Abstract
Histones, by packaging and organizing the DNA into chromatin, serve as essential building blocks for eukaryotic life. The basic structure of the chromatin is established by four canonical histones (H2A, H2B, H3 and H4), while histone variants are more commonly utilized to alter the properties of specific chromatin domains. H3.3, a variant of histone H3, was found to have diverse localization patterns and functions across species but has been rather poorly studied in protists. Here we present the first genome-wide analysis of H3.3 in the malaria-causing, apicomplexan parasite, P. falciparum, which revealed a complex occupancy profile consisting of conserved and parasite-specific features. In contrast to other histone variants, PfH3.3 primarily demarcates euchromatic coding and subtelomeric repetitive sequences. Stable occupancy of PfH3.3 in these regions is largely uncoupled from the transcriptional activity and appears to be primarily dependent on the GC-content of the underlying DNA. Importantly, PfH3.3 specifically marks the promoter region of an active and poised, but not inactive antigenic variation (var) gene, thereby potentially contributing to immune evasion. Collectively, our data suggest that PfH3.3, together with other histone variants, indexes the P. falciparum genome to functionally distinct domains and contribute to a key survival strategy of this deadly pathogen.
In all eukaryotes, chromatin enables high-degree compaction of the genetic material while allowing access to the DNA for essential cellular processes such as transcription, replication, DNA damage response and repair. Canonical histones, namely H2A, H2B, H3 and H4, are mainly synthesized and assembled into nucleosomes in a replication-coupled manner to form the basic layout of the newly synthetized chromatin1,2,3. In contrast, histone variants (non-allelic histone isoforms1,3), and most posttranslational histone modifications (e.g. acetylation, methylation4) are locally incorporated into chromatin in a replication-independent fashion to alter its properties and function.
Given their essential function, histones are among the most conserved proteins and their evolutionary origins can be traced back to archaea, where one or two histone like proteins are present5. In Eukarya, the histone family has expanded into canonical histones, the linker histone H1 and a great variety of histone variants for H2A, H2B and H36. Strikingly, the amino acid sequences of the four eukaryotic canonical histones and of many histone variants are extremely similar among distantly related species, although histone variants have emerged by convergent evolution7,8,9. The requirement of convergent evolution of histone variants indicates a universal theme in chromatin regulation; and indeed some histone variants display universal functions. Histone H3 variant CenH3, for instance, demarcates the centromere and functions in chromosome segregation10,11,12,13. The function of H3.3, another member of the H3 histone family is however more diverse among the species.
Phylogenetically earlier organisms, such as Saccharomyces cerevisiae and the algal protist, Cyanidioschyzon merolae, possess only one form of non-centromeric H3 that is needed for both replication-coupled and replication-independent assembly11,14. This single form corresponds to variant H3.3 of higher eukaryotes, which suggests that canonical forms of H3 evolved recurrently in eukaryotic evolution, most likely by divergence from H3.3-like forms15,16. In most eukaryotes H3.3 differs from H3 only by few amino acid substitutions; for example by four in most mammals and plants and by 16 in the ciliate Tetrahymena thermophila (Superphylum Alveolata)11,17. Yet, these few amino acid substitutions are sufficient to determine H3.3 and H3 chaperone selectivity and their respective nucleosome deposition pathways18,19,20,21.
Genome-wide studies performed mainly in animals, but also in Arabidopsis thaliana revealed that H3.3 is mainly deposited into the coding sequence of transcribed genes, promoters of active and inactive genes, transcription start and end sites as well as further gene regulatory elements in euchromatic regions8,16,22,23. H3.3 is incorporated into transcription start sites and coding regions of genes in a transcription-coupled manner24,25. However, enrichment of H3.3 has also been observed in regions of the genome that are presumed to be transcriptionally silent26,27,28,29,30. For example, in embryonic stem cells H3.3 contributes to the maintenance of the condensed chromatin state of telomeres26,28,31,32, while it appears to be absent from the telomeres of A. thaliana33,34. Surprisingly, deletions of H3.3 genes are not always lethal, probably due to compensation by other H3 genes. For instance, increased H3 expression compensates for H3.3 lacking in D. melanogaster35 and overexpression of H3.3 makes canonical H3 dispensable in T. thermophila17. In contrast, H3.3 disruption in D. melanogaster and in T. thermophila results in meiotic defects17,36,37. Furthermore, in mammals, H3.3 is not only required for reproduction but also for early development32,38 and mutation in H3.3 can lead to various pediatric cancer types39,40. Hence, the function of H3 variants has been extensively studied in animals, fungi and plants - where it appears to have several conserved, but also specialized function.
In this article, we focus on the localization and role of H3.3 in the apicomplexan parasite, Plasmodium falciparum. Besides the canonical histones four histone variants are encoded in the Plasmodium genome, i.e. PfH2A.Z, PfH2B.Z, PfCenH3 and PfH3.341,42. Interestingly, P. falciparum has an extremely AT-rich genome (on average ~80% adenine and thymine bases) where different genomic regions have distinctly different base composition. Earlier we observed an intriguing correlation between the base composition of these genomic regions and the incorporation of various histone variants41. Centromeres are formed via incorporation of PfCenH3 into a ~2kb long extremely AT-rich (~97% AT) sequence present once on every chromosome13. On the other end of the spectrum are H3K9me3 and HP1-marked heterochromatic regions with distinctly higher GC content (~73% AT). These regions are present at the chromosome ends and some chromosome internal islands, and so far have found to contain mainly canonical nucleosomes43,44,45,46. Within the euchromatic domain intergenic regions having distinctly higher AT-content (~87% AT) compared to coding sequences (~78% AT), are demarcated by double-variant nucleosomes containing PfH2A.Z and the Plasmodium-specific variant PfH2B.Z46,47,48,49. Collectively these observations support a model in which histone variants help indexing the Plasmodium epigenome into functionally distinct domains and the rough blueprint of the epigenome is sketched by the AT-content of the underlying sequence. Here, in further support of this model, we show that PfH3.3 demarcates euchromatic coding sequences and subtelomeric repeat regions in a GC-content coupled-manner. Furthermore, we observe that PfH3.3 specifically associates with the promoter region of the single expressed or poised - but not inactive - antigenic variation gene (var gene). This suggests that PfH3.3 might contribute to the epigenetic memory of var gene expression and hence could underlie a major defense mechanism of this deadly human pathogen.
Results
PfH3.3 primarily localizes to GC-rich euchromatic coding sequences and subtelomeric repetitive regions
Plasmodium falciparum’s histone variant PfH3.3 (PF3D7_0617900) and its single copy canonical counterpart PfH3 (PF3D7_0610400) both encode for a 136 amino-acid protein. They only differ in eight amino acids including an Apicomplexan specific feature for PfH3.3, namely the substitution of the conserved amino acids RY with KF at position 54 and 5542,50. With an identity of 94% they are hence difficult to investigate individually employing antibodies.
We circumvented this problem by generating a P. falciparum 3D7 parasite line that expresses an episomal Ty1-tagged copy of PfH3.3 under the control of the chloroquine resistance transporter promoter (similar to pARL-1a-Ty1-H2A in ref. 47). Additionally, to avoid any artefacts due to episomal (over)expression of the tagged protein, we engineered a construct (pHH1-PfH3.3-Ty1) that, after single crossover integration, resulted in a P. falciparum NF54-DCJ parasite line51 expressing PfH3.3-Ty1 under control of the endogenous promoter (Fig. S1). Notably, these tagged lines show a very similar multiplication rate (data not shown) and expression pattern (Fig. S2) to wild type parasites suggesting that the tag does not interfere with normal function of PfH3.3.
To determine the genome-wide localization of PfH3.3, we performed chromatin immunoprecipitation of mono-nucleosomes using an anti-Ty1 antibody followed by next generation sequencing of the immunoprecipitated DNA (ChIP-seq). First we performed a test ChIP-seq on native chromatin isolated from the episomal Ty1-PfH3.3 line for two intraerythrocytic stages (30 and 40 hours post-invasion) in duplicates (Fig. S3). The respective ChIP input samples were sequenced as control. For detailed analysis we performed ChIP-seq on formaldehyde cross-linked chromatin from the endogenously PfH3.3-Ty1 line for four intraerythrocytic stages (10, 20, 30 and 40 hpi). Additionally, we performed ChIP-seq using an anti-H3 antibody that recognizes the C-terminal end of both PfH3.3 and PfH3 (referred to as H3core control) and RNA-seq on the same four parasite populations. The resulting PfH3.3 ChIP-seq data were normalized either to input or H3core ChIP-seq data to correct for any biases due to differential cross-linking, PCR amplification efficiency and reduced mapability of the highly AT-rich sequences, typical to the P. falciparum genome47. The normalized PfH3.3 ChIP-seq data from episomal and endogenously PfH3.3-tagged parasites displayed a highly similar localization pattern (Fig. S3A) and good genome-wide correlation (Fig. S3B; r = 0.71–0.86), despite numerous technical differences between the two experiments (N- versus C-terminal tagging, cross-linked versus native ChIP, different parasite lines, different library preparation methods) (Fig. S3). Hence, our ChIP-seq analysis provides a robust measure of PfH3.3 localization, which we analyzed in detail using the data from the endogenously tagged line.
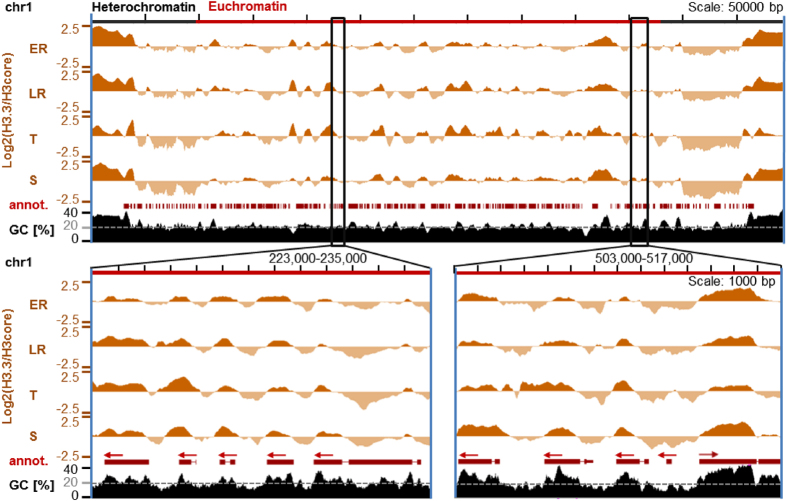
PfH3.3 is mainly enriched in euchromatic coding sequences and is generally absent in the heterochromatic domain with the exception of its enrichment over GC-rich subtelomeric repetitive sequences (Fig. 1). In these regions the occupancy of PfH3.3 is largely invariable through the intraerythrocytic stages. Additionally, PfH3.3 occupancy shows dynamic changes in euchromatic intergenic regions (Fig. 1). Centromeres are depleted of PfH3.3 and PfH3 (Fig. S4), as they are occupied by the third H3 variant, PfCenH313.
Figure 1. PfH3.3-Ty1 ChIP-seq profiles from different stages of intraerythrocytic development.
ChIP-seq ratio tracks displaying relative enrichment of the endogenous PfH3.3-Ty1 compared to H3core (antibody capturing histone PfH3 and PfH3.3) over entire chromosome 1 (upper panel) or two smaller sections of it (lower panels). Red arrows indicate the directionality of the respective gene (“annot.”). Stages: ER: early rings (10 hpi), LR: late rings (20 hpi), T: trophozoites (30 hpi), S: schizonts (40 hpi). Heterochromatic regions are defined based on HP1 occupancy in an earlier study43.
PfH3.3 is demarcating the P. falciparum genome and exogenous DNA sequences in a GC-content coupled manner
Visual inspection of the genome-wide PfH3.3 profiles led to the observation that PfH3.3 is mainly incorporated into GC-rich sequences (Fig. 1), including euchromatic coding sequences with an average GC-content of ~22% and the subtelomeric repetitive sequences with an GC-content ~32%. While at these GC-rich regions PfH3.3 occupancy is invariable across the stages, AT-rich intergenic regions (average 14% GC) are only temporarily marked or completely depleted of PfH3.3 (Fig. 1).
To further investigate the correlation between GC-content and PfH3.3 occupancy on a genome-wide level, we in silico subdivided the genome into 150 bp windows and calculated the GC percentage and PfH3.3 coverage for each window. In line with our visual observation, GC-poor intergenic regions show the lowest PfH3.3 coverage; whereas in euchromatic coding sequences PfH3.3 levels exhibited clear correlation with the GC-content (Fig. 2A and Fig. S5A for data from all four stages). Importantly, similar correlation was also observed in the ChIP-seq data obtained from the episomally tagged line (Fig. S5B). Subtelomeric GC-rich repetitive sequences display the highest PfH3.3 levels. Thus, our genome-wide computational analysis is consistent with the initial observation.
Figure 2. PfH3.3 localizes to GC-rich regions within the P. falciparum genome and exogenous DNA.
(A) Density plots depicting PfH3.3 levels in relation to GC-content. PfH3.3 levels and GC-content were calculated genome-wide per 150 bp windows. Data depicted are generated from schizont stages. IG: intergenic region, CS: coding sequence, Sub. Rep.: subtelomeric repetitive region. Pearson correlation values are displayed at the lower right corner of the graphs. (B) Screen shot from PfH3.3 ChIP-seq ratio tracks encompassing the integrated pHH1-PfH3.3-Ty1 plasmid sequence (left) and the sequence of two nearby genes (right). Grey blocks indicate non-unique P. falciparum sequences within the pHH1-PfH3.3-Ty1 plasmid, which were excluded from the analysis in (C). (C) Scatter plot of GC percentage and PfH3.3 occupancy over unique 75 bp windows of the integrated pHH1-PfH3.3-Ty1 plasmid. Stages: ER: early rings (10 hpi); LR: late rings (20 hpi), T: trophozoites (30 hpi), S: schizonts (40 hpi). Pearson correlation values are displayed at the lower right corner of the graphs.
Additionally, we tested whether PfH3.3 is also incorporated into an exogenous DNA in a GC-dependent manner. Therefore, the non-P. falciparum-specific sequences within the integrated pHH1-Ty1-PfH3.3 plasmid were examined for their GC-content and PfH3.3 occupancy. Also here a clear correlation of incorporated PfH3.3 and GC-content can be observed (Fig. 2B,C). As these exogenous sequences are unlikely to contain sequence-element for recruitment or detainment of PfH3.3 or its chaperone, these findings support a base-content-driven genome marking by PfH3.3.
PfH3.3 dynamically occupies intergenic regions predominantly during trophozoite development
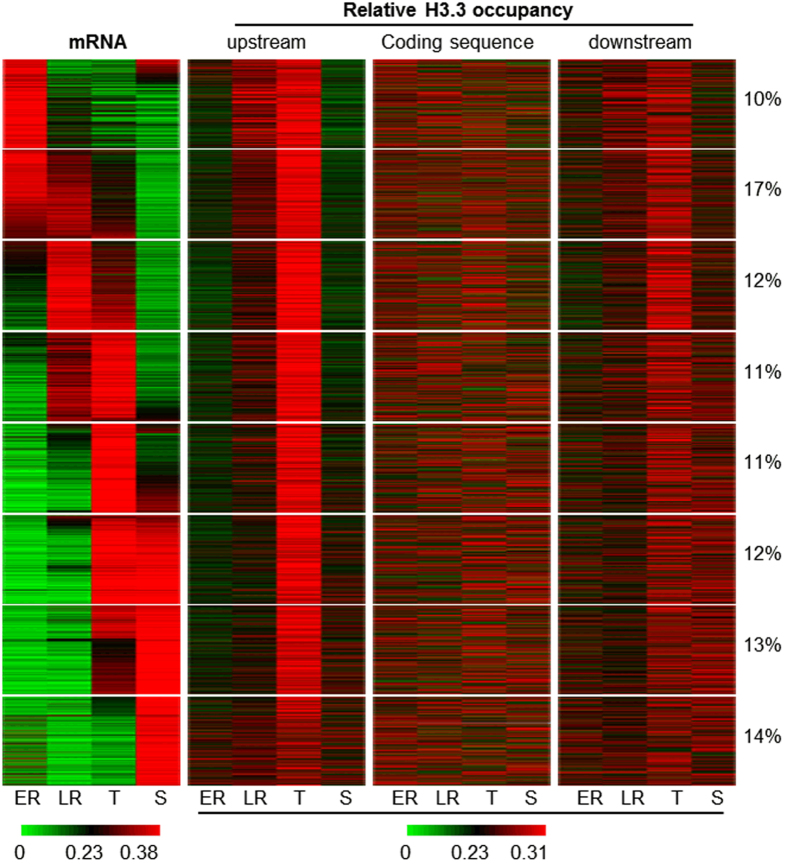
The dynamic occupancy of H3.3 in some intergenic regions prompted us to test whether correlation exists between temporal gene activity and the dynamics of PfH3.3 occupancy. Genes were clustered according to their relative mRNA abundance (matching RNA-seq data) during the intraerythrocytic cycle and matched with the respective PfH3.3 occupancy values (in proportion of the sum of the occupancy values) for upstream regions, coding sequences and downstream regions (Fig. 3). Whereas PfH3.3 coverage in coding sequences showed no obvious stage-dependent or transcription-coupled dynamics, mainly upstream regions but to some extent also downstream regions showed dynamic PfH3.3 enrichment during the intraerythrocytic cycle. Interestingly, most intergenic regions have the highest relative PfH3.3 occupancy during the trophozoite stage largely independent of their transcriptional dynamics.
Figure 3. PfH3.3 dynamically occupies intergenic regions predominantly during trophozoite stage.
Heatmap represents relative mRNA abundance of 3800 genes (heterochromatic and genes with low mRNA abundance have been excluded) and relative PfH3.3 occupancy in upstream, coding and downstream region throughout intraerythrocytic development. Color-scale depicts the relative mRNA abundance in relation to the sum of the RPKM values in all four stages or the relative PfH3.3 occupancy in proportion to the sum of occupancy values in all four stages (green – low, red – high). 8 groups of co-expressed genes were identified using k-means clustering. The percentage of genes belonging to each cluster is displayed next to the heatmap. Stages: ER: early rings (10 hpi), LR: late rings (20 hpi), T: trophozoites (30 hpi), S: schizonts (40 hpi).
To investigate PfH3.3 enrichment in relation to gene expression level, we plotted the mRNA abundance (RPKM values) for each gene against PfH3.3 occupancy in coding, upstream or downstream regions for each stage separately (Fig. S6). This analysis also did not reveal clear correlations between transcription and PfH3.3 occupancy. Only in 5′ intergenic regions and in particular at the trophozoite stage were moderate correlations observed.
In summary, PfH3.3 is temporarily enriched in AT-rich, 5′ intergenic regions mainly at the trophozoite stage. While this enrichment is somewhat more apparent for very highly expressed genes, and least apparent for (nearly) silent genes (Fig. S6), it does not exhibit clear correlation to gene expression dynamics (Fig. 3).
PfH3.3 marks the promoter of the active and poised var gene
Persistence of P. falciparum within the human bloodstream primarily depends on the var multigene family, encoding for a highly variable cytoadherence protein called P. falciparum erythrocyte membrane protein 1 (PfEMP1)52,53,54,55. Only one of the ~60 var gene family members is expressed in any given parasite while other family members remain transcriptionally silenced. Importantly, var genes are expressed in a clonally variant fashion56 as the same var gene is reactivated over several generation by a yet elusive mechanisms of epigenetic memory57,58.
In order to investigate the role of PfH3.3 in var gene regulation, we specifically looked at the var2CSA locus of CSA-selected (10, 20, 30, 40 hpi) (Fig. S7A) and non-selected (mix of trophozoites and schizonts) parasites. Interestingly, PfH3.3 is enriched in the promoter as well as in the coding region of the active as well as poised var2CSA gene (Fig. 4A) (Note that the var gene is only transcribed during rings stages while at trophozoites and schizonts stages it is poised, yet marked for reactivation in the next generation). The var2CSA gene in non-CSA selected parasites, however, is clearly less enriched in PfH3.3 and only shows PfH3.3 coverage at the var2CSA intron and the second exon (regions which are known to be transcribed from the var intronic promoter55,59,60,61,62,63) (Fig. 4A). To ensure that this observation was not an artefact due to the comparison of two independently generated data sets (as the CSA-selected and non-selected parasites originate from independent experiments) we performed a ChIP-qPCR experiment that enabled us to directly compare the var2CSA locus of CSA-selected and non-selected parasites. First, a culture of endogenously PfH3.3-Ty1 expressing NF54-DCJ P. falciparum parasites was selected for var2CSA expression via chondroitin sulfate affinity purification (var2CSA positive line). Subsequently, half of the culture was treated with blasticidin (2.5 mM), which forced a switch to another var promoter (PF3D7_0223500) driving expression of the BSD resistance cassette51. Successful selection for parasites expressing var2CSA or PF3D7_0223500 was confirmed by RT-qPCR (Fig. S7). Subsequently, ChIP-qPCR was performed for the active histone modification H3K9ac, the inactive histone modification H3K9me3, the histone variant PfH2A.Z and for PfH3.3 for three different time points (15, 25 and 40 hpi). The switch from var2CSA expression to PF3D7_0223500 was confirmed at the epigenetic level: CSA-selected parasites showed high H3K9ac recoveries for the whole promoter region of the var2CSA gene and low H3K9me3 recoveries for the promoter as well as coding sequences (Fig. 4B). BSD-selected parasites, however, showed the opposite pattern (Fig. 4B). As also reported earlier, only the CSA-selected parasite line showed high occupancy of PfH2A.Z close to the promoter start site 1500 bp upstream of the ATG49. Incorporation of PfH3.3 into the promoter region of the var2CSA gene was clearly increased when the gene was active compared to its silenced state validating the observation made based on the ChIP-seq data. In summary, PfH3.3 stably occupies the promoter region and coding sequence of the active var gene but is evidently less incorporated into the promoter and coding sequence of silenced var genes.
Figure 4. PfH3.3 marks the promoter of the active and poised, but not the inactive var gene.
(A) RNA-seq and ChIP-seq ratio tracks displaying PfH3.3 enrichment over H3core over the var2CSA locus. Data was obtained from var2CSA-selected parasites at four intraerythrocytic stages (ER, LR, T, S) as well as from unselected mixed stage parasites (mainly containing trophozoites and schizonts, T/S). Arrows indicate the approximate position of primers used in (B). (B) Bar graphs displaying ChIP-qPCR data for H3K9ac (euchromatic mark) and H3K9me3 (heterochromatic mark) marking as well as PfH3.3 and PfH2A.Z occupancy over the var2CSA locus in CSA-selected (red) or - BSD-selected (grey) parasites. After affinity-based selection for var2CSA-expressing parasites half of the parasite population was forced to express PF3D7_0223500 by blasticidin treatment (NF54-DCJ strain: first exon of PF3D7_0223500 is replaced by a blasticidin resistance cassette). Numbers indicate the position of the forward primer relative to the ATG. Stages (R: rings (15 hpi); T: trophozoites (25 hpi); S: schizonts (40 hpi)) are depicted by different shades.
Discussion
Here we present the first genome-wide analysis of histone variant H3.3 in the apicomplexan parasite, P. falciparum. Interestingly, PfH3.3 has a distinct localization compared to other variants. While PfH2A.Z and PfH2B.Z48 as well as PfCenH313 appear to occupy AT-rich sequences, PfH3.3 is most commonly found in GC-rich coding sequences and the subtelomeric repetitive regions (Fig. 1) and only temporarily occupies AT-richer intergenic regions (Fig. 3). In many animals, H3.3 is also found in coding regions, where it is incorporated during transcriptional elongation24,25. In P. falciparum, however, we could not observe a clear correlation between PfH3.3 occupancy in euchromatic coding sequences and the steady-state mRNA level of the corresponding genes (Fig. 3, Fig. S6). Instead we find that most coding sequences are constantly marked by PfH3.3 throughout the intraerythrocytic cycle and we observed a positive correlation between PfH3.3 occupancy and the GC-content of the underlying DNA (Fig. 2A). This correlation is even more striking over exogenous DNA sequences that do not contain any P. falciparum “compatible” gene regulatory elements (Fig. 2B/C). Furthermore, although transcriptionally silent heterochromatic regions appear to have a generally lower PfH3.3 occupancy, GC-richer coding sequences are yet more likely to contain some PfH3.3 (Fig. 2A). These observations together suggest that PfH3.3 incorporation to coding regions does not (necessarily) require transcriptional activity and PfH3.3 occupancy is rather influenced by the GC-content of DNA sequence.
Next to constant occupancy of PfH3.3 at euchromatic intergenic regions we observe clear PfH3.3 marking at subtelomeric repeat sequences. In mammals, PfH3.3 is also deposited at repressive chromatin regions and in mouse embryonic cells additionally at the telomeres26,28,31,64, suggesting that PfH3.3 might serve a conserved telomere associated function. However, unlike in mouse embryonic stem cells31,64, PfH3.3 occupancy seems to be uncoupled from K9me3 marking. In fact, PfH3.3 appears to be specifically depleted from most heterochromatic regions - marked by K9me3 and HP1 in P. falciparum - except for the subtelomeric repetitive regions (Figs 1 and 2). Notably, these latter regions have the highest density of G quadruplex motifs in P. falciparum genome65. Since, earlier work in mouse established a potential link between G quadruplex sequences and H3.3 occupancy at the telomeres66, it is conceivable that a similar link might underlie the subtelomeric H3.3 localization in P. falciparum. However, it is even more straightforward to think these regions have high PfH3.3 occupancy simply because they have a distinctly higher GC content (Fig. 2A), providing further support for the above model.
In addition to the GC-based correlation of Pf H3.3 localization to coding and subtelomeric sequences, we observe dynamic incorporation of PfH3.3 into AT-rich intergenic regions, almost exclusively at the trophozoite stages. This stage is peculiar in two ways: i) this is the stage when extensive replication of the parasites genome begins; ii) this is the transcriptionally most active stage. Notably, in contrast to many other eukaryotic cells, cell division during the intraerythrocytic cycle of P. falciparum does not only double the DNA content, but rather results in 8 to 32 genome copies. Given that an enormous amount of histones needed to support the synthesis of the new chromatin it is conceivable that both H3 variants (PfH3 and PfH3.3) need to be incorporated during replication to the newly synthetized chromatin, after which PfH3.3 might be preferentially retained at GC-rich sequences. A similar phenomenon is observed in mouse spermatocytes, when almost all histones are replaced by protamine to form a highly compact paternal chromatin, nucleosomes that are retained at unmethylated CpG-rich sequences, primarily contain H3.367. Alternatively, the stage-specific incorporation of PfH3.3 to promoter regions could be explained by temporal transcriptional activity, typical to intraerythrocytic stages of malaria parasites (Fig. 3). In fact in many other organisms H2A.Z and H3.3 containing nucleosomes are located around the transcriptional start site (for review see ref. 68). However, while our analysis revealed a moderate correlation between mRNA abundance and PfH3.3 enrichment in promoter regions in particular at the trophozoite stage (Fig. S6) PfH3.3 occupancy profiles in promoter regions are largely uncoupled from the dynamics of their transcriptional activity (Fig. 3). Accordingly, while we cannot completely exclude the possibility that PfH3.3 is incorporated to some promoter regions in a transcription coupled manner, the extensive incorporation of PfH3.3 to intergenic regions at the trophozoite stage is perhaps better explained by the replicative activity of this stage.
In mammalian cells, the different distribution patterns of H3.3 compared to H3 is achieved by histone- and site-specific chaperones. While H3.3 is deposited at euchromatic regions by the chaperone Histone regulator A (HIRA) replication-independently20,69, the death-associated protein (DAXX) together with the α-thalassemia and/or mental retardation X-linked syndrome protein (ATRX) mediates H3.3 deposition in pericentric heterochromatin and telomeric regions26,27. H3 deposition, however, is mediated by the chaperone CAF-1 replication-dependently20. Recognition of H3 and H3.3 by these chaperons is mediated by amino acid residues at positions such as 87 and 9018,19,70 which also differ in PfH3 and PfH3.318,19,42,50,70 and might support differential recognition by different chaperon proteins. Thus far, however, only one P. falciparum protein with weak homology to both CAF1 and HIRA has been identified (PF3D7_0501800). Therefore, additional experiments will have to be performed to determine whether this protein supports the incorporation of both PfH3 and PfH3.3 during replication or yet unidentified PfH3/PfH3.3-specific chaperons exist that ensure differential genomic incorporation of PfH3 and PfH3.3.
Of particular interest is the specific PfH3.3 incorporation to the promoter region of the active, but not inactive var gene (Fig. 4). This is similar to the earlier reported specific incorporation of the PfH2A.Z/PfH2B.Z double-variant nucleosomes around the transcription start site of the active var gene during the ring stage, when this gene is being actively transcribed49. Furthermore, the active var promoter is marked by two active marks (H3K9ac and H3K4me3), while the poised var gene carries H3K9ac and H3K4me258,71,72. Interestingly, unlike PfH2A.Z/PfH2B.Z, PfH3.3 is retained at this AT-rich var promoter even after active transcription has ceased (so called poised state). This raises the exciting possibility that PfH3.3 may contribute to the epigenetic memory of var gene expression57,58, and enables the parasites to express the same var gene over several generations. In line with such hypothesis, nuclear transplantation experiments in Xenopus laevis showed that H3.3 mediates the epigenetic memory of certain genes; and H3.3 overexpression in nuclear transplant embryos even enhances this effect73,74,75. Furthermore, H3.3 enrichment at promoters has also been observed for inactive genes, which are potentially in a poised state25,76,77 as we observe it for the poised var2CSA gene during the second half of the intraerythrocytic cycle. All in all, these observations made in other organisms support the idea of a potential role of PfH3.3 in epigenetic memory in P. falciparum. In order to investigate the functional relevance of PfH3.3 in parasite biology in general or in var gene regulation in particular, we made several attempts to tag PfH3.3 with a AID-GFP-glmS78,79 or GFP-glmS79 tag. Unfortunately, these attempts failed to result in viable parasite lines using these rather bulky conditional knock-down tags, hinting towards an essential function of PfH3.3. Accordingly, whether PfH3.3 is critical for “epigenetic memory” of var gene expression and how it could excerpt such function will be the subject of future studies.
In summary, we show that PfH3.3 has a complex localization pattern that consists of conserved, but also parasite-specific features. It demarcates euchromatic coding and subtelomeric repetitive sequences in a GC-content coupled manner. Within the coding sequences it might promote the process of transcription by forming labile nucleosomes, as has been reported for other organisms. The strong marking of subtelomeric repetitive sequences resembles findings from higher eukaryotes, which could elude to a conserved, yet uncharacterized function. Excitingly, PfH3.3 specifically marks the promoter region of the var gene in its active and poised, but not inactive state and could contribute to the process of antigenic variation which is critical to immune evasion of this deadly pathogen.
Methods
Generation of Ty1-tagged PfH3.3 parasites
In order to profile PfH3.3, 3D7 P. falciparum parasites were transfected with a pARL-1a- plasmid80 encoding a N-terminally Ty1-tagged version of PfH3.3 (PF3D7_0617900) under the control of the chloroquine resistance transporter promoter (similar to pARL-1a-Ty1-H2A in ref. 47) and cultured in presence of 40 nM WR99210. To generate PfH3.3 profiles from endogenously expressed PfH3.3, NF54-DCJ P. falciparum parasites51 were transfected with a pHH1-plasmid81 encoding a C-terminally Ty1-tagged version of PfH3.3 (PF3D7_0617900) and a CTG instead of an ATG as “start codon” (Fig. S1A). Parasites were cultured in presence of 2.5 nM WR99210. Single crossover integration of the plasmid resulted in parasites that express a fully functional PfH3.3-Ty1 under the endogenous promoter (ATG as start codon) and a non-functional untagged PfH3.3 (CTG as start codon) (Fig. S1B). A clonal parasite line was obtained by limiting dilution. Correct integration of the pHH1-plasmid and absence of wild type parasites was confirmed by performing Southern Blot analysis (Fig. S1B,C). Expression of episomal and endogenously-expressed tagged PfH3.3 was confirmed using the monoclonal anti-Ty1 antibody (BB2)82 that exclusively recognizes the tagged PfH3.3 protein on Western blot (Fig. S1D).
Parasite culture and collection of parasites
Parasites were cultured in standard RPMI medium supplemented with 10% human serum (AB), 0.2% NaHCO3 and 2.5% human O+ red blood cells. Culturing occurred in 250 ml tissue culture flasks placed in candle jars or in an incubator with a gas composition of 3% O2, 4% CO2 and 93% N2 and incubated at 37 °C. Parasites were synchronized with sorbitol treatments and Percoll gradient centrifugations as described previously47. During collections, medium was changed every 10 hours, but not less than 10 hours before collection to ensure optimal parasite growth. After 20 hours post invasion (hpi), medium volume was doubled. During each medium change parasite cultures were mixed to minimize growth differences within single culture flasks. For the generation of genome-wide PfH3.3 profiles early ring (10 hpi), late ring (20 hpi), trophozoite (30 hpi) and schizont stages (40 hpi) were collected. For the ChIP-qPCR investigation of PfH3.3 var2CSA- or PF3D7_0223500-selected parasites, ring, trophozoite and schizont stages were collected at 15, 25 and 40 hpi. Parasite cultures used for RNA extraction or native ChIP were immediately placed and processed on ice (4 °C). Parasite cultures used for cross-linked ChIP were immediately treated with 1% formaldehyde (final concentration) and incubated at 37 °C for 10 min while shaking. Cross-linking reaction was quenched by adding 0.125 M glycine (final concentration). All subsequent steps were performed at 4 °C unless noted otherwise. To remove contaminating human white blood cells that would cause considerable background upon sequencing, all blood used for collection of sequence samples was filtered through Plasmodipure filters (EuroProxima).
Etic statement
Blood and serum was obtained from the local blood bank (Sanquin) with the full consent of healthy donors to use this material for malaria research.
var2CSA (PF3D7_1200600) and PF3D7_0223500 selection
Endogenously PfH3.3-Ty1 expressing NF54-DCJ P. falciparum parasites were selected for var2CSA expression by affinity purification as in ref. 83. For this purpose, petri dishes (150 × 15 mm, BD biosciences Falcon 351058) were coated with Chondroitin sulfate A (0.05% CSA in PBS) overnight and subsequently blocked with 1% Casein/PBS solution for at least one hour and rinsed with RPMI twice. In the meanwhile parasite cultures were centrifuged, resuspended in RPMI containing 10% human serum and transferred to the petri dishes. Subsequently, petri dishes containing the parasite culture were incubated for 30 min at 37 °C in a candle jar. Afterwards, unbound parasites and non-infected erythrocytes were removed by gentle RPMI washes. Bound parasites were extensively resuspended in complete medium, fresh blood was added, and parasites were cultured as described above. var2CSA panning was repeated several times to achieve an increasing proportion of var2CSA-expressing parasites. Subsequent switching to PF3D7_0223500 expression, was enforced by culturing the parasites with 2.5 mM Blasticidin51. Successful var gene selection was confirmed via RNA extraction (RNeasy mini kit, Qiagen) followed by cDNA synthesis and subsequent quantitative real time PCR (qPCR) using var gene specific primers71,84 (Fig. S7A,B).
cDNA synthesis and qPCR
To synthesize the cDNA 0.5–5 ug of total RNA were mixed with random hexamer primers (0.5 ug, Roche), OligodT12-18 (0.5 ug, Invitrogen) and dNTPs (0.5 mM in the final volume of 20 ul, Invitrogen) and incubated for 5 min at 70 °C. Then first strand synthesis was performed for 1 hour at 42 °C in First Strand Buffer supplemented with DTT (10 mM), Superscript III (200 units) and RNasin Plus RNase inhibitor (40 units, Promega), after which superscript III was inactivated by incubation at 70 °C for 15 minutes. RT “minus” reactions (where Superscript III was replaced by water) were performed in all cases as negative control. Subsequent qPCR was performed using the CFX96 Real Time Systems C1000 Touch Thermal Cycler. P. falciparum genomic DNA served as standard (500 pg, 50 pg, 5 pg). SYBRgreen supermix (BioRad) and primers were mixed according to manufacturer’s instructions. qPCR was performed in 96 well plate format (BioRad) using the following protocol: 95 °C for 3 min, (94 °C for 10 sec, 52 °C for 30 sec, 68 °C for 30 sec) 39x, 95 °C 1 min, 65 °C 1 min and a gradient from 65 °C to 94.5 °C with a 0.5 °C increase every 10 sec.
Western Blot analysis
Nuclear extracts from the 3D7 mother line (~40 ug), the 3D7 parasites expressing Ty1-PfH3.3 episomally (~60 ug) and the NF54-DCJ parasites expressing PfH3.3-Ty1 under the endogenous promoter respectively, were separated on 18% SDS-PAGE and transferred to a protean nitrocellulose membrane (Whatman, 0.45 mm). The membrane was probed with mouse anti-Ty1 antibody (1:2000, BB2 antibody in ref. 82) and rabbit anti-histone H3 antibody (1:3000, Abcam ab1791) and secondary goat anti-mouse IRDye680 (1:10000, LI-COR Biosciences 926-32220) and goat anti-rabbit IRDye 800CW (1:10000, LI-COR 926-32211). Fluorescence was measured on the Odyssey system (LI-COR Biosciences).
Southern Blot analysis
Genomic DNA was isolated from native nuclei using phenol-chloroform extraction as described previously47. 2.5 μg of genomic DNA and 0.5 ng of plasmid DNA were digested with EcoRI and XbaI (NEB) (Fig. S1B). A PfH3.3 sequence specific hybridization probe was generated by PCR labelling using Digoxigenin-11-dUTP (DIG) (alkali-labile, Roche) according to manufacturer’s instructions. The PCR reaction was performed using the Advantage® cDNA PCR Kit and Polymerase Mix (Clonetech) and a DIG-dUTP to dTTP ratio of 1:3 under the following PCR conditions: 5 min 95 °C; 30 s 95 °C, 30 s 52 °C, 90 s 68 °C (35 cycles); 5 min 68 °C (Fig. S1B). Gel preparation, subsequent DNA transfer to a positively charged Nylon membrane (Hybond-N+, GE Healthcare Amersham), prehybridization (DIG Easy Hyb Kit from Roche, 65 °C) and probe-DNA hybridization (DIG Easy Hyb Kit from Roche, 65 °C) were all performed according to manufacturer’s recommendations. The detection of the hybridized probe was performed using the DIG Wash and Block Buffer Set, the anti-Digoxigenin antibody (anti-Digoxigenin-AB conjugate Fab frag; 1:10 000 dilution) and the CDP-Star Kit (ready-to-use) from Roche according to manufacturer’s instructions.
Chromatin immunoprecipitation
After erythrocyte and parasite lysis using 0.05% saponin and a hypotonic buffer (10 mM Tris pH 8.0, 3 mM MgCl2, 0.2% NP-40, Roche Protease Inhibitor Cocktail) respectively, native nuclei (ChIP-seq of episomal Ty1-PfH3.3 and var gene switch experiment, see below) or formaldehyde-cross-linked nuclei (ChIP-seq experiment) were separated from cell debris using a 0.25 M sucrose buffer cushion as described before47. Subsequent, cross-linked chromatin was prepared as described in ref. 85 with a few adjustments. In short, for generation of genome-wide PfH3.3 profiles, enzymatic MNase digestion of cross-linked chromatin was performed using digestion buffer (50 mM Tris pH7.4, 4 mM MgCl2, 1 mM CaCl2, 0.075% NP40, Roche Protease Inhibitor Cocktail) with 0.5 U MNase (Worthington Biochemicals Corporation) in 150 μl aliquots for 6 min (10 hpi), 12 min (20 hpi) or 13 min (30 hpi & 40 hpi) at 37 °C. Optimal digestion times were empirically determined for each stage by test-digestion of a nuclear aliquot for varying times, to obtain primarily mono-nucleosomal DNA fragments. Digestion reactions were stopped by adding 150 μl quenching solution (2% Triton X100, 0.6% SDS, 300 mM NaCl, 6 mM EDTA, Roche Protease Inhibitor Cocktail) and placing the samples on ice. Subsequently, digested chromatin was sonicated for 6 × 10 sec (setting low, BioruptorTM Next Gen, Diagenode) to free cross-linked chromatin from nuclear membranes. Approximately 200 ng of digested DNA-containing cross-linked chromatin was incubated in ChIP buffer (150 mM NaCl, 20 mM Tris pH 8.0, 2 mM EDTA, 1% Triton X-100, 0.15% SDS, Roche Protease Inhibitor Cocktail) with 2 μg of anti-Ty1, 1 ug of anti-histone H3 (recognizes PfH3 and PfH3.3, Abcam Ab1791) or 1 μg polyclonal anti-IgG (Upstate 12-370) overnight at 4 °C while rotating, followed by the addition of 10 μl ProtA and 10 ul ProtG dynabeads (Life Technologies, 10008D and 10009D) and further incubation at 4 °C for 2 h. For the H3core antibody (capturing histone PfH3 and PfH3.3) one ChIP reaction was performed, for PfH3.3 (anti-Ty1) four ChIP reactions were performed in parallel per life cycle stage to ensure sufficient amount of DNA for subsequent ChIP-seq. After washing with three different buffers (buffer1, 1x: 20 mM Tris pH 8.0, 2 mM EDTA, 1% Triton-X100, 0.1% SDS, 150 mM NaCl; buffer 2, 2x: 20 mM Tris pH 8.0, 2 mM EDTA, 1% Triton-X100, 0.1% SDS, 500 mM NaCl and buffer 3, 2x: 10 mM Tris pH 8.0, 1 mM EDTA) immunoprecipitated chromatin was eluted using 1% SDS and 0.1 M NaHCO3. Immunoprecipitated chromatin was decross-linked (1% SDS, 0.1 M NaHCO3, 1 M NaCl) overnight at 45 °C shaking, followed by DNA isolation via QIAquick column purification (Qiagen).
To investigate PfH3.3 and PfH2A.Z occupancy as well as H3K9ac and H3K9me3 marking of the var2CSA locus of var2CSA- or PF3D7_0223500-selected parasites, chromatin was extracted and native ChIPs were performed as described previously47. Native chromatin was digested with 0.5 U MNase in 150 μl aliquots for 4 min (15 hpi var2CSA-selected parasites), 7 min (15 hpi, 25 hpi, 40 hpi PF3D7_0223500-selected parasites and 25 hpi var2CSA-selected parasites) and 10 min (40 hpi Var2CSA-selected parasites) at 37 °C in a waterbath and regular tapping of the tubes (again optimal digestion times to primarily mono- nucleosomal fragments were determined empirically by a test-digestion of an nuclear-aliquot from each sample). Subsequent chromatin extraction was performed in low-salt buffer (buffer1: 50 mM Tris pH 7.4, 4 mM MgCl2, 1 mM CaCl2, Roche Protease Inhibitor Cocktail; buffer2: 10 mM Tris pH 7.4, 1 mM EDTA, Roche Protease Inhibitor Cocktail; buffer3: 1 mM Tris pH 7.4, 0.2 mM EDTA, Roche Protease Inhibitor Cocktail) and incubated with 1 volume ChIP incubation buffer (100 mM NaCl, 19.7 mM Tris pH 7.4, 6.25 mM EDTA, 1% Triton X-100, 0.1% SDS). For native ChIP reactions, native chromatin (containing about 200 ng DNA) from each time point of var2CSA- and PF3D7_0223500-selected parasites was incubated with 2 μg of anti-Ty1 (PfH3.3), 0.5 μl polyclonal H2A.Z47, 1 μg of polyclonal anti-H3K9/14ac antibody (Diagenode, C15410200), 1 μg of polyclonal H3K9me3 (in house, corresponds to Merck Millipore 07-442) or 1 μg polyclonal IgG (Upstate 12-370) overnight at 4 °C followed by the addition of 20 μl ProtA/G dynabeads and further incubation at 4 °C for 2 h. 200 ng of native chromatin were kept as input. After washing with buffers three different buffers (buffer 1, 1x: 50 mM Tris pH 7.5, 10 mM EDTA, 100 mM NaCl, 0.5% Triton X-100, 0.05% SDS; buffer 2, 2x: 50 mM Tris pH 7.5, 10 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, 0.05% SDS and buffer 3, 2x: 50 mM Tris pH 7.5; 10 mM EDTA, 250 mM NaCl) immunoprecipitated DNA was eluted and purified using PCR purification columns.
ChIP-qPCR
To test relative enrichment of PfH3.3-Ty1, H2A.Z, H3K9ac and H3K9me3 xlChIP was performed as described above. qPCR (Bio-Rad MyIQ) on each ChIP sample was performed alongside with a 1:10 dilution series of the corresponding input DNA to calculate recovery as a percentage of input. Recovery values for the var2CSA locus where normalized across the different samples using primer sets for regions that are invariably marked with the histone mark/variant. For H3K9ac, PfH2A.Z and PfH3.3 primers for actin (PF3D7_1246200, coding sequence and -1100bp promoter region) and seryl-tRNA synthetase (PF3D7_0717700, coding sequence), HSP70 (PF3D7_0818900, coding sequence and -1000 promoter regions) and 18s RNA (PF3D7_1148600, coding sequence) were chosen. For H3K9me3 primers for bp the telomere repetitive regions, a rifin (PF3D7_0324500, - 400bp promoter regions) and AP2G (PF3D7_1222600, coding sequence). Normalization factor was calculated by dividing the average of the recoveries from the control primers within each sample with the average of the average recovery values across the samples. Finally, all recovery values from the var2CSA specific primers have been multiplied by the normalization factors. The normalization factor scaled between 0.75 and 1.8.
ChIP-seq library preparation
For each sequencing library generated from endogenously PfH3.3-Ty1 expressing NF54-DCJ P. falciparum parasites 3 ng of ChIP DNA were end repaired, extended with 3′ A-overhangs, and ligated to barcoded NextFlex adapters (Bio Scientific)86. Subsequent library amplification was performed under similar conditions as described previously85 with a few adjustments: First, 2x KAPA HiFi HotStart ready-mix (KAPA Biosystems) and NextFlex primers were used for 4 cycles of PCR amplification (98 °C for 2 min; 4 cycles of 98 °C for 20 sec, 62 °C for 3 min; 62 °C for 5 min). Subsequently, amplified libraries were size selected for 270 bp (mono-nucleosomes + ~125 bp NextFlex adapter) using 2% E-Gel Size Select Agarose Gels (Thermo Fisher Scientific) and again amplified as described above for 9 cycles. To deplete adapter dimers and clean-up the DNA, libraries were subsequently purified with Agencourt AMPure XP bead (Beckman Coulter) (1:1 library beads ratio) before sequencing. Sequencing libraries generated from episomally Ty1-PfH3.3-expressing parasites were generated as described in refs 47,86.
RNA-seq library preparation
The preparation of strand-specific RNA-seq libraries was performed as in ref. 85. In short, after total RNA isolation (with on-column DNase treatment) and oligo-dT-selection87 PolyA-selected RNA was further treated with DNase (TURBO DNase, Ambion) to remove remaining genomic DNA. Subsequently, 1.2 μg PolyA-selected total RNA equivalent (see ref. 87 for details) for 10 hpi and 2 μg for 20 hpi, 30 hpi and 40 hpi were fragmented by means of alkaline hydrolysis (5x fragmentation buffer: 200 mM Tris acetate pH 8.2, 500 mM potassium acetate, 150 mM magnesium acetate) for 2 min at 85 °C in 250 μl volume. RNA was precipitated overnight and checked for genomic DNA contamination using qPCR, followed by - if needed - another DNase treatment and RNA precipitation for clean-up. Remaining material was processed for strand-specific RNA-seq as in ref. 85. To prevent unwanted DNA-dependent second strand cDNA synthesis - a major source of artificial antisense ‘transcripts’88 −0.2 μg Actinomycin D was included into the first strand synthesis reaction, which was followed by a 15 min 70 °C enzyme deactivation step and QIAquick MinElute purification (Qiagen). During second strand synthesis replacement of dTTP with dUTPs resulted in incorporation of U-bases into the second cDNA strand. Subsequently, 2 ng double stranded cDNA were used for each sequencing library which were prepared as described above with the exception that after adaptor ligation samples were treated with USER enzyme (New England Biolabs) for 15 min at 37 °C that resulted in dUTP-dependent second strand-specific degradation, followed by a first amplification cycle (4 cycles), size-selection for 300–400 bp fragments, a second amplification cycle (9 cycles) and Ampure Beads purification (see above).
High throughput sequencing
ChIP-seq and strand-specific RNA-seq libraries generated from endogenously PfH3.3-Ty1 expressing NF54-DCJ parasites were sequenced on a HiSeq 2000 system (Illumina) to obtain 100 bp single-end reads (TruSeq SE Cluster Kit v2). ChIP-seq libraries generated from episomal Ty1-PfH3.3 expressing parasites were sequenced on a GAIIx system (Illumina) to obtain 75 bp single-end reads (Standard Cluster Generation KitV4). 100 bp and 75 bp reads respectively were mapped against the Plasmodium falciparum genome assembly (PlasmoDB v6.1) for ChIP samples or against the Plasmodium falciparum annotated transcripts (PlasmoDB v6.1) for mRNA samples using BWA89. Single-end ChIP and RNA reads were filtered to mapping quality ≥15 and only uniquely mapped reads (between 13.5 and 23.5 million reads for ChIP-seq and 15.2 and 26.8 million reads for RNA-seq) were used for further analysis.
High-throughput sequencing data analysis
To visualize ChIP-seq and RNA-seq data in the UCSC browser, all libraries were normalized to the amount of mapped reads per million (RPM). For log2 ratio tracks PfH3.3 values were divided through the respective PfH3core values and log2 was calculated (“unmappable” regions containing the value 0 were discarded). Bedgraph.gz files were generated using bedtools v2.20.1. Genome browser tracks were set to ‘mean’ as windowing function and ‘16’ as smoothing window function.
To investigate the correlation of PfH3.3 with the GC-content of the Plasmodium falciparum genome the genome was binned into 150 bp windows and the GC-content as well as PfH3.3 and PfH3core tags were calculated in each window. PfH3.3 tags were divided through PfH3core tags (windows containing the value 0 were discarded), log2 transformed and plotted over the GC-content as a density plot using the python package seaborn 0.6.0 in ipython notebook (Enthought Canopy 1.5.5.3123). For more detailed analysis the 150 bp windows were intersected with different chromatin domains e.g. heterochromatic or euchromatic regions (at least 50% overlap) using bedtools v2.20.1 and depicted as described above.
In order to investigate the correlation of PfH3.3 in upstream regions, downstream regions or coding sequences with mRNA abundance, the genome was subdivided as described85. In short, intergenic regions between convergent and divergent genes were split in half and each half was assigned to the nearest gene. Intergenic regions between genes with the same orientation were split in 3/4 and 1/4. The longer part was assigned to the upstream region of the nearest gene and the shorter part to the downstream region of the other gene. All upstream and downstream regions were shortened to a maximum of 3 kb as this covers the vast majority of upstream regions in full length, but excludes some extremely long intergenic regions. PfH3.3 and PfH3core tags were calculated within each upstream region, downstream region and coding sequence, normalized to the amount of reads per kb per million mapped reads (RPKM) and PfH3.3/PfH3core ratios (PfH3.3 occupancy) were determined. To assess RNA abundance of each gene, RNA-seq data were mapped against the annotated P. falciparum transcriptome, tags were counted for each transcript (excluding tRNAs, rRNAs, mitchochondrial and apicoplast RNA) and RPKM values were calculated. To illustrate the dynamics of RNA expression during intraerythrocytic development, the relative RNA abundance was calculated by dividing the amount of tags per transcript of each intraerythrocytic stage through the sum of tags per transcript of all four investigated intraerythrocytic stages (Heterochromatic genes and 25% of euchromatic genes with the lowest expression has been excluded from this analysis). Subsequently, RNAs were clustered into eight groups according to their expression pattern throughout the intraerythrocytic cycle using k-means clustering (Multi Experiment Viewer 4.9.0). Clusters were visualized in a heatmap. After calculating the relative PfH3.3 occupancy in upstream regions for all four intraerythrocytic stages as described above, the profile of PfH3.3 occupancy in upstream regions was matched to the respective RNA expression profile. The same method was applied to coding sequences and downstream regions.
To illustrate the relationship between PfH3.3 enrichment and gene expression level, RPKM values were log10 transformed and plotted against log2 transformed PfH3.3 occupancy levels of matching upstream region, downstream region and coding sequence.
Data Availability
ChIP-seq and RNA-seq datasets will be available via GEO (accession number GSE80466) and PlasmoDB.
Additional Information
How to cite this article: Fraschka, S. A.-K. et al. H3.3 demarcates GC-rich coding and subtelomeric regions and serves as potential memory mark for virulence gene expression in Plasmodium falciparum. Sci. Rep. 6, 31965; doi: 10.1038/srep31965 (2016).
Supplementary Material
Acknowledgments
We are very grateful to: i) Hendrik G. Stunnenberg for advice and support at an early stage of this project; ii) Moritz Treeck and Tim-Wolf Gilberger for transfecting the Ty1-PfH3.3 episomal construct to P. falciparum 3D7 parasites; iii) Sash Lopaticki and Alan Cowman for providing us with protocol and hands on experience for CSA selection of parasites; iv) Ron Dzikowski for providing us the NF54-DCJ parasite line and v) Andy Waters, Christine Clayton and David R. Westhead for fruitful advice throughout the project. We would also like to thank Wieteke Hoeijmakers and Christa Toenhake for lively discussions and critical reading of the manuscript. SF received her PhD fellowship from the European Community’s Seventh Framework Program [grant number FP7/2007–2013] under grant agreements No 242095 and No ParaMet 290080. This work also received support from The Netherlands Organization for Scientific Research [NWO-Vidi 864.11.007] and the National Institutes of Health [EuPathDB-Driving Biological Project, subaward no 553539].
Footnotes
Author Contributions S.A.-K.F. and R.B. designed and conceived the project. R.W.M.H. generated transgenic knock-down parasite lines and provided technical help. S.A.-K.F. with R.B. analyzed the data and wrote the manuscript.
References
- Ramachandran S. & Henikoff S. Replicating Nucleosomes. Sci Adv 1, 10.1126/sciadv.1500587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A., Paik J. & Verreault A. Regulation of histone synthesis and nucleosome assembly. Biochimie 87, 625–635, 10.1016/j.biochi.2005.02.008 (2005). [DOI] [PubMed] [Google Scholar]
- Melters D. P., Nye J., Zhao H. & Dalal Y. Chromatin Dynamics in vivo: A Game of Musical Chairs. Genes (Basel) 6, 751–776, 10.3390/genes6030751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Daujat S. & Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet 32, 42–56, 10.1016/j.tig.2015.10.007 (2016). [DOI] [PubMed] [Google Scholar]
- Ammar R. et al. Chromatin is an ancient innovation conserved between Archaea and Eukarya. Elife 1, e00078, 10.7554/eLife.00078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B. et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5, 7, 10.1186/1756-8935-5-7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg J. H. Evolution of histone H3: emergence of variants and conservation of post-translational modification sites. Biochem Cell Biol 90, 79–95, 10.1139/o11-036 (2012). [DOI] [PubMed] [Google Scholar]
- Elsaesser S. J., Goldberg A. D. & Allis C. D. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev 20, 110–117, 10.1016/j.gde.2010.01.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. et al. Genome-wide identification, evolutionary, and expression analyses of histone H3 variants in plants. Biomed Res Int 2015, 341598, 10.1155/2015/341598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S. J. & Black B. E. Centromeric chromatin and the pathway that drives its propagation. Biochim Biophys Acta 1819, 313–321 (2013). [PubMed] [Google Scholar]
- Talbert P. B. & Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11, 264–275, 10.1038/nrm2861 (2010). [DOI] [PubMed] [Google Scholar]
- McKinley K. L. & Cheeseman I. M. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17, 16–29, 10.1038/nrm.2015.5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers W. A. et al. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol 14, 1391–1401, 10.1111/j.1462-5822.2012.01803.x (2012). [DOI] [PubMed] [Google Scholar]
- Altheim B. A. & Schultz M. C. Histone modification governs the cell cycle regulation of a replication-independent chromatin assembly pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96, 1345–1350 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S. & Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol 10, 882–891, 10.1038/nsb996 (2003). [DOI] [PubMed] [Google Scholar]
- Szenker E., Ray-Gallet D. & Almouzni G. The double face of the histone variant H3.3. Cell Res 21, 421–434, 10.1038/cr.2011.14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B., Liu Y. & Gorovsky M. A. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol Cell Biol 26, 7719–7730, 10.1128/MCB.01139-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. P. et al. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 19, 1287–1292, 10.1038/nsmb.2439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer S. J. et al. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 491, 560–565, 10.1038/nature11608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G. & Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). [DOI] [PubMed] [Google Scholar]
- Ahmad K. & Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9, 1191–1200 (2002). [DOI] [PubMed] [Google Scholar]
- Shu H. et al. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol 15, R62, 10.1186/gb-2014-15-4-r62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H. et al. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci USA 109, 5370–5375, 10.1073/pnas.1203145109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. E. & Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19, 804–814, 10.1101/gad.1259805 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T. et al. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem 284, 12217–12225, 10.1074/jbc.M805651200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691, 10.1016/j.cell.2010.01.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P., Ouararhni K., Depaux A., Shuaib M. & Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 24, 1253–1265, 10.1101/gad.566910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A. et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 12, 853–862, 10.1038/ncb2089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H. et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res 19, 404–414, 10.1101/gr.084947.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S. B. et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA 102, 6344–6349, 10.1073/pnas.0502413102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M. et al. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res, 10.1093/nar/gkv847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C. W., Shibata Y., Starmer J., Yee D. & Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev 29, 1377–1392, 10.1101/gad.264150.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H. et al. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet 8, e1002658, 10.1371/journal.pgen.1002658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y. et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253, 10.1126/science.1248357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Schwartz B. E., Goldstein S. & Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol 19, 1816–1820, 10.1016/j.cub.2009.09.021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C. & Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21, 1519–1529, 10.1101/gad.1547707 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hödl M. & Basler K. Transcription in the absence of histone H3.3. Curr Biol 19, 1221–1226, 10.1016/j.cub.2009.05.048 (2009). [DOI] [PubMed] [Google Scholar]
- Couldrey C., Carlton M. B., Nolan P. M., Colledge W. H. & Evans M. J. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet 8, 2489–2495 (1999). [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231, 10.1038/nature10833 (2012). [DOI] [PubMed] [Google Scholar]
- Lan F. & Shi Y. Histone H3.3 and cancer: A potential reader connection. Proc Natl Acad Sci USA 112, 6814–6819, 10.1073/pnas.1418996111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers W. A., Stunnenberg H. G. & Bártfai R. Placing the Plasmodium falciparum epigenome on the map. Trends Parasitol 28, 486–495, 10.1016/j.pt.2012.08.006 (2012). [DOI] [PubMed] [Google Scholar]
- Miao J., Fan Q., Cui L. & Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369, 53–65, 10.1016/j.gene.2005.10.022 (2006). [DOI] [PubMed] [Google Scholar]
- Flueck C. et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog 5, e1000569, 10.1371/journal.ppat.1000569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio J. J., Mancio-Silva L. & Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5, 179–190, 10.1016/j.chom.2008.12.012 (2009). [DOI] [PubMed] [Google Scholar]
- Pérez-Toledo K. et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res 37, 2596–2606, 10.1093/nar/gkp115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511, 10.1038/nature01097 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártfai R. et al. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog 6, e1001223, 10.1371/journal.ppat.1001223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers W. A. et al. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol Microbiol 87, 1061–1073, 10.1111/mmi.12151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M. et al. H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum. Mol Microbiol 87, 1167–1182, 10.1111/mmi.12154 (2013). [DOI] [PubMed] [Google Scholar]
- Sullivan W. J. Histone H3 and H3.3 variants in the protozoan pathogens Plasmodium falciparum and Toxoplasma gondii. DNA Seq 14, 227–231 (2003). [DOI] [PubMed] [Google Scholar]
- Dzikowski R., Frank M. & Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog 2, e22, 10.1371/journal.ppat.0020022 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Lopez-Rubio J. J. & Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62, 445–470, 10.1146/annurev.micro.61.080706.093134 (2008). [DOI] [PubMed] [Google Scholar]
- Baruch D. I. et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82, 77–87 (1995). [DOI] [PubMed] [Google Scholar]
- Smith J. D. et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Z. et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 (1995). [DOI] [PubMed] [Google Scholar]
- Cortés A., Crowley V. M., Vaquero A. & Voss T. S. A view on the role of epigenetics in the biology of malaria parasites. PLoS Pathog 8, e1002943, 10.1371/journal.ppat.1002943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss T. S., Bozdech Z. & Bártfai R. Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol 20, 88–95, 10.1016/j.mib.2014.05.007 (2014). [DOI] [PubMed] [Google Scholar]
- Chookajorn T. et al. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA 104, 899–902, 10.1073/pnas.0609084103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp C., Li F., Howitt C. A., Chookajorn T. & Deitsch K. W. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15, 116–127, 10.1261/rna.1080109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit-Avraham I. et al. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA 112, E982–E991, 10.1073/pnas.1420855112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood M. S., Gannoun-Zaki L., Wellems T. E. & Deitsch K. W. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem 278, 34125–34132, 10.1074/jbc.M213065200 (2003). [DOI] [PubMed] [Google Scholar]
- Broadbent K. M. et al. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12, R56, 10.1186/gb-2011-12-6-r56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel T. N. et al. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics 15, 150, 10.1186/1471-2164-15-150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon H. P. & Wong L. H. New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res, 10.1093/nar/gkw012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. M. & Merrick C. J. G-quadruplexes in pathogens: a common route to virulence control? PLoS Pathog 11, e1004562, 10.1371/journal.ppat.1004562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. & Gray L. T. The G4 genome. PLoS Genet 9, e1003468, 10.1371/journal.pgen.1003468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S. et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20, 868–875, 10.1038/nsmb.2599 (2013). [DOI] [PubMed] [Google Scholar]
- Huang C. & Zhu B. H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin? Bioessays 36, 579–584, 10.1002/bies.201400005 (2014). [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell 9, 1091–1100 (2002). [DOI] [PubMed] [Google Scholar]
- DeNizio J. E., Elsässer S. J. & Black B. E. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res 42, 4318–4331, 10.1093/nar/gku090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio J. J. et al. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol 66, 1296–1305, 10.1111/j.1365-2958.2007.06009.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz J. et al. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol 40, 109–121, 10.1016/j.ijpara.2009.09.002 (2010). [DOI] [PubMed] [Google Scholar]
- Ng R. K. & Gurdon J. B. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci USA 102, 1957–1962, 10.1073/pnas.0409813102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R. K. & Gurdon J. B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10, 102–109, 10.1038/ncb1674 (2008). [DOI] [PubMed] [Google Scholar]
- Ng R. K. & Gurdon J. B. Epigenetic inheritance of cell differentiation status. Cell Cycle 7, 1173–1177 (2008). [DOI] [PubMed] [Google Scholar]
- Jin C. & Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci USA 103, 574–579, 10.1073/pnas.0509974103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J. G. & Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411, 10.1126/science.1134004 (2007). [DOI] [PubMed] [Google Scholar]
- Philip N. & Waters A. P. Conditional Degradation of Plasmodium Calcineurin Reveals Functions in Parasite Colonization of both Host and Vector. Cell Host Microbe 18, 122–131, 10.1016/j.chom.2015.05.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prommana P. et al. Inducible knockdown of Plasmodium gene expression using the glmS ribozyme. PLoS One 8, e73783, 10.1371/journal.pone.0073783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb B. S. et al. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol Biol 270, 263–276, 10.1385/1-59259-793-9:263 (2004). [DOI] [PubMed] [Google Scholar]
- Reed M. B., Saliba K. J., Caruana S. R., Kirk K. & Cowman A. F. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403, 906–909, 10.1038/35002615 (2000). [DOI] [PubMed] [Google Scholar]
- Brookman J. L. et al. An immunological analysis of Ty1 virus-like particle structure. Virology 207, 59–67, 10.1006/viro.1995.1051 (1995). [DOI] [PubMed] [Google Scholar]
- Noviyanti R. et al. Multiple var gene transcripts are expressed in Plasmodium falciparum infected erythrocytes selected for adhesion. Mol Biochem Parasitol 114, 227–237 (2001). [DOI] [PubMed] [Google Scholar]
- Salanti A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49, 179–191 (2003). [DOI] [PubMed] [Google Scholar]
- Kensche P. R. et al. The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences. Nucleic Acids Res, 10.1093/nar/gkv1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers W. A., Bártfai R., Françoijs K. J. & Stunnenberg H. G. Linear amplification for deep sequencing. Nat Protoc 6, 1026–1036, 10.1038/nprot.2011.345 (2011). [DOI] [PubMed] [Google Scholar]
- Hoeijmakers W. A., Bártfai R. & Stunnenberg H. G. Transcriptome analysis using RNA-Seq. Methods Mol Biol 923, 221–239, 10.1007/978-1-62703-026-7_15 (2013). [DOI] [PubMed] [Google Scholar]
- Perocchi F., Xu Z., Clauder-Münster S. & Steinmetz L. M. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res 35, e128, 10.1093/nar/gkm683 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq datasets will be available via GEO (accession number GSE80466) and PlasmoDB.