Abstract
It has been hypothesized that nitrogen fixation occurs in the human gut. However, whether the gut microbiota truly has this potential remains unclear. We investigated the nitrogen-fixing activity and diversity of the nitrogenase reductase (NifH) genes in the faecal microbiota of humans, focusing on Papua New Guinean and Japanese individuals with low to high habitual nitrogen intake. A 15N2 incorporation assay showed significant enrichment of 15N in all faecal samples, irrespective of the host nitrogen intake, which was also supported by an acetylene reduction assay. The fixed nitrogen corresponded to 0.01% of the standard nitrogen requirement for humans, although our data implied that the contribution in the gut in vivo might be higher than this value. The nifH genes recovered in cloning and metagenomic analyses were classified in two clusters: one comprising sequences almost identical to Klebsiella sequences and the other related to sequences of Clostridiales members. These results are consistent with an analysis of databases of faecal metagenomes from other human populations. Collectively, the human gut microbiota has a potential for nitrogen fixation, which may be attributable to Klebsiella and Clostridiales strains, although no evidence was found that the nitrogen-fixing activity substantially contributes to the host nitrogen balance.
Nitrogen fixation, the reduction of atmospheric dinitrogen to ammonium, is a process that changes a biologically inactive form of nitrogen to a compound that can be readily used by organisms1. Diverse microbial communities use this process to fulfil their nitrogen demands. For example, the microbiota associated with invertebrates living in nitrogen-poor environments occasionally include diazotrophs (nitrogen-fixing prokaryotes). Nitrogen fixation in these animals, including termites2, wood-boring beetles3, shipworms4, sponges5, and corals6, has been tested by the measurement of 15N2 incorporation and of reduction of acetylene, an analogue of dinitrogen, and recently also by nanoscale secondary ion mass spectrometry (NanoSIMS). The phylogenetic diversity of the nitrogenase reductase (NifH) gene, a molecular marker of nitrogen fixation, has also been extensively examined in the symbiotic microbiota of various invertebrates2,3,5,7.
In contrast, there have been few studies of nitrogen fixation in vertebrates. Several reports have been published, but almost exclusively in the 1970s. No or only slight acetylene-reducing activity was detected in the rumen contents or faeces of herbivorous mammals, such as sheep8,9,10, goats8,10, cows11,12, rabbits8, and reindeer8. In situ examinations of nitrogen fixation were conducted in a sheep13 and a guinea pig14. In the sheep, acetylene gas was injected into the rumen of a live animal, and acetylene-reduction rates of 0–5.1 nmol ethylene/ml rumen gas/h were detected. The live guinea pig was maintained in an atmosphere containing acetylene, and 75 nmol of ethylene was detected in the rearing chamber after 22 h.
The human distal gut is an anaerobic environment, like those of other mammals, where dinitrogen accounts for 60–80% of the gaseous content15. In a study of people with a low-nitrogen diet in Papua New Guinea (PNG), Bergersen and Hipsley (1970) hypothesized that nitrogen fixation occurs in the human distal gut14. The habitual nitrogen intake of the study subjects was estimated to be 49–74 mg/kg body weight/day in 196716, which is much lower than the estimated average requirement for humans (105 mg/kg body weight/day)17. Nevertheless, these individuals displayed no disorders associated with nitrogen (protein) deficiency18. Bergersen and Hipsley collected faeces from an unspecified number of PNG individuals and five Europeans, and analysed them with an acetylene reduction assay. They detected 0.75 nmol/g/h acetylene-reducing activity in one of the European faecal samples and ‘slight but significant’ activities in all the PNG samples. However, the details were not described, and the samples were supplemented with glucose, which may have altered their acetylene-reducing activity. Because there have been no subsequent studies, whether the human gut microbiota truly has a capacity for nitrogen fixation, and if it does, whether it is specific to individuals with low nitrogen intake, remain unclear.
The diet of certain PNG individuals is still composed mostly of sweet potatoes today, resulting in a below-standard nitrogen intake19, even though their average nitrogen intake has been increasing since the 1970s. Therefore, we collected faecal samples from PNG individuals with low to sufficient nitrogen intake, and from Japanese individuals, with much higher nitrogen intake. To detect the nitrogen-fixing activity of the faecal microbiota and evaluate the effects of the habitual nitrogen intake, the 15N2 incorporation and acetylene reduction by the faecal samples were tested. We also examined the phylogenetic diversity of the nifH gene in the faecal samples by analysing clone libraries of nifH genes and transcripts and with metagenomic sequencing. In addition to nifH, the nitrogenase (NifDK) genes were identified in the PNG and Japanese metagenomes and in publicly available databases of human faecal metagenomes from other populations. This study demonstrates the nitrogen-fixing activity of the human faecal microbiota and presents a list of candidate nitrogen-fixing bacteria in the human gut.
Results
Nitrogen-fixing activity
Table 1 lists the faecal samples used in this study, with information on the PNG and Japanese host individuals, including their habitual nitrogen intake. Most samples were collected from males, because it has been suggested that nitrogen requirements fluctuate during the menstrual cycle and pregnancy in females20,21. To measure nitrogen-fixing activity, we selected six PNG (V001–035) and two Japanese (J001 and 002) individuals, so that the host nitrogen intake ranged from below to above (67.3–209.1 mg/kg body weight/day) the standard requirement (105 mg/kg body weight/day)17 (Table 1). Frozen faecal samples from these individuals were subjected to a 15N2 incorporation assay.
Table 1. PNG and Japanese sample information.
| Sampling area and year | ID | Sexa | Age | BMIb | N intakec | Sample typed | Analysese |
|
|---|---|---|---|---|---|---|---|---|
| Activity | nifH | |||||||
| PNG | ||||||||
| Levani (2012) | L005 | M | 23 | 22.9 | 85.2 | R | ✓ | |
| Levani (2012) | L006 | M | 35 | 22.8 | 89.6 | R | ✓ | |
| Levani (2012) | L015 | M | 20 | 21.2 | 69.6 | R | ✓ | |
| Levani (2013) | V001 | M | 39 | 22.9 | 103.4 | Fro, R | ✓ | |
| Levani (2013) | V004 | M | 22 | 22.7 | 109.6 | Fro, R | ✓ | ✓ |
| Levani (2013) | V009 | M | 35 | 24.3 | 117.2 | Fro, R | ✓ | |
| Levani (2013) | V014 | M | 22 | 26.2 | 67.3 | Fro, R | ✓ | ✓ |
| Levani (2013) | V022 | F | 35 | 27.2 | 87.3 | Fro, R | ✓ | |
| Levani (2013) | V035 | M | 40 | 22.9 | 148.0 | Fro, R | ✓ | ✓ |
| Japanese | ||||||||
| Tokyo (2013) | J001 | M | 44 | 25.1 | 209.1 | Fro, R | ✓ | ✓ |
| Tokyo (2013) | J002 | M | 40 | 21.6 | 183.2 | Fro, R | ✓ | ✓ |
| Tokyo (2014) | J004 | M | 31 | 22.9 | 273.3 | R | ✓ | |
| Tokyo (2014) | J005 | M | 24 | 22.2 | 210.3 | R | ✓ | |
| Tokyo (2016) | N001 | M | 25 | 24.4 | 170.9 | Unfrozen | ✓ | |
| Tokyo (2016) | N002 | M | 23 | 22.7 | 176.4 | Unfrozen | ✓ | |
| Tokyo (2016) | N003 | F | 41 | 19.2 | 250.0 | Unfrozen | ✓ | |
| Tokyo (2016) | N004 | M | 39 | 24.6 | 177.1 | Unfrozen | ✓ | |
| Tokyo (2016) | N005 | F | 38 | 19.7 | 197.6 | Unfrozen | ✓ | |
aM, male; F, female.
bBMI, body mass index, calculated as body weight (kg) per body height (m)2.
cValues are given as total N (mg) per body weight (kg) per day.
dR, preserved in RNAlater; Fro, frozen.
eActivity, 15N2-incorporation and/or acetylene reduction assays; nifH, cloning analysis.
All the faecal samples from both the PNG and Japanese individuals incorporated 15N2 gas, whereas faecal samples sterilized by autoclaving did not (Table 2). The increases in the δ15N values (Δδ15N) ranged from 0.68‰ to 1.26‰, and were statistically significant (Student’s t test or Welch’s t test: p < 0.05, n = 3). These values corresponded to nitrogen fixation rates of 0.065–0.147 nmol/g wet faeces/h (Supplementary Table S1). There was no correlation between Δδ15N and the nitrogen intake of the host (Pearson’s product-moment correlation: r = 0.22; Spearman’s rank correlation: ρ = 0.21). The acetylene reduction assay supported the occurrence of nitrogen fixation in the two Japanese samples with high nitrogen intake (209.1 and 183.2 mg/kg body weight/day). The emission of ethylene was observed in the presence of acetylene (Supplementary Figure S1), and the nitrogen fixation rates were estimated to be 0.008 and 0.143 nmol/g/h, when the theoretical reduction ratio C2H2:N2 ≈ 322 was used.
Table 2. Incorporation of 15N2 gas by PNG and Japanese faecal samples.
| Sample ID | δ15N valuea(‰) |
Δδ15Nb | p valuec | |
|---|---|---|---|---|
| 15N2 gas (+) | 15N2 gas (−) | |||
| PNG | ||||
| V001 | 5.90 ± 0.32 | 4.83 ± 0.10 | 1.07 | 0.0025 |
| V004 | 4.26 ± 0.22 | 3.09 ± 0.38 | 1.17 | 0.0049 |
| V009 | 6.17 ± 0.26 | 5.33 ± 0.04 | 0.84 | 0.0139 |
| V014 | 6.44 ± 0.60 | 5.32 ± 0.05 | 1.11 | 0.0429 |
| V022 | 6.32 ± 0.60 | 5.64 ± 0.05 | 0.68 | 0.0283 |
| V035 | 3.74 | 2.66 | 1.08 | N. A. |
| Japanese | ||||
| J001 | 7.48 | 6.22 ± 0.33 | 1.26 | N. A. |
| J002 | 5.58 ± 0.18 | 4.78 ± 0.21 | 0.80 | 0.0041 |
| Sterile control | ||||
| V004 | 3.22 | 3.09 ± 0.38 | 0.13 | N. A. |
| V009 | 4.86 | 5.33 ± 0.04 | − 0.47 | N. A. |
| J002 | 4.66 ± 0.08 | 4.78 ± 0.21 | − 0.12 | 0.2066 |
aAverages and SD of triplicate samples are shown. For duplicate samples, only average values are shown.
bΔδ15N = δ15N[15N2 (+)] − δ15N [15N2 (−)].
cStudent’s t test for V001 and V004; Welch’s t test for the other samples. N. A., not applicable.
To further confirm the occurrence of nitrogen fixation in the faecal samples of human individuals with high nitrogen intake, we additionally examined the acetylene-reducing activities in six faecal samples from five Japanese individuals (N001–005) (Table 1). In this assay, each faecal sample was divided into two: one was analysed directly (‘unfrozen sample’) and the other was frozen before analysis (‘frozen sample’). By comparing these two types of sample, we also evaluated the effect of freezing on the nitrogen-fixing activity. All the unfrozen samples showed acetylene-reducing activities, ranging 0.055–1.608 nmol/g/h, which were 1.6–9.5 times higher than the values for the corresponding frozen samples (Table 3). The differences were statistically significant for samples N001-1 and N003 (Welch’s t test: p < 0.01, n = 3). We also observed that the acetylene-reducing activity declined within 24 h of sampling (defecation) (Fig. 1). Furthermore, the acetylene-reduction rates correlated negatively with the time after defecation in the unfrozen samples (Pearson’s product-moment correlation: r = −0.86), although the host difference might have affected the data (Table 3).
Table 3. Acetylene-reduction rates in Japanese faecal samples.
| Sample ID | Time between sampling and assaya (h) | Acetylene reduction rates (nmol/g/h) |
|
|---|---|---|---|
| Unfrozen | Frozen | ||
| N001-1b | 8 | 1.390 ± 0.237 | 0.472 ± 0.109 |
| N001-2b | 6 | 1.608 ± 0.422 | 0.954 ± 0.010 |
| N002 | 8 | 0.660 ± 0.403 | N. A. |
| N003 | 15.5 | 0.466 ± 0.117 | 0.049 ± 0.034 |
| N004 | 16.5 | 0.331 ± 0.127 | 0.082 ± 0.055 |
| N005 | 27 | 0.055 ± 0.010 | N. A. |
aTime between defecation and the addition of acetylene to the unfrozen samples or freezing (the frozen samples).
bN001-1 and N001-2 were sampled from the same individual (N001) on different days.
Figure 1. Decline in the acetylene-reduction rates in unfrozen Japanese faecal samples, N001-1 and N003.
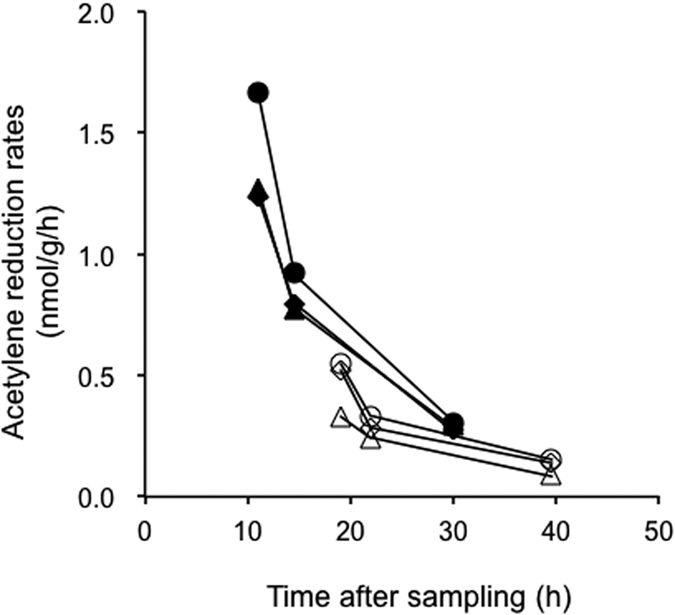
The experiments were performed in triplicate. Closed symbols, N001-1; open symbols, N003.
PCR and reverse transcription (RT)–PCR amplification of nifH
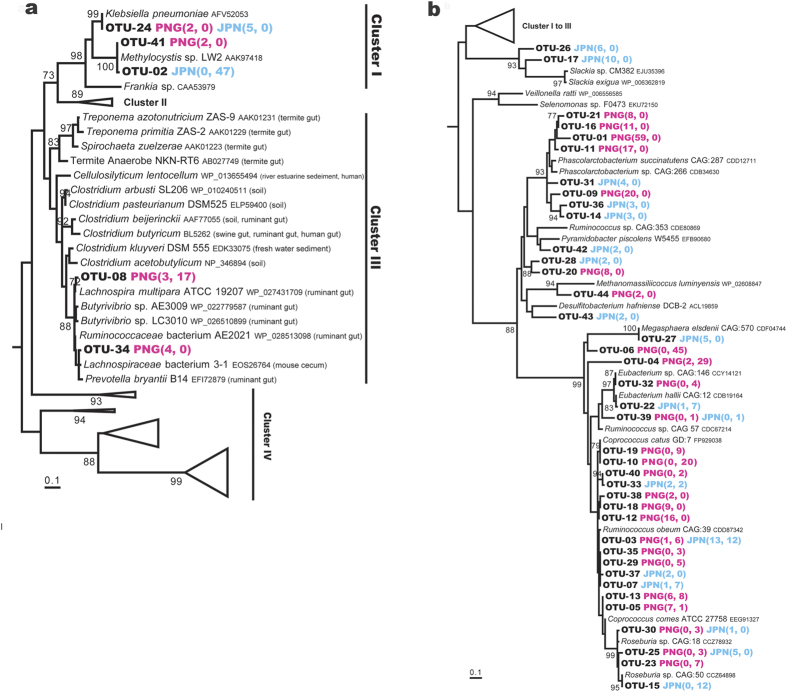
DNA and RNA were extracted from each of the RNAlater®-preserved faecal samples from six PNG individuals and four Japanese individuals (Table 1). PCR and RT–PCR were performed with universal primers for nifH23. Genes and transcripts of the predicted size, around 360 base pairs (bp), were detected in all the samples. A total of 259 genes and 265 transcripts encoding NifH homologues were detected and sorted to 44 operational taxonomic units (OTUs) (Supplementary Table S2). These were phylogenetically affiliated with nifH Cluster I (3 OTUs, 56 clones), Cluster III (2 OTUs, 24 clones), or Cluster IV (39 OTUs, 444 clones) (Fig. 2), according to the classification of nifH sequences by Zehr et al.24
Figure 2. Phylogenetic positions of nifH recovered from Papua New Guinean and Japanese faecal samples, based on the deduced amino acid sequences.
(a) Sequences affiliated with nifH Clusters I and III. (b) Sequences affiliated with nifH Cluster IV. Numbers in parentheses indicate the numbers of clones of genes and transcripts affiliated with the OTUs. A total of 113 amino acid positions were used, corresponding to positions 45–157 of the Klebsiella pneumoniae NifH sequence (AFV52053). 100 bootstrap resamplings were performed. Chlorophyllide reductase subunit BchX of Rhodobacter sphaeroides (CAB38747) was used as an outgroup. Only bootstrap confidence levels ≥70% are shown. OTU-02 and OTU-41 are most probably sequences contaminating in reagents.
Cluster I is composed of nifH sequences from diverse proteobacteria and cyanobacteria24. The OTU-24 sequence was recovered as gene clones from the PNG and Japanese samples (Fig. 2a). The nucleotide sequences were nearly identical (99%) to the nifH sequences of Klebsiella species, such as Klebsiella pneumoniae. The OTU-02 and OTU-41 sequences were almost identical to known contaminants frequently detected in PCR reagents25, and were not detected in the metagenomic datasets described below. Therefore, it is likely that these were not true constituents of the nifH community in the faecal samples.
Cluster III is known to contain nifH sequences of anaerobes from various environments. OTU-08 and OTU-34 showed high amino acid sequence similarities to NiH of anaerobic bacteria isolated from mammalian intestinal tracts (Fig. 2a). OTU-08 was recovered as both genes and transcripts from a PNG faecal sample. This OTU shared 98% amino acid sequence identity with NifH of Lachnospira multipara and also 97% identity with those of Butyrivibrio species and Ruminococcaceae bacterium AE2021. These bacteria have been isolated from ruminant foreguts, according to the description in the sequence database. OTU-34 was recovered as gene clones from two PNG samples. This OTU shared more than 97% amino acid sequence identity with NifH of Lachnospiraceae bacterium 3-1 isolated from a mouse cecum and Ruminococcaceae bacterium AE2021 and Prevotella bryantii isolated from ruminant foreguts.
Diverse and abundant sequences were assigned to Cluster IV, which is also called the ‘NifH-like cluster’. The function of Cluster IV NifH is largely unknown, but it has been suggested to play a role other than in nitrogen fixation24,26. Most of the 39 OTUs showed high amino acid sequence similarities to NifH-like sequences of members of the order Clostridiales, belonging to genera such as Ruminococcus, Coprococcus, and Eubacterium (Fig. 2b).
Detection of nifH Cluster I and III sequences with quantitative PCR (qPCR)
Because the abundant Cluster IV nifH sequences probably interfered with the detection of Cluster I and III sequences in the clone analysis discussed above, we also performed a qPCR analysis of nine PNG and four Japanese faecal samples (Table 4) to specifically detect sequences of OTU-24, OTU-08, and OTU-34. The detection limit was 1.25 × 104 copies per one gram of faecal sample. Of the 13 samples tested, 10 showed the presence of at least one of these OTUs.
Table 4. Detection of Cluster I and Cluster III nifH genes with qPCR.
| ID | Target copy number (copies/wet faeces g) |
||
|---|---|---|---|
| OTU-24 | OTU-08 | OTU-34 | |
| PNG | |||
| L005 | 8.4 × 104 | 2.4 × 106 | 3.1 × 107 |
| L006 | UD | UD | 1.4 × 106 |
| L015 | UD | 6.1 × 107 | 1.4 × 108 |
| V001 | 1.1 × 105 | UD | UD |
| V004 | 5.1 × 106 | DNQ | UD |
| V009 | UD | UD | UD |
| V014 | UD | 4.0 × 106 | 3.9 × 107 |
| V022 | UD | DNQ | 1.4 × 106 |
| V035 | UD | UD | 9.6 × 107 |
| Japanese | |||
| J001 | UD | UD | DNQ |
| J002 | 1.6 × 109 | UD | 1.4 × 106 |
| J004 | UD | UD | UD |
| J005 | UD | UD | 1.8 × 105 |
Underlining indicates OTUs detected in the cloning analysis of the corresponding samples. Each reaction was conducted in triplicate, and standards (101–107 plasmid copies) and negative controls were measured in duplicate. Quantification was performed when the target sequences were detected at ≥5 copies in at least two of the triplicate reactions. Abbreviations: UD, under detection limit; DNQ, detected but not quantified.
OTU-24 of Cluster I was detected in samples V004 and J002, as in the clone analysis, and also in another two PNG samples, L005 and V001. The copy number of the OTU-24 sequence ranged from 105 to 106 per gram wet faeces in the three PNG samples, whereas the Japanese sample J002 contained 109 copies per gram wet faeces (Table 4). OTU-08 of Cluster III was detected in five PNG samples, including L005, in which the OTU was found with the clone analysis. OTU-34 was detected more widely, in six PNG and three Japanese samples, including samples L005 and V014, in both of which the OTU was also found in the clone analysis. The copy numbers of the OTU-08 and OTU-34 sequences ranged from 106 to 108 per gram wet faeces in the PNG samples, while 105 to 106 in the Japanese samples (Table 4).
Metagenomic analysis
The faecal DNA samples from 16 PNG and two Japanese individuals, including the six PNG and two Japanese individuals tested in the 15N2 incorporation assay, were subjected to whole-genome shotgun sequencing (Supplementary Table S3). To facilitate homology searches, non-redundant gene sets were prepared from the metagenomic contigs from the PNG and Japanese samples. A published metagenomic dataset27 was incorporated into the Japanese set.
In the PNG non-redundant gene set, four nifH homologues were identified and designated PNG-1–4 (Supplementary Table S4). These were affiliated with nifH Cluster III, sharing 94–98% amino acid sequence identities with NifH of Lachnospira and Butyrivibrio species. PNG-1 and PNG-3 showed 100% amino acid sequence similarity to OTU-08 and OTU-34, respectively (Fig. 3). The PNG-2 and PNG-4 sequence regions did not overlap most regions of the OTU sequences, and are therefore not shown in Fig. 3. In the Japanese gene set, two NifH homologues, designated JPN-1 and JPN-2, were identified and affiliated with nifH Clusters I and III, respectively (Fig. 3). JPN-1 shared 98% amino acid sequence identity with OTU-24, and JPN-2 shared 99% with OTU-34.
Figure 3. Phylogenetic positions of nifH from the human faecal metagenomes from six countries, based on deduced amino acid sequences.
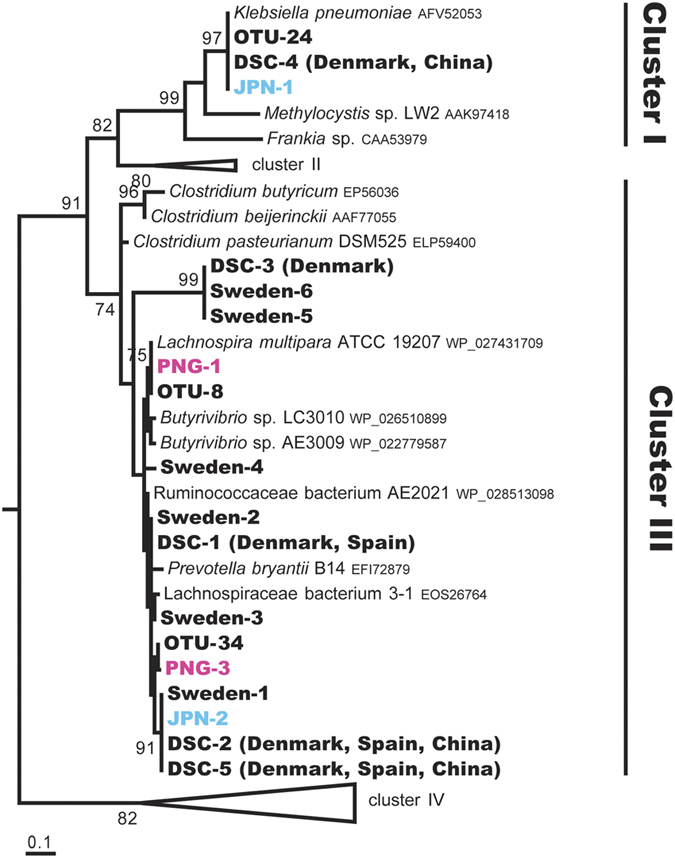
A total of 112 amino acid positions were used, corresponding to positions 45–157 of the Klebsiella pneumoniae NifH sequence (AFV52053). Of the four nifH genes from Papua New Guinean individuals, PNG-2 and -4 were not included because they corresponded to positions 138–283 and 124–264, respectively. ‘DSC’ indicates a combined non-redundant gene set of Danish, Spanish, and Chinese populations. See also the legend to Fig. 2.
We also searched for nifH sequences in non-redundant gene sets constructed from publicly available databases of faecal metagenomes from other human populations, each consisting of 291 Danish28,29, 39 Spanish28, 145 Swedish30, or 363 Chinese31 individuals. In total, 11 NifH homologues were identified. Ten were assigned to nifH Cluster III, sharing 83–100% amino acid sequence identity with the amino acid sequences of Clostridiales species (Lachnospira and Butyrivibrio species) and also of OTU-08 and OTU-34 (Fig. 3 and Supplementary Table S4). The remaining homologue shared an identical amino acid sequence with NifH of Klebsiella species and OTU-24 in nifH Cluster I. Cluster III sequences were detected in 31.7%, 21.6%, 17.9%, and 10.7% of the Swedish, Danish, Spanish, and Chinese individuals, respectively, when 106 sequencing reads were randomly chosen from each population. Klebsiella-related Cluster I sequences were detected in 0.7% and 6.6% of Danish and Chinese individuals, respectively.
The sequencing reads of the faecal metagenomes of the PNG and Japanese individuals who were tested with the 15N2 incorporation assay were further searched for nifH, using MG-RAST v3.232. The results were consistent with the data presented above (Supplementary Table S5). We also searched for genes encoding the nitrogenase Mo-Fe protein subunits NifD and NifK in all the non-redundant gene sets as well as the sequencing reads of the PNG and Japanese, used in the search for NifH. NifD and NifK were encoded in all the datasets and shared high amino acid sequence identities with those of Klebsiella and members of the Clostridiales (Supplementary Tables S6 and S7).
Discussion
We have demonstrated that the human gut microbiota truly has a capacity for nitrogen fixation, using 15N2 incorporation and acetylene reduction assays. The nitrogen-fixing activities were detected irrespective of the host habitual nitrogen intake; even the samples from Japanese individuals with nitrogen intake much higher than the standard requirement exhibited the activity. In addition, nifHDK genes were detected in the faecal metagenomes of PNG, Japanese, Chinese, and European individuals. Collectively, we suggest that a capacity for nitrogen fixation is maintained in various human populations.
The acetylene-reduction rates in the human faecal samples were up to 1.6 nmol/g/h, which is comparable to those detected in herbivorous mammals, such as sheep (0–1.4 nmol/g/h in the rumen)8,9,10, a rabbit (0.38 nmol/g/h in the cecum)8, and a reindeer (0–0.77 nmol/g/h in faeces)8. The nitrogen fixed in the human faecal samples corresponded to 0.36 mg/kg gut content/day, and the contribution to the host nitrogen balance was estimated to be 0.01%, at most. Thus, the nutritional impact of nitrogen fixation should be low in humans. However, caution is needed. Our data suggest that the nitrogen fixation by the human faecal microbiota is sensitive to the time after sampling (Fig. 1 and Table 3), as seen in termites. The nitrogen-fixing activity in termites declines within several hours of sampling33,34,35,36. Therefore, the nitrogen-fixing activity in the human gut in vivo might be higher than the values estimated in this study. In addition, freezing considerably reduced the nitrogen-fixing activity; this activity of the PNG faecal samples was possibly underestimated, because the sampling required a long-distance transportation and freezing.
It is generally believed that nitrogen fixation is suppressed when available nitrogen sources, such as ammonia and nitrate, are sufficient in the environment. Therefore, it has been assumed that the concentration of nitrogen compounds in the human gut is too high to allow biological nitrogen fixation26. However, previous studies have reported plasticity in the regulation of nitrogen-fixing activity in natural environments. In aquatic environments, such as marine sediments and salt marshes, nitrogen fixation (acetylene reduction) has been detected under high concentrations of nitrogen compounds (e.g., millimolar levels of ammonia)37,38. A high demand for nitrogen and balance between nitrogen fixation and nitrification have been suggested to explain these examples. In another case, certain plant-associated diazotrophs are less sensitive to ammonia in the symbiotic phase than in the free-living phase, and contribute to the nutrition of the host1. Our data suggest that the human gut microbiota also contains members capable of fixing nitrogen, even when the nitrogen input is sufficient.
In both of the PNG and Japanese faecal samples, the nifH genes identified (except the Cluster IV nifH-like genes) were affiliated with two clades: those in one set were almost identical to the nifH genes of Klebsiella species, and these genes in the other set were closely related to those of Clostridiales members. This result is consistent between the cloning results and metagenomic analyses, and the nifH genes found in the European and Chinese faecal metagenomes also fell within these two phylogenetic groups. Thus, Klebsiella and Clostridiales species are the candidate nitrogen-fixers in the human gut, although the possibility that other bacterial lineages possess the nif-genes via horizontal gene transfer cannot be excluded. The total copy numbers of these Klebsiella- and Clostridiales-related nifH genes were 105–109 copies per gram wet faeces (Table 4) and not correlated with the host nitrogen intake (Table 1) or with the nitrogen-fixing activity (Table 2). These nifH copy numbers are comparable to those in other diazotrophic environments, such as corals (107 copies per gram tissue)7 and the rhizosphere (105–107 copies per gram soil)39,40.
Many Klebsiella strains have been identified as active diazotrophs in various environments, including soil, water, and plants41. Klebsiella species are generally not predominant, but are widely distributed in the gut microbiota of humans28,42,43. Bergersen and Hipsley (1970) predicted that Klebsiella species are one of the nitrogen fixers present in the human gut microbiota14. They isolated three bacterial strains, provisionally identified as Klebsiella aerogenes, on a nitrogen-free medium, from human faecal samples and determined their 15N2-incorporating activities.
The nitrogen-fixing activities of various Clostridiales strains containing Cluster III NifH were demonstrated with an acetylene reduction or 15N2 incorporation assay44. However, the bacterial strains, such as Lachnospira and Butyrivibrio species, of which NifH sequences constitute a clade with those from the human faecal microbiota, were not tested for nitrogen-fixing activity. A computational study suggested that certain bacteria expressing Cluster III NifH are likely to reduce acetylene and azide, but not dinitrogen, because they lack a histidine residue in the NifD protein (His422 in Azotobacter vinelandii)45. However, 15N2 fixation has recently been reported in Endomicrobium proavitum, which lacks His422 but retains all cysteine residues in NifD46. Among the relevant members of the Clostridiales and Bacteroidales (Figs 2a and 3), L. multipara ATCC 19207, Ruminococcaceae bacterium AE2021, and P. bryantii B14 lack His422 in NifD, but all the essential cysteine residues are conserved, as in E. proavitum. The other members of Clostridiales in this clade retain both the histidine and cysteine residues. In the NifD homologues recovered from the human faecal metagenomes, both of those with and without the histidine residue were identified. None of the NifD homologues lacked the cysteine residues. Therefore, there is currently no reason to consider that these bacteria cannot fix dinitrogen.
This study has shown that the human faecal microbiota has a potential capacity for nitrogen fixation, and that this capacity may exist in a wide range of human populations. However, evidence that this capacity substantially contributes to the host nitrogen balance was not obtained; the ecological meaning of nitrogen fixation in the human gut remains to be clarified.
Materials and Methods
Ethics statement
The study, involving human participants, was approved by the Institutional Review Board at the Papua New Guinea Institute of Medical Research (1025), the Papua New Guinea Medical Research Advisory Committee (11.25), and the Research Ethics Committee at the Graduate School of Medicine, The University of Tokyo (3391). All the participants provided their written informed consent. All experiments were performed in accordance with the approved study protocols.
Study population
PNG participants were recruited in the Levani area (Levani Valley, Hela Province, Papua New Guinea) in March 2012 and March 2013, and in the Maprik area (East Maprik, East Sepik Province) in August 2012. Dietary data were obtained with a semi-quantitative food frequency questionnaire developed in our previous study19, including demographic and anthropometric data. Nineteen healthy PNG individuals, aged between 15 and 40 years, with body mass indices (BMIs) ranging from 19.2 to 27.2 kg/m2, were selected for the study (Table 1 and Supplementary Table S3). Their nitrogen intake was calculated based on their protein intake using a nitrogen-to-protein conversion factor of 6.25 (nitrogen weight = protein weight/6.25)47.
The Japanese participants were recruited in Tokyo, Japan, between June 2013 and April 2016. Their dietary information was obtained with a brief self-administered diet history questionnaire48, and their age, height, and weight were self-reported. Nine individuals, with ages and BMIs similar to those of the PNG participants, were selected (Table 1).
Sample collection in PNG
In Levani in 2012, faecal samples were collected by each participant in a plastic container, and a portion of approximately 500 mg of the faeces was immediately suspended in 2 ml of RNAlater® (Ambion) (Table 1). In Maprik in 2012 and in Levani in 2013, faecal samples were collected and immediately enclosed in an AnaeroPack™ (Mitsubishi Gas Chemical), placed in a cooler container, and brought to our research base in town (Table 1 and Supplementary Table S3), which took 7–8 h to drive and/or walk from the rural communities in Maprik and Levani. At the base, a portion of the faeces was suspended in RNAlater® as described above, and another portion of approximately 2.0 g was frozen in a liquid nitrogen dry vapor shipper. These samples were transported to our laboratory at The University of Tokyo, Japan, and stored at −80 °C until analysis.
Sample collection in Japan
Faecal samples were collected in 2012 and 2013 in Tokyo (Table 1), and suspended in RNAlater® immediately after defecation by each participant. Frozen samples from two individuals were also prepared. The samples were enclosed anaerobically in an AnaeroPack™ by each participant, brought to the laboratory at the ambient temperature (around 15 °C), and stored at −80 °C until analysis. In 2016, additional faecal samples were collected from five individuals without freezing or treatment with RNAlater® (Table 1). The samples were enclosed in an AnaeroPack™ by each participant and were stored at 4 °C until analysis.
15N2 incorporation assay
The frozen faeces were divided (cracked) into pieces without thawing. The faecal pieces were placed in a vacuum desiccator (7 l), as duplicate or triplicate samples, at the ambient temperature. The gas phase was immediately replaced with argon (Ar) and subsequently adjusted to (60% [v/v] 15N2 [99.7% atoms] and 40% [v/v] Ar) (SI Science, Saitama, Japan). The samples were incubated at 37 °C for 48 h. Control experiments without 15N2 gas were also performed in duplicate or triplicate. Sterile controls were prepared by autoclaving the samples at 120 °C for 20 min and incubating them with 15N2 gas under the same conditions. The 15N abundance (δ15N) was measured with the DELTA V Advantage ConFlo IV system (Thermo Fisher Scientific) at SI Science. The total nitrogen mass concentration (%N) was measured with a Flash 2000 elemental analyser (Thermo Fisher Scientific) in the Isotope Ecology Laboratory at The University of Tokyo. The analytical standard deviation (SD) was approximately <0.2‰ for δ15N and <0.77% for %N. For samples analysed in triplicate, Student’s t test or Welch’s t test was performed based on the results of an F-test. The correlation between Δδ15N and the host nitrogen intake was determined with Pearson’s product–moment correlation coefficient (r) and Spearman’s rank correlation coefficient (ρ). The calculation of the 15N mass is shown in Supplementary Table S1.
Acetylene reduction assay
Frozen faeces from samples J001 and J002 were cracked into approximately 1.0–2.0 g pieces and divided into glass vials (net capacity 8.7 ml). The headspace was replaced with nitrogen gas followed by the injection of acetylene gas at a final concentration of 15% (v/v). The vials were placed at 37 °C. Faecal samples incubated without acetylene gas and empty vials with only acetylene gas were also prepared as controls. An aliquot (0.2 ml) of the headspace gas was analysed to measure the ethylene concentrations from day 0 to day 3 at 40 °C using a GC7A gas chromatograph (Shimadzu) equipped with a Propack N column (80/100 mesh, 2 m in length, 2.2 mm in diameter; Shimadzu) and a flame ionization detector.
Unfrozen faeces from samples N001 to N005 were divided into approximately 1.0–2.0 g portions in glass vials in an anaerobic chamber. Half the vials were frozen at −80 °C for 10–12 h for comparisons with the corresponding unfrozen samples. Aliquots (0.125 ml) of the headspace gas from both the unfrozen and frozen samples were analysed at 3, 6, and 21 h after the addition of acetylene gas for samples N001-1 and N003, and at 1 and 3 h after the addition of acetylene gas for the other samples.
DNA extraction
DNA was extracted from the faecal samples preserved in RNAlater® or from frozen samples, according to the previously described methods49,50, with modifications. The faecal samples in RNAlater® were diluted and washed twice with phosphate-buffered saline (PBS). Faecal aliquots of 4 mg were prepared and suspended in 300 μl of Tris-SDS (250 μl of 200 mM Tris-HCl, 80 mM EDTA, pH 9.0, and 50 μl of 10% SDS). To this suspension, 300 mg of glass beads (0.1 mm diameter) and 500 μl of Tris-EDTA (TE)-saturated phenol (Wako, Japan) were added, and the cells disrupted with a Shakemaster Auto (Biomedical Science, Tokyo, Japan) for 15 min. After centrifugation, the supernatant was subjected to phenol/chloroform/isoamyl alcohol extraction and isopropanol precipitation. The extracted DNA was suspended in 100 μl of TE.
A frozen faecal sample was treated with 15 mg/ml lysozyme at 37 °C for 1 h, with 60 units/ml purified achromopeptidase (Wako 015-09951, Japan) at 37 °C for 30 min, and then with 1% SDS and 1 mg/ml proteinase K at 55 °C for 1 h. The sample was subjected to phenol/chloroform/isoamyl alcohol extraction and isopropanol precipitation. The extracted DNA was treated with RNase A, and purified with polyethylene glycol precipitation to remove any residual protein. The purified DNA was suspended in 300 μl of TE.
RNA extraction and cDNA synthesis
RNA was extracted from faecal samples preserved in RNAlater®, using the RNeasy® Mini Kit (Qiagen). The samples were washed in PBS, and 20 mg aliquots of faeces were combined with a mixture of 600 μl of the RLT buffer contained in the kit, 7 μl of β-mercaptoethanol, 100 μl of TE buffer, and 300 mg of glass beads, and disrupted with the Shakemaster Auto for 5 min. The subsequent procedure was performed according to the manufacturer’s guidelines. DNA was removed from the extracted RNA with the Turbo DNA-free Kit (Ambion) at 37 °C for 1 h. Reverse transcription was performed at 55 °C using the SuperScript III First Strand Synthesis System (Invitrogen) with 0.5 μM nifH3 primer23. Negative controls without reverse transcription were prepared for each sample.
PCR and RT–PCR amplification, cloning, and sequencing
Genes and transcripts encoding NifH were amplified with PCR and RT–PCR, respectively, using primers specific for nifH under previously described conditions51, with modifications (Supplementary Methods). The PCR products were separated by agarose gel electrophoresis and those of approximately 360 bp in length were purified with the QIAquick Gel Extraction Kit (Qiagen), cloned into the pGEM®-T Easy Vector System (Promega), and used to transform Escherichia coli DH5α cells. Randomly chosen clones were sequenced with the T7 primer using the BigDye Terminator Cycle Sequencing Kit V3.1 (Applied Biosystems) on an ABI 3730 Genetic Analyzer (Applied Biosystems).
Identification and phylogenetic analysis of nifH
The sequences obtained with PCR and RT–PCR were examined with BLASTX searches of the National Center for Biotechnology Information (NCBI) non-redundant database. The sequences that showed high similarities (E-value ≤ 10–50) to known nifH sequences and encoded the amino acid residues conserved in NifH23 were subjected to subsequent screening. The selected sequences were aligned with ClustalX252, and a distance matrix was calculated with the DNADIST program in the PHYLIP package53. The nucleotide sequences were sorted to OTUs with a 96% similarity cut-off, using the mothur program54. A phylogenetic analysis was performed based on the deduced amino acid sequences, using a maximum likelihood method with the Le and Gascuel substitution model in MEGA 6.055.
Quantitative PCR
Specific PCR primers and TaqMan® probes were designed for the nifH phylotypes OTU-08, OTU-24, and OTU-34 (Supplementary Table S8). Standard curves were constructed in triplicate using serial dilutions of linearized plasmids containing each target sequence. Quantitative PCR amplification was performed with the FastStart Essential DNA Probes Master (Roche Diagnostics) on the LightCycler Nano System (Roche) under cycling conditions: 95 °C for 10 min, and 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Specific amplification was verified by cloning and sequencing the PCR products.
Metagenomic analysis
Whole-genome shotgun sequencing was performed on an Ion PGM™ System with the Ion 318™ Chip Kit and Ion PGM™ 400 Sequencing Kit (Life Technologies). A total of 51,633,321 and 7,099,635 reads (average 3,227,083 and 3,549,818) were obtained for the 16 PNG and two Japanese samples (Supplementary Table S3), respectively, and a non-redundant gene set was constructed for each population. The method is described in detail in the Supplementary Methods.
Faecal metagenomic sequences for Danish28,29, Spanish28, Swedish30, and Chinese31 populations were retrieved from the NCBI archives. A combined non-redundant gene set for the Danish, Spanish, and Chinese populations was obtained at the GigaDB database (http://gigadb.org), and the non-redundant gene set for the Swedish population was provided by Fredrik Karlsson (Chalmers University of Technology, Sweden).
The amino acid sequences deduced from the non-redundant gene sets were searched for NifH homologues using BLASTP with a cut-off level of E-value ≤ 10–5, against nifH (K02588) genes from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. A supercomputer at the Human Genome Center, The University of Tokyo (http://sc.hgc.jp/shirokane.html) was used for the analysis. Sequences of more than 120 amino acids were incorporated into the nifH database in the ARB format created by Zehr’s laboratory (www.jzehrlab.com/#!nifh-database/c1coj) to classify the sequences into nifH Clusters24. The BLASTX search results for all the amino acid sequences containing ≤ 120 amino acids were also manually checked to eliminate Cluster IV NifH. The frequencies of the retrieved NifH homologues in each metagenomic dataset were calculated by mapping 106 randomly chosen reads to the corresponding non-redundant gene sets, using the Bowtie2 tool56, with a criterion of 95% nucleotide sequence identity. The NifD and NifK sequences were identified with BLASTP using a cut-off of E-value ≤ 10–5 against NifD (K02586) and NifK (K02591) in the KEGG database, respectively. NifD and NifK sequences with more than 80% amino acid identity to those of bacteria containing Cluster I or Cluster III NifH were selected.
Additional searches for nifH, nifD, and nifK were made in the metagenomic sequence reads from the samples used for the 15N incorporation assay, using MG-RAST31. The method is described in detail in the Supplementary Methods.
Additional Information
Accession numbers: The nifH sequences of the 41 OTUs were deposited at GenBank/EMBL/DDBJ under accession numbers, LC097013–56.
How to cite this article: Igai, K. et al. Nitrogen fixation and nifH diversity in human gut microbiota. Sci. Rep. 6, 31942; doi: 10.1038/srep31942 (2016).
Supplementary Material
Acknowledgments
We thank Hirokazu Tsuji and Koji Nomoto of Yakult Honsha Co., Ltd. for their helpful advice on sampling and preparing nucleic acids. We also thank Erika Iioka, Misa Kiuchi, Rina Kurokawa, Keiko Komiya, Naoko Yamashita, Chie Shindo, and Yasue Hattori for technical assistance with DNA sequencing, and Takayuki Wada for advising on data analysis. This study was supported by the Next Generation World-Leading Researchers from Japan Society for the Promotion of Science LS024 to M.U. and partially a Grant-in-Aid for Scientific Research 26292047 to M.O. and 23117003 to Y.H., and also supported by an incubation study supported by the Research Institute for Humanity and Nature, Japan, to T.Y. This work was partly conducted at the Joint Usage/Research Center on Tropical Disease, Institute of Tropical Medicine, Nagasaki University.
Footnotes
Author Contributions M.U. conceived the study. K.I. and K.M. designed the experiments. M.Y., M.O., Y.H., T.Y., M.H., K.M. and M.U. provided the reagents and equipment. K.T., J.B., S.O., K.N., A.M., A.R.G., P.F.H., P.M.S. and M.U. conducted field studies. K.I., M.I., H.T., T.T., E.T., J.I. and K.M. performed experiments and data analyses. K.I., S.N., W.S. and M.H. performed metagenomic analyses. K.I. and Y.H. wrote the manuscript with significant contributions from A.R.G., P.F.H., M.O., T.Y., K.M. and M.U. and input from all the other authors.
References
- Dixon R. & Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2, 621–631 (2004). [DOI] [PubMed] [Google Scholar]
- Brune A. & Ohkuma M. Role of the termite gut microbiota in symbiotic digestion in Biology of Termites: a Modern Synthesis (eds Bignell D. E., Roisin Y., & Lo N.) 439–475 (Springer, Dordrecht, 2011). [Google Scholar]
- Ceja-Navarro J. A. et al. Compartmentalized microbial composition, oxygen gradients and nitrogen fixation in the gut of Odontotaenius disjunctus. ISME J 8, 6–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechene C. P., Luyten Y., McMahon G. & Distel D. L. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 317, 1563–1566 (2007). [DOI] [PubMed] [Google Scholar]
- Mohamed N. M., Colman A. S., Tal Y. & Hill R. T. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ Microbiol 10, 2910–2921 (2008). [DOI] [PubMed] [Google Scholar]
- Lesser M. P. et al. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar Ecol Prog Ser 346, 143–152 (2007). [Google Scholar]
- Olson N. D., Ainsworth T. D., Gates R. D. & Takabayashi M. Diazotrophic bacteria associated with Hawaiian Montipora corals: Diversity and abundance in correlation with symbiotic dinoflagellates. J Exp Mar Biol Ecol 371, 140–146 (2009). [Google Scholar]
- Granhall U. & Ciszuk P. Nitrogen fixation in rumen contents indicated by the acetylene reduction test. J Gen Microbiol 65, 91–93 (1971). [DOI] [PubMed] [Google Scholar]
- Jones K. & Thomas J. G. Nitrogen fixation by the rumen contents of sheep. J Gen Microbiol 85, 97–101 (1974). [DOI] [PubMed] [Google Scholar]
- Li Pun H. H. & Satter L. D. Nitrogen fixation in ruminants. J Anim Sci 41, 1161–1163 (1975). [DOI] [PubMed] [Google Scholar]
- Hardy R. W., Holsten R. D., Jackson E. K. & Burns R. C. The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol 43, 1185–1207 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleway R. F., Sabine J. R. & Nicholas D. J. Acetylene reduction by rumen microflora. Arch Mikrobiol 76, 277–291 (1971). [DOI] [PubMed] [Google Scholar]
- Hobson P. N., Summers R., Postgate J. R. & Ware D. A. Nitrogen fixation in the rumen of a living sheep. J Gen Microbiol 77, 225–226 (1973). [DOI] [PubMed] [Google Scholar]
- Bergersen F. J. & Hipsley E. H. The presence of N2-fixing bacteria in the intestines of man and animals. J Gen Microbiol 60, 61–65 (1970). [DOI] [PubMed] [Google Scholar]
- Levitt M. D., Bond, John H., Levitt & David G. Gastrointestinal gas in Physiology of the Gastrointestinal Tract (ed. Johnson L. R.) 1301–1316 (Raven Press, 1981). [Google Scholar]
- Oomen H. A. Interrelationship of the human intestinal flora and protein utilization. Proc Nutr Soc 29, 197–206 (1970). [DOI] [PubMed] [Google Scholar]
- Rand W. M., Pellett P. L. & Young V. R. Meta-analysis of nitrogen balance studies for estimating protein requirements in healthy adults. Am J Clin Nutr 77, 109–127 (2003). [DOI] [PubMed] [Google Scholar]
- Luyken R., Luyken-Koning F. W. & Pikaar N. A. Nutrition studies in New Guinea. Am J Clin Nutr 14, 13–27 (1964). [DOI] [PubMed] [Google Scholar]
- Morita A. et al. Development, validation, and use of a semi-quantitative food frequency questionnaire for assessing protein intake in Papua New Guinean Highlanders. Am J Hum Biol 27, 349–357 (2015). [DOI] [PubMed] [Google Scholar]
- Stephens T. V., Payne M., Ball R. O., Pencharz P. B. & Elango R. Protein requirements of healthy pregnant women during early and late gestation are higher than current recommendations. J Nutr 145, 73–78 (2015). [DOI] [PubMed] [Google Scholar]
- Egun G. N. & Atinmo T. Protein requirement of young adult Nigerian females on habitual Nigerian diet at the usual level of energy intake. Br J Nutr 70, 439–448 (1993). [DOI] [PubMed] [Google Scholar]
- Montoya J. P., Voss M., Kahler P. & Capone D. G. A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl Environ Microbiol 62, 986–993 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J. P. & McReynolds L. A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55, 2522–2526 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J. P., Jenkins B. D., Short S. M. & Steward G. F. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5, 539–554 (2003). [DOI] [PubMed] [Google Scholar]
- Goto M., Ando S., Hachisuka Y. & Yoneyama T. Contamination of diverse nifH and nifH-like DNA into commercial PCR primers. FEMS Microbiol Lett 246, 33–38 (2005). [DOI] [PubMed] [Google Scholar]
- Raymond J., Siefert J. L., Staples C. R. & Blankenship R. E. The natural history of nitrogen fixation. Mol Biol Evol 21, 541–554 (2003). [DOI] [PubMed] [Google Scholar]
- Nishijima S. et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res 2, 125–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013). [DOI] [PubMed] [Google Scholar]
- Karlsson F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- Qin J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- Meyer F. et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwich G. D., Bentley B. L. & Carpenter E. J. Nitrogen-sources for neotropical Nasute termites - fixation and selective foraging. Oecologia 46, 397–401 (1980). [DOI] [PubMed] [Google Scholar]
- Lovelock M., Obrien R. W. & Slaytor M. Effect of laboratory containment on the nitrogen-metabolism of termites. Insect Biochem 15, 503–509 (1985). [Google Scholar]
- Yamada A. et al. Nitrogen fixation by termites in tropical forests, Thailand. Ecosystems 9, 75–83 (2006). [Google Scholar]
- Pandey S. Waller, D. A. & Gordon A. S. Variation in acetylene-reductoin (nitrogen-fixation) rates in Reticulitermes spp. (Isoptera; Rhinotermitidae). Virginia J Sci 43, 333–338 (1992). [Google Scholar]
- Dekas A. E., Chadwick G. L., Bowles M. W., Joye S. B. & Orphan V. J. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ Microbiol 16, 2012–2029 (2013). [DOI] [PubMed] [Google Scholar]
- Dicker H. J. & Smith D. W. Physiological ecology of acetylene reduction (nitrogen fixation) in a Delaware salt marsh. Microb Ecol 6, 161–171 (1980). [DOI] [PubMed] [Google Scholar]
- Hai B. et al. Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol 75, 4993–5000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf B., Kumar R., Mishra A. & Jha B. Differential distribution and abundance of diazotrophic bacterial communities across different soil niches using a gene-targeted clone library approach. FEMS Microbiol Lett 360, 117–125 (2014). [DOI] [PubMed] [Google Scholar]
- Brisse S. G. F. & Grimont P. A. D. The Genus Klebsiella in Proteobacteria: Gamma Subclass (eds Dworkin M., Falkow S., Rosenberg E., Schleifer K. & Stackebrandt E.) 159–196 (Springer, 2006). [Google Scholar]
- Martinez I. et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11, 527–538 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J 9, 1979–1990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum E. D. & Wilson P. W. Fixation of isotopic nitrogen by Clostridium. J Bacteriol 57, 413 (1949). [DOI] [PubMed] [Google Scholar]
- Dos Santos P. C., Fang Z., Mason S. W., Setubal J. C. & Dixon R. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13, 162–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Dietrich C., Radek R. & Brune A. Endomicrobium proavitum, the first isolate of Endomicrobia class. nov. (phylum Elusimicrobia) - an ultramicrobacterium with an unusual cell cycle that fixes nitrogen with a Group IV nitrogenase. Environ Microbiol 18 (2015). [DOI] [PubMed] [Google Scholar]
- Jones D. B. Factors for converting percentages of nitrogen in foods and feeds into percentages of protein. Report No. 183, (US Dept. Agric. Cir., Washington, D.C., 1931).
- Kobayashi S. et al. Comparison of relative validity of food group intakes estimated by comprehensive and brief-type self-administered diet history questionnaires against 16 d dietary records in Japanese adults. Public Health Nutr 14, 1200–1211 (2011). [DOI] [PubMed] [Google Scholar]
- Matsuki T. et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70, 167–173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M. et al. Assessment and Improvement of Methods for Microbial DNA Preparation from Fecal Samples in Handbook of Molecular Microbial Ecology II (ed. Bruijn F. J. D.) 191–198 (John Wiley & Sons, Inc., 2011). [Google Scholar]
- Moisander P. H., Beinart R. A., Voss M. & Zehr J. P. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J 2, 954–967 (2008). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 (1989). [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.