Abstract
Lithium is the mainstay prophylactic treatment for bipolar disorder (BD), but treatment response varies considerably across individuals. Patients who respond well to lithium treatment might represent a relatively homogeneous subtype of this genetically and phenotypically diverse disorder. Here, we performed genome-wide association studies (GWAS) to identify (i) specific genetic variations influencing lithium response and (ii) genetic variants associated with risk for lithium-responsive BD. Patients with BD and controls were recruited from Sweden and the United Kingdom. GWAS were performed on 2698 patients with subjectively defined (self-reported) lithium response and 1176 patients with objectively defined (clinically documented) lithium response. We next conducted GWAS comparing lithium responders with healthy controls (1639 subjective responders and 8899 controls; 323 objective responders and 6684 controls). Meta-analyses of Swedish and UK results revealed no significant associations with lithium response within the bipolar subjects. However, when comparing lithium-responsive patients with controls, two imputed markers attained genome-wide significant associations, among which one was validated in confirmatory genotyping (rs116323614, P=2.74 × 10−8). It is an intronic single-nucleotide polymorphism (SNP) on chromosome 2q31.2 in the gene SEC14 and spectrin domains 1 (SESTD1), which encodes a protein involved in regulation of phospholipids. Phospholipids have been strongly implicated as lithium treatment targets. Furthermore, we estimated the proportion of variance for lithium-responsive BD explained by common variants (‘SNP heritability') as 0.25 and 0.29 using two definitions of lithium response. Our results revealed a genetic variant in SESTD1 associated with risk for lithium-responsive BD, suggesting that the understanding of BD etiology could be furthered by focusing on this subtype of BD.
Introduction
Bipolar disorder (BD) is a serious psychiatric illness characterized by recurrent episodes of depression and mania.1 The mood stabilizer lithium effectively reduces episode recurrences and remains a first-line option for maintenance treatment of BD.2 However, treatment responses vary considerably across individuals and a number of persons with BD show only partial or no response.3, 4 An understanding of which patients with BD are most likely to respond to lithium would enable tailored treatment.
Genetic factors may mediate variation in response to lithium, and several genetic association studies have been performed to assess this possibility. Most have been candidate gene studies based on putative pharmacological mechanisms of lithium or the pathophysiology of BD.5 Although a minority of these findings have been observed in more than one study (for example, the CREB family of genes6, 7 and synapse-related genes8, 9), the majority of reported associations have not been reproduced. For example, studies of single-nucleotide polymorphisms (SNPs) in the brain-derived neurotrophic factor (BDNF) gene have reported different findings between ethnicities.10, 11, 12, 13 Also, two Japanese groups have examined the X-box binding protein 1 gene and found that -116C allele carriers seem to have better lithium response, but each was in a small number of patients (N<70).14, 15 Small sample sizes in conjunction with the limited understanding of not only the pathophysiology of BD but also lithium's pharmacodynamics to guide candidate gene selection are likely reasons for these equivocal findings.
Hypothesis-free genome-wide association studies (GWAS) of larger samples therefore offer a promising approach. One GWAS of lithium response performed in a Han Chinese population showed a strong association with variants in the gene GADL1 (glutamate decarboxylase-like protein 1),16 but not only are the susceptibility alleles rare in European populations, the findings were not replicated in other Asian samples.17 Other GWAS of lithium response found no SNPs that reached genome-wide significance, but reported suggestive findings for the gene DGKH (diacylglycerol kinase eta),18 the GRIA2 gene coding for the glutamate receptor,19 the gene ACCN1 (amiloride-sensitive cation channel 1 neuronal)20 and clock genes.21
Another impetus to explore the genetic variability underlying lithium response is the possibility for new insights into the genetic basis of BD. The pathophysiology of BD is likely to be heterogeneous with multiple genetic mechanisms related to distinct biochemical pathway abnormalities giving rise to illnesses with similar clinical manifestations.5 The group of patients responding to lithium could suffer from a more etiologically homogeneous illness subtype caused by abnormal pathways that are direct or indirect targets of lithium.22 Indeed, based on longitudinal stability and familial clustering,23, 24 lithium response has been suggested to define a distinct genetically based subtype of BD. Since genetic heterogeneity reduces the power to identify significantly associated variants, GWAS focusing on a homogeneous subgroup such as lithium responders may be more fruitful.
Here, we utilized samples from Sweden and the United Kingdom to identify common genetic variants associated with lithium response in BD patients. Furthermore, we conducted analyses comparing lithium responders with controls to identify common genetic variants that increase susceptibility for the lithium-responsive subtype of BD. Finally, we estimated the extent of genetic contribution to lithium-responsive BD (that is, SNP heritability).
Materials and methods
Subjects
Cases from Sweden were collected through two recruitment streams: the Stanley study and the St Göran Bipolar Project. Most cases were recruited from the Stanley study that identified cases through the Swedish Quality Register for BD (BipoläR), which has been described in detail previously.25, 26 In brief, BipoläR contains individualized data on diagnoses (that is, BD type 1, type 2, not otherwise specified, or schizoaffective disorder bipolar type according to the DSM-IV-TR), medical intervention and outcomes. It also captures basic clinical epidemiological data as well as longitudinal data on the natural history and clinical course of the disease. Genotyping has been completed for 1591 BD patients (passing quality control (QC)) enrolled in the Stanley study. The other recruitment stream for cases was from the St Göran Bipolar Project (N=231), which provides assessment, treatment and follow-up of BD patients within the Northern Stockholm Mental Health Service and the Affective Unit in Mölndal. The work-up procedures have been previously described in detail.27, 28, 29 Both projects were approved by the Regional Ethical Review Board in Stockholm, Sweden, and all participants provided written informed consent.
Control subjects (N=3486) were randomly selected from Swedish population registers, ascertained on a national basis. The exclusion criterion was any hospitalization with schizophrenia or BD. DNA collection procedures have been previously described.30 A small number of controls (N=56) were from St Göran Bipolar Project as described previously.27
Cases from the United Kingdom were drawn from the Bipolar Disorder Research Network (BDRN) study, a large ongoing program of genetic and clinical research into the causes of BD and related mood disorders. Subjects were recruited via both systematic and non-systematic methods. Information was gathered retrospectively by semi-structured interview and case-notes review. More detailed description can be found in prior publications.31, 32 Controls were from the Wellcome Trust Case Control Consortium 2 (WTCCC2) common control set, which comprised 2675 healthy blood donors recruited from the UK Blood Service and 2742 samples from the 1958 British Birth Cohort.33 This study received approval from the Multi-Region and Local Research Ethics Committee in the United Kingdom, and all participants provided written informed consent.
Phenotype definition and assessment
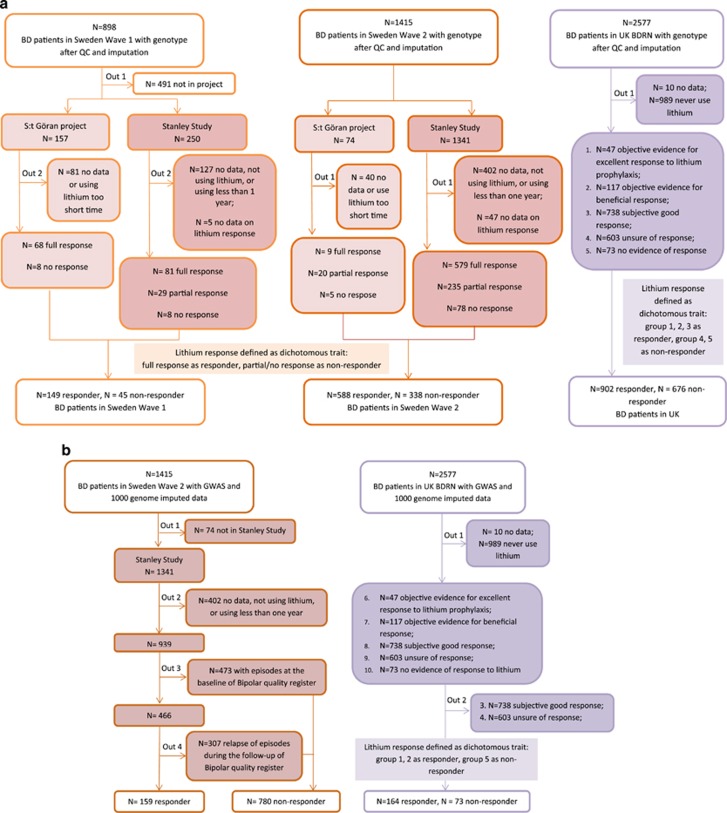
A summary of phenotype assessments and the corresponding numbers of Swedish and UK samples is shown in Table 1 and Figure 1. The detailed phenotype definitions and assessments of lithium response were described in Supplementary Methods.
Table 1. Sample sizes by group and study.
| Phenotype assessment |
Swedish sample |
UK BDRN | Meta-analysis | |
|---|---|---|---|---|
| Wave 1 | Wave 2 | |||
| Subjective measurement | ||||
| Responders | 149 | 588 | 902 | 1639 |
| Non-responders | 45 | 338 | 676 | 1059 |
| Controls | 2215 | 1271 | 5413 | 8899 |
| Objective measurement | ||||
| Responders | — | 159 | 164 | 323 |
| Non-responders | — | 780 | 73 | 853 |
| Controls | — | 1271 | 5413 | 6684 |
Abbreviation: BDRN, Bipolar Disorder Research Network.
The numbers are subjects with: (1) available assessments of lithium response according to our definition (for details, see Supplementary Methods) and (2) passed genotyping quality control. Too few subjects in Sweden wave 1 had objective measures of lithium response. Therefore, a GWAS for that sample and measure was not able to be performed.
Figure 1.
Sample ascertainment flow chart. (a) Subjectively defined lithium assessment. (b) Objectively defined lithium assessment.
In brief, lithium response in Swedish participants was assessed through both interviews (subjective assessment) and register data (objective assessment). The extent to which subjective assessment and objective assessment were in line with each other is shown in Supplementary Table S2. For participants from United Kingdom, lithium response information was collected by interviews and reviews of clinical notes. The inter-rater reliability is not applicable in Swedish samples but has been assessed in UK samples, with a kappa score of 0.76 calculated from a random selection of cases. We treat lithium response as a dichotomous trait based on the subjective and objective measurements, to maximize the sample size and to arrive at a narrower phenotype definition, respectively.
Genotyping, QC, imputation and validation
DNA from Swedish subjects was extracted from whole blood samples at the Karolinska Institutet Biobank. Genotyping was conducted at the Broad Institute of Harvard and MIT using Affymetrix 6.0 (wave 1) (Affymetrix, Santa Clara, CA, USA) and Illumina OmniExpress (wave 2) chips (Illumina, San Diego, CA, USA). Blood samples from UK BDRN cases underwent DNA extraction at the neuropsychiatric genetics laboratory at Cardiff University and were genotyped using Illumina OmniExpress and Illumina ComboChip. UK control samples were genotyped using Illumina 1.2M Custom Chip (Illumina), as described in the WTCCC GWAS.33 The detailed genotyping QC for Swedish and UK samples is described in Supplementary Methods. Following all QC steps, the final analysis data set consisted of 898 bipolar cases, 2215 controls with 744 932 genotyped SNPs in Swedish wave 1, 1415 bipolar cases, 1271 controls with 598 894 genotyped SNPs in Swedish wave 2, and 2577 bipolar cases, 5413 controls with 393 635 SNPs genotyped in UK sample.
Data sets were then pre-phased using SHAPEIT34 and imputed with the full 1000 Genomes Project integrated variant set (Phase 1 integrated data version 3, released March 2012) as the reference panel35 using IMPUTE2.36 After imputation, the three waves shared more than 8 million SNPs.
To confirm the accuracy of imputed SNPs with GWAS significance, DNA from Swedish subjects putatively carrying the minor alleles for rs116323614 and rs146727601 and additional non-carriers were sent for confirmatory genotyping (N=85). TaqMan assays (Applied Biosystems, Foster City, CA, USA) were used for genotyping and were performed by LGC Genomics (Teddington, Middlesex, UK).
Association analyses
Association analyses were conducted using imputed SNP dosages and logistic regression in PLINK version 1.07.37 Multidimensional scaling (MDS) was performed on each collection wave and MDS components were included as covariates (first six MDS components for Swedish wave 2 and first four MDS components for others) to control for population substructure. We retained only SNP dosages imputed with high confidence (Info >0.6) and with minor allele frequency >1%. The genome-wide significance threshold was set at P<5 × 10−8.
We first compared lithium responders with non-responders. Both subjective and objective assessments were used for association analysis separately for each wave, except for Sweden wave 1 due to an inadequate number of subjects with information regarding objective lithium measures. Results were then combined by meta-analysis using a fixed-effects model in PLINK for each measure of lithium response, respectively. Second, we conducted a GWAS comparing lithium responders, subjectively and objectively defined, respectively, with controls without history of schizophrenia and BD. Multiple testing correction was not attempted since the samples and phenotype definition were not independent (Supplementary Table S2). To define regions of association, we conducted PLINK linkage disequilibrium (LD) based ‘clumping' to group SNPs with association P<0.0001 and near-by SNPs with nominal associations (P<0.05) and LD (r2>0.1) within 500-kb windows. We also identified genes in these and the 20-kb flanking regions.
SNP-heritability estimation
Furthermore, we estimated the heritability of lithium-responsive BD using the GCTA (genome-wide complex trait analysis) method.38 We first merged Sweden wave 2 and UK data sets, which were used for both subjective assessment and objective assessment of phenotype. We then used GCTA version 1.24 to calculate the pair-wise genetic relationship between individuals and created the genetic relationship matrix.38 SNP heritability of lithium-responsive BD phenotype was then estimated using the restricted maximum likelihood method in GCTA, with sex, different batches and the first four eigenvectors calculated from principal component analysis included as covariates to control for population structure. Details about the analysis procedure are provided in the Supplementary Methods.
Results
Demographic characteristics for the Swedish and UK samples can be found in Supplementary Table S1.
Lithium responders vs non-responders
For each genome-wide association analysis, and for meta-analyses combining each GWAS with comparable assessments of lithium response, the genomic inflation factor λ showed little or no evidence for inflation (λ=0.97–1.02, Supplementary Table S3). The regions of association with the lowest P-values for each analysis can be found in Supplementary Table S4, while the top results for each meta-analysis are summarized in Table 2 (corresponding results for analyses of each sample separately are in Supplementary Table S7). The quantile-quantile and Manhattan plots for all analyses are shown in Supplementary Figure S1.
Table 2. Top regions of genetic association for each meta-analysis.
| Chr | Index SNP | A1/A2 | Freq | OR | P-value | N | Position | KB | Genes |
|---|---|---|---|---|---|---|---|---|---|
| Responders vs non-responders, subjective assessments | |||||||||
| 20 | rs73918339 | T/C | 0.91 | 0.56 | 3.80 × 10−7 | 60 | 61150190–61213367 | 63 | MIR133A2, MIR1-1, C20orf200, C20orf166 |
| 18 | rs7240206 | C/G | 0.09 | 0.60 | 3.86 × 10−7 | 67 | 74393681–74447380 | 54 | LOC105372213 |
| 7 | rs116927879 | G/A | 0.85 | 1.53 | 4.99 × 10−7 | 267 | 45588502–46011988 | 423 | SEPT7P2, IGFBP3, IGFBP1, ADCY1 |
| 4 | rs78295376 | T/C | 0.90 | 0.57 | 6.77 × 10−7 | 26 | 76480371–76729937 | 250 | USO1, TMSL3, G3BP2, FRG2, CDKL2, C4orf26 |
| Responders vs non-responders, objective assessments | |||||||||
| 6 | rs438475 | G/A | 0.88 | 0.49 | 2.13 × 10−6 | 165 | 31 770265–32625494 | 855 | NOTCH4, many genes in MHC region |
| 11 | rs113262272 | A/− | 0.71 | 1.93 | 3.77 × 10−6 | 99 | 99298992–99587113 | 288 | CNTN5 |
| 14 | rs809 | C/T | 0.53 | 0.61 | 4.08 × 10−6 | 39 | 78502019–78558331 | 56 | NRXN3 |
| 6 | rs181812561 | G/A | 0.98 | 0.13 | 6.39 × 10−6 | 3 | 75079066–75231552 | 152 | LOC101928516 |
| Responders vs controls, subjective assessments | |||||||||
| 1 | rs12144699 | G/A | 0.96 | 0.60 | 6.05 × 10−7 | 7 | 59605197–59863936 | 259 | OR4F16, OR4F29, LOC729467, LOC100133331, LOC100132287, FGGY |
| 3 | rs9834970 | T/C | 0.50 | 0.82 | 7.88 × 10−7 | 153 | 36834099–37285522 | 451 | TRANK1, MLH1, MIR4273, LRRFIP2, GOLGA4, FRG2C, EPM2AIP1 |
| 3 | rs12493050 | G/A | 0.20 | 1.27 | 8.78 × 10−7 | 276 | 182478533–182909924 | 431 | MCF2L2, MCCC1, LAMP3, DCUN1D1, ATP11B |
| 7 | rs4947962 | G/C | 0.11 | 1.35 | 1.35 × 10−6 | 85 | 54906722–55161372 | 255 | EGFR |
| Responders vs controls, objective assessments | |||||||||
| 11 | rs146727601 | −/TA | 0.01 | 3.98 | 1.33 × 10−8 | 2 | 112118590–112343856 | 225 | PTS, PLET1 |
| 2 | rs116323614 | A/G | 0.03 | 3.14 | 2.74 × 10−8 | 58 | 179859406–180139219 | 280 | SESTD1, CCDC141 |
| 19 | rs77866734 | C/T | 0.98 | 0.27 | 1.49 × 10−7 | 11 | 1633923–1642221 | 8 | TCF3, KIR3DP1, KIR2DL4 |
| 17 | rs142643109 | T/G | 0.98 | 0.28 | 3.93 × 10−7 | 3 | 60086587–60497572 | 411 | TBC1D3P2, MIR4315-2, MIR4315-1, METTL2A, MED13, EFCAB3 |
Abbreviations: A1/A2, reference and alternate allele; Chr, chromosome; Freq, frequency of reference allele; Index SNP, the single-nucleotide polymorphism with the strongest association in the genomic region; MHC, major histocompatibility complex; N, number of SNPs in the reported region; OR, odds ratio. We used LD clumping to define regions of association. Positions are given in UCSC hg19 coordinates. Lines in bold indicate associations that are genome-wide statistically significant.
No SNP reached genome-wide significance in any sample separately, except for one imputed SNP in GWAS for Sweden wave 2 comparing lithium responders and non-responders with subjective assessment (588 responders vs 338 non-responders, rs56177802, P=2.03 × 10−9, OR=2.14, Supplementary Table S4). However, this marker failed to meet the threshold for genome-wide significance following meta-analysis (1639 responders vs 1059 non-responders, P=1.81 × 10−4, OR=1.31).
No SNP achieved genome-wide significance in meta-analyses of association with lithium response. In the analyses of objective lithium response, the SNPs with the greatest evidence for association were located in an intron of the NOTCH4 gene and all in high LD (r2>0.8).
Lithium responders vs controls
For these GWAS and meta-analyses, the resulting genomic inflation factors again showed little evidence of inflation (λ=1.01–1.05, Supplementary Table S5). The regions of association with the lowest P-values for each analysis are shown in Supplementary Table S6, while the top results for each meta-analysis are shown in Table 2 (corresponding results for analyses of each sample separately are in Supplementary Table S7). The quantile-quantile and Manhattan plots for all analyses are in Supplementary Figure S2.
In GWAS for each sample comparing lithium responders (subjective assessment) and controls, one imputed SNP achieved genome-wide significance in Sweden wave 1 (149 responders vs 2215 controls, rs10979017, P=1.08 × 10−8, OR=0.20), and one imputed SNP approached genome-wide significance in Sweden wave 2 (588 responders vs 1271 controls, rs1442378, P=5.19 × 10−8, OR=1.55) (Supplementary Table S6). However, the meta-analysis combining Swedish and UK samples together showed no genome-wide significant associations for these SNPs.
In the subsequent meta-analyses comparing lithium responders (objective assessment) with controls, two imputed variants reached genome-wide significance (323 responders vs 6684 controls, rs146727601, P=1.33 × 10−8, OR=3.98; and rs116323614, P=2.74 × 10−8, OR=3.14; Table 2). Rs146727601 is a two-base deletion on chromosome 11q22.4 in the gene PTS (6-pyruvoyltetrahydropterin synthase) and also about 500 base pair (bp) downstream of the gene placenta expressed transcript 1 (PLET1). Rs116323614 is an intronic marker on chromosome 2q31.2 in the gene SEC14 and spectrin domains 1 (SESTD1). Direct genotyping was successful in 82 Swedish samples confirming the imputed rs116323614 alleles (Pearson correlation coefficient=0.92). However, three distinct primer designs failed to genotype rs146727601. The region plots depicting association signals surrounding rs116323614 and rs146727601 are in Supplementary Figure S3.
We calculated the predicted genotype frequencies for imputed rs116323614 and performed the exact test of Hardy–Weinberg Equilibrium (details are provided in the Supplementary Methods). There was no evidence of departure from Hardy–Weinberg Equilibrium for this variant in either Swedish or the UK population (Supplementary Table S8).
Noticing the sex differences in the ratio of responders to non-responders in the UK sample (Supplementary Table S1), we performed association tests and meta-analysis for male and female samples separately. The ORs for rs116323614 by sex were not significantly different (P=0.55) and including sex as a covariate in the association analyses did not alter the significance (P=3.93 × 10–8) (Supplementary Table S9).
SNP-heritability estimates
SNP heritability (h2) for lithium-responsive BD was 29% for the subjective definition (95% CI 23–36, P<0.0001) and 25% for the objective definition (95% CI 0–51, P=0.03 for the null hypothesis of being non-heritable) (Supplementary Table S10).
Discussion
By using two large samples of persons with BD treated with lithium and healthy controls, we found that (i) no markers met the threshold for genome-wide association with lithium response; (ii) variation in SESTD1 demonstrated association in the case–control analysis, where we used lithium response to define a BD subtype, with the proportion of variance explained by common genetic variants (SNP heritability) estimated at 0.25–0.29. Interestingly, SESTD1 encodes a protein involved in the regulation of phospholipids.39 Inositol phospholipids have been implicated not only in the pathophysiology of BD but also are among the most studied molecular targets of lithium.40, 41 These results suggest that using pharmacoresponse to define subgroups within complex disorders might be a complementary approach for identifying disease risk loci in case–control association studies.
Lithium-responsive BD
Lithium response has been suggested to be a distinct phenotype of BD for decades.42, 43, 44, 45, 46, 47 A number of studies have identified specific clinical features in patients responsive to lithium, such as fewer hospitalizations preceding treatment, later age of onset and an episodic pattern of mania followed by depression.48 In addition, lithium response is suggested to have a genetic basis, based on evidence such as longitudinal stability23 and familial clustering.24 Moreover, both clinical observational and neurobiological studies show that lithium responders differ from responders to other mood stabilizers.47, 49 Therefore, the present study utilizes this distinct subtype of BD to investigate the mechanisms behind it.
Lithium responders vs non-responders
GWAS for each sample separately yielded one SNP (Sweden wave 2) showing genome-wide significant association (Supplementary Figure S1-II, Supplementary Table S4), but it failed to remain significant in the ensuing meta-analysis. The variants ranking highly in prior GWAS18, 19, 20, 21 also showed little evidence of association in our results, and although the alleles previously reported to be strongly associated with lithium response in GADL1 did not exist in our sample,16 other common variants in GADL1 did not support an association.
Our findings with strong, but not significant, associations might nevertheless be informative for future genetic and protein-level studies. For example, the SNPs with the lowest P-values for objectively defined lithium response are located in the NOTCH4 gene within the major histocompatibility complex. Several studies have associated NOTCH4 with schizophrenia and BD,50, 51 and it was found to be upregulated in BD.52 The mechanisms might be related to neurodevelopment and inflammation.52 Thus, our results are in line with the evidence implicating NOTCH4 in the pathophysiology of BD and potentially lithium response.
Lithium responders vs controls
By comparing the subgroup of BD patients who were lithium responders with healthy controls, we identified two variants, rs146727601 and rs116323614, reaching genome-wide significance. When we performed GWAS comparing all patients with BD with healthy controls, the results for rs146727601 and rs116323614 were OR=1.28, P=0.05 and OR=1.31, P=0.003, respectively, which supports the contention of an association in a subset of BD cases. Moreover, the minor allele frequencies in these two variants in lithium responders were markedly higher than that in non-responders (with objective response definition meta-analysis, rs146727601, OR=2.18, P=0.04; rs116323614, OR=2.25, P=0.01). The different allele distributions suggest that these variants are unlikely to be general BD-associated markers, but specifically associated with lithium-responsive BD. Furthermore, the estimate of SNP heritability in our sample showed that lithium-responsive BD is modestly heritable. Although the largest published GWAS for BD did not include the significant markers in our study,53 we hope that future studies incorporating them will offer the possibility of replication.
For rs146727601, it is noteworthy that this variant is located in the gene PTS, a catalyst involved in the regulation of serotonin biosynthesis and nitric oxide synthase activity. These processes are suggested to be potential therapeutic targets for lithium.54, 55, 56 However, validation attempts failed, and no reasonable proxy marker exists.
By contrast, we successfully validated the rs116323614 association. This is an intronic SNP in the gene SESTD1, which shows relatively high expression in brain and is also significantly expressed in vascular cells.39 Importantly, SESTD1 Ca2+ dependently binds phospholipids (for example, phosphatidylinositol monophosphates (PIP) and diphosphates (PIP2)) that are involved in the phosphoinositide signaling pathway. This pathway, activated by G protein-coupled receptors, leads to the hydrolysis of PIP2 and produces inositol 1,4,5-trisphosphate (IP3) via the enzyme phospholipase C.57 IP3 regulates calcium signaling58 and is used to produce inositol, which is utilized to form PIP2, directing the initial step of this pathway.57 Decades of research into the therapeutic effects of lithium suggest40, 41, 59, 60, 61 that they are mediated through the inhibition of inositol monophosphatase and inositol polyphosphate 1-phosphatase.59, 61, 62 Both enzymes are important in the process of recycling inositol to PIP2.62, 63 In fact, Soares et al.40, 64 showed that lithium at therapeutic levels significantly decreases platelet membrane PIP2 levels in vivo. Lithium thereby modulates the phosphoinositide pathway, and indirectly influences the actions of downstream neurotransmitter receptors.65 The fact that the phosphoinositide pathway is the most validated lithium target confers biological plausibility to our finding of an association between SESTD1 and risk for lithium-responsive BD.5, 66
Additionally, SESTD1 is proposed to be involved in phospholipid regulation of transient receptor potential channels TRPC4 and TRPC5.39, 67 Both TRPC4 and TRPC5 are Ca2+ permeable channels and have an important role in functions of neurotransmitter release68 and in regulation of neurite growth.69, 70 Interestingly, TRPC4 and TRPC5 are assumed to be activated by stimulation of phospholipase C, through IP3-induced intracellular Ca2+ store depletion.71 Moreover, recent GWAS for BD identified a novel variant located in the gene TRPC4 Associated Protein (TRPC4AP).72 The relationships between the TRPC families and the phosphoinositide signaling pathway further imply that SESTD1 is associated, possibly indirectly, with BD as well as lithium targets.
The A allele frequency for SNP rs116323614 is ~3% in European populations (1000Genome phase 1) with an odds ratio of 3.14 (95% confidence interval 2.10–4.70) in our study, suggesting that this variant alone is not well suited for clinical use. Moreover, it is likely that response to lithium is polygenically mediated, and only by examining multiple variants simultaneously will genetic screening be sufficiently powerful to guide decisions regarding mood stabilizers.5
Strengths and limitations
Our samples comprise two distinct populations, and the total number of patients with available assessment of lithium response in our study exceeds that of other reported studies to date. Nevertheless, the sample size is still small by GWAS standards, and results should be interpreted cautiously pending replication. To some extent, our results identifying associated variants in the responders/controls comparison but not in the responders/non-responders comparison could reflect reduced power rather than underlying biology.
The evaluation of genetic heterogeneity is important for meta-analyses, and our relatively homogeneous samples from Sweden and the United Kingdom offer greater power compared with studies involving populations that are more diverse. The markers demonstrating the greatest association in our results showed no evidence of heterogeneity using Q-tests (P=0.73) and I2 heterogeneity index (I2=0.00).73
A key issue for pharmocogenetic studies is reliable assessment of treatment response. The irregular clinical course of BD74 and the difficulty in accurately measuring treatment adherence75 complicate the assessment of lithium response. Furthermore, whether a person is lithium responsive takes years to reliably establish. In this study, the harmonization of lithium response measures across sites is a potential concern as the assessments were conducted differently in Sweden and United Kingdom. Even though we carefully attempted to harmonize measures across sites, potential phenotypic misclassification cannot be excluded, and is a possible reason why significantly associated loci in the respective samples did not hold in the meta-analysis.
In addition to measuring the extent of clinical improvement, a refined evaluation could also consider additional factors (for example, the duration and severity of illness before and during lithium treatment, the duration and compliance of lithium treatment and additional medications) to determine whether the reduction of BD symptoms is actually due to the treatment.24 However, in the trade-off between certainty of response and sufficient sample size, the rate of missing data hampered inclusion of such variables. Hence, it cannot be excluded that a more elaborate measurement of lithium response might yield positive results.
Conclusion
In conclusion, the present study suggests that rs116323614, located in an intron of SESTD1, is associated with a lithium-responsive subtype of BD. This illustrates that defining homogeneous subgroups of complex genetic traits by response to treatment might be a productive strategy for identifying susceptibility alleles. Although the specific genetic variants identified in this study are not clinically applicable due to rare frequency, the findings provide new insights into the pathophysiology of BD and lithium's mechanism of action.
Acknowledgments
We would like to thank all of the participants who have kindly given their time and DNA to participate in our research, the Swedish quality register for bipolar disorders (BipoläR), data collectors at the Department of Medical Epidemiology and Biostatistics (MEB) at Karolinska Institutet, Bipolar UK, BDRN clinical collaborators and facilitators, and the Mental Health Research Network (MHRN) for help with recruitment of participants. The Swedish part was funded by the Stanley Center for Psychiatric Research, Broad Institute from a grant from Stanley Medical Research Institute, the Swedish Medical Research Council (K2014-62X-14647-12-51 and K2010-61P-21568-01-4), the Söderström-Königska Foundation (SLS-472751), the Swedish foundation for Strategic Research (KF10-0039) and the China Scholarship Council. The UK collection was supported by grants from the Wellcome Trust and the Stanley Medical Research Institute. The International Cohort Collection for Bipolar Disorder (ICCBD) was funded by NIMH grant RO1MH085542.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Craddock N, Sklar P. Genetics of bipolar disorder. Lancet 2013; 381: 1654–1662. [DOI] [PubMed] [Google Scholar]
- Geddes JR, Miklowitz DJ. Treatment of bipolar disorder. Lancet 2013; 381: 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham J, Munro A, Slaney C, Macdougall M, Passmore M, Duffy A et al. Prophylactic treatment response in bipolar disorder: results of a naturalistic observation study. J Affect Disord 2007; 104: 185–190. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK. Lithium in neuropsychiatry: a 2010 update. World J Biol Psychiatry 2011; 12: 340–348. [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, Leckband SG, Kelsoe JR. Pharmacogenetics of lithium response in bipolar disorder. Pharmacogenomics 2010; 11: 1439–1465. [DOI] [PubMed] [Google Scholar]
- Mamdani F, Alda M, Grof P, Young LT, Rouleau G, Turecki G. Lithium response and genetic variation in the CREB family of genes. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alda M, Shao L, Wang JF, Lopez de Lara C, Jaitovich-Groisman I, Lebel V et al. Alterations in phosphorylated cAMP response element-binding protein (pCREB) signaling: an endophenotype of lithium-responsive bipolar disorder? Bipolar Disord 2013; 15: 824–831. [DOI] [PubMed] [Google Scholar]
- Lopez de Lara C, Jaitovich-Groisman I, Cruceanu C, Mamdani F, Lebel V, Yerko V et al. Implication of synapse-related genes in bipolar disorder by linkage and gene expression analyses. Int J Neuropsychopharmacol 2010; 13: 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu C, Alda M, Grof P, Rouleau GA, Turecki G. Synapsin II is involved in the molecular pathway of lithium treatment in bipolar disorder. PLoS One 2012; 7: e32680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrzak-Weglarz M, Rybakowski JK, Suwalska A, Skibinska M, Leszczynska-Rodziewicz A, Szczepankiewicz A et al. Association studies of the BDNF and the NTRK2 gene polymorphisms with prophylactic lithium response in bipolar patients. Pharmacogenomics 2008; 9: 1595–1603. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK, Suwalska A, Skibinska M, Szczepankiewicz A, Leszczynska-Rodziewicz A, Permoda A et al. Prophylactic lithium response and polymorphism of the brain-derived neurotrophic factor gene. Pharmacopsychiatry 2005; 38: 166–170. [DOI] [PubMed] [Google Scholar]
- Masui T, Hashimoto R, Kusumi I, Suzuki K, Tanaka T, Nakagawa S et al. Lithium response and Val66Met polymorphism of the brain-derived neurotrophic factor gene in Japanese patients with bipolar disorder. Psychiatr Genet 2006; 16: 49–50. [DOI] [PubMed] [Google Scholar]
- Michelon L, Meira-Lima I, Cordeiro Q, Miguita K, Breen G, Collier D et al. Association study of the INPP1, 5HTT, BDNF, AP-2beta and GSK-3beta GENE variants and restrospectively scored response to lithium prophylaxis in bipolar disorder. Neurosci Lett 2006; 403: 288–293. [DOI] [PubMed] [Google Scholar]
- Masui T, Hashimoto R, Kusumi I, Suzuki K, Tanaka T, Nakagawa S et al. A possible association between the -116C/G single nucleotide polymorphism of the XBP1 gene and lithium prophylaxis in bipolar disorder. Int J Neuropsychopharmacol 2006; 9: 83–88. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Kato T. Lithium response and -116C/G polymorphism of XBP1 in Japanese patients with bipolar disorder. Int J Neuropsychopharmacol 2005; 8: 631–632. [DOI] [PubMed] [Google Scholar]
- Chen CH, Lee CS, Lee MT, Ouyang WC, Chen CC, Chong MY et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. N Engl J Med 2014; 370: 119–128. [DOI] [PubMed] [Google Scholar]
- Consortium on Lithium Genetics, Hou L, Heilbronner U, Rietschel M, Kato T, Kuo PH et al. Variant GADL1 and response to lithium in bipolar I disorder. N Engl J Med 2014; 370: 1857–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 2008; 13: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Ferreira MA, McQuillin A, Bass N, Lawrence J et al. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry 2009; 166: 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squassina A, Manchia M, Borg J, Congiu D, Costa M, Georgitsi M et al. Evidence for association of an ACCN1 gene variant with response to lithium treatment in Sardinian patients with bipolar disorder. Pharmacogenomics 2011; 12: 1559–1569. [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS One 2012; 7: e32091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu C, Alda M, Turecki G. Lithium: a key to the genetics of bipolar disorder. Genome Med 2009; 1: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghofer A, Alda M, Adli M, Baethge C, Bauer M, Bschor T et al. Long-term effectiveness of lithium in bipolar disorder: a multicenter investigation of patients with typical and atypical features. J Clin Psychiatry 2008; 69: 1860–1868. [DOI] [PubMed] [Google Scholar]
- Grof P, Duffy A, Cavazzoni P, Grof E, Garnham J, MacDougall M et al. Is response to prophylactic lithium a familial trait? J Clin Psychiatry 2002; 63: 942–947. [DOI] [PubMed] [Google Scholar]
- Karanti A, Bobeck C, Osterman M, Kardell M, Tidemalm D, Runeson B et al. Gender differences in the treatment of patients with bipolar disorder: a study of 7354 patients. J Affect Disord 2015; 174: 303–309. [DOI] [PubMed] [Google Scholar]
- Tidemalm D, Haglund A, Karanti A, Landen M, Runeson B. Attempted suicide in bipolar disorder: risk factors in a cohort of 6086 patients. PLoS One 2014; 9: e94097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson J, Zetterberg H, Blennow K, Johan Ekman C, Johansson AG, Landen M. Altered concentrations of amyloid precursor protein metabolites in the cerebrospinal fluid of patients with bipolar disorder. Neuropsychopharmacology 2013; 38: 664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden E, Thase ME, Straht D, Aberg-Wistedt A, Bejerot S, Landen M. A history of childhood attention-deficit hyperactivity disorder (ADHD) impacts clinical outcome in adult bipolar patients regardless of current ADHD. Acta Psychiatr Scand 2009; 120: 239–246. [DOI] [PubMed] [Google Scholar]
- Ekman CJ, Lind J, Ryden E, Ingvar M, Landen M. Manic episodes are associated with grey matter volume reduction - a voxel-based morphometry brain analysis. Acta Psychiatr Scand 2010; 122: 507–515. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H, Forty L, Jones L, Gordon-Smith K, Jones I, Craddock N et al. Improving the psychometric utility of the hypomania checklist (HCL-32): a Rasch analysis approach. J Affect Disord 2014; 152-154: 448–453. [DOI] [PubMed] [Google Scholar]
- Gordon-Smith K, Forty L, Chan C, Knott S, Jones I, Craddock N et al. Rapid cycling as a feature of bipolar disorder and comorbid migraine. J Affect Disord 2015; 175C: 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK IBD Genetics Consortium, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 2009; 41: 1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Coulonges C, Zagury JF. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics 2008; 9: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 2011; 1: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miehe S, Bieberstein A, Arnould I, Ihdene O, Rutten H, Strubing C. The phospholipid-binding protein SESTD1 is a novel regulator of the transient receptor potential channels TRPC4 and TRPC5. J Biol Chem 2010; 285: 12426–12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares JC, Mallinger AG, Dippold CS, Frank E, Kupfer DJ. Platelet membrane phospholipids in euthymic bipolar disorder patients: are they affected by lithium treatment? Biol Psychiatry 1999; 45: 453–457. [DOI] [PubMed] [Google Scholar]
- Niciu MJ, Ionescu DF, Mathews DC, Richards EM, Zarate CA Jr. Second messenger/signal transduction pathways in major mood disorders: moving from membrane to mechanism of action, part II: bipolar disorder. CNS Spectr 2013; 18: 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grof P, Alda M, Grof E, Fox D, Cameron P. The challenge of predicting response to stabilising lithium treatment. The importance of patient selection. Br J Psychiatry Suppl 1993; 21: 16–19. [PubMed] [Google Scholar]
- Calabrese JR, Fatemi SH, Kujawa M, Woyshville MJ. Predictors of response to mood stabilizers. J Clin Psychopharmacol 1996; 16: 24S–31S. [DOI] [PubMed] [Google Scholar]
- Greil W, Ludwig-Mayerhofer W, Erazo N, Schochlin C, Schmidt S, Engel RR et al. Lithium versus carbamazepine in the maintenance treatment of bipolar disorders—a randomised study. J Affect Disord 1997; 43: 151–161. [DOI] [PubMed] [Google Scholar]
- Baldessarini RJ, Tondo L, Floris G, Hennen J. Effects of rapid cycling on response to lithium maintenance treatment in 360 bipolar I and II disorder patients. J Affect Disord 2000; 61: 13–22. [DOI] [PubMed] [Google Scholar]
- Bowden CL. Clinical correlates of therapeutic response in bipolar disorder. J Affect Disord 2001; 67: 257–265. [DOI] [PubMed] [Google Scholar]
- Passmore MJ, Garnham J, Duffy A, MacDougall M, Munro A, Slaney C et al. Phenotypic spectra of bipolar disorder in responders to lithium versus lamotrigine. Bipolar Disord 2003; 5: 110–114. [DOI] [PubMed] [Google Scholar]
- Kleindienst N, Engel R, Greil W. Which clinical factors predict response to prophylactic lithium? A systematic review for bipolar disorders. Bipolar Disord 2005; 7: 404–417. [DOI] [PubMed] [Google Scholar]
- Kruger S, Alda M, Young LT, Goldapple K, Parikh S, Mayberg HS. Risk and esilience markers in bipolar disorder: brain responses to emotional challenge in bipolar patients and their healthy siblings. Am J Psychiatry 2006; 163: 257–264. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S et al. Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum Mol Genet 2011; 20: 387–391. [DOI] [PubMed] [Google Scholar]
- Dieset I, Djurovic S, Tesli M, Hope S, Mattingsdal M, Michelsen A et al. Up-regulation of NOTCH4 gene expression in bipolar disorder. Am J Psychiatry 2012; 169: 1292–1300. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GN, Pandey SC, Ren X, Dwivedi Y, Janicak PG. Serotonin receptors in platelets of bipolar and schizoaffective patients: effect of lithium treatment. Psychopharmacology 2003; 170: 115–123. [DOI] [PubMed] [Google Scholar]
- Ghasemi M, Dehpour AR. The NMDA receptor/nitric oxide pathway: a target for the therapeutic and toxic effects of lithium. Trends Pharmacol Sci 2011; 32: 420–434. [DOI] [PubMed] [Google Scholar]
- Lenox RH, Hahn CG. Overview of the mechanism of action of lithium in the brain: fifty-year update. J Clin Psychiatry 2000; 61: 5–15. [PubMed] [Google Scholar]
- Hilgemann DW, Feng S, Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE 2001; 2001: re19. [DOI] [PubMed] [Google Scholar]
- Gill DL, Ghosh TK, Mullaney JM. Calcium signalling mechanisms in endoplasmic reticulum activated by inositol 1,4,5-trisphosphate and GTP. Cell Calcium 1989; 10: 363–374. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Downes CP, Hanley MR. Lithium amplifies agonist-dependent phosphatidylinositol responses in brain and salivary glands. Biochem J 1982; 206: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EM, Detwiler TC. The effects of lithium on platelet phosphoinositide metabolism. Biochem J 1986; 236: 895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Cheng L, Mudge AW, Harwood AJ. A common mechanism of action for three mood-stabilizing drugs. Nature 2002; 417: 292–295. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell 1989; 59: 411–419. [DOI] [PubMed] [Google Scholar]
- Hallcher LM, Sherman WR. The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J Biol Chem 1980; 255: 10896–10901. [PubMed] [Google Scholar]
- Soares JC, Mallinger AG, Dippold CS, Forster Wells K, Frank E, Kupfer DJ. Effects of lithium on platelet membrane phosphoinositides in bipolar disorder patients: a pilot study. Psychopharmacology 2000; 149: 12–16. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta 2009; 1793: 933–940. [DOI] [PubMed] [Google Scholar]
- Pollack SJ, Atack JR, Knowles MR, McAllister G, Ragan CI, Baker R et al. Mechanism of inositol monophosphatase, the putative target of lithium therapy. Proc Natl Acad Sci USA 1994; 91: 5766–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JP, Hong C, Park EJ, Jeon JH, Cho NH, Kim IG et al. Selective Galphai subunits as novel direct activators of transient receptor potential canonical (TRPC)4 and TRPC5 channels. J Biol Chem 2012; 287: 17029–17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsch T, Freichel M, Flockerzi V, Pape HC. Contribution of transient receptor potential channels to the control of GABA release from dendrites. Proc Natl Acad Sci USA 2003; 100: 16065–16070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greka A, Navarro B, Oancea E, Duggan A, Clapham DE. TRPC5 is a regulator of hippocampal neurite length and growth cone morphology. Nat Neurosci 2003; 6: 837–845. [DOI] [PubMed] [Google Scholar]
- Hui H, McHugh D, Hannan M, Zeng F, Xu SZ, Khan SU et al. Calcium-sensing mechanism in TRPC5 channels contributing to retardation of neurite outgrowth. J Physiol 2006; 572: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. The TRP superfamily of cation channels. Sci STKE 2005; 2005: re3. [DOI] [PubMed] [Google Scholar]
- Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry 2013; 18: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Interpretation of tests of heterogeneity and bias in meta-analysis. J Eval Clin Pract 2008; 14: 951–957. [DOI] [PubMed] [Google Scholar]
- Angst J, Sellaro R. Historical perspectives and natural history of bipolar disorder. Biol Psychiatry 2000; 48: 445–457. [DOI] [PubMed] [Google Scholar]
- Sachs GS, Rush AJ. Response, remission, and recovery in bipolar disorders: what are the realistic treatment goals? J Clin Psychiatry 2003; 64: 18–22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.