or without open ring structures, have been described as potent electron donors. Cyclic‐3‐hydroxymelatonin, N1‐acetyl‐5‐methoxykynuramine (secondary metabolite) (AMK, tertiary metabolite) and N‐acetyl‐N‐formyl‐5‐methoxykynuramine (AFMK, quaternary metabolite) scavenge free radicals neutralizing reactive oxygen and nitrogen species (Ressmeyer et al., 2003; Tan et al., 2007; Zavala‐Oseguera et al., 2014). Hence, one melatonin molecule and its associated metabolites could scavenge a large number of reactive species, and thus, the overall antioxidant capacity of melatonin is believed to be greater than that of other well‐known antioxidants, such as vitamin C and vitamin E, under in vitro or in vivo conditions (Gitto et al., 2001; Sharma and Haldar, 2006; Ortiz et al., 2013). However, the ability of melatonin in reducing oxidative stress does not only rely on donating electrons. Indeed, by acting on MT1 and MT2 receptors, low pM and low nM concentrations of melatonin increased the expression or activity of enzymes such as superoxide dismutase, catalase and glutathione peroxidase, which are involved in oxygen detoxification (Rosen et al., 2009). Thus, depending on the dose of exogenous or endogenous melatonin, receptor‐dependent or ‐independent mechanisms may be involved. A further complexity in the interplay between receptor‐dependent and ‐independent processes could arise from the fact that melatonin, by changing the redox state of the cell, might influence receptor‐mediated functions. Indeed, the function of several GPCRs has been shown to be sensitive to the cellular redox state. Whether this is also the case for MT receptors has to be addressed in future studies. Although endogenous melatonin levels are typically considered to range from low pM to low nM concentrations, much higher concentrations may be reached locally in the brain (Legros et al., 2014) and in activated immune cells (Conti et al., 2000). In addition, melatonin can be actively taken up through the GLUT1 glucose transporter (Hevia et al., 2015). In conclusion, the role of the MT3 binding site is still not fully understood and warrants further attention. Concerning the free radical scavenging properties of melatonin, it is surprising that opposing opinions are still in the literature. Overall, the antioxidant effects of melatonin appear to be complex, relying on a mixture of MT receptor‐dependent and ‐independent processes.
Conflict of interests
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Agence Nationale de la Recherche (ANR 2011 ‐BSV1‐012‐01 ‘MLT2D’ and ANR‐2011‐META ‘MELA‐BETES’, ANR‐12‐RPIB‐0016 ‘MED‐HET‐REC‐2’), the Fondation de la Recherche Médicale (Equipe FRM DEQ20130326503), Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS) and the ‘Who am I?’ laboratory of excellence no.ANR‐11‐LABX‐0071 funded by the French Government through its ‘Investments for the Future’ programme operated by The French National Research Agency (ANR) under grant no.ANR‐11‐IDEX‐0005‐01 (to R. J.); USPHS grants MH‐42922, MH 52685, DA021870 and NS 061068 (to M. L. D); the National Institutes of Health grants EY022216 and EY020821 and by 5U54NS083932, S21MD000101, G12‐RR03034 and U54RR026137 (to G. T.), the São Paulo Research Foundation (FAPESP – Thematic Project – 2013/13691‐1) and Brazilian National Council of Research and Innovation (CNPq – 480097) (to R. P. M.). PD is an employee of the pharmaceutical company Institut de Recherches Servier. NC‐IUPHAR receives financial support from the Wellcome Trust.
Jockers, R. , Delagrange, P. , Dubocovich, M. L. , Markus, R. P. , Renault, N. , Tosini, G. , Cecon, E. , and Zlotos, D. P. (2016) Update on melatonin receptors: IUPHAR Review 20. British Journal of Pharmacology, 173: 2702–2725. doi: 10.1111/bph.13536.
This article is an NC‐IUPHAR review contributed by members of the International Union of Basic and Clinical Pharmacology Committee on Receptor Nomenclature and Drug Classification (NC‐IUPHAR) subcommittee for the melatonin receptors.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi A, Natesan AK, Whitfield‐Rucker MG, Weigum SE, Cassone VM (2002). Functional melatonin receptors and metabolic coupling in cultured chick astrocytes. Glia 39: 268–278. [DOI] [PubMed] [Google Scholar]
- Adamah‐Biassi EB, Stepien I, Hudson RL, Dubocovich ML (2013). Automated video analysis system reveals distinct diurnal behaviors in C57BL/6 and C3H/HeN mice. Behav Brain Res 243: 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adi N, Mash DC, Ali Y, Singer C, Shehadeh L, Papapetropoulos S (2010). Melatonin MT1 and MT2 receptor expression in Parkinson's disease. Med Sci Monit 16: BR61–BR67. [PubMed] [Google Scholar]
- Alarma‐Estrany P, Crooke A, Pintor J (2009). 5‐MCA‐NAT does not act through NQO2 to reduce intraocular pressure in New‐Zealand white rabbit. J Pineal Res 47: 201–209. [DOI] [PubMed] [Google Scholar]
- Alcantara‐Contreras S, Baba K, Tosini G (2011). Removal of melatonin receptor type 1 increases intraocular pressure and retinal ganglion cells death in the mouse. Neurosci Lett 494: 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Davenport AP, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015a). The Concise Guide to PHARMACOLOGY 2015/16: G Protein‐Coupled Receptors. Br J Pharmacol 172: 5744–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Catterall WA, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Voltage‐gated ion channels. Br J Pharmacol 172: 5904–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E et al. (2015c). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. Br J Pharmacol 172: 6110–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015d). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson EA, Holst B, Sparso T, Grarup N, Banasik K, Holmkvist J et al. (2010). MTNR1B G24E variant associates with BMI and fasting plasma glucose in the general population in studies of 22,142 Europeans. Diabetes 59: 1539–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audinot V, Mailliet F, Lahaye‐Brasseur C, Bonnaud A, Le Gall A, Amosse C et al. (2003). New selective ligands of human cloned melatonin MT1 and MT2 receptors. Naunyn Schmiedebergs Arch Pharmacol 367: 553–561. [DOI] [PubMed] [Google Scholar]
- Ayoub MA, Couturier C, Lucas‐Meunier E, Angers S, Fossier P, Bouvier M et al. (2002). Monitoring of ligand‐independent dimerization and ligand‐induced conformational changes of melatonin receptors in living cells by bioluminescence resonance energy transfer. J Biol Chem 277: 21522–21528. [DOI] [PubMed] [Google Scholar]
- Ayoub MA, Levoye A, Delagrange P, Jockers R (2004). Preferential formation of MT1/MT2 melatonin receptor heterodimers with distinct ligand interaction properties compared with MT2 homodimers. Mol Pharmacol 66: 312–321. [DOI] [PubMed] [Google Scholar]
- Baba K, Benleulmi‐Chaachoua A, Journe AS, Kamal M, Guillaume JL, Dussaud S et al. (2013). Heteromeric MT1/MT2 melatonin receptors modulate photoreceptor function. Sci Signal 6: ra89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Davidson AJ, Tosini G (2015). Melatonin Entrains PER2::LUC Bioluminescence Circadian Rhythm in the Mouse Cornea. Invest Ophthalmol Vis Sci 56: 4753–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Pozdeyev N, Mazzoni F, Contreras‐Alcantara S, Liu C, Kasamatsu M et al. (2009). Melatonin modulates visual function and cell viability in the mouse retina via the MT1 melatonin receptor. Proc Natl Acad Sci U S A 106: 15043–15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benleulmi‐Chaachoua A, Chen L, Sokolina K, Wong V, Jurisica I, Emerit MB et al. (2016). Protein interactome mining defines melatonin MT1 receptors as integral component of presynaptic protein complexes of neurons. J Pineal Res 60: 95–108. [DOI] [PubMed] [Google Scholar]
- Bonnefond A, Clement N, Fawcett K, Yengo L, Vaillant E, Guillaume JL et al. (2012). Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 44: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatia‐Naji N, Bonnefond A, Cavalcanti‐Proenca C, Sparso T, Holmkvist J, Marchand M et al. (2009). A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41: 89–94. [DOI] [PubMed] [Google Scholar]
- Browning C, Beresford I, Fraser N, Giles H (2000). Pharmacological characterization of human recombinant melatonin mt(1) and MT2 receptors. Br J Pharmacol 129: 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali DP, Furio AM, Brusco LI (2010). Clinical aspects of melatonin intervention in Alzheimer's disease progression. Curr Neuropharmacol 8: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo VA, Calvo JR, Abreu P, Lardone PJ, Garcia MS, Reiter RJ et al. (2004). Evidence of melatonin synthesis by human lymphocytes and its physiological significance: possible role as intracrine, autocrine, and/or paracrine substance. FASEB J 18: 537–539. [DOI] [PubMed] [Google Scholar]
- Carrillo‐Vico A, Lardone PJ, Alvarez‐Sanchez N, Rodriguez‐Rodriguez A, Guerrero JM (2013). Melatonin: buffering the immune system. Int J Mol Sci 14: 8638–8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecon E, Chen M, Marcola M, Fernandes PA, Jockers R, Markus RP (2015). Amyloid beta peptide directly impairs pineal gland melatonin synthesis and melatonin receptor signaling through the ERK pathway. FASEB J 29: 2566–2582. [DOI] [PubMed] [Google Scholar]
- Cernysiov V, Mauricas M, Girkontaite I (2015). Melatonin inhibits granulocyte adhesion to ICAM via MT3/QR2 and MT2 receptors. Int Immunol 27: 599–608. [DOI] [PubMed] [Google Scholar]
- Chan KH, Hu Y, Ho MK, Wong YH (2013). Characterization of substituted phenylpropylamides as highly selective agonists at the melatonin MT2 receptor. Curr Med Chem 20: 289–300. [DOI] [PubMed] [Google Scholar]
- Chaste P, Clement N, Botros HG, Guillaume JL, Konyukh M, Pagan C et al. (2011). Genetic variations of the melatonin pathway in patients with attention‐deficit and hyperactivity disorders. J Pineal Res 51: 394–399. [DOI] [PubMed] [Google Scholar]
- Chaste P, Clement N, Mercati O, Guillaume JL, Delorme R, Botros HG et al. (2010). Identification of pathway‐biased and deleterious melatonin receptor mutants in autism spectrum disorders and in the general population. PLoS One 5: e11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li X, Du Z, Shi W, Yao Y, Wang C et al. (2014). Melatonin promotes the acquisition of neural identity through extracellular‐signal‐regulated kinases 1/2 activation. J Pineal Res 57: 168–176. [DOI] [PubMed] [Google Scholar]
- Chugunov AO, Farce A, Chavatte P, Efremov RG (2006). Differences in binding sites of two melatonin receptors help to explain their selectivity to some melatonin analogs: a molecular modeling study. J Biomol Struct Dyn 24: 91–107. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Hutchinson AJ, Hudson RL, Dubocovich ML (2014). Genetic deletion of the MT1 or MT2 melatonin receptors abrogates methamphetamine‐induced reward in C3H/HeN mice. Physiol Behav 132: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai S, Ochoa‐Sanchez R, Gobbi G (2013). Sleep–wake characterization of double MT(1)/MT(2) receptor knockout mice and comparison with MT(1) and MT(2) receptor knockout mice. Behav Brain Res 243: 231–238. [DOI] [PubMed] [Google Scholar]
- Conti A, Conconi S, Hertens E, Skwarlo‐Sonta K, Markowska M, Maestroni JM (2000). Evidence for melatonin synthesis in mouse and human bone marrow cells. J Pineal Res 28: 193–202. [DOI] [PubMed] [Google Scholar]
- Contreras‐Alcantara S, Baba K, Tosini G (2010). Removal of melatonin receptor type 1 induces insulin resistance in the mouse. Obesity (Silver Spring) 18: 1861–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway S, Mowat ES, Drew JE, Barrett P, Delagrange P, Morgan PJ (2001). Serine residues 110 and 114 are required for agonist binding but not antagonist binding to the melatonin MT(1) receptor. Biochem Biophys Res Commun 282: 1229–1236. [DOI] [PubMed] [Google Scholar]
- Daulat AM, Maurice P, Froment C, Guillaume JL, Broussard C, Monsarrat B et al. (2007). Purification and identification of G protein‐coupled receptor protein complexes under native conditions. Mol Cell Proteomics 6: 835–844. [DOI] [PubMed] [Google Scholar]
- de Bodinat C, Guardiola‐Lemaitre B, Mocaer E, Renard P, Munoz C, Millan MJ (2010). Agomelatine, the first melatonergic antidepressant: discovery, characterization and development. Nat Rev Drug Discov 9: 628–642. [DOI] [PubMed] [Google Scholar]
- de la Fuente Revenga M, Fernandez‐Saez N, Herrera‐Arozamena C, Morales‐Garcia JA, Alonso‐Gil S, Perez‐Castillo A et al. (2015). Novel N‐Acetyl Bioisosteres of Melatonin: Melatonergic Receptor Pharmacology, Physicochemical Studies, and Phenotypic Assessment of Their Neurogenic Potential. J Med Chem 58: 4998–5014. [DOI] [PubMed] [Google Scholar]
- Descamps‐Francois C, Yous S, Chavatte P, Audinot V, Bonnaud A, Boutin JA et al. (2003). Design and Synthesis of Naphthalenic Dimers as Selective MT1 Melatoninergic Ligands. J Med Chem 46: 1127–1129. [DOI] [PubMed] [Google Scholar]
- Dominguez‐Alonso A, Valdes‐Tovar M, Solis‐Chagoyan H, Benitez‐King G (2015). Melatonin stimulates dendrite formation and complexity in the hilar zone of the rat hippocampus: participation of the Ca++/Calmodulin complex. Int J Mol Sci 16: 1907–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC et al. (2013). Structural basis for modulation of a G‐protein‐coupled receptor by allosteric drugs. Nature 503: 295–299. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML (1985). Characterization of a retinal melatonin receptor. J Pharmacol Exp Ther 234: 395–401. [PubMed] [Google Scholar]
- Dubocovich ML (1988). Luzindole (N‐0774): a novel melatonin receptor antagonist. J Pharmacol Exp Ther 246: 902–910. [PubMed] [Google Scholar]
- Dubocovich ML (1983). Melatonin is a potent modulator of dopamine release in the retina. Nature 306: 782–784. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML (1995). Melatonin receptors: Are there multiple subtypes? Trends Pharmacol Sci 16: 50–56. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML (2007). Melatonin receptors: role on sleep and circadian rhythm regulation. Sleep Med 8 (Suppl 3): 34–42. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Delagrange P, Krause DN, Sugden D, Cardinali DP, Olcese J (2010). International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein‐coupled melatonin receptors. Pharmacol Rev 62: 343–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubocovich ML, Hudson RL, Sumaya IC, Masana MI, Manna E (2005). Effect of MT1 melatonin receptor deletion on melatonin‐mediated phase shift of circadian rhythms in the C57BL/6 mouse. J Pineal Res 39: 113–120. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Masana MI, Iacob S, Sauri DM (1997). Melatonin receptor antagonists that differentiate between the human Mel1a and Mel1b recombinant subtypes are used to assess the pharmacological profile of the rabbit retina ML1 presynaptic heteroreceptor. Naunyn Schmiedebergs Arch Pharmacol 355: 365–375. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Rivera‐Bermudez MA, Gerdin MJ, Masana MI (2003). Molecular pharmacology, regulation and function of mammalian melatonin receptors. Front Biosci 8: d1093–d1108. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Takahashi JS (1987). Use of 2‐[125I]iodomelatonin to characterize melatonin binding sites in chicken retina. Proc Natl Acad Sci U S A 84: 3916–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubocovich ML, Yun K, AlGhoul WM, Benloucif S, Masana MI (1998). Selective MT2 melatonin receptor antagonists block melatonin‐mediated phase advances of circadian rhythms. FASEB J 12: 1211–1220. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Kajimura N, Uchiyama M, Katoh M, Sekimoto M, Watanabe T et al. (1999). Alleic variants of human melatonin 1a receptor: Function and prevalence in subjects with circadian rhythm sleep disorders. Biochem Biophys Res Commun 262: 832–837. [DOI] [PubMed] [Google Scholar]
- Escribano BM, Colin‐Gonzalez AL, Santamaria A, Tunez I (2014). The role of melatonin in multiple sclerosis, Huntington's disease and cerebral ischemia. CNS Neurol Disord Drug Targets 13: 1096–1119. [DOI] [PubMed] [Google Scholar]
- Ettaoussi M, Sabaouni A, Rami M, Boutin JA, Delagrange P, Renard P et al. (2012). Design, synthesis and pharmacological evaluation of new series of naphthalenic analogues as melatoninergic (MT1/MT2) and serotoninergic 5‐HT2C dual ligands (I). Eur J Med Chem 49: 310–323. [DOI] [PubMed] [Google Scholar]
- Evely KM, Hudson RL, Dubocovich ML, Haj‐Dahmane S (2016). Melatonin receptor activation increases glutamatergic synaptic transmission in the rat medial lateral habenula. Synapse 70: 181–186. [DOI] [PubMed] [Google Scholar]
- Fang JM, Dubocovich ML (1990). Activation of melatonin receptor sites retarded the depletion of norepinephrine following inhibition of synthesis in the C3H/HeN mouse hypothalamus. J Neurochem 55: 76–82. [DOI] [PubMed] [Google Scholar]
- Farce A, Chugunov AO, Loge C, Sabaouni A, Yous S, Dilly S et al. (2008). Homology modeling of MT1 and MT2 receptors. Eur J Med Chem 43: 1926–1944. [DOI] [PubMed] [Google Scholar]
- Farez MF, Mascanfroni ID, Mendez‐Huergo SP, Yeste A, Murugaiyan G, Garo LP et al. (2015). Melatonin Contributes to the Seasonality of Multiple Sclerosis Relapses. Cell 162: 1338–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust R, Garratt PJ, Jones R, Yeh LK, Tsotinis A, Panoussopoulou M et al. (2000). Mapping the melatonin receptor. 6. Melatonin agonists and antagonists derived from 6H‐isoindolo[2,1‐a]indoles, 5,6‐dihydroindolo[2,1‐a]isoquinolines, and 6,7‐dihydro‐5H‐benzo[c]azepino[2,1‐a]indoles. J Med Chem 43: 1050–1061. [DOI] [PubMed] [Google Scholar]
- Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ et al. (2014). G protein‐coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev 66: 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SP, Sugden D (2009). Sleep‐promoting action of IIK7, a selective MT2 melatonin receptor agonist in the rat. Neurosci Lett 457: 93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdin MJ, Masana MI, Rivera‐Bermudez MA, Hudson RL, Earnest DJ, Gillette MU et al. (2004). Melatonin desensitizes endogenous MT2 melatonin receptors in the rat suprachiasmatic nucleus: relevance for defining the periods of sensitivity of the mammalian circadian clock to melatonin. FASEB J 18: 1646–1656. [DOI] [PubMed] [Google Scholar]
- Gerdin MJ, Mseeh F, Dubocovich ML (2003). Mutagenesis studies of the human MT2 melatonin receptor. Biochem Pharmacol 66: 315–320. [DOI] [PubMed] [Google Scholar]
- Gianesini C, Hiragaki S, Laurent V, Hicks D, Tosini G (2016). Cone Viability Is Affected by Disruption of Melatonin Receptors Signaling. Invest Ophthalmol Vis Sci 57: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitto E, Karbownik M, Reiter RJ, Tan DX, Cuzzocrea S, Chiurazzi P et al. (2001). Effects of melatonin treatment in septic newborns. Pediatr Res 50: 756–760. [DOI] [PubMed] [Google Scholar]
- Green NH, Jackson CR, Iwamoto H, Tackenberg MC, McMahon DG (2015). Photoperiod programs dorsal raphe serotonergic neurons and affective behaviors. Curr Biol 25: 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman D, Attia MI, Behnam MAM, Mohsen AMY, Markl C, Julius J et al. (2011). 2‐[(1,3‐Dihydro‐2H‐isoindol‐2yl)methyl]‐melatonin‐ a novel MT2‐selective melatonin receptor antagonist. Med Chem Commun 2: 991–994. [Google Scholar]
- Hevia D, Gonzalez‐Menendez P, Quiros‐Gonzalez I, Miar A, Rodriguez‐Garcia A, Tan DX et al. (2015). Melatonin uptake through glucose transporters: a new target for melatonin inhibition of cancer. J Pineal Res 58: 234–250. [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gazarini ML, Beraldo FH, Varotti FP, Lopes C, Markus RP et al. (2000). Calcium‐dependent modulation by melatonin of the circadian rhythm in malarial parasites. Nat Cell Biol 2: 466–468. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ho MKC, Chan KH, New DC, Wong YH (2010). Synthesis of substituted N‐[3‐(3‐methoxyphenyl)propyl] amides as highly potent MT2‐selective melatonin ligands. Bioorg Med Chem Lett 20: 2582–2585. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhu J, Chan KH, Wong YH (2013). Development of substituted N‐[3‐(3‐methoxylphenyl)propyl] amides as MT(2)‐selective melatonin agonists: improving metabolic stability. Bioorg Med Chem 21: 547–552. [DOI] [PubMed] [Google Scholar]
- Hunt AE, AlGhoul WM, Gillette MU, Dubocovich ML (2001). Activation of MT2 melatonin receptors in rat suprachiasmatic nucleus phase advances the circadian clock. Am J Physiol Cell Physiol 280: C110–C118. [DOI] [PubMed] [Google Scholar]
- Hutchinson AJ, Hudson RL, Dubocovich ML (2012). Genetic deletion of MT(1) and MT(2) melatonin receptors differentially abrogates the development and expression of methamphetamine‐induced locomotor sensitization during the day and the night in C3H/HeN mice. J Pineal Res 53: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg A, Moek J, Weaver DR, Korf HW, Stehle JH, von Gall C (2005). Rhythms in clock proteins in the mouse pars tuberalis depend on MT1 melatonin receptor signalling. Eur J Neurosci 22: 2845–2854. [DOI] [PubMed] [Google Scholar]
- Jin XW, von Gall C, Pieschl RL, Gribkoff VK, Stehle JH, Reppert SM et al. (2003). Targeted disruption of the mouse Mel(1b) melatonin receptor. Mol Cell Biol 23: 1054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockers R, Maurice P, Boutin JA, Delagrange P (2008). Melatonin receptors, heterodimerization, signal transduction and binding sites: what's new? Br J Pharmacol 154: 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JD, Skene DJ (2015). 60 YEARS OF NEUROENDOCRINOLOGY: Regulation of mammalian neuroendocrine physiology and rhythms by melatonin. J Endocrinol 226: T187–T198. [DOI] [PubMed] [Google Scholar]
- Johnston JM, Filizola M (2014). Beyond standard molecular dynamics: investigating the molecular mechanisms of G protein‐coupled receptors with enhanced molecular dynamics methods. Adv Exp Med Biol 796: 95–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Biswas J, Nath C, Singh S (2015). Promising Role of Melatonin as Neuroprotectant in Neurodegenerative Pathology. Mol Neurobiol 52: 330–340. [DOI] [PubMed] [Google Scholar]
- Journe AS, Habib SAM, Dodda BR, Morcos MNF, Sadek MS, Tadros SAA et al. (2014). N1‐linked melatonin dimers as bivalent ligands targeting dimeric melatonin receptors. Med Chem Commun 5: 792–796. [Google Scholar]
- Kamal M, Gbahou F, Guillaume JL, Daulat AM, Benleulmi‐Chaachoua A, Luka M et al. (2015). Convergence of melatonin and serotonin (5‐HT) signaling at MT2/5‐HT2C receptor heteromers. J Biol Chem 290: 11537–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A et al. (2015). Crystal structure of rhodopsin bound to arrestin by femtosecond X‐ray laser. Nature 523: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamitri A, Jockers R (2014). Exon Sequencing of G Protein‐Coupled Receptor Genes and Perspectives for Disease Treatment In: Stefens CW. (ed). Methods in Pharmacology and Toxicology series, G Protein‐Coupled Receptor Genetics: Research and Methods in the Post‐Genomic Era. Springer Science: New York, pp. 313–332. [Google Scholar]
- Karamitri A, Renault N, Clement N, Guillaume JL, Jockers R (2013). Minireview: Toward the Establishment of a Link between Melatonin and Glucose Homeostasis: Association of Melatonin MT2 Receptor Variants with Type 2 Diabetes. Mol Endocrinol 27: 1217–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Hirai K, Nishiyama K, Uchikawa O, Fukatsu K, Ohkawa S et al. (2005). Neurochemical properties of ramelteon (TAK‐375), a selective MT1/MT2 receptor agonist. Neuropharmacology 48: 301–310. [DOI] [PubMed] [Google Scholar]
- Koike T, Hoashi Y, Takai T, Nakayama M, Yukuhiro N, Ishikawa T et al. (2011). 1,6‐Dihydro‐2H‐indeno[5,4‐b]furan derivatives: design, synthesis, and pharmacological characterization of a novel class of highly potent MT(2)‐selective agonists. J Med Chem 54: 3436–3444. [DOI] [PubMed] [Google Scholar]
- Kokkola T, Foord SM, Watson MA, Vakkuri O, Laitinen JT (2003). Important amino acids for the function of the human MT1 melatonin receptor. Biochem Pharmacol 65: 1463–1471. [DOI] [PubMed] [Google Scholar]
- Kong X, Li X, Cai Z, Yang N, Liu Y, Shu J et al. (2008). Melatonin regulates the viability and differentiation of rat midbrain neural stem cells. Cell Mol Neurobiol 28: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DN, Siuciak JA, Dubocovich ML (1992). Optic nerve transection decreases 2‐[125I]iodomelatonin binding in the chick optic tectum. Brain Res 590: 325–328. [DOI] [PubMed] [Google Scholar]
- Krause DN, Siuciak JA, Dubocovich ML (1994). Unilateral optic nerve transection decreases 2‐[125I]‐iodomelatonin binding in retinorecipient areas and visual pathways of chick brain. Brain Res 654: 63–74. [DOI] [PubMed] [Google Scholar]
- Lahiri S, Haldar C (2009). Response of melatonin receptor MT1 in spleen of a tropical Indian rodent, Funambulus pennanti, to natural solar insolation and different photoperiodic conditions. Chronobiol Int 26: 1559–1574. [DOI] [PubMed] [Google Scholar]
- Landolt HP, Wehrle R (2009). Antagonism of serotonergic 5‐HT2A/2C receptors: mutual improvement of sleep, cognition and mood? Eur J Neurosci 29: 1795–1809. [DOI] [PubMed] [Google Scholar]
- Laranjeira‐Silva MF, Zampieri RA, Muxel SM, Floeter‐Winter LM, Markus RP (2015). Melatonin attenuates Leishmania (L.) amazonensis infection by modulating arginine metabolism. J Pineal Res 59: 478–487. [DOI] [PubMed] [Google Scholar]
- Lardone PJ, Carrillo‐Vico A, Naranjo MC, De Felipe B, Vallejo A, Karasek M et al. (2006). Melatonin synthesized by Jurkat human leukemic T cell line is implicated in IL‐2 production. J Cell Physiol 206: 273–279. [DOI] [PubMed] [Google Scholar]
- Lardone PJ, Rubio A, Cerrillo I, Gomez‐Corvera A, Carrillo‐Vico A, Sanchez‐Hidalgo M et al. (2010). Blocking of melatonin synthesis and MT(1) receptor impairs the activation of Jurkat T cells. Cell Mol Life Sci 67: 3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J, Jessen RE, Uz T, Arslan AD, Kurtuncu M, Imbesi M et al. (2006). Impaired hippocampal long‐term potentiation in melatonin MT2 receptor‐deficient mice. Neurosci Lett 393: 23–26. [DOI] [PubMed] [Google Scholar]
- Lavedan C, Forsberg M, Gentile AJ (2015). Tasimelteon: a selective and unique receptor binding profile. Neuropharmacology 91: 142–147. [DOI] [PubMed] [Google Scholar]
- Legros C, Brasseur C, Delagrange P, Ducrot P, Nosjean O, Boutin JA (2016). Alternative Radioligands for Investigating the Molecular Pharmacology of Melatonin Receptors. J Pharmacol Exp Ther 356: 681–692. [DOI] [PubMed] [Google Scholar]
- Legros C, Chesneau D, Boutin JA, Barc C, Malpaux B (2014). Melatonin from cerebrospinal fluid but not from blood reaches sheep cerebral tissues under physiological conditions. J Neuroendocrinol 26: 151–163. [DOI] [PubMed] [Google Scholar]
- Legros C, Matthey U, Grelak T, Pedragona‐Moreau S, Hassler W, Yous S et al. (2013). New Radioligands for Describing the Molecular Pharmacology of MT1 and MT2 Melatonin Receptors. Int J Mol Sci 14: 8948–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Dam J, Ayoub MA, Guillaume JL, Couturier C, Delagrange P et al. (2006a). The orphan GPR50 receptor specifically inhibits MT(1) melatonin receptor function through heterodimerization. EMBO J 25: 3012–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Dam J, Ayoub MA, Guillaume JL, Jockers R (2006b). Do orphan G‐protein‐coupled receptors have ligand‐independent functions? New insights from receptor heterodimers. EMBO Rep 7: 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Williams‐Ashman HG (1961). Enzymatic oxidation of some non‐phosphorylated derivatives of dihydronicotinamide. Biochem Biophys Res Commun 4: 208–213. [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Jin X, Shearman LP, Pieschl RL, Gribkoff VK et al. (1997). Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron 19: 91–102. [DOI] [PubMed] [Google Scholar]
- Liu D, Wei N, Man HY, Lu Y, Zhu LQ, Wang JZ (2015). The MT2 receptor stimulates axonogenesis and enhances synaptic transmission by activating Akt signaling. Cell Death Differ 22: 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Clough SJ, Hutchinson AJ, Adamah‐Biassi EB, Popovska‐Gorevski M, Dubocovich ML (2016). MT1 and MT2 Melatonin Receptors: A Therapeutic Perspective. Annu Rev Pharmacol Toxicol 56: 361–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotufo CM, Lopes C, Dubocovich ML, Farsky SH, Markus RP (2001). Melatonin and N‐acetylserotonin inhibit leukocyte rolling and adhesion to rat microcirculation. Eur J Pharmacol 430: 351–357. [DOI] [PubMed] [Google Scholar]
- Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P et al. (2009). Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 41: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestroni G (1993). Th immunoneuroendocrine role of melatonin. J Pineal Res 14: 1–10. [DOI] [PubMed] [Google Scholar]
- Markl C, Clafshenkel WP, Attia MI, Sethi S, Witt‐Enderby PA, Zlotos DP (2011). N‐Acetyl‐5‐arylalkoxytryptamine analogs: probing the melatonin receptors for MT(1) ‐selectivity. Arch Pharm (Weinheim) 344: 666–674. [DOI] [PubMed] [Google Scholar]
- Markus RP, Cecon E, Pires‐Lapa MA (2013). Immune‐Pineal Axis: Nuclear Factor Œ∫B (NF‐kB) Mediates the Shift in the Melatonin Source from Pinealocytes to Immune Competent Cells. Int J Mol Sci 14: 10979–10997 %! Immune‐Pineal Axis: Nuclear Factor Œ∫B (NF‐kB) Mediates the Shift in the Melatonin Source from Pinealocytes to Immune Competent Cells %@ 11422‐10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice P, Daulat AM, Broussard C, Mozo J, Clary G, Hotellier F et al. (2008). A generic approach for the purification of signaling complexes that specifically interact with the carboxy‐terminal domain of G protein‐coupled receptors. Mol Cell Proteomics 7: 1556–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice P, Kamal M, Jockers R (2011). Asymmetry of GPCR oligomers supports their functional relevance. Trends Pharmacol Sci 32: 514–520. [DOI] [PubMed] [Google Scholar]
- Mazna P, Obsilova V, Jelinkova I, Balik A, Berka K, Sovova Z et al. (2004). Molecular modeling of human MT2 melatonin receptor: the role of Val204, Leu272 and Tyr298 in ligand binding. J Neurochem 91: 836–842. [DOI] [PubMed] [Google Scholar]
- McArthur A, Hunt A, Gillette M (1997). Melatonin action and signal transduction in the rat suprachiasmatic circadian clock: Activation of protein kinase C at dusk and dawn. Endocrinology 138: 627–634. [DOI] [PubMed] [Google Scholar]
- Medeiros CA, Carvalhedo de Bruin PF, Lopes LA, Magalhaes MC, de Lourdes Seabra M, de Bruin VM (2007). Effect of exogenous melatonin on sleep and motor dysfunction in Parkinson's disease. A randomized, double blind, placebo‐controlled study. J Neurol 254: 459–464. [DOI] [PubMed] [Google Scholar]
- Mesangeau C, Peres B, Descamps‐Francois C, Chavatte P, Audinot V, Coumailleau S et al. (2010). Design, synthesis and pharmacological evaluation of novel naphthalenic derivatives as selective MT(1) melatoninergic ligands. Bioorg Med Chem 18: 3426–3436. [DOI] [PubMed] [Google Scholar]
- Millan MJ, Gobert A, Lejeune F, Dekeyne A, Newman‐Tancredi A, Pasteau V et al. (2003). The novel melatonin agonist agomelatine (S20098) is an antagonist at 5‐hydroxytryptamine2C receptors, blockade of which enhances the activity of frontocortical dopaminergic and adrenergic pathways. J Pharmacol Exp Ther 306: 954–964. [DOI] [PubMed] [Google Scholar]
- Mini LJ, Wang‐Weigand S, Zhang J (2007). Self‐reported efficacy and tolerability of ramelteon 8 mg in older adults experiencing severe sleep‐onset difficulty. Am J Geriatr Pharmacother 5: 177–184. [DOI] [PubMed] [Google Scholar]
- Molinari EJ, North PC, Dubocovich ML (1996). 2‐[I‐125]iodo‐5‐methoxycarbonylamino‐N‐acetyltryptamine: A selective radioligand for the characterization of melatonin ML(2) binding sites. Eur J Pharmacol 301: 159–168. [DOI] [PubMed] [Google Scholar]
- Morellato L, Lefas‐Le Gall M, Langlois M, Caignard DH, Renard P, Delagrange P et al. (2013). Synthesis of new N‐(arylcyclopropyl)acetamides and N‐(arylvinyl)acetamides as conformationally‐restricted ligands for melatonin receptors. Bioorg Med Chem Lett 23: 430–434. [DOI] [PubMed] [Google Scholar]
- Muhlbauer E, Gross E, Labucay K, Wolgast S, Peschke E (2009). Loss of melatonin signalling and its impact on circadian rhythms in mouse organs regulating blood glucose. Eur J Pharmacol 606: 61–71. [DOI] [PubMed] [Google Scholar]
- Mulchahey JJ, Goldwater DR, Zemlan FP (2004). A single blind, placebo controlled, across groups dose escalation study of the safety, tolerability, pharmacokinetics and pharmacodynamics of the melatonin analog beta‐methyl‐6‐chloromelatonin. Life Sci 75: 1843–1856. [DOI] [PubMed] [Google Scholar]
- Muxel SM, Laranjeira‐Silva MF, de Carvalho Sousa CE, Floeter‐Winter LM, Markus RP (2016). The RelA/cRel nuclear factor‐kappaB (NF‐kappaB) dimer, crucial for inflammation resolution, mediates the transcription of the key enzyme in melatonin synthesis in RAW 264.7 macrophages. J Pineal Res 60: 394–404. [DOI] [PubMed] [Google Scholar]
- Muxel SM, Pires‐Lapa MA, Monteiro AW, Cecon E, Tamura EK, Floeter‐Winter LM et al. (2012). NF‐kappaB drives the synthesis of melatonin in RAW 264.7 macrophages by inducing the transcription of the arylalkylamine‐N‐acetyltransferase (AA‐NAT) gene. PLoS One 7: e52010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naskar A, Prabhakar V, Singh R, Dutta D, Mohanakumar KP (2015). Melatonin enhances L‐DOPA therapeutic effects, helps to reduce its dose, and protects dopaminergic neurons in 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine‐induced Parkinsonism in mice. J Pineal Res 58: 262–274. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C et al. (2012). An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonno R, Lucini V, Spadoni G, Pannacci M, Croce A, Esposti D et al. (2000). A new melatonin receptor ligand with mt(1)‐agonist and MT2‐antagonist properties. J Pineal Res 29: 234–240. [DOI] [PubMed] [Google Scholar]
- Nosjean O, Ferro M, Cogé F, Beauverger P, Henlin JM, Lefoulon F et al. (2000). Identification of the melatonin‐binding site MT3 as the quinone reductase 2. J Biol Chem 275: 31311–31317. [DOI] [PubMed] [Google Scholar]
- O'Neal‐Moffitt G, Delic V, Bradshaw PC, Olcese J (2015). Prophylactic melatonin significantly reduces Alzheimer's neuropathology and associated cognitive deficits independent of antioxidant pathways in AbetaPP(swe)/PS1 mice. Mol Neurodegener 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal‐Moffitt G, Pilli J, Kumar SS, Olcese J (2014). Genetic deletion of MT1/MT2 melatonin receptors enhances murine cognitive and motor performance. Neuroscience 277: 506–521. [DOI] [PubMed] [Google Scholar]
- Ochoa‐Sanchez R, Comai S, Lacoste B, Bambico FR, Dominguez‐Lopez S, Spadoni G et al. (2011). Promotion of non‐rapid eye movement sleep and activation of reticular thalamic neurons by a novel MT2 melatonin receptor ligand. J Neurosci 31: 18439–18452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y (2002). Functional role of internal water molecules in rhodopsin revealed by X‐ray crystallography. Proc Natl Acad Sci U S A 99: 5982–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V (2004). The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol 342: 571–583. [DOI] [PubMed] [Google Scholar]
- Olcese JM, Cao C, Mori T, Mamcarz MB, Maxwell A, Runfeldt MJ et al. (2009). Protection against cognitive deficits and markers of neurodegeneration by long‐term oral administration of melatonin in a transgenic model of Alzheimer disease. J Pineal Res 47: 82–96. [DOI] [PubMed] [Google Scholar]
- Ortiz GG, Pacheco‐Moises FP, Gomez‐Rodriguez VM, Gonzalez‐Renovato ED, Torres‐Sanchez ED, Ramirez‐Anguiano AC (2013). Fish oil, melatonin and vitamin E attenuates midbrain cyclooxygenase‐2 activity and oxidative stress after the administration of 1‐methyl‐4‐phenyl‐1,2,3,6‐ tetrahydropyridine. Metab Brain Dis 28: 705–709. [DOI] [PubMed] [Google Scholar]
- Owino S, Contreras‐Alcantara S, Baba K, Tosini G (2016). Melatonin Signaling Controls the Daily Rhythm in Blood Glucose Levels Independent of Peripheral Clocks. PLoS One 11: e0148214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala D, Beuming T, Sherman W, Lodola A, Rivara S, Mor M (2013). Structure‐based virtual screening of MT2 melatonin receptor: influence of template choice and structural refinement. J Chem Inf Model 53: 821–835. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA et al. (2000). Crystal structure of rhodopsin: A G protein‐coupled receptor. Science 289: 739–745. [DOI] [PubMed] [Google Scholar]
- Peng CX, Hu J, Liu D, Hong XP, Wu YY, Zhu LQ et al. (2013). Disease‐modified glycogen synthase kinase‐3beta intervention by melatonin arrests the pathology and memory deficits in an Alzheimer's animal model. Neurobiol Aging 34: 1555–1563. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Rauch A, Korf HW, von Gall C (2012). The endogenous melatonin (MT) signal facilitates reentrainment of the circadian system to light‐induced phase advances by acting upon MT2 receptors. Chronobiol Int 29: 415–429. [DOI] [PubMed] [Google Scholar]
- Pinato L, da Silveira Cruz‐Machado S, Franco DG, Campos LM, Cecon E, Fernandes PA et al. (2015). Selective protection of the cerebellum against intracerebroventricular LPS is mediated by local melatonin synthesis. Brain Struct Funct 220: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires‐Lapa MA, Tamura EK, Salustiano EM, Markus RP (2013). Melatonin synthesis in human colostrum mononuclear cells enhances dectin‐1‐mediated phagocytosis by mononuclear cells. J Pineal Res 55: 240–246. [DOI] [PubMed] [Google Scholar]
- Poissonnier‐Durieux S, Ettaoussi M, Peres B, Boutin JA, Audinot V, Bennejean C et al. (2008). Synthesis of 3‐phenylnaphthalenic derivatives as new selective MT(2) melatoninergic ligands. Bioorg Med Chem 16: 8339–8348. [DOI] [PubMed] [Google Scholar]
- Pontes GN, Cardoso EC, Carneiro‐Sampaio MM, Markus RP (2007). Pineal melatonin and the innate immune response: the TNF‐alpha increase after cesarean section suppresses nocturnal melatonin production. J Pineal Res 43: 365–371. [DOI] [PubMed] [Google Scholar]
- Prada C, Udin SB, Wiechmann AF, Zhdanova IV (2005). Stimulation of melatonin receptors decreases calcium levels in xenopus tectal cells by activating GABA(C) receptors. J Neurophysiol 94: 968–978. [DOI] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G et al. (2009). Variants in MTNR1B influence fasting glucose levels. Nat Genet 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasi D, Artacho MC, Negri A, Mobarec JC, Filizola M (2011). Ligand‐induced modulation of the free‐energy landscape of G protein‐coupled receptors explored by adaptive biasing techniques. PLoS Comput Biol 7: e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racagni G, Riva MA, Molteni R, Musazzi L, Calabrese F, Popoli M et al. (2011). Mode of action of agomelatine: Synergy between melatonergic and 5‐HT(2C) receptors. World J Biol Psychiatry 12: 574–587. [DOI] [PubMed] [Google Scholar]
- Rajaratnam SM, Polymeropoulos MH, Fisher DM, Roth T, Scott C, Birznieks G et al. (2009). Melatonin agonist tasimelteon (VEC‐162) for transient insomnia after sleep‐time shift: two randomised controlled multicentre trials. Lancet 373: 482–491. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS et al. (2011a). Structure of a nanobody‐stabilized active state of the beta(2) adrenoceptor. Nature 469: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS et al. (2011b). Crystal structure of the beta2 adrenergic receptor‐Gs protein complex. Nature 477: 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawashdeh O, Hudson RL, Stepien I, Dubocovich ML (2011). Circadian periods of sensitivity for ramelteon on the onset of running‐wheel activity and the peak of suprachiasmatic nucleus neuronal firing rhythms in C3H/HeN mice. Chronobiol Int 28: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressmeyer AR, Mayo JC, Zelosko V, Sainz RM, Tan DX, Poeggeler B et al. (2003). Antioxidant properties of the melatonin metabolite N1‐acetyl‐5‐methoxykynuramine (AMK): scavenging of free radicals and prevention of protein destruction. Redox Rep 8: 205–213. [DOI] [PubMed] [Google Scholar]
- Rivara S, Lodola A, Mor M, Bedini A, Spadoni G, Lucini V et al. (2007). N‐(substituted‐anilinoethyl)amides: design, synthesis, and pharmacological characterization of a new class of melatonin receptor ligands. J Med Chem 50: 6618–6626. [DOI] [PubMed] [Google Scholar]
- Rivara S, Lorenzi S, Mor M, Plazzi PV, Spadoni G, Bedini A et al. (2005). Analysis of structure–activity relationships for MT2 selective antagonists by melatonin MT1 and MT2 receptor models. J Med Chem 48: 4049–4060. [DOI] [PubMed] [Google Scholar]
- Rivara S, Pala D, Lodola A, Mor M, Lucini V, Dugnani S et al. (2012). MT1‐selective melatonin receptor ligands: synthesis, pharmacological evaluation, and molecular dynamics investigation of N‐{[(3‐O‐substituted)anilino]alkyl}amides. ChemMedChem 7: 1954–1964. [DOI] [PubMed] [Google Scholar]
- Rosen R, Hu DN, Perez V, Tai K, Yu GP, Chen M et al. (2009). Urinary 6‐sulfatoxymelatonin level in age‐related macular degeneration patients. Mol Vis 15: 1673–1679. [PMC free article] [PubMed] [Google Scholar]
- Sampaio Lde F, Mesquita FP, de Sousa PR, Silva JL, Alves CN (2014). The melatonin analog 5‐MCA‐NAT increases endogenous dopamine levels by binding NRH:quinone reductase enzyme in the developing chick retina. Int J Dev Neurosci 38: 119–126. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Olivieri G, Meier F, Brydon L, Jockers R, Ravid R et al. (2002). Increased melatonin 1a‐receptor immunoreactivity in the hippocampus of Alzheimer's disease patients. J Pineal Res 32: 59–62. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Schnitzler C, Schroder C, Cajochen C, Muller‐Spahn F, Wirz‐Justice A (2006). Treatment of behavioural, cognitive and circadian rest‐activity cycle disturbances in Alzheimer's disease: haloperidol vs. quetiapine. Int J Neuropsychopharmacol 9: 507–516. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW et al. (2008). Crystal structure of opsin in its G‐protein‐interacting conformation. Nature 455: 497–502. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Baba K, Mazzoni F, Pozdeyev NV, Strettoi E, Iuvone PM et al. (2011). Localization of melatonin receptor 1 in mouse retina and its role in the circadian regulation of the electroretinogram and dopamine levels. PLoS One 6: e24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Haldar C (2006). Melatonin prevents X‐ray irradiation induced oxidative damagein peripheral blood and spleen of the seasonally breeding rodent, Funambulus pennanti during reproductively active phase. Int J Radiat Biol 82: 411–419. [DOI] [PubMed] [Google Scholar]
- Sheynzon P, Korf HW (2006). Targeted deletions of Mel1a and Mel1b melatonin receptors affect pCREB levels in lactotroph and pars intermedia cells of mice. Neurosci Lett 407: 48–52. [DOI] [PubMed] [Google Scholar]
- Shin IS, Shin NR, Park JW, Jeon CM, Hong JM, Kwon OK et al. (2015). Melatonin attenuates neutrophil inflammation and mucus secretion in cigarette smoke‐induced chronic obstructive pulmonary diseases via the suppression of Erk‐Sp1 signaling. J Pineal Res 58: 50–60. [DOI] [PubMed] [Google Scholar]
- Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA et al. (2008). Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha‐subunits. Proc Natl Acad Sci U S A 105: 6457–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH et al. (2016). The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 44 (Database Issue): D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadoni G, Bedini A, Lucarini S, Mari M, Caignard DH, Boutin JA et al. (2015). Highly Potent and Selective MT2 Melatonin Receptor Full Agonists from Conformational Analysis of 1‐Benzyl‐2‐acylaminomethyl‐tetrahydroquinolines. J Med Chem 58: 7512–7525. [DOI] [PubMed] [Google Scholar]
- Spadoni G, Bedini A, Orlando P, Lucarini S, Tarzia G, Mor M et al. (2011). Bivalent ligand approach on N‐{2‐[(3‐methoxyphenyl)methylamino]ethyl}acetamide: synthesis, binding affinity and intrinsic activity for MT(1) and MT(2) melatonin receptors. Bioorg Med Chem 19: 4910–4916. [DOI] [PubMed] [Google Scholar]
- Stumpf I, Muhlbauer E, Peschke E (2008). Involvement of the cGMP pathway in mediating the insulin‐inhibitory effect of melatonin in pancreatic beta‐cells. J Pineal Res 45: 318–327. [DOI] [PubMed] [Google Scholar]
- Sugden D, Chong NW, Lewis DF (1995). Structural requirements at the melatonin receptor. Br J Pharmacol 114: 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden D, Yeh LK, Teh MT (1999). Design of subtype selective melatonin receptor agonists and antagonists. Reprod Nutr Dev 39: 335–344. [DOI] [PubMed] [Google Scholar]
- Tamura EK, Fernandes PA, Marcola M, da Silveira Cruz‐Machado S, Markus RP (2010). Long‐lasting priming of endothelial cells by plasma melatonin levels. PLoS One 5: e13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DX, Manchester LC, Esteban‐Zubero E, Zhou Z, Reiter RJ (2015). Melatonin as a Potent and Inducible Endogenous Antioxidant: Synthesis and Metabolism. Molecules 20: 18886–18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DX, Manchester LC, Terron MP, Flores LJ, Reiter RJ (2007). One molecule, many derivatives: a never‐ending interaction of melatonin with reactive oxygen and nitrogen species? J Pineal Res 42: 28–42. [DOI] [PubMed] [Google Scholar]
- Tchekalarova J, Nenchovska Z, Atanasova D, Atanasova M, Kortenska L, Stefanova M et al. (2016). Consequences of long‐term treatment with agomelatine on depressive‐like behavior and neurobiological abnormalities in pinealectomized rats. Behav Brain Res 302: 11–28. [DOI] [PubMed] [Google Scholar]
- Tosini G, Baba K, Hwang CK, Iuvone PM (2012). Melatonin: an underappreciated player in retinal physiology and pathophysiology. Exp Eye Res 103: 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosini G, Owino S, Guillaume JL, Jockers R (2014). Understanding melatonin receptor pharmacology: Latest insights from mouse models, and their relevance to human disease. Bioessays 36: 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa O, Fukatsu K, Tokunoh R, Kawada M, Matsumoto K, Imai Y et al. (2002). Synthesis of a novel series of tricyclic indan derivatives as melatonin receptor agonists. J Med Chem 45: 4222–4239. [DOI] [PubMed] [Google Scholar]
- Vakkuri O, Leppaluoto J, Vuolteenaho O (1984). Development and validation of a melatonin radioimmunoassay using radioiodinated melatonin as a tracer. Acta Endocrinol 106: 152–157. [DOI] [PubMed] [Google Scholar]
- van Wamelen DJ, Aziz NA, Anink JJ, van Steenhoven R, Angeloni D, Fraschini F et al. (2013). Suprachiasmatic nucleus neuropeptide expression in patients with Huntington's Disease. Sleep 36: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wamelen DJ, Roos RA, Aziz NA (2015). Therapeutic strategies for circadian rhythm and sleep disturbances in Huntington disease. Neurodegener Dis Manag 5: 549–559. [DOI] [PubMed] [Google Scholar]
- Vanecek J (1988). Melatonin binding sites. J Neurochem 51: 1436–1440. [DOI] [PubMed] [Google Scholar]
- Vincent L, Cohen W, Delagrange P, Boutin JA, Nosjean O (2010). Molecular and cellular pharmacological properties of 5‐methoxycarbonylamino‐N‐acetyltryptamine (MCA‐NAT): a nonspecific MT3 ligand. J Pineal Res 48: 222–229. [DOI] [PubMed] [Google Scholar]
- von Gall C, Garabette ML, Kell CA, Frenzel S, Dehghani F, Schumm DP et al. (2002). Rhythmic gene expression in pituitary depends on heterologous sensitization by the neurohormone melatonin. Nat Neurosci 5: 234–238. [DOI] [PubMed] [Google Scholar]
- Voronkov AE, Ivanov AA, Baskin II, Palyulin VA, Zefirov NS (2005). Molecular modeling study of the mechanism of ligand binding to human melatonin receptors. Dokl Biochem Biophys 403: 284–288. [DOI] [PubMed] [Google Scholar]
- Wade AG, Farmer M, Harari G, Fund N, Laudon M, Nir T et al. (2014). Add‐on prolonged‐release melatonin for cognitive function and sleep in mild to moderate Alzheimer's disease: a 6‐month, randomized, placebo‐controlled, multicenter trial. Clin Interv Aging 9: 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LM, Suthana NA, Chaudhury D, Weaver DR, Colwell CS (2005). Melatonin inhibits hippocampal long‐term potentiation. Eur J Neurosci 22: 2231–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sirianni A, Pei Z, Cormier K, Smith K, Jiang J et al. (2011). The melatonin MT1 receptor axis modulates mutant Huntingtin‐mediated toxicity. J Neurosci 31: 14496–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil ZM, Borniger JC, Cisse YM, Abi Salloum BA, Nelson RJ (2015). Neuroendocrine control of photoperiodic changes in immune function. Front Neuroendocrinol 37: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weishaupt JH, Bartels C, Polking E, Dietrich J, Rohde G, Poeggeler B et al. (2006). Reduced oxidative damage in ALS by high‐dose enteral melatonin treatment. J Pineal Res 41: 313–323. [DOI] [PubMed] [Google Scholar]
- Wu YH, Zhou JN, Van Heerikhuize J, Jockers R, Swaab DF (2007). Decreased MT1 melatonin receptor expression in the suprachiasmatic nucleus in aging and Alzheimer's disease. Neurobiol Aging 28: 1239–1247. [DOI] [PubMed] [Google Scholar]
- Yadav SK, Haldar C (2013). Reciprocal interaction between melatonin receptors (Mel(1a), Mel(1b), and Mel(1c)) and androgen receptor (AR) expression in immunoregulation of a seasonally breeding bird, Perdicula asiatica: role of photoperiod. J Photochem Photobiol B 122: 52–60. [DOI] [PubMed] [Google Scholar]
- Yasuo S, Yoshimura T, Ebihara S, Korf HW (2009). Melatonin transmits photoperiodic signals through the MT1 melatonin receptor. J Neurosci 29: 2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala‐Oseguera C, Galano A, Merino G (2014). Computational study on the kinetics and mechanism of the carbaryl + OH reaction. J Phys Chem A 118: 7776–7781. [DOI] [PubMed] [Google Scholar]
- Zefirova ON, Baranova TY, Ivanova AA, Ivanov AA, Zefirov NS (2011). Application of the bridgehead fragments for the design of conformationally restricted melatonin analogues. Bioorg Chem 39: 67–72. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen XY, Su SW, Jia QZ, Ding T, Zhu ZN et al. (2016). Exogenous melatonin for sleep disorders in neurodegenerative diseases: a meta‐analysis of randomized clinical trials. Neurol Sci 37: 57–65. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cook A, Kim J, Baranov SV, Jiang J, Smith K et al. (2013). Melatonin inhibits the caspase‐1/cytochrome c/caspase‐3 cell death pathway, inhibits MT1 receptor loss and delays disease progression in a mouse model of amyotrophic lateral sclerosis. Neurobiol Dis 55: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JN, Liu RY, Kamphorst W, Hofman MA, Swaab DF (2003). Early neuropathological Alzheimer's changes in aged individuals are accompanied by decreased cerebrospinal fluid melatonin levels. J Pineal Res 35: 125–130. [DOI] [PubMed] [Google Scholar]
- Zisapel N, Egozi Y, Laudon M (1985). Circadian variations in the inhibition of dopamine release from adult and newborn rat hypothalamus by melatonin. Neuroendocrinology 40: 102–108. [DOI] [PubMed] [Google Scholar]
- Zisapel N, Laudon M (1982). Dopamine release induced by electrical field stimulation of rat hypothalamus in vitro: inhibition by melatonin. Biochem Biophys Res Commun 104: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Zlotos DP (2012). Recent progress in the development of agonists and antagonists for melatonin receptors. Curr Med Chem 19: 3532–3549. [DOI] [PubMed] [Google Scholar]
- Zlotos DP, Attia MI, Julius J, Sethi S, Witt‐Enderby PA (2009). 2‐[(2,3‐dihydro‐1H‐indol‐1‐yl)methyl]melatonin analogues: a novel class of MT2‐selective melatonin receptor antagonists. J Med Chem 52: 826–833. [DOI] [PubMed] [Google Scholar]
- Zlotos DP, Jockers R, Cecon E, Rivara S, Witt‐Enderby PA (2014). MT1 and MT2 Melatonin Receptors: Ligands, Models, Oligomers, and Therapeutic Potential. J Med Chem 57: 3161–3185. [DOI] [PubMed] [Google Scholar]
- Zlotos DP, Riad NM, Osman MB, Dodda BR, Witt‐Enderby PA (2015). Novel difluoroacetamide analogues of agomelatine and melatonin: probing the melatonin receptors for MT1 selectivity. Med Chem Commun 6: 1340–1344. [Google Scholar]


