Abstract
N-terminal acetylation is a highly abundant protein modification catalyzed by N-terminal acetyltransferases (NATs) NatA-NatG. The Saccharomyces cerevisiae protein Arl3 depends on interaction with Sys1 for its localization to the Golgi and this targeting strictly requires NatC-mediated N-terminal acetylation of Arl3. We utilized the Arl3 acetylation-dependent localization phenotype as a model system for assessing the functional conservation and in vivo redundancy of several human NATs. The catalytic subunit of human NatC, hNaa30 (Mak3), restored Arl3 localization in the absence of yNaa30, but only in the presence of either yeast or human Naa35 subunit (Mak10). In contrast, hNaa35 was not able to replace its yeast orthologue without the co-expression of hNaa30, suggesting co-evolution of the two NatC subunits. The most recently discovered and organellar human NAT, NatF/Naa60, restored the Golgi localization of Arl3 in the absence of yNaa30. Interestingly, this was also true for hNaa60 lacking its membrane-binding domain whereas hNaa50 did not complement NatC function. This in vivo redundancy reflects NatC and NatF´s overlapping in vitro substrate specificities. The yeast model presented here provides a robust and rapid readout of NatC and NatF activity in vivo, and revealed evolutionary conservation of the NatC complex and redundancy between NatC and NatF.
N-terminal (Nt) acetylation is a prevalent protein modification catalyzed by N-terminal acetyltransferases (NATs). It involves the transfer of an acetyl moiety from acetyl-coenzyme A (Ac-CoA) to the α-amino group of the first amino acid residue of a polypeptide. This is mainly considered to be a co-translational event although evidence exist that Nt-acetylation also occurs post-translationally1,2.
In the yeast S. cerevisiae more than 50% of the soluble proteome is subjected to Nt-acetylation, whereas in humans this number exceeds 70% 3,4,5. There is an increasing number of substrates uncovered for which Nt-acetylation has significant functional roles, including global protein folding6, protein-protein interaction7, protein stabilization or degradation8,9,10 and subcellular protein targeting11,12,13.
Five NATs have been identified in yeast, NatA-NatE, all of which are conserved to multicellular eukaryotes, which additionally contain NatF4,14. NatG was recently identified in the plant kingdom15. NatA and NatD target Met-cleaved N-termini3,16,17,18 whereas NatB, NatC, NatE and NatF acetylate Met-starting N-termini4,5,16,19,20,21. NatG is found in chloroplasts of plant cells and its activity is towards Ala-, Met-, Ser-, and Thr-starting N-termini15. NatC, NatE and NatF display overlapping substrate specificities in vitro, targeting Met-hydrophobic and Met-amphipathic N-termini4,22,23, whereas their in vivo substrates have been proposed to be distinct5,24. For instance, NatF localizes to the cytosolic surface of the Golgi apparatus and specifically Nt-acetylates membrane proteins with their N-termini facing the cytosol5.
NatC is a complex consisting of the catalytic subunit Naa30 (Mak3) and the auxiliary subunits Naa35 (Mak10) and Naa38 (Mak31)22,25,26. In yeast, the individual deletions of NatC subunits cause growth defects on non-fermentable carbon sources (e.g. glycerol) at elevated temperatures25. NatC knockdown in human cells results in reduced cell viability and p53-dependent apoptosis22. One of the verified NatC substrates is the yeast ADP-ribosylation factor-like 3 (Arl3) protein whose interaction with Golgi-anchored Sys1 strictly depends on NatC-mediated Nt-acetylation. In the absence of either Naa30 or Naa35, Arl3 remains unmodified and as a consequence is unable to associate with the Golgi12,13.
Although NatC has been deemed evolutionarily conserved in several aspects - such as in vitro substrate specificity, ribosome association and subunit composition22,27 - its conservation in terms of in vivo protein function remains unknown. In this study, we applied the yeast Arl3 localization phenotype as an in vivo system to study the functional conservation between yeast and human NatC. We found that the human Naa30-Naa35 NatC complex could functionally complement yeast lacking NatC activity, as indicated by its ability to restore the punctate Golgi localization of Arl3. Moreover, the possible NatC-redundancy of NatE and NatF was challenged in the Arl3 complementation assay to enlighten a potential in vivo redundancy. The organelle associated human NatF/Naa60, but not NatE/Naa50, was able to restore normal Arl3 distribution pattern in yeast. Surprisingly, also a truncated cytosolic hNaa60 complemented the Arl3 localization phenotype in the absence of active NatC complex. These results enlightened a functional difference in vivo among NatC, NatE and NatF, which are known to have partially overlapping in vitro substrate specificities.
Results and Discussion
Functional conservation of Naa30 revealed by Arl3 localization complementation assay
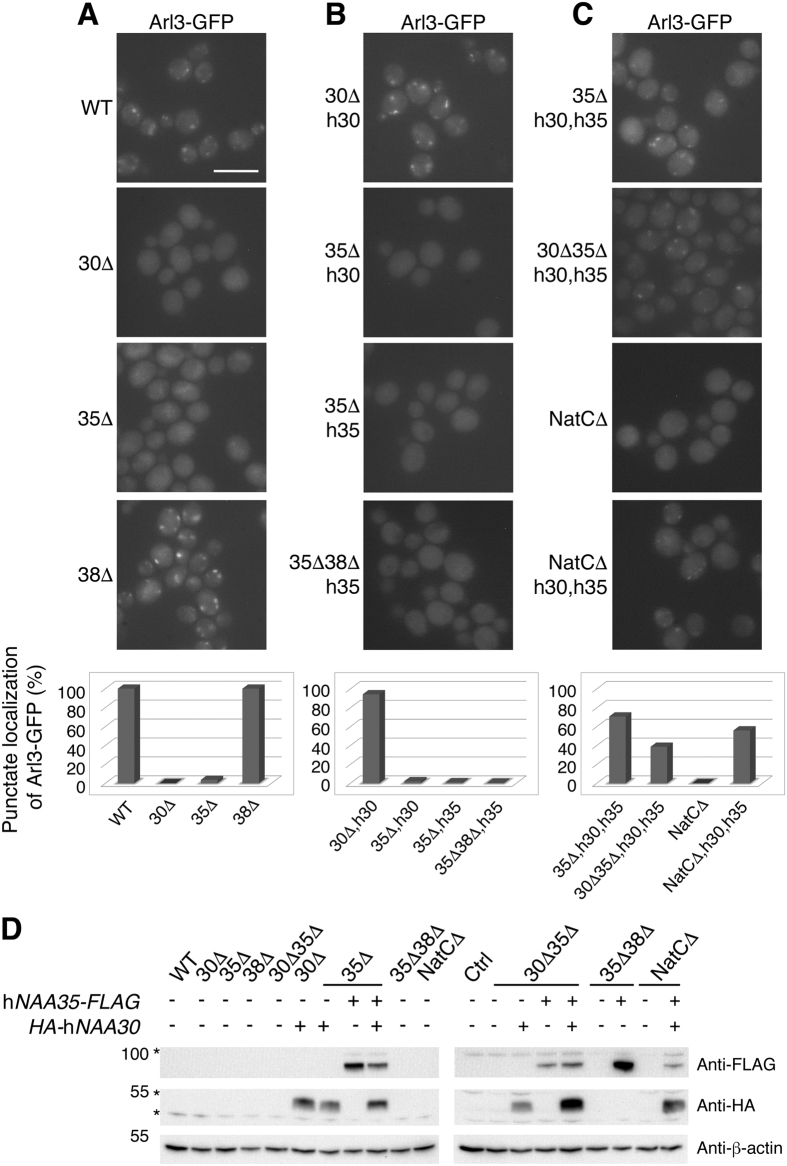
The dependency of NatC-mediated Nt-acetylation of Arl3 for correct subcellular localization has previously been shown by fluorescence microscopy and mass spectrometry12,13. Here, we utilized the Arl3 localization as a model for NatC activity in vivo (Fig. 1). In WT cells, Arl3-GFP localized to the Golgi whereas in naa30Δ and naa35Δ cells the punctate localization was lost (Fig. 1A), as previously reported12,13. Yeast Naa38 was shown to be dispensable for Nt-acetylation of Arl3 by mass spectrometry12 and we accordingly observed the subcellular localization of naa38Δ yeast to be similar to the WT (Fig. 1A).
Figure 1. Arl3-GFP punctate localization as a model for active NatC revealed functional conservation of the NatC complex.
(A–C) Live-cell microscopy of endogenous Arl3-GFP in yeast cells with the indicated genotypes. A corresponding bar graph below the fluorescent images shows the number of cells (n > 500 cells) harbouring a punctate localization pattern of Arl3-GFP. Scale bar, 5 μm. (A) WT, naa30Δ, naa35Δ and naa38Δ yeast. (B) Human Naa30 exogenously expressed in naa30Δ and naa35Δ cells, whereas hNaa35 was exogenously expressed in naa35Δ and naa35Δnaa38Δ yeast. (C) Exogenous expression of both human Naa30 and Naa35 in naa35Δ, naa30Δnaa35Δ and NatCΔ cells. (D) Western blot analysis using anti-HA and anti-FLAG antibody verified the exogenous expression of hNaa30 and hNaa35, respectively, in different yeast strains. Asterisks represent undetermined band detected by anti-FLAG or anti-HA (Sigma-Aldrich) antibodies. Abbreviations: 30Δ, naa30Δ; 35Δ, naa35Δ; 38Δ, naa38Δ; NatCΔ, naa30Δnaa35Δnaa38Δ; h30, HA-hNaa30; h35, hNaa35-FLAG.
Next, we used the Arl3 localization model in various rescue assays and found the human orthologue of yNaa30 to be functionally conserved, as it was able to restore the mislocalization of Arl3-GFP in naa30Δ yeast (Fig. 1B). Previously, hNaa30 was reported to restore subcellular localization of Imh1, downstream of Arl3 function13. To investigate whether the complementation ability of hNaa30 was due to the catalytic subunit performing Nt-acetylation on its own or in a human-yeast interspecies complex with the other essential NatC subunit, yNaa35, we expressed HA-hNaa30 (hereafter referred to as hNaa30) in naa35Δ cells. In this yeast background, exogenously expressed hNaa30 failed to restore Arl3 localization (Fig. 1B), thus suggesting interspecies NatC functionality of hNaa30-yNaa35.
Human Naa35 is unable to complement yeast Naa35 for Arl3 Nt-acetylation
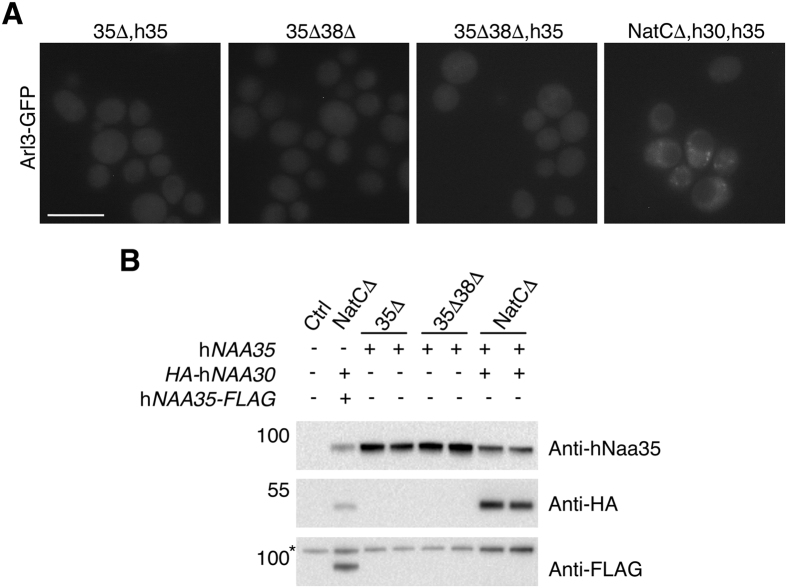
The functional conservation of hNaa35 was then addressed by expressing hNaa35-FLAG (hereafter referred to as hNaa35) in naa35Δ cells, but this did not restore Arl3 localization. To exclude the possibility of yNaa38 hindering a necessary binding between hNaa35 and yNaa30 we further expressed hNaa35 in naa35Δnaa38Δ yeast. No rescue of Arl3 localization was seen in this condition either (Fig. 1B). Additionally, we performed the complementation assay using hNaa35 without the C-terminal FLAG-tag, which was also negative in terms of restoring Arl3 punctate localization (Fig. 2).
Figure 2. Untagged human Naa35 behaved similarly to hNaa35-FLAG.
(A) Fluorescence microscopy of Arl3-GFP in different yeast strains. Untagged hNaa35 was exogenously expressed in naa35Δ and naa35Δnaa38Δ cells as well as together with hNaa30 in NatCΔ yeast. The Arl3 mislocalization phenotype was demonstrated in naa35Δnaa38Δ cells. Scale bar, 5 μm. (B) Western blot analysis of cell lysates revealed the presence of hNaa30 and hNaa35 (untagged) in the three yeast strains, represented by two individual clones. Asterisk indicates an undetermined band achieved by anti-FLAG. Abbreviations: 35Δ, naa35Δ; 38Δ, naa38Δ; NatCΔ, naa30Δnaa35Δnaa38Δ; h30, HA-hNaa30; h35, hNaa35.
Taken together, these data demonstrate that hNaa30, but not hNaa35, can replace the corresponding yeast orthologue. Whereas hNaa30 functions in the presence of yNaa35, hNaa35 cannot function despite the presence of yNaa30, thus suggesting the formation of a functional hNaa30-yNaa35 interspecies complex, but not yNaa30-hNaa35.
Human Naa30 and Naa35 cooperate to perform NatC activity in yeast
Based on the above results showing that co-expression of hNaa30 and yNaa35 restore NatC function and the inability of yNaa30 and hNaa35 to do so, we proceeded by studying both human NatC subunits in yeast. Human Naa30 and Naa35 were exogenously co-expressed in three different yeast strains: naa35Δ, naa30Δnaa35Δ, and naa30Δnaa35Δnaa38Δ (NatCΔ) (Fig. 1D). For all deletion strains, a clear rescue of Arl3 localization was observed, indicating that NatC activity was present (Fig. 1C). Thus, the two human subunits Naa30 and Naa35 cooperated and performed NatC activity on a yeast substrate protein in vivo. From this, we suggest a functional conservation of the Naa30-Naa35 NatC complex between S. cerevisiae and humans, and that there has been a co-evolution of these subunits since heterologous expression of yNaa30 and hNaa35 is non-functional. Our observation that human Naa30-Naa35 NatC complex is functionally active in yeast and complements yNatC, is analogous to earlier complementation studies of NatA and NatB3,21, and suggests a large degree of conservation of these major NATs from yeast to humans. However, the yeast/human Naa30 complementation observed here, differs from those of NatA and NatB studies in which the human catalytic subunits alone (Naa10 and Naa20, respectively) cannot phenotypically complement a yeast strain lacking its yeast orthologue3,21.
The punctate localization pattern of Arl3 corresponds to Golgi structures
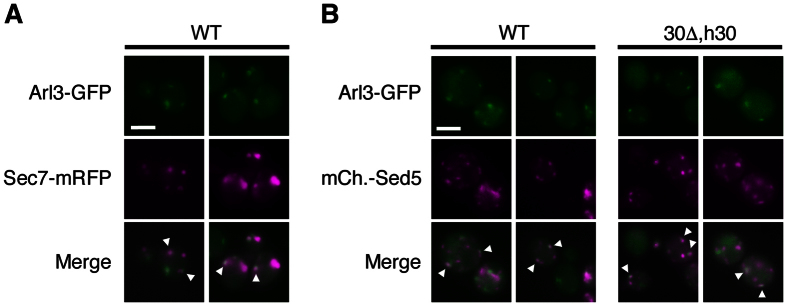
To verify that the observed localization pattern of Arl3 in the rescue assays did indeed represent restoration of the Arl3 WT localization, the trans-Golgi Sec7-mRFP and cis-Golgi mCherry-Sed5 marker proteins were individually expressed in Arl3-GFP cells. Both Golgi markers partially co-localized with Arl3-GFP in the WT, apparently with equal amount of overlap (Fig. 3). We further examined the restored punctate Arl3-GFP localization in naa30Δ cells expressing hNaa30 and detected similar co-localization with mCherry-Sed5 as for the WT yeast (Fig. 3B). This confirmed that the reappeared punctate structures detected represents restored Arl3 Golgi localization.
Figure 3. Partial co-localization was observed between Arl3 and two individual Golgi marker proteins.
(A) Imaging of live WT yeast expressing endogenous Arl3-GFP and exogenous Sec7-mRFP. (B) Endogenous Arl3-GFP and exogenous mCherry-Sed5 were imaged in live WT cells and in naa30Δ cells exogenously expressing HA-hNaa30. Scale bars, 2 μm. Abbreviations: 30Δ, naa30Δ; h30, HA-hNaa30.
Overlapping in vivo activities of human NatC and NatF in yeast
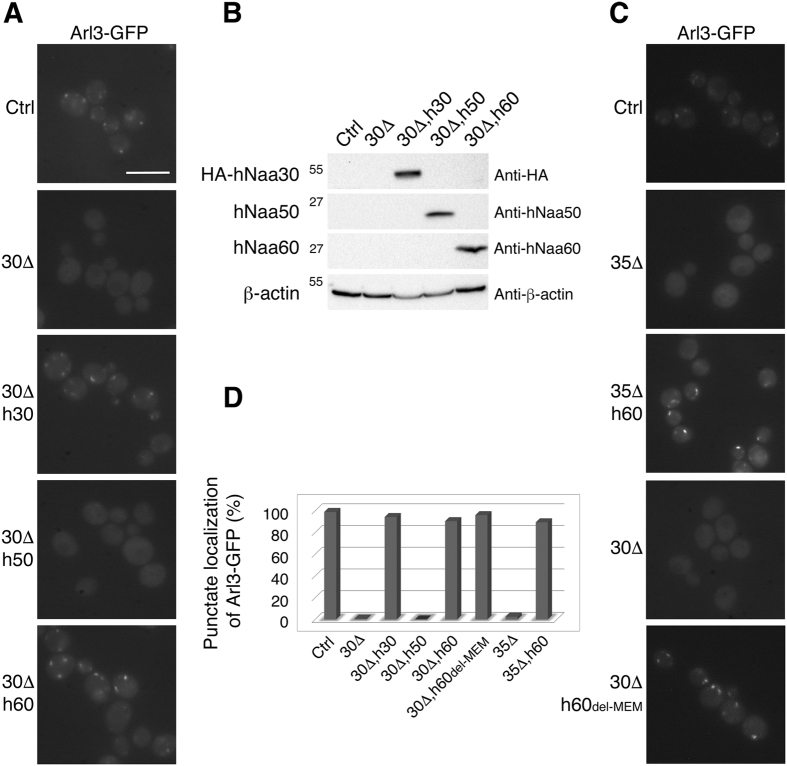
Human NatC, NatE and NatF display overlapping in vitro substrate specificities4,22,23. Furthermore, human NatE and NatF exhibit overlapping in vivo substrate profiles when expressed in yeast4,24. All three enzymes recognize Met-starting N-termini containing a hydrophobic or amphipathic amino acid residue in the second position. Arl3 has a Met-Phe-starting N-terminus and thus may be a putative target of these enzymes. To challenge the suggested substrate redundancy between human NatC, NatE and NatF in vivo, we again utilized the Arl3 complementation model (Fig. 4). Here, the human catalytic subunits Naa30, Naa50 and Naa60 were individually expressed in Arl3-GFP naa30Δ cells. Human Naa50, which in human cells is cytosolic and partially ribosome-associated23,28, was not able to complement yNaa30 whereas hNaa60 almost completely reinstated the punctate Arl3 localization, similar to hNaa30 (Fig. 4A). The same was true for hNaa60 expressed in naa35Δ cells (Fig. 4C), indicating that hNaa60 may operate independently of any of the yNatC subunits, unlike hNaa30, which requires the assumed ribosome binding subunit Naa35.
Figure 4. Human Naa60 has in vivo redundancy with Naa30 that is independent of its organellar localization.
(A) Live-cell microscopy of Arl3-GFP in yeast genetically modified as indicated. Restoration of Arl3-GFP localization by hNaa30 is shown as a positive control. (A,C) Scale bar, 5 μm. (B) Western blot analysis of cell lysates verified the expression of exogenous human Naa30, Naa50 or Naa60 in yeast strains shown in (A). (C) Detection of endogenous Arl3-GFP in live yeast. Human Naa60 and a C-terminally truncated variant Naa601−184 (h60del-MEM) were exogenously expressed in naa35Δ and naa30Δ cells, respectively. (D) Amount of cells for which the Arl3-GFP punctate localization was observed in the indicated yeast strains. The number from naa30Δ yeast expressing hNaa30 is a re-representation of data in Fig. 1B as a positive control reference (n > 500 cells). Abbreviations: 30Δ, naa30Δ; 35Δ, naa35Δ; h30, HA-hNaa30; h50, hNaa50; h60, hNaa60; h60del-MEM, hNaa60 without membrane binding region.
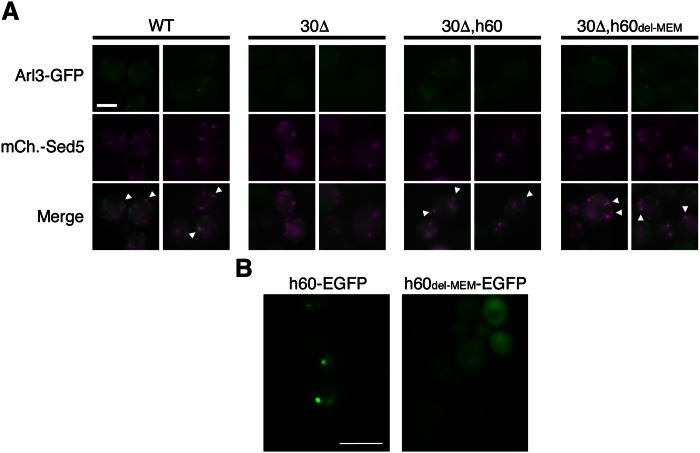
Naa60 was recently described in human cells as the first organellar NAT and its catalytic activity is facing towards the cytosol5. It might be possible that the ability of human Naa60 to complement the NatC activity on Arl3 relates to Naa60´s association with the yeast Golgi membrane. However, upon expression of truncated Naa60del-MEM, without the C-terminal end that mediates Naa60 membrane-association in mammalian cells, (ref. 5), the Arl3 localization pattern was again represented as punctate structures that partially co-localized with the cis-Golgi marker Sed5 (Figs 4C and 5A). Human Naa60 and Naa60del-MEM fused to a C-terminal EGFP tag mainly localized to punctate structures and the cytosol, respectively (Fig. 5B). Taken together these data may indicate a post-translational activity of NatF/Naa60 towards Arl3-GFP at the Golgi or freely in the cytosol. In any case, the data verify Met-Phe as an in vivo N-terminal target sequence of NatF and demonstrate the ability of human Naa30 and Naa60 to act on the same substrate in vivo4.
Figure 5. Human Naa60-rescued Arl3 partially co-localizes with the cis-Golgi marker Sed5.
Yeast cells of the indicated genotypes were imaged for the detection of endogenous Arl3-GFP and exogenous mCherry-Sed5. Scale bar, 2 μm. (B) Subcellular localization of C-terminally EGFP-tagged hNaa60 and hNaa60del-MEM in live yeast cells. Scale bar, 5 μm. Abbreviations: 30Δ, naa30Δ; h60, hNaa60; h60del-MEM, hNaa60 without membrane binding region.
From these results, it is clear that human Naa60 and Naa60del-MEM, but not hNaa50, are able to rescue the Arl3 localization phenotype, thus enlightening an interesting Nt-acetylation performance of NatF as well as functional redundancy with NatC. Moreover, human Naa50 acts as a negative control in the assay for Arl3 localization, excluding mere NAT overexpression to be sufficient for phenotype rescue.
Conclusions
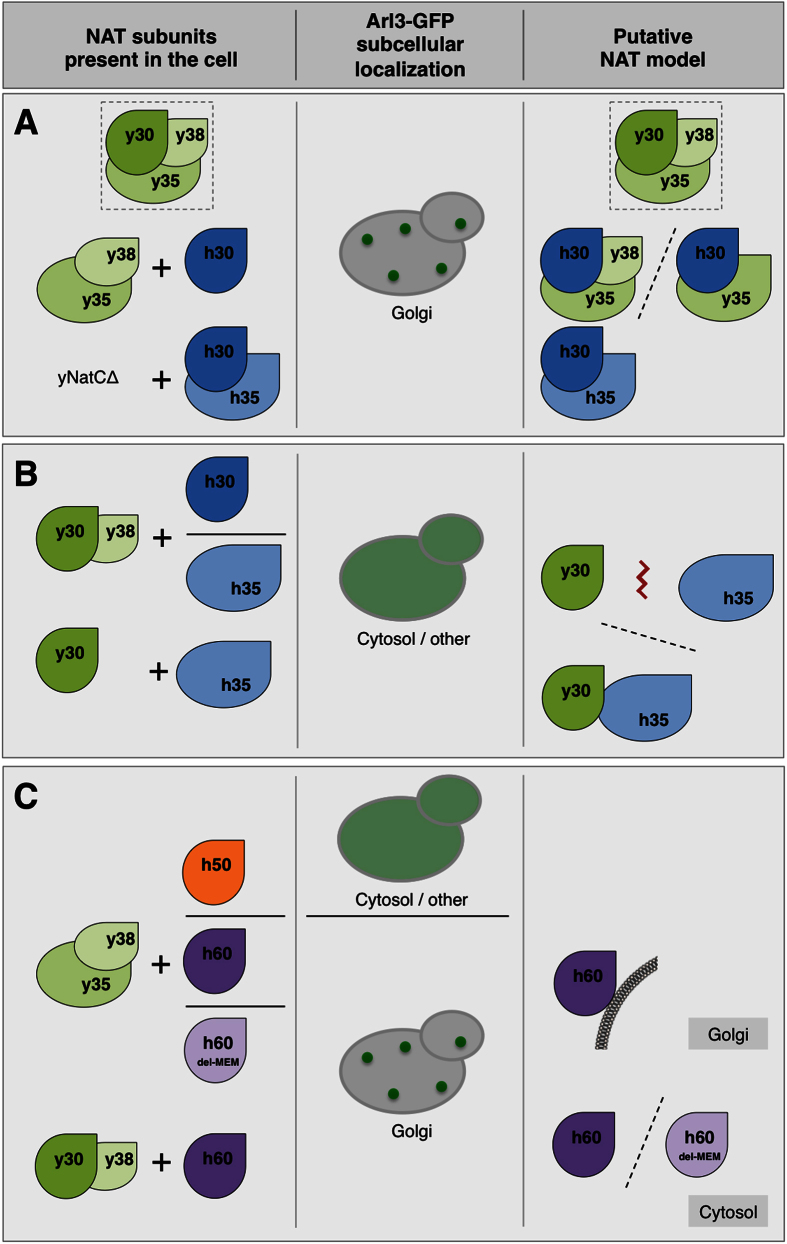
We applied S. cerevisiae as a model system to study the in vivo function of human NATs. The NatC-dependent subcellular localization of yeast Arl3 proved to be a suitable model for studying functional conservation among NatC orthologues as well as diverging in vivo function among NatC, NatE and NatF harbouring overlapping in vitro substrate specificities (Fig. 6). Naa30 was defined as functionally conserved between yeast and humans through its ability to complement Arl3 mislocalization, which interestingly required the presence of either yeast or human Naa35; accordingly we propose that NatC complex formation occurred (Fig. 6A). Human Naa35 on the other hand, could not complement its yeast orthologue and we suggest this to be due to insufficient binding between yNaa30 and hNaa35 or formation of an impaired complex (Fig. 6B). Taken together, these results indicate that the two subunits Naa30 and Naa35 have co-evolved.
Figure 6. Putative model of the NatC, NatE and NatF subunits and their effect on Arl3 localization.
In WT yeast cells Arl3 is N-terminally acetylated by the NatC complex (Naa30 and Naa35, whereas Naa38 is dispensable for Arl3-acetylation) and targeted to the Golgi transmembrane protein Sys1, mediating Arl3 punctate Golgi localization. However, in the absence of a functional NatC complex Arl3 remains unacetylated after protein synthesis and the targeting is abrogated, leading to Arl3 cytosolic mislocalization12,13. (A) Human Naa30 is able to function in complex with yNaa35 (and possibly yNaa38) or hNaa35, indicating a functionally conserved catalytic subunit between yeast and humans. WT yNatC is depicted framed in dotted line. (B) No Nt-acetylation activity of hNaa30 is observed in the absence of yeast or human Naa35, which is thought to be the ribosome-binding subunit. Human Naa35 cannot replace its yeast orthologue and function together with yNaa30, neither in the presence nor absence of yNaa38. (C) Naa30 and Naa60 exhibit in vivo redundancy. The Arl3 localization phenotype is complemented by human Naa60 and Naa60del-MEM in both naa30Δ and naa35Δ yeast. Based on the subcellular localizations of human Naa60 and Naa60del-MEM, a possible post-translational NAT activity at the cytosolic side of Golgi or freely in the cytosol is suggested. Abbreviations: y30, yNaa30; y35, yNaa35; y38, yNaa38; h30, hNaa30; h35, hNaa35; h50, hNaa50; h60, hNaa60; h60del-MEM, hNaa60 without membrane binding region; Ac-CoA, acetyl-coenzyme A.
Despite human Naa30, Naa50 and Naa60’s shared catalytic activity towards specific N-terminal sequences, we found opposite complementation patterns of NatE/Naa50 and NatF/Naa60 for the NatC-deficient phenotype of Arl3. This clearly hints to unique in vivo functions of the two. Furthermore, in light of the different subcellular localizations of Naa60 and Naa60del-MEM in human cells5 and in yeast cells, both being able to rescue the Arl3 localization (Fig. 6C), the difference between Naa50 and Naa60 may reflect differences in the substrate binding site rather than differences in subcellular localization. This assumption is also based on the crystal structures and catalytic efficiencies of human Naa5029 and Naa6030 suggesting that Naa60 may accommodate larger and more bulky substrate side chains as compared to Naa50.
The yeast model presented here may act as a useful and rapid in vivo setup to assess the functionality of various NAT subunits, meaning isoforms of human Naa30, Naa35 and Naa60. For a Naa10 mutant causing the Ogden syndrome31,32, a yeast model provided useful insights into the impaired functionality of NatA33 Similarly, the yeast model described here may be used in any future studies of pathological mutants of NatC or NatF.
Methods
Yeast Cultivation
The yeast strains used in this study (see Supplementary Table S1) are derived from one of the two parental strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). According to their genotype and required selection, the cells were either grown in YPD (Sigma-Aldrich, YPD Broth #Y1375 and YPD Agar #Y1500) or appropriate synthetic defined dropout media (Sunrise Science Products, SD-URA #1703-500, SD-LEU #1707-500, SD-LEU-URA #1721-500, Agar #1910-500). Protein expression of mCherry-Sed5 under the control of the MET25 promoter required growth medium with maximum 10 μg/ml methionine.
Plasmid construction
A record of all plasmids and primers used is found in Supplementary Table S2 and Table S3, respectively. Human NAA30 (Gene ID 122830) was sub-cloned from pRS315-HA-hMAK3, which was a kind gift from Professor Sean Munro, MRC, Cambridge13, into the pBEVY-U yeast vector34 downstream of the ADH1 promoter using restriction enzyme sites XmaI (5´) and EcoRI (3´). The N-terminal HA-tag was retained from the original gene construct.
Human NAA35 (Gene ID 60560) was amplified from pCMV6-AC-NAA35-GFP (OriGene, #RG213022) with the insertion of XbaI (5´) and PstI (3´) restriction enzyme sites as well as a C-terminal FLAG-tag and ligated into pBEVY-L after the GPD promoter. Additionally, pBEVY-L-hNAA35 was generated by introducing a stop codon downstream of the NAA35 ORF by mutagenesis (Agilent Technologies, #210514).
A plasmid containing human Naa601-184 (Naa60del-MEM) was generated by mutagenesis of pBEVY-U-hNAA60 4, introducing a stop codon after the codon representing amino acid 184 of Naa60. Moreover, pBEVY-U-hNAA60-EGFP and pBEVY-U-hNAA601−182-EGFP (Naa60del-MEM) were generated for subcellular localization studies by subcloning from mammalian pEGFP-N1 vectors in which the respective NAA60 sequences were inserted between NheI and KpnI sites. The C-terminally EGFP-tagged NAA60 constructs were inserted between XbaI (5´) and SalI (3´) restriction enzymes sites in the pBEVY-U vector after the GPD promoter.
Plasmid p415MET25-mRFP-SED5 was kindly provided by Professor Blanche Schwappach, UMG, Göttingen35 and pRS316-SEC7-mRFP was from RIKEN BRC DNA BANK (#RDB08663)36.
Yeast transformation
Yeast transformation was based on a method developed by Gietz and Schiestl37. Exponentially growing yeast cells (10 ml culture, OD600 1.0) were harvested, washed twice with water and resuspended in 240 μl 50% PEG-3350 before addition of 36 μl 1 M lithium acetate, 100 μg salmon sperm ssDNA, DNA template (10 μg gene deletion cassette or 5 μg plasmid), and water up to 360 μl. After a 40 min heat-shock at 42 °C the cells were washed four times with water, before plating or incubation in YPD, depending on selection.
Yeast gene deletions were performed by homologous recombination and a complete overview of the primers used to amplify the respective deletion cassettes as well as the PCR-based screen for positive mutants are listed in Supplementary Table S3.
Protein extraction and Western blot analysis
Protein extraction was done according to Kushnirov38 with modifications. Exponentially growing yeast cells (4 ml yeast culture, OD600 1.0) were harvested for 10 min at 5,000 × g. The pellet was washed twice with water, resuspended in 200 μl water before adding 200 μl 0.2 M NaOH for 5 min. Cells were harvested for 1 min at 12,000 × g, resuspended and boiled for 3 min in 100 μl 1x Sample buffer (10% glycerol, 2% SDS, 50 mM Tris-HCl, pH 6.8, 0.0025% bromophenol blue, 50 mM dithiothreitol), before pelleting cell debris. Typically, 6 μl supernatant was loaded for SDS-PAGE on 4–20% Mini-PROTEAN® gels (Bio-Rad, #456–8093).
Exogenous expression of HA-hNaa30 and hNaa35-FLAG were verified by Western blotting using anti-HA (1:400, rabbit (Rb), polyclonal, Sigma-Aldrich, #H6908) or anti-HA (1:1000, Rb, polyclonal, Abcam, #ab9110) and anti-FLAG (1:1000, Ms, M2 monoclonal, Sigma-Aldrich, #F1804) or anti-hNaa35 (1:1000, Rb, polyclonal, Sigma-Aldrich, #HPA021547), respectively. Beta-actin served as loading control and was detected by anti-β-actin (1:4000, Ms, monoclonal, Abcam, #8224). Additionally, protein expression of human Naa50 and Naa60 were verified by anti-hNaa50 (1:500, Rb, polyclonal, custom made raised against a peptide representing the amino acids 150–163 of hNaa50, BioGenes) and anti-hNaa60 (1:500, Rb, polyclonal, custom made raised against a protein representing the amino acids 1–184 of hNaa60, BioGenes), respectively.
Fluorescence microscopy
Cells in exponential growth phase (OD600 0.8–1.2) were pelleted and washed three times in 1x PBS, 2% glucose. Fluorescence images were acquired in the washing solution on a Leica DMI6000 B widefield microscope as previously described39. Recorded images were processed using Photoshop CS5 (Adobe Systems, San Jose, CA, USA). Arl3-GFP subcellular localization was characterized as normal (punctate) or mislocalization (other). More than 500 cells were counted per strain.
Additional Information
How to cite this article: Osberg, C. et al. Microscopy-based Saccharomyces cerevisiae complementation model reveals functional conservation and redundancy of N-terminal acetyltransferases. Sci. Rep. 6, 31627; doi: 10.1038/srep31627 (2016).
Supplementary Material
Acknowledgments
We thank N. Glomnes for technical assistance and Professor S. Munro and Professor B. Schwappach for kindly providing plasmids. This work was supported by the Research Council of Norway (grants 197136 and 230865 to TA), the Western Norway Regional Health Authority (grant 911883 to TA and grant 911761 to CO), the Norwegian Cancer Society (to TA), and the Bergen Research Foundation (to TA).
Footnotes
The authors declare no competing financial interests.
Author Contributions C.O. wrote the manuscript, generated most of the plasmids and yeast strains for this study, conceived and performed most of the experimental work; H.A. generated plasmids and yeast strains, supervised the project and worked on the manuscript; S.N. generated yeast strains and performed fluorescence microscopy; M.M. generated Figures 3 and 5A based on data from C.O. and worked on the manuscript; T.A. initiated, conceived and supervised the project as well as prepared the manuscript.
References
- Helsens K. et al. Bioinformatics analysis of a Saccharomyces cerevisiae N-terminal proteome provides evidence of alternative translation initiation and post-translational N-terminal acetylation. J Proteome Res. 10, 3578–3589 (2011). [DOI] [PubMed] [Google Scholar]
- Varland S., Osberg C. & Arnesen T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T. et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA 106, 8157–8162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P. et al. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. Plos Genet 7, e1002169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksnes H. et al. An organellar Nα-acetyltransferase, Naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep. 10, 1362–1374 (2015). [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Mannakee B. K., Gutenkunst R. N. & Serio T. R. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 5, 4383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. C., Monda J. K., Bennett E. J., Harper J. W. & Schulman B. A. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C. S., Shemorry A. & Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K. et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemorry A., Hwang C. S. & Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Molecular cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A. & Hopper A. K. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: roles for N-acetylation and an integral membrane protein. Genetics 170, 1553–1560 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S. R., Strochlic T. I., Tong A. H., Boone C. & Burd C. G. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 6, 414–419 (2004). [DOI] [PubMed] [Google Scholar]
- Behnia R., Panic B., Whyte J. R. & Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 6, 405–413 (2004). [DOI] [PubMed] [Google Scholar]
- Aksnes H., Hole K. & Arnesen T. Molecular, cellular, and physiological significance of N-terminal acetylation. Int Rev Cell Mol Biol. 316, 267–305 (2015). [DOI] [PubMed] [Google Scholar]
- Dinh T. V. et al. Molecular identification and functional characterization of the first Nalpha-acetyltransferase in plastids by global acetylome profiling. Proteomics 15, 2426–2435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Norbeck J., Takakura H., Blomberg A. & Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J 18, 6155–6168 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole K. et al. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. Plos One 6, e24713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song O. K., Wang X. R., Waterborg J. H. & Sternglanz R. An N-alpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. Journal of Biological Chemistry 278, 38109–38112 (2003). [DOI] [PubMed] [Google Scholar]
- Evjenth R. et al. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J Biol Chem. 284, 31122–31129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. C. & Wickner R. B. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J Biol Chem. 267, 20277–20281 (1992). [PubMed] [Google Scholar]
- Van Damme P. et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. P Natl Acad Sci USA 109, 12449–12454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starheim K. K. et al. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol. 29, 3569–3581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P. et al. Proteome-derived Peptide Libraries Allow Detailed Analysis of the Substrate Specificities of N-alpha-acetyltransferases and Point to hNaa10p as the Post-translational Actin N-alpha-acetyltransferase. Molecular & Cellular Proteomics 10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P., Hole K., Gevaert K. & Arnesen T. N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics 15, 2436–2446 (2015). [DOI] [PubMed] [Google Scholar]
- Polevoda B. & Sherman F. NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J Biol Chem. 276, 20154–20159 (2001). [DOI] [PubMed] [Google Scholar]
- Rigaut G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 17, 1030–1032 (1999). [DOI] [PubMed] [Google Scholar]
- Polevoda B., Brown S., Cardillo T. S., Rigby S. & Sherman F. Yeast N-alpha-terminal acetyltransferases are associated with ribosomes. Journal of Cellular Biochemistry 103, 492–508 (2008). [DOI] [PubMed] [Google Scholar]
- Arnesen T. et al. Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein N-alpha-acetyltransferase complex. Gene 371, 291–295 (2006). [DOI] [PubMed] [Google Scholar]
- Liszczak G., Arnesen T. & Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J Biol Chem. 286, 37002–37010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Støve S. I. et al. Crystal structure of the Golgi-associated human Nα-acetyltransferase 60 reveals the molecular determinants for substrate-specific acetylation. Structure 24, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rope A. F. et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet 89, 28–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myklebust L. M. et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Human molecular genetics 24, 1956–1976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P., Stove S. I., Glomnes N., Gevaert K. & Arnesen T. A Saccharomyces cerevisiae model reveals in vivo functional impairment of the Ogden syndrome N-terminal acetyltransferase NAA10 Ser37Pro mutant. Mol Cell Proteomics 13, 2031–2041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Martinat M. A. & Hyman L. E. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Research 26, 3577–3583 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K. et al. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. Journal of Cell Science 126, 473–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Okamoto M. & Nakano A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat Commun 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D. & Schiestl R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 (2000). [DOI] [PubMed] [Google Scholar]
- Aksnes H., Osberg C. & Arnesen T. N-terminal acetylation by NatC is not a general determinant for substrate subcellular localization in Saccharomyces cerevisiae. PLoS One 8, e61012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.