Abstract
Neuroblastoma is a pediatric cancer of the developing sympathoadrenal lineage. The tumors are known to develop from the adrenal gland or paraspinal ganglia and have molecular and cellular features of sympathetic neurons such as dense core vesicles and catecholamine production. Here we present the detailed molecular, cellular, genetic and epigenetic characterization of an orthotopic xenograft derived from a high-risk stage 4 neuroblastoma patient. Overall, the xenografted tumor retained the high risk features of the primary tumor and showed aggressive growth and metastasis in the mouse. Also, the genome was preserved with no additional copy number variations, structural variations or aneuploidy. There were 13 missense mutations identified in the xenograft that were not present in the patient’s primary tumor and there were no new nonsense mutations. None of the missense mutations acquired in the xenograft were in known cancer genes. We also demonstrate the feasibility of using the orthotopic neuroblastoma xenograft to test standard of care chemotherapy and molecular targeted therapeutics. Finally, we optimized a new approach to produce primary cultures of the neuroblastoma xenografts for high-throughput drug screening which can be used to test new combinations of therapeutic agents for neuroblastoma.
1. Introduction
Neuroblastoma is the most common extracranial solid tumor and the most common tumor in infants (Maris et al., 2007). The course of the disease is variable with some tumors regressing spontaneously and others progressing rapidly despite intense therapeutic intervention (Cheung and Dyer, 2013). Current treatment includes chemotherapy, surgical resection, radiotherapy, stem cell transplantation, and immunotherapy (Maris, 2010). While low-risk patients may be cured with surgery alone, patients with high-risk neuroblastoma have a poor prognosis despite this aggressive therapy (Maris, 2010).
Neuroblastoma typically arises in the retroperitoneum from the adrenal glands and paravertebral sympathetic plexus. It is a highly vascular tumor that wraps around major abdominal vessels, developing extensive tumor neovasculature (Chlenski et al., 2003; Maris et al., 2007). This suggests activation of angiogenic signaling pathways, many of which are regulated through receptor tyrosine kinase (RTK) signaling cascades. For example, the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) (PI3K/AKT/mTOR) pathway is a pro-survival signaling cascade that is aberrantly activated in neuroblastoma and activation correlates with a poor prognosis (Opel et al., 2007). mTOR regulates cell growth and autophagy, and its activity in neuroblastoma cells has been shown to affect cell differentiation by coordinating macromolecule synthesis and degradation (Zeng and Zhou, 2008). Targeted agents including monoclonal antibodies, small molecule tyrosine kinase inhibitors and serine-threonine kinase inhibitors can block this pathway at multiple regulatory nodes (Segerstrom et al., 2011). Indeed, therapeutics targeting PI3K/AKT/mTOR have shown efficacy in preclinical models of neuroblastoma (Chanthery et al., 2012) and may be a promising approach for patients with high-risk disease.
We previously developed an ultrasound guided, minimally invasive method for injecting neuroblastoma cell lines into the adrenal gland or para-adrenal space of immunocompromised mice (Teitz et al., 2011). This model was used to test a standard-of-care induction chemotherapy regimen, which includes cyclophosphamide, doxorubicin, and etoposide with alternating courses of cisplatin and etoposide (Teitz et al., 2011). Tumor growth and response was monitored using ultrasound, MRI and bioluminescence imaging. 30% of the mice treated with induction chemotherapy demonstrated a clinical response with complete response (CR) or stable disease (SD). Animals with SD or CR could be differentiated by the 5th course. This was reminiscent of observations in clinical trials, which supported reduction of the number of cycles of intensive induction chemotherapy previously used for high-risk neuroblastoma (Kushner et al., 2004).
A major challenge in oncology research is the low success of translating preclinical results to effective outcomes in patients. This discrepancy can result from many different factors including the dose, schedule and combination of drugs used in preclinical testing, the method for monitoring and reporting tumor response and the relevance of the animal models used in preclinical trials. Orthotopic neuroblastoma xenografts have previously been shown to be more likely to develop distant metastasis compared to subcutaneous tumors (Khanna et al., 2002), and others have adopted similar techniques to create patient-derived xenografts that retained the major characteristics seen in the patient (Braekeveldt et al., 2015). Indeed, Braekeveldt et al. (2015) have shown that patient-derived neuroblastoma xenografts retain the genetic and histopathologic features of the patient tumor they were derived from. In this study, we report the development and characterization of an orthotopic neuroblastoma xenograft from a high-risk primary patient tumor. We performed comprehensive genomic characterization of the primary tumor and the xenograft. As shown previously (Braekeveldt et al., 2015), our patient-derived neuroblastoma xenograft retained the genetic features of the primary tumor. To establish a baseline response, we tested standard of care induction chemotherapy (Matthay et al., 1999) and compared those data to molecular targeted therapy directed at the PI3K/AKT/mTOR pathway. We developed a double blind, randomized preclinical trial design along with an integrated database using OpenClinica for storing and sharing all preclinical data. Having validated the xenograft and establishing the value for preclinical testing, we then developed culture conditions for high throughput screening of the primary xenograft cells to identify novel therapeutics. This example serves as the foundation for a larger more comprehensive effort to establish and characterize orthotopic xenografts for pediatric solid tumors as a tool for translational research.
2. Results
2.1. Characterization of a high-risk primary neuroblastoma tumor sample
We have previously described a method for orthotopically implanting neuroblastoma cell lines into immunocompromised mice (Teitz et al., 2011). One disadvantage of using cell lines to model neuroblastoma in vivo is that they may undergo selection in culture and the genomic landscape may deviate from the primary patient tumor (Sausen et al., 2013). Therefore, to more accurately model neuroblastoma in preclinical models, we developed a biology protocol called Molecular Analysis of Solid Tumors (MAST) to obtain excess tumor from surgical resection or biopsy for orthotopic implantation in immunocompromised mice.
The first neuroblastoma orthotopic xenograft from the MAST protocol came from a 2 year old patient (SJNBL046_D) that originally presented with a history of intermittent fevers, abdominal distention, fatigue and a limp. Imaging revealed a large adrenal mass consistent with neuroblastoma. Urine catecholamines were elevated and there was evidence of metastatic lesions in the lymph nodes and bones. Biopsy confirmed neuroblastoma with MYCN amplification and unfavorable histology (INSS stage 4) (Fig. 1A–C). The patient had a poor response to the first two courses of induction chemotherapy with cyclophosphamide/doxorubicin/etoposide alternating with cisplatin/etoposide. Therefore, the patient was switched to a cyclophosphamide/topotecan regimen. The tumor was resected after 2 cycles of these agents and a small specimen was dissociated, suspended in matrigel and injected into the para-adrenal space of NOD scid gamma (NSG) mice as described previously (Teitz et al., 2011). The patient tumor specimen used for orthotopic implantation had cellular features of neuroblastoma including expression of synaptophysin and chromogranin (Fig. 1D–F) as well as abundant dense core vesicles in electron micrographs, which are a hallmark of catecholamine storage (Fig. 1G and H).
Fig. 1.
Characterization of the primary SJNBL046 tumor. (A) Micrograph of hematoxylin and eosin (H&E) stained needle biopsy of the diagnostic tumor prior to treatment. (B and C) Synaptophysin and chromogranin immunohistochemical staining (brown) of the needle biopsy specimen shown in (A). (D–F) Micrographs of H&E, synaptophysin and chromograning immunohistochemical staining of the patient tumor at the time of surgical resection (SJNBL046_D). (G and H) Electron micrographs of SJNBL046_D with dense core vesicles (DCV, arrows). (I) Circos plot of whole genome sequencing of SJNBL046_D. Loss of heterozygosity (orange), copy number gain (red), and losses (blue) are shown. Intrachromosomal translocations (green lines) and interchromosomal translocations (purple lines) are indicated. Sequence mutations in Refseq genes included silent single nucleotide variants (SNVs, green), nonsense and missense SNVs (brown), splice-site and UTR mutations (dark blue), and insertion/deletion mutations (red). (J) Principle component analysis of gene expression arrays for the SJNBL046_D sample and 3 xenografts (SJNBL046_X) derived from SJNBL046_D. This includes an early passage, a late passage and a matched metastatic lesion of SJNBL046_X. Abbreviations: H&E, hematoxlyin and eosin; DCV, dense core vesicles; m, mitochondria. Scale bars: A–F: 50 μm.
We characterized the genomic landscape of the patient’s tumor (SJNBL046_D) and germline by whole genome sequencing as described previously (Cheung et al., 2012). The genome of SJNBL046_D was typical of high-risk stage 4 MYCN amplified neuroblastoma (Fig. 1I). Gene expression array analysis of SJNBL046_D and the xenografts derived from the patient tumor (see below) was similar to other MYCN amplified neuroblastomas (Fig. 1J).
2.2. Characterization of a neuroblastoma orthotopic xenograft
Six months after initial implantation into the para-adrenal space of NSG mice (Fig. 2A–C), we identified a mass consistent with neuroblastoma, which was confirmed by ultrasound. The animal was euthanized and the primary tumor at the site of injection was removed (Fig. 2D). We also identified metastatic lesions in the liver with extensive infiltration (Fig. 2E). Histopathological and transmission electron microscopic analyses confirmed that the orthotopic xenograft (SJNBL046_X) had cellular features of the patient’s tumor (SJNBL046_D) (Fig. 2F–J). The tumor cells on the liver were infiltrating and invading the normal liver tissue (Fig. 2K–M). The dissociated tumors were cryopreserved and also injected into an additional cohort of NSG mice to expand and propagate the xenograft. With subsequent passage, the tumors grew more rapidly (4–6 weeks) than the original diagnostic tumor.
Fig. 2.
Establishing an orthotopic xenograft of SJNBL046. (A) Drawing of method for para-adrenal injections. The injection procedure is a closed procedure using ultrasound guidance. (B) Ultrasound image of needle immediately after deposition of SJNBL046_D cells in matrigel (red) adjacent to the adrenal gland (blue). (C) 3D reconstruction of the kidney (green), adrenal gland (blue) and deposited tumor cells in matrigel (red). (D) Photograph of a large primary SJNBL046_X mass surrounding the adrenal and kidney and expanding into the abdominal cavity. (F and G) Low and high power micrographs of H&E stained sections of the SJNBL046_X tumor. (H and I) Micrographs of chromogranin and synaptophysin immunohistochemical staining of sections of SJNBL046_X. (J) Transmission electron micrograph of SJNBL046_X showing dense core vesicles (inset arrows). (K and L) Micrographs of chromogranin and synaptophysin staining of metastatic lesions from SJNBL046_X in the liver. (M) Electron micrograph of metastatic SjNBL046_X cells invading the liver (red shading). Scale bars: F, H, I, K, L: 50 μm; G: 10 μm.
In addition to the cellular and histological characterization, we also performed detailed molecular and genomic profiling of SJNBL046_X. SNP 6.0 analysis of the patient’s germline DNA, diagnostic tumor (SJNBL046_D), primary and metastatic xenografts (SJNBL046_X) showed that they retained MYCN amplification and the other chromosomal lesions (chromosomes 1, 15 and 17) found in the diagnostic tumor (Fig. 3A). More importantly, SJNBL046_X did not acquire any additional large chromosomal gains or losses. Spectral karyotype analysis (SKY) confirmed these results and provided additional information on the chromosomal lesions (Fig. 3B). In particular, fragments of chromosome 17 that were gained in SJNBL046 were fused to chromosome 13 and 22. The gain on chromosome 1 was fused to chromosome 7 and portions of chromosome 7 and 15 were fused to chromosome 1. The MYCN amplification was a double minute as validated by fluorescence in situ hybridization (FISH) (Fig. 3C). A comparison of the gene expression array data of the diagnostic tumor (SJNBL046_D) and the orthotopic xenograft (SJNBL046_X) showed a correlation coefficient of 0.92 (Fig. 3D). The most significant upregulated genes in the xenograft were associated with cell cycle and the most downregulated genes were associated with extracellular matrix organization and cell adhesion (Supplemental information).
Fig. 3.
Characterization of the SJNBL046 xenograft. (A) Diagram of copy number variations from SNP 6.0 analysis of the SJNBL046_D tumor and a sample from the primary site of engraftment (para-adrenal) and a metastatic site (liver). Copy number gains are shown in red and losses are shown in blue. The MYCN amplification is indicated with an arrow. (B) Spectral karyotype analysis of SJNBL046_X showing the MYCN amplification on a double minute fragment and several translocations, losses and gains. The SKY analysis agrees with the copy number analysis in (A). (C) Fluorescence in situ hybridization for MYCN in a metaphase SJNBL046_D cell. The DNA fragments that hybridized to the MYCN probe are shown in green and the metaphase chromosomes are stained with DAPI (blue). (D) Scatterplot of correlation in gene expression arrays for SJNBL046_D and SJNBL046_X with a 0.92 correlation coefficient. (E) Circos plot showing the coding region single nucleotide variations (SNV) across SJNBL046_D and SJNBL046_X. Those genes with SNVs in both the SJNBL046_D and SJNBL046_X samples are shown in black and those unique to SJNBL046_X are in blue. (F) Heat map of correlation of DNA methylation (Illumina 450K) across SJNBL046_D and two SJNBL046_X samples (primary and metastatic). Abbreviations: SNV, single nucleotide variation.
We also performed whole exome sequencing of the orthotopic xenograft and compared the exonic single nucleotide variations (SNVs) in the xenograft to those identified in the diagnostic tumor by whole genome sequencing (Cheung et al., 2012). We identified a series of missense and nonsense mutations in genes that arose in the xenograft and were not present in the diagnostic tumor (Fig. 3E and Table S1) but very few were in known cancer genes or have been found to be recurrently mutated in neuroblastoma. We also performed DNA methylation analysis using the Illumina 450k beadplex platform on the diagnostic tumor, and the xenografts from the primary and metastatic sites. There was good agreement between the primary tumor and each xenograft with 93% and 91% correlation (Fig. 3F and Table S2). There was 98% correlation between the two xenograft samples from the primary and metastatic sites (Fig. 3F and Table S2).
2.3. Targeting the PI3K/AKT/mTOR pathway in SJNBL046_X
Neuroblastomas are highly vascular tumors that often wrap around major abdominal vessels. Activation of the PI3K/AKT/mTOR pathway frequently occurs in neuroblastoma and correlates with a poor prognosis (Opel et al., 2007). mTOR is an important regulator of cell growth and autophagy, and its activity in neuroblastoma cells has been shown to impact cell differentiation by regulating the balance between macromolecule synthesis and degradation (Zeng and Zhou, 2008). Moreover, preclinical studies have shown that small molecule inhibitors of this pathway can have an anti-angiogenic effect on neuroblastoma in vivo (Chanthery et al., 2012). To establish a standardized procedure for preclinical testing of novel therapeutics, we developed an integrated database using OpenClinica. We performed a randomized, double blind study comparing standard of care chemotherapy and molecular targeted therapy for the PI3K/AKT/mTOR pathway. For standard of care (SOC), the first course of chemotherapy was cyclophosphamide, doxorubicin and etoposide (CAE) and the second course was etoposide and cisplatin (CE) (Supplemental information). Alternating CAE/CE treatments were repeated for a total of 4 courses over 12 weeks (Fig. 4A and B). To target the PI3K/AKT/mTOR pathway, we treated mice for 2 courses with SOC followed by OSI-906 with BEZ235 or OSI-906 with BKM120 on a dailyx5 schedule (Fig. 4C and D). OSI-906 is a small molecule inhibitor of the IGFR1 receptor, BEZ235 is a small molecule inhibitor of PI3K and mTOR and BKM120 is a pan-PI3K inhibitor. The IGFR1 receptor signals through the PI3K/AKT/mTOR pathway. All three drugs are currently in clinical development for cancer patients and we used plasma pharmacokinetics to achieve clinically relevant doses in mice (data not shown).
Fig. 4.
Preclinical testing using SJNBL046_X. (A–D) Diagram of the dosing schedule for standard of care chemotherapy (CAE, CE) and therapy targeting the PI3K pathway. (C and D) Drug doses were AUC guided based on pharmacokinetic studies performed on mice and humans. (E) Kaplan–Meier curves for each of the treatment groups. All mice in treatment groups received standard of care (SOC) for the first 2 courses (black shading). The placebo group received no treatment during these first 2 courses (red line). After the first 2 courses, one cohort continued on with SOC treatment and the other groups received the specified PI3K targeted therapy. (F–I) Estimated doubling time from the analysis of tumor volume and immunhistochemical staining for each treatment group at each course of treatment. Statistical analysis is presented in the Supplemental information. Representative ultrasound image and 3D reconstruction of a tumor with complete response (J) and progressive disease (K).
Initially, 120 mice received para-adrenal injections of 10 μl SJNBL_046X in matrigel (20,000 cells/μl). The mice were monitored weekly by ultrasound and when the tumor volume was 6–8 mm3, they were assigned a mouse medical record number (mMRN) and enrolled on study (Supplemental information). A total of 80 mice were enrolled on the study (20 per treatment group) and we used a randomized double blind preclinical trial design (Supplemental information). The mice were randomized to 4 treatment groups (placebo, SOC, OSI-906+BEZ235 and OSI-906+BKM120) and were treated for 4 courses (12 weeks) (Fig. 4A–D). The tumor volume was measured every 3 weeks by ultrasound and animals were euthanized when tumor burden reached 20% of body weight. MRI was also used to validate the ultrasound volumetric data (Supplemental information). When the animals were euthanized at the end of study or due to progressive disease, the tumors were processed and embedded for histopathological analysis.
Survival times among the four groups showed significant differences using the log-rank test (p=0.001) or the Gehan–Wilcoxon test (p<0.001) and each of the 3 treatment arms were significantly different from the placebo group (Fig. 4E). However, pairwise comparison of the survival times between the treatment arms were not statistically significant (Supplemental information). The OSI-906+BKM120 treatment arm had greater mean and median survival time compared to all other treatment arms (Fig. 4E and Supplemental information). We quantitated the rate of change in tumor volume by calculating the doubling time between two successive pairs of volumetric measurements using ultrasound (Fig. 4F–K). In the SOC treatment arm, 23% of the tumors diminished in size, 7% remained stable and 70% showed slower growth compared to placebo (placebo: 4 ± 1 days vs. SOC: 16 ± 11 days, p<0.001). None of the tumors in the placebo group decreased in size or regressed during the study. There was no significant difference in growth doubling times among any of the treatment groups (Fig. 4G–I).
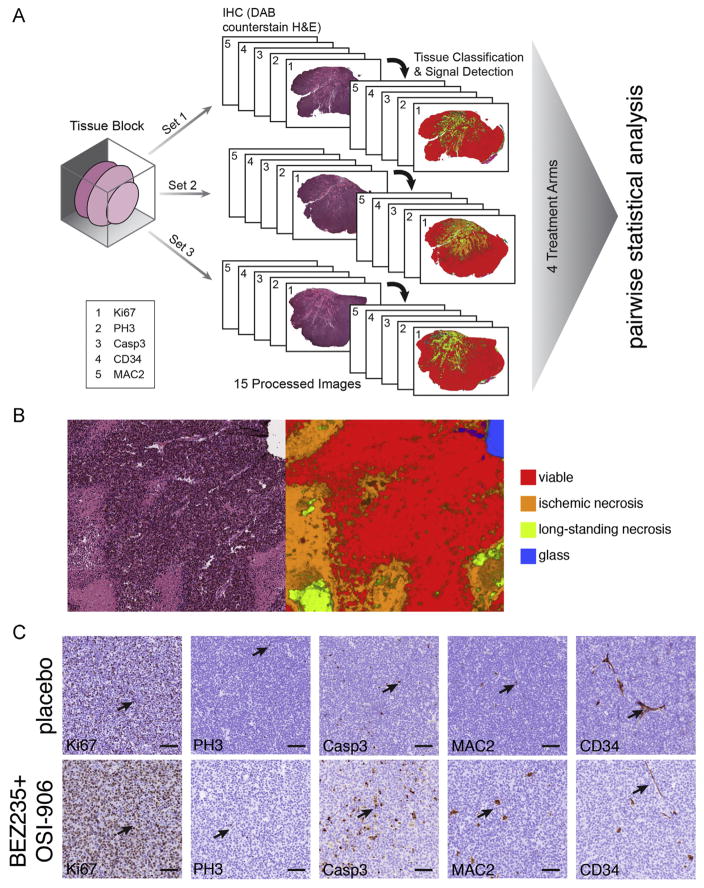
To relate the tumor response across treatment arms to cellular mechanisms of each drug combination, we performed histopathological analysis of the tumors. From the 80 mice enrolled on study, there were 51 tumors available for analysis. For each tumor, 4 μm sections were cut to prepare 115 slides for each tissue block for a total of 5865 slides for this study. We used three sets of 5 serial slides per tumor with 200 μm between each set. Sequential slides in each set were stained with Ki67 to monitor proliferation, phospho-histone H3 to monitor M-phase, activated caspase 3 to monitor cell death, Mac2 to monitor macrophage infiltration and CD34 to monitor tumor vasculature (Fig. 5A). Each slide was digitally scanned and then linked to the individual mouse data in OpenClinica. An image processing pipeline was developed to quantitate proliferation, apoptosis, angiogenesis, macrophage infiltration and necrosis in each of the 51 tumors (Supplemental information).
Fig. 5.
Histological analysis of SJNBL046_X tumors after treatment. (A) Drawing of the scheme used to sample the tumor tissue isolated from mice that came off of study. Three regions of tumor were processed separately and serial sections were collected to produce 3 distinct sets for each tumor. After staining, the slides were scanned and images were segmented and scored using the automated system described in the Supplemental information. (B) Representative H&E stained section of a SJNBL046_X tumor and the matched segmentation and automated scoring of the tissue for viable (red), ischemic necrotic (orange) and long-standing necrotic tissue (yellow). (C) Representative micrographs of immunohistochemical staining of a tumor from a mouse who received placebo and one that received BEZ-235+OSI-906. Arrows indicate immunopositive cells for each antibody. Scale bars: 50 μm. Abbreviations: Ki67 labels the nuclei of proliferating cells in any stage of the cell cycle. PH3 labels cells in S-phase. Casp3 labels cells undergoing cell death. CD34 labels vascular endothelial cells and MAC2 labels macrophages.
Our automated image processing pipeline was used to segment the tumor tissue scans and measure the area of viable tumor, ischemic necrosis, long standing necrosis, extracellular matrix/stroma/fat, tissue folds, liver, kidney, spleen, muscle and glass based on the texture and H&E staining properties (Fig. 5B–D and Supplemental information). The algorithm achieved greater than 95% accuracy per region in cross-validation tests (Supplemental information). SJNBL_046X is a fast growing tumor that outgrows its vasculature and has large areas of necrotic tissue in untreated mice. The viable-to-total-necrotic tissue ratio was significantly higher in the SOC and OSI-906+BEZ235 treatment groups (p<0.001) (Fig. 5E) and this is consistent with previous reports that apoptosis and necrosis are inversely correlated in rapidly growing tumors such as SJNBL_046X (Arends et al., 1994). Both the SOC and OSI-906+BKM120 treatment arms showed increased levels of pH3 and Ki67 relative to placebo when normalized to the viable tumor area (Fig. 5E). In addition to inhibiting PI3K, it has been reported that BKM120 can induce G2/M-phase arrest (Brachmann et al., 2012). Consistent with this observation, the pH3/Ki67 ratio was higher in OSI-906+BKM120 treated tumors compared to placebo or OSI-906+BEZ235 (Fig. 5F). The proportion of apoptotic cells (activated caspase 3 immunopositive) was significantly higher in the viable tumor regions for the OSI-906+BKM120 treatment arms compared to placebo (Fig. 5G). The other treatment arms did not reach statistical significance. It has been previously reported that targeting the PI3K/AKT/mTOR pathway in tumor models in vivo can have anti-angiogenic effects (Chanthery et al., 2012; Cho et al., 2010; Guba et al., 2002). Consistent with these previous reports, the OSI-906+BEZ235 and OSI-906+BKM120 treatment arms showed significant reduction in CD34 immunostaining for tumor vasculature in the viable regions of the tumor (Fig. 5H). Finally, we analyzed macrophage infiltration in the viable and necrotic regions of the tumor. There was a significant reduction in macrophages in both viable and necrotic regions of tumors treated with OSI-906+BEZ235 and OSI-906+BKM120 (Fig. 5I).
2.4. High throughput screening of primary neuroblastoma xenograft
To determine if the human neuroblastoma orthotopic xenograft described here (SJNBL046_X) could be used for testing drug sensitivity, we developed a method for dissociating and plating tumor cells in 384 well plates. Briefly, the tumor is dissociated into a single cell suspension with trypsin and filtered through a cells strainer to remove debris (Fig. 6A, Day 0). The contaminating red blood cells are eliminated and the tumor cells are plated in 384 well dishes in matrigel. Once the matrigel polymerizes, the cells are overlaid with cell culture medium (Supplemental information) and placed in a cell culture incubator (Fig. 6A). Twenty four hours later drugs are added in triplicate dose response using a pin tool (Fig. 6A, Day 1). Seventy two hours later, cell titer glow is added to the plate and the ATP levels are quantitated on a plate reader (Fig. 6A, Day 4). Positive and negative control compounds are included in each experiment for data normalization. Validated data are then entered into a central database and curve fitting is performed to generate an EC50 for each drug (Fig. 6B and C). Overall, the SJNBL046_X cells are much less sensitive to chemotherapy than neuroblastoma cell lines. The most sensitive cell line analyzed in this study was IMR32 and SK-N-AS was the least sensitive. These data demonstrate the feasibility of performing high throughput drug screening with a primary orthotopic neuroblastoma xenograft.
Fig. 6.
High-throughput screening of SJNBL046_X primary cultures. (A) Drawing of the scheme used for primary cultures of orthotopic xenografts in the screening platform. On day 0, the tumor is isolated and processed into a single cell suspension that is then plated into 384 well dishes. The next day (Day 1) the dishes are drugged in dose response in triplicate and then incubated for 72 h. Cell-titer glo is used to measure the level of ATP in the cultures as a proxy for the number of viable cells. (B) Representative dose response curves for some of the active drugs against neuroblastoma cell lines and the SJNBL046_X xenograft. (C) Heatmap of the entire drug library tested in this study.
3. Materials and methods
3.1. Mast protocol and patient samples
The study, Molecular Analysis of Solid Tumors (MAST), is a biology study at St. Jude Children’s Research Hospital designed to prospectively characterize the molecular, cellular and genetic properties of primary and metastatic pediatric solid tumors. Additionally, if a participant agrees, it allows for the establishment of cell lines and xenograft models. Patients are eligible to participate if they have a solid malignancy and are less than or equal to 25 years old. The MAST protocol was approved by the institutional review board of St. Jude Children’s Research Hospital and the parents of SJNB046 provided written informed consent in accordance with institutional and federal guidance.
3.2. Immunohistochemistry and TEM analysis
The formalin-fixed-paraffin-embedded block of the xenograft tissue was cut at 4μm sections and processed for immunohistochemistry using heat-induced epitope retrieval with ER1 for 20–30 min. Mouse monoclonal antibodies against synaptophysin (27G12; Leica Microsystems) and chromogranin A (LK2H10(3); Leica Microsystems) were applied at 1:200 dilution with 15-min incubation at room temperature using the Leica Bond (Leica Microsystems) staining platform and the Bond Refine Polymer detection kit.
3.3. WGS and WES
Whole-genome sequencing (WGS) and SNP or gene expression profiling by array were performed as previously described (Zhang et al., 2012a, 2012b). For WGS, paired-end sequencing was performed using the Illumina GAIIx or HighSeq platform with 100 bp read length. The WGS data are deposited at the European Bioinformatics Institute (EBI) with accession number EGAS00001000256. WGS mapping, coverage and quality assessment, single nucleotide variation (SNV)/indel detection, tier annotation for sequence mutations, prediction of deleterious effects of missense mutations, and identification of loss of heterozygosity (LOH) have been described previously (Zhang et al., 2012b) Structural variations (SVs) were analyzed using CREST and annotated as previously described (Wang et al., 2011; Zhang et al., 2012b). The reference human genome assembly NCBI Build 36 was used for mapping 5 samples (SJRHB001–005) and Build 37 was used for mapping other samples. Genomic coordinates for variations in samples mapped to Build 36 were lifted over to Build 37. Copy number variations (CNVs) were identified by evaluating the difference of read depth for each tumor and its matching normal using a novel algorithm, COpy Number SEgmentation by Regression Tree In Next-Gen sequencing (CONSERTING).
For whole exome sequencing (WES), DNA libraries were prepared from 1 μg of whole genome amplified (WGA) material from matched samples using the Illumnia TruSeq DNA library prep kit following the recommended manufacturer’s protocol. Libraries were analyzed on an Agilent Bioanalyzer to inspect quality of each library construction. Germline and diagnostic library samples were independently pooled and applied for exome capture using the Illumina TruSeq Exome Enrichment kit as described by the manufacturer. Captured libraries were then clustered on the Illumina c-bot and were sequenced on an Illumina HiSeq 2000 platform with 100 base pair end multiplexed reads at an equivalent of 3 samples per lane.
For enrichment and validation of the regions containing putative alterations, genomic coordinates of the putative WGS targets were used to order either Nimbelgen Seqcap EZ solution bait sets (Roche), or Nimblegen Seqcap 2.1M arrays (Roche). The library construction and target enrichment was performed per manufacturer’s instructions (Roche) using repli-G (Qiagen) WGA DNA. Enriched targets were sequenced on the illumina platform using paired end 100 cycle sequencing. The resulting data was converted to FASTQ files using CASAVA 1.8.2 (Illumina), and mapped with BWA prior to pipeline analysis.
Putative SNVs from exon sequencing were validated by NGS amplicon sequencing. Briefly, primers were designed to genomic regions (hg19) flanking the SNV no closer than 100 base pairs to the detected SNV. PCR was performed using 20–30 ng of whole genome amplified (WGA) DNA from each patient sample. DNA from the tumor (diagnostic sample) and a matched germline sample were used for each primer set to confirm the presence of the SNV in the diagnostic sample. Standard PCR was performed in 25 μl reactions using Accuprime GC-rich DNA polymerase (Invitrogen) with the following parameters: 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min followed by a 72 °C 10 min extension with cooling to 4 °C. All PCR amplicons were checked on a 2% E-gel (Invitrogen) to ensure single amplified products.
PCR amplified products were collected into diagnostic or germ-line pools and purified using a Qiagen PCR purification kit. One nanogram of pooled sample was treated for sequencing using the Nextera XT kit (Illumina) following the manufacturer’s protocol. Samples were normalized and denatured prior to the MiSeq run using the 2 ×150 base pair run (version 1) reagent kit.
3.4. Gene expression array analysis
RNA from the primary tumor (SJNBL046_D) and xenograft (SJNBL046_X) were assayed with the Affymetrix HT HG-U133+ PM Array Plate (NIH GEO Platform GPL13158). The data was then RMA summarized (Irizarry et al., 2003) using Partek Genomics Suite 6.12 (St Louis, MO). Scatterplots, log2ratios and correlations were produced using STATA/MP 11.2 (College Station, TX). SJNB046_D/X data was joined by probeset sequence to 125 public human neuroblastoma microarrays analyzed as previously described (Teitz et al., 2011). After platform corrected the joined data set was visualized by principal component analysis (PCA) using Partek 6.6.
3.5. SNP 6.0 analysis
SJNBL046 germline, primary tumor, xenograft and metastasis sample DNA was assayed on GenomeWideSNP_6 Affymetrix arrays. The data was normalized and logratio and copy number were estimated using the Partek Genomics Suites 6.3 workflow. Data was then visualized in Spotfire DecisionSite.
3.6. Ultrasound guided para-adrenal injection
All ultrasound procedures were performed using the VEVO-770, fitted with a RMV-706 probe. Cells were suspended in Matrigel (BD Worldwide, Cat#354234) at a concentration of 2 ×104 cells per microliter and placed on ice. Anesthetized recipient CD1 nude mice (isoflurane 1.5% in O2 delivered at 2 l/min) were placed laterally on the imaging bed such that the left flank faced upward. In order to provide a channel for delivery of the implant, a 22 gauge catheter (BD Worldwide, Cat#381423) was gently inserted through the skin and back muscle into the para-adrenal region and the hub was removed. A chilled Hamilton syringe fitted with a 27 gauge needle (1.5 in.) and loaded with 10 μl of the cell suspension was guided steretoactically through the catheter and positioned between the kidney and adrenal gland as visualized using ultrasound. The cells were injected into the region and the needle was left in place for 30 s to permit the matrigel component to set. The needle was then slowly removed, followed by gentle removal of the catheter.
3.7. Neuroblastoma xenograft dissociation and banking
Tumor was harvested from SJNB046_X tumor bearing NSG mice. Tumor was minced with sterile scalpels and rinsed in phosphate buffered saline without calcium or magnesium (PBS). Tumor suspension was transferred to a 50 ml conical tube and filled with PBS. Dissociation was done by adding 600 μl of trypsin (10 mg/ml, Sigma Cat#T9935) and the tube was incubated 37 °C for 10 min. Dissociation was stopped by adding 600 μl of Soybean Trypsin Inhibitor (10 mg/ml, Sigma Cat#T6522). Deoxyribonuclease I (2 mg/ml, Sigma Cat#D4513) and magnesium chloride (1 M) were added in equal amounts of 60 μL increments until tumor fragments easily settle at the bottom of the tube. The dissociated tumor cell suspension was filtered with a 40 μm cell strainer and then centrifuged at 450g (G=RCF) for 5 min. Supernatant was discarded and 10 ml of red blood cell lysis solution (5 Prime Cat#2301310) added and incubated at room temperature for 10 min. PBS with 10% FBS (Biowest Cat#SO1520) was added to fill 50 ml conical tube and cell suspension was centrifuged at 450g (G=RCF) for 5 min. The supernatant was discarded and the cell pellet was resuspended in PBS/10% FBS for counting.
3.8. SKY
After a 4 h colcemid incubation, cells from this sample were harvested using routine cytogenetic methods. A commercially prepared SKY probe from Applied Spectral Imaging (Carlsbad, CA) was used as the probe for this analysis. Applied Spectral Imaging protocols were followed for the hybridization and detection steps. Sixteen metaphase cells were analyzed which were determined to be male in origin. 69% of analyzed metaphase cells were found to have the following karyotype determined by SKY as 45,XY,t(1;7)(q?21;p?13)der(1)t(1;15)(p?31;?),der(13)t(13;17)(p? 13;?),-17,der(22)t(17;22)(?;p?13),dmin(2). The remaining 31% of analyzed metaphase cells were found to have the following karyotype determined by SKY analysis as 46,XY,t(1;7)t(q?21;p? 13)der(1)t(1;15)(p?31;?),der(13)t(13;17)(p?13;?),der(22)t(17;22) (?;p?13),dmin(2).
4. Discussion
In this study, we describe the detailed characterization of a patient derived orthotopic xenograft of a high risk pediatric neuroblastoma tumor. This is one example of a broader collection of more than 50 orthotopic xenografts of pediatric solid tumors that are made available to the research community free of charge through the childhood solid tumor network (CSTN). Not only are these preclinical models of pediatric cancer useful for studying the basic biology of their respective diseases, but they can be used for preclinical testing and high-throughput drug screening of primary cultures derived from the xenografts. Moreover, they provide a well-validated and renewable source of cells for future epigenetic and proteomic studies that are difficult to carry out on primary patient tumors.
4.1. Orthotopic xenografts preserve features of primary tumor
Many key features of the high-risk neuroblastoma sample described here in the patient tumor were preserved in the orthotopic xenograft. The histopathological features of the patient tumor were preserved in the xenograft with robust expression of antigens typically found in neuroblastoma including synaptophysin and chromogranin. Importantly, key subcellular features of neuroblastoma tumors including dense core vesicles were found in the primary patient tumor and the xenograft. The gene expression profile and DNA methylation profile of the xenograft were similar to the patient’s primary tumor. The differences that were observed were associated with normal cells found within the tumor such as macrophages and vascular endothelial cells. The xenografted tumor does have vasculature and infiltration of macrophages but those are contributed by the mouse and are therefore not detected in the human array platforms. As in the patient, the xenografted tumor was aggressive with metastasis to the liver, spleen, lymph nodes and other sites in over 70% of mice. While some of the metastatic lesions could have been the result of seeding associated with ultrasound guided para-adrenal injections, the cells in lymph node and lung were likely the result of dissemination from the primary tumor. Also, even in the liver, the cells infiltrated into the tissue suggesting that they retained the ability to disrupt the cell–cell junctions and invade normal organs associated with neuroblastoma metastasis. One notable exception for metastasis was the bone marrow, which is a common site of metastasis in human neuroblastoma. The xenografted tumors did not metastasis as efficiently to the bone marrow as noted in other orthotopic neuroblastoma xenografts (Braekeveldt et al., 2015). It is possible that the immunocompromised status of these mice prevents the tumors from metastasizing to the bone marrow because the microenvironment is altered. Future studies with humanized mice will be useful to test this hypothesis.
Exome sequencing identified a series of SNVs that were unique to the xenograft. They were not present in the whole genome sequence data from the original patient’s tumor. It is possible that they were from a minor clone in a distinct region of the primary patient tumor that had a growth advantage in the xenograft or that they were acquired in the xenograft with passage. It is not known if those SNVs contribute to tumor progression or if they are simply passenger mutations. At the chromosomal level, there were no significant changes in copy number variations, structural variations or aneuploidy in the xenograft compared to the primary tumor. These data show the value of comprehensive molecular, cellular and genetic characterization of orthotopic xenografts to ensure that they faithfully recapitulate the patient tumor. All of the tumors described in the companion paper titled “The Childhood Solid Tumor Network: A New Resource for the Developmental Biology and Oncology Research Communities” are currently undergoing similar characterization.
Importantly, in a previous study published by Braekeveldt, they also showed that patient-derived neuroblastoma xenografts retained molecular, cellular, genetic and histopathologic features of the primary tumor (Braekeveldt et al., 2015). Thus, in two independent laboratories, neuroblastoma xenografts have been generated that retain the genetic and phenotypic features of the patient tumor and can be passaged in vivo and in culture. These patient derived xenografts have many advantages over cell lines that have been maintained in culture for many years and will provide a useful resource for the biomedical research community.
4.2. Preclincial testing and drug screening using orthotopic xenografts
In order to demonstrate the feasibility of using the orthotopic xenografts for preclinical studies, we tested standard of care induction chemotherapy and molecular targeted therapeutic combinations previously shown to have activity for neuroblastoma in mouse models. Key features of our study are the use of AUC guided dosing and clinically relevant schedules of drug combinations. Also, the mice were randomized into different treatment arms and the study was performed double blind to minimize any experimental bias. We also entered all the primary data from each mouse and the histopathological analysis of the individual tumors into a central database using OpenClinica. This allows us to share the raw data with other investigators and provides a convenient platform to test hypotheses and identify biologically relevant correlations. Finally, we showed that the orthotopic xenografts can be cultured as primary acute cultures and used to test drug sensitivity in vitro. While this does not recapitulate the complexity of the tumor microenvironment in vivo, it does allow researchers to test a large number of individual drugs quickly. In addition, this technique will also allow for more sophisticated in vitro studies of drug combinations as novel agents become available. Promising new therapeutic agents can then be efficiently moved in vivo using our standard procedure for preclinical testing. This is also important because it suggests that the agents tested here (OSI-906, BEZ235 and BKM120) have very limited efficacy in this high-risk orthotopic neuroblastoma xenograft model when combined with induction chemotherapy.
Supplementary Material
Acknowledgments
This work was supported, in part, by Cancer Center Support (CA21765) from the NCI, grants to M.A.D. from the NIH (EY014867 and EY018599 and CA168875), and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. was also supported by a grant from Alex’s Lemonade Stand Foundation for Childhood Cancer and HHMI. Finally, we thank John and Andra Tully and the Tully Family Foundation for generous support of Pediatric Solid Tumor Research at St. Jude Children’s Research Hospital. All animal pathology support was provided by the Veterinary Pathology Core Laboratory of St. Jude Hospital.
Appendix A. Supplementary information
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.ydbio.2015.02.002.
References
- Arends MJ, McGregor AH, Wyllie AH. Apoptosis is inversely related to necrosis and determines net growth in tumors bearing constitutively expressed myc, ras, and HPV oncogenes. Am J Pathol. 1994;144:1045–1057. [PMC free article] [PubMed] [Google Scholar]
- Brachmann SM, Kleylein-Sohn J, Gaulis S, Kauffmann A, Blommers MJ, Kazic-Legueux M, Laborde L, Hattenberger M, Stauffer F, Vaxelaire J, et al. Characterization of the mechanism of action of the pan class I PI3K inhibitor NVP-BKM120 across a broad range of concentrations. Mol Cancer Ther. 2012;11:1747–1757. doi: 10.1158/1535-7163.MCT-11-1021. [DOI] [PubMed] [Google Scholar]
- Braekeveldt N, Wigerup C, Gisselsson D, Mohlin S, Merselius M, Beckman S, Jonson T, Borjesson A, Backman T, Tadeo I, et al. Neuroblastoma patient-derived orthotopic xenografts retain metastatic patterns and geno- and phenotypes of patient tumours. Int J Cancer. 2015;136:E252–E261. doi: 10.1002/ijc.29217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanthery YH, Gustafson WC, Itsara M, Persson A, Hackett CS, Grimmer M, Charron E, Yakovenko S, Kim G, Matthay KK, Weiss WA. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci Transl Med. 2012;4:115ra113. doi: 10.1126/scitranslmed.3002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. J Am Med Assoc. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlenski A, Liu S, Cohn SL. The regulation of angiogenesis in neuroblastoma. Cancer Lett. 2003;197:47–52. doi: 10.1016/s0304-3835(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Cho DC, Cohen MB, Panka DJ, Collins M, Ghebremichael M, Atkins MB, Signoretti S, Mier JW. The efficacy of the novel dual PI3-kinase/mTOR inhibitor NVP-BEZ235 compared with rapamycin in renal cell carcinoma. Clin Cancer Res. 2010;16:3628–3638. doi: 10.1158/1078-0432.CCR-09-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guba M, von Breitenbuch P, Steinbauer M, Koehl G, Flegel S, Hornung M, Bruns CJ, Zuelke C, Farkas S, Anthuber M, et al. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat Med. 2002;8:128–135. doi: 10.1038/nm0202-128. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Khanna C, Jaboin JJ, Drakos E, Tsokos M, Thiele CJ. Biologically relevant orthotopic neuroblastoma xenograft models: primary adrenal tumor growth and spontaneous distant metastasis. In Vivo. 2002;16:77–85. [PubMed] [Google Scholar]
- Kushner BH, Kramer K, LaQuaglia MP, Modak S, Yataghene K, Cheung NK. Reduction from seven to five cycles of intensive induction chemotherapy in children with high-risk neuroblastoma. J Clin Oncol. 2004;22:4888–4892. doi: 10.1200/JCO.2004.02.101. [DOI] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- Opel D, Poremba C, Simon T, Debatin KM, Fulda S. Activation of Akt predicts poor outcome in neuroblastoma. Cancer Res. 2007;67:735–745. doi: 10.1158/0008-5472.CAN-06-2201. [DOI] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr, Papadopoulos N, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstrom L, Baryawno N, Sveinbjornsson B, Wickstrom M, Elfman L, Kogner P, Johnsen JI. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int J Cancer. 2011;129:2958–2965. doi: 10.1002/ijc.26268. [DOI] [PubMed] [Google Scholar]
- Teitz T, Stanke JJ, Federico S, Bradley CL, Brennan R, Zhang J, Johnson MD, Sedlacik J, Inoue M, Zhang ZM, et al. Preclinical models for neuroblastoma: establishing a baseline for treatment. PLoS One. 2011;6:e19133. doi: 10.1371/journal.pone.0019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Zhou JN. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell Signal. 2008;20:659–665. doi: 10.1016/j.cellsig.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, Ulyanov A, Wu G, Wilson M, Wang J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012a;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012b;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.