Abstract
We report the main characteristics of ‘Lachnoclostridium massiliosenegalense’ strain mt23T (=CSUR P299 =DSM 102084), a new bacterial species isolated from the gut microbiota of a healthy young girl from Senegal.
Keywords: anaerobe, culturomics, gut microbiota, Lachnoclostridium massiliosenegalense, taxono-genomics
As part of the ‘culturomics’ exploration of the gut microbiota [1], [2], a stool sample was collected from a 38-month-old healthy girl from Senegal. Her weight-for-height z-score was −0.12. Oral consent was obtained from the parents, and the ethics committee of the Institut Federatif de Recherche IFR48 validated this study. After a 7-day anaerobic pre-incubation of the stool sample in a marine broth and seeding on 5% sheep-blood-enriched Colombia agar (bioMérieux, La Balme-les-Grottes, France), strain mt23T was isolated in October 2015. Colonies were translucent with a mean diameter of 2 mm on 5% sheep-blood-enriched Colombia agar. Cells were Gram-positive rods and showed a mean length of 2.5 μm and a mean diameter of 0.5 μm on electron microscopy. Strain mt23T was catalase and oxidase negative. Routine identification of colonies was carried out using matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3], [4], which failed to identify strain mt23T. Therefore, the 16S rRNA gene was sequenced with fD1-rP2 primers as previously described [5] using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). The resulting sequence showed a 95.1% similarity with the 16S rRNA gene of Lachnoclostridium phytofermentans strain ISDg (GenBank Accession number NR_074652.1), the phylogenetically closest species (Fig. 1) [6]. According to the 16S rRNA gene sequence similarity for species delineation of prokaryotes [7], [8], strain mt23T is representative of a new species within the recently described Lachnoclostridium genus. We then propose the name of ‘Lachnoclostridium massiliosenegalense’ (mas.si.li.o.se.ne.gal.en′sis. L. gen. masc. n. massiliosenegalense contraction of the Latin name of Marseille where strain mt23T was isolated and Senegal where the stool sample was collected) for which strain mt23 is the type strain.
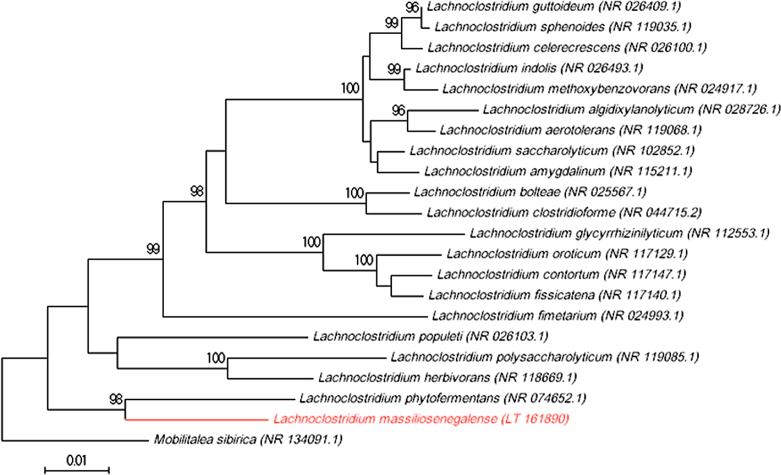
Fig. 1.
Phylogenetic tree showing the position of ‘Lachnoclostridium massiliosenegalense’ sp. nov. strain mt23T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum-likelihood method within the MEGA software. Numbers at the nodes are percentages of bootstrap values (>95%) obtained by repeating the analysis 500 times to generate a majority consensus tree. Mobilitalea sibirica was used as an outgroup. The scale bar indicates a 1% nucleotide sequence divergence.
MALDI-TOF-MS spectrum. The MALDI-TOF-MS spectrum of ‘L. massiliosenegalense’ is available at http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database.
Nucleotide sequence accession number. The 16S rRNA gene sequence was deposited in GenBank under Accession number LT161890.
Deposit in a culture collection. Strain mt23T was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P299 and in the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) under number DSM 102084.
Conflict of interest
None to declare.
Funding
This work was funded by the Mediterrannée-Infection Foundation.
References
- 1.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.-E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 4.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.-C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yutin N., Galperin M.Y. A genomic update on clostridial phylogeny: gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152. [Google Scholar]
- 8.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]