Abstract
The nonheterocystous filamentous cyanobacterium, strain ESFC-1, is a recently described member of the order Oscillatoriales within the Cyanobacteria. ESFC-1 has been shown to be a major diazotroph in the intertidal microbial mat system at Elkhorn Slough, CA, USA. Based on phylogenetic analyses of the 16S RNA gene, ESFC-1 appears to belong to a unique, genus-level divergence; the draft genome sequence of this strain has now been determined. Here we report features of this genome as they relate to the ecological functions and capabilities of strain ESFC-1. The 5,632,035 bp genome sequence encodes 4914 protein-coding genes and 92 RNA genes. One striking feature of this cyanobacterium is the apparent lack of either uptake or bi-directional hydrogenases typically expected within a diazotroph. Additionally, a large genomic island is found that contains numerous low GC-content genes and genes related to extracellular polysaccharide production and cell wall synthesis and maintenance.
Keywords: Cyanobacteria, Nitrogen fixation, Hydrogenase, Intertidal microbial mat
Introduction
Microbial mats played a key role in the evolution of the early Earth and today provide a model system for exploring relationships between evolution, ecology, and biogeochemical cycles. In many mats, nitrogen-fixing filamentous Cyanobacteria are often central components with important roles in carbon, nitrogen and sulfur cycling [1, 2]. Recently, a previously unknown lineage of filamentous nitrogen-fixing Cyanobacteria was described in intertidal microbial mats from Elkhorn Slough, Moss Landing, California [3]. The type strain of this organism, ESFC-1, lacks both heterocysts and an extracellular sheath and has been shown to be an important cyanobacterial diazotroph in the Elkhorn Slough system [3]. At Elkhorn Slough this strain is often a dominant cyanobacterial member of the community (along with Cyanobacteria closely related to Coleofasciculus chthonoplastesPCC 7420); the sequence abundance of ESFC-1 in 16S rRNA libraries based on DNA and cDNA has been observed to reach up to 5 % (based on pyrosequencing) and 33–36 % (based on clone libraries and pyrosequencing), respectively [3, 4]. Although it is not always dominant, ESFC-1 is highly active, based on nifH transcript abundance and rRNA transcript to rRNA gene ratios [3, 5]. Recent work has shown that ESFC-1 produces a considerable external carbon pool as an EPS; this EPS is managed by means of an active exoproteome, and provides a source of organic carbon for the cyanobacterium and other community members [6]. Previous phenetic analyses using full-length 16S rRNA gene sequences have indicated this organism shares only a moderate identity with other identified Cyanobacteria; its best cultured BLAST hit is the marine Aphanocapsa sp. HBC6 at 93.6 % similarity [7, 8]. Given the importance of ESFC-1 in the Elkhorn Slough mat system and its evolutionarily divergent 16S rRNA, the genomic sequence was determined [3, 8]. Here we report a detailed description of the genome of ESFC-1 as it relates to the ecology of this important mat community organism.
Organism information
Classification and features
Strain ESFC-1 was isolated by L. Prufert-Bebout at NASA Ames Research Center in Moffett Field, California from the top 2 mm of intertidal microbial mat samples collected at Elkhorn Slough, California, USA. Fresh microbial mat was repeatedly streaked onto plates of a modified version of ASN artificial seawater medium, until a unialgal culture was obtained [3, 9]. Strain ESFC-1 is a motile, Gram negative, non-heterocystous filament (Fig. 1). Trichomes are cylindrical in shape and straight to slightly curved, with rounded to slightly conical ends. Individual cells are approximately 1.8 μm across, and cells are typically longer than wide, up to 3.5 μm in length, slightly longer than reported previously [3]. Constrictions between cells are shallow but clearly visible. Hormogonia and akinetes have not been observed. Heterocysts have not been observed in cells, even when actively fixing N2. Morphologically, ESFC-1 appears most similar to isolates of the form-genus Geitlerinema, but with a cell size more typical of the form-genus Leptolyngbya [10].
Fig. 1.
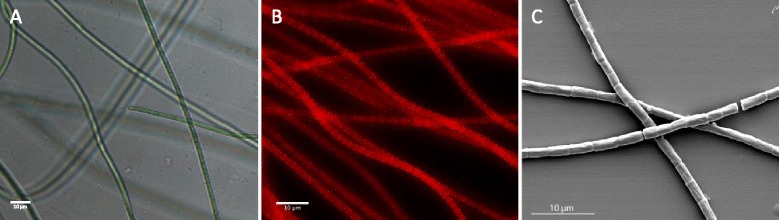
a Photomicrograph (400×) showing the filament morphology and size of Cyanobacterium ESFC-1, b Epifluorescent image of an ESFC-1 biofilm. Image is a calculated maximum intensity projection of a 50 μm z-stack. Red is autofluorescent ESFC-1 trichomes. Cells were fixed with 10 % formaldehyde prior to imaging. c Scanning electron microscopy image of ESFC-1 trichomes. ESFC-1 samples were fixed with 10 % formaldehyde, rinsed with sterile water, spotted onto a silicon wafer, air-dried and coated with ~5 nm of gold. Imaged with an FEI Inspect F SEM (Hillsboro, OR). For all panels, scale bar represents 10 μm
General features of ESFC-1 and project information are presented in Tables 1 and 2. In previous similarity and phylogenetic analyses based on the 16S rRNA locus, strain ESFC-1 did not show a close similarity (<94 %) with any other cyanobacterial sequence, and its phylogenetic placement within the cyanobacterial radiation was ambiguous [3, 8]. A 31-marker gene phylogenomic analysis of the cyanobacterial radiation, including ESFC-1, is presented in Fig. 2. This analysis places ESFC-1 with strong support in a clade with two non-diazotrophic Spirulina strains, PCC 6313 and PCC 9445 [10].
Table 1.
Classification and general features of strain ESFC-1 according to the MIGS recommendations [38]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [39] | |
| Phylum Cyanobacteria | TAS [40] | ||
| Class Cyanobacteria | TAS [10] | ||
| Order Oscillatoriales | TAS [10] | ||
| Family Oscillatoriaceae | TAS [10] | ||
| Genus Unclassified | TAS [3] | ||
| Species Unclassified | TAS [3] | ||
| strain: ESFC-1 | |||
| Gram stain | Negative | NAS | |
| Cell shape | Cylindrical cells in filaments | TAS [3] | |
| Motility | Motile | IDA | |
| Sporulation | Not reported | NAS | |
| Temperature range | Mesophilic | NAS | |
| Optimum temperature | Unknown | NAS | |
| pH range; Optimum | Unknown | NAS | |
| Carbon source | Photoautotroph | TAS [3] | |
| MIGS-6 | Habitat | Marine/Intertidal | TAS [3] |
| MIGS-6.3 | Salinity | Euryhaline; 3.5 % NaCl (w/v) | TAS [3] |
| MIGS-22 | Oxygen requirement | Aerobic | NAS |
| MIGS-15 | Biotic relationship | free-living | TAS [3] |
| MIGS-14 | Pathogenicity | Non-pathogenic | NAS |
| MIGS-4 | Geographic location | Elkhorn Slough, California, USA | TAS [3] |
| MIGS-5 | Sample collection | October, 2009 | TAS [3] |
| MIGS-4.1 | Latitude | 36°48′47″N | TAS [3] |
| MIGS-4.2 | Longitude | 121°47′5″W | TAS [3] |
| MIGS-4.4 | Altitude | 1.5 m | TAS [3] |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly ob-served for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [41]
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Improved High-Quality Draft |
| MIGS-28 | Libraries used | Two Illumina PE libraries: 222 bp avg. insert, and 7791 bp avg. insert |
| MIGS-29 | Sequencing platforms | Illumina |
| MIGS-31.2 | Fold coverage | 719× |
| MIGS-30 | Assemblers | Velvet v. 1.1.05; ALLPATHS v. r38445; Phrap v. 4.24 |
| MIGS-32 | Gene calling method | Prodigal 2.5 |
| Genbank ID | ARCP00000000 | |
| Genbank Date of Release | December 25, 2014 | |
| GOLD ID | Gi14129 | |
| BIOPROJECT | PRJNA165547 | |
| NCBI taxon ID | 1128427 | |
| MIGS-13 | Source Material Identifier | ESFC-1 |
| Project relevance | Cyanobacterial ecology |
Fig. 2.

Maximum likelihood (ML) phylogenomic analysis of the cyanobacterial radiation based on a concatenated amino acid sequences for 31 conserved loci [35, 42] showing the phylogenomic affiliation of ESFC-1 with two species of Spirulina (PCC 6313 and PCC 9445; clade in red). Only the portion of the larger tree corresponding to lineage B2 (sensu [42]) is shown. The full 126-taxon ML tree was built using PHYML using the LG protein substitution matrix, and was rooted with Chloroflexus auranticus J-10, Rhodobacter sphaeroides 2.4.1, Heliobacterium modesticaldum Ice1, and Chlorobium tepidum TLS [43–45]. ML bootstrap values for nodes >50 are shown; black boxes at the nodes denote bootstrap values of 100. Strain ESFC-1 and the nearest neighbors are highlighted in red
Genome sequencing information
Genome project history
Strain ESFC-1 was selected for sequencing because of its recent discovery as a major diazotroph in intertidal mat communities and its unique taxonomic position within the cyanobacteria. The genome project is deposited in the Genome On Line Database (GOLD Legacy ID Gi14129) and the complete genome sequence is deposited in GenBank (accession ARCP00000000). Sequencing, finishing and annotation were performed by the DOE-JGI. A summary of the project information is shown in Table 2.
Growth conditions and genomic DNA preparation
ESFC-1 was maintained in culture in liquid modified ASN at 25 °C on a 14:10 L:D cycle under cool fluorescent lamps at approximately 50 μmol photons · m-2 · s-1. High molecular weight genomic DNA was isolated based on the “JGI Bacterial DNA isolation CTAB” protocol from JGI [11], including an RNA digestion step according to the protocol. 50 μg of gDNA was provided to the JGI for sequencing.
Genome sequencing and assembly
The high-quality draft genome of strain ESFC-1 was generated by the DOE-JGI using the Illumina GAIIx platform [12]. An Illumina standard short-insert paired-end library with an average insert size of 222 bp +/− 50 bp generated 15,283,374 reads. An Illumina CLIP-PE long-insert paired-end library with an average insert size of 7791 +/− 660 bp generated 18,062,354 reads [13]. In total, 4099 Mbp of Ilumina data were generated.
The Illumina draft data was assembled with Allpaths, version r38445 [14], and contained 117 contigs in 14 scaffolds. The consensus was computationally shredded into 10 Kbp overlapping fake reads (shreds). The draft data was also assembled with Velvet, version 1.1.05 [15], and the consensus sequences were computationally shredded into 1.5 Kbp overlapping shreds. The Illumina draft data was reassembled with Velvet using the shreds from the first Velvet assembly to guide the reassembly. The consensus from this second Velvet assembly was shredded into 1.5 Kbp overlapping fake reads. Fake reads from the Allpaths and both Velvet assemblies were assembled using parallel phrap, version 4.24 (High Performance Software, LLC) with a subset of the Illumina CLIP-PE reads [16, 17]. Possible misassemblies were checked and manually corrected in Consed [18]. Gap closure was accomplished using repeat resolution software (Wei Gu, unpublished). The final assembly is based on 4099 Mbp of Illumina draft data, with an average genome coverage of 719×.
Genome annotation
The genome was annotated automatically with Prodigal 2.5 [19] by IMG [20], locally using the RAST server [21], and by GeneMarkS+ [22] by the NCBI annotation pipeline. Pathways of interest were mapped to the KEGG maps through both IMG and RAST.
Genome properties
The high quality draft genome of cyanobacterium ESFC-1 was resolved to 3 scaffolds consisting of 5,431,811, 135,349 and 64,875 bp, for a total of 5,632,035 bp. GC content was 46.47 %. The genome sequence is predicted to encode 5006 total genes, with 92 RNA genes, and 4914 protein-encoding genes. A majority (79.0 %) of genes were assigned putative functions, and the remainder were annotated as hypothetical proteins. The properties of the ESFC-1 genome, and the distribution of genes into COG functional groups are presented in Tables 3, 4, and Fig. 3. 16S rRNA gene sequence similarity to closely related cultured cyanobacteria, as determined by the phylogeny in Fig. 2, is summarized in Table 5 for the two 16S rRNA genes found in this genome.
Table 3.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 5,632,035 | 100.00 |
| DNA coding (bp) | 4,742,170 | 84.20 |
| DNA G+C (bp) | 2,617,048 | 46.47 |
| DNA scaffolds | 3 | 100.00 |
| Total genes | 5006 | 100.00 |
| Protein coding genes | 4914 | 98.16 |
| RNA genes | 92 | 1.84 |
| Pseudo genes | 146 | 2.92 |
| Genes in internal clusters | 880 | 17.58 |
| Genes with function prediction | 3505 | 70.02 |
| Genes assigned to COGs | 2625 | 52.44 |
| Genes with Pfam domains | 3742 | 74.75 |
| Genes with signal peptides | 253 | 5.05 |
| Genes with transmembrane helices | 1157 | 23.11 |
| CRISPR repeats | 19 | 0.25 |
Table 4.
Number of genes associated with the general COG functional categories
| Code | Value | % of total (2895) | Description |
|---|---|---|---|
| J | 194 | 6.70 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 106 | 3.66 | Transcription |
| L | 94 | 3.25 | Replication, recombination and repair |
| B | 2 | 0.07 | Chromatin structure and dynamics |
| D | 27 | 0.93 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.00 | Nuclear Structure |
| V | 96 | 3.32 | Defense mechanisms |
| T | 230 | 7.94 | Signal transduction mechanisms |
| M | 223 | 7.70 | Cell wall/membrane biogenesis |
| N | 37 | 1.28 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 12 | 0.41 | Extracellular Structures |
| U | 33 | 1.14 | Intracellular trafficking and secretion |
| O | 145 | 5.01 | Posttranslational modification, protein turnover, chaperones |
| C | 139 | 4.8 | Energy production and conversion |
| G | 143 | 4.94 | Carbohydrate transport and metabolism |
| E | 201 | 6.94 | Amino acid transport and metabolism |
| F | 73 | 2.52 | Nucleotide transport and metabolism |
| H | 187 | 6.46 | Coenzyme transport and metabolism |
| I | 74 | 2.56 | Lipid transport and metabolism |
| P | 167 | 5.77 | Inorganic ion transport and metabolism |
| Q | 51 | 1.76 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 398 | 13.75 | General function prediction only |
| S | 205 | 7.08 | Function unknown |
| - | 2381 | 47.56 | Not in COGs |
Fig. 3.
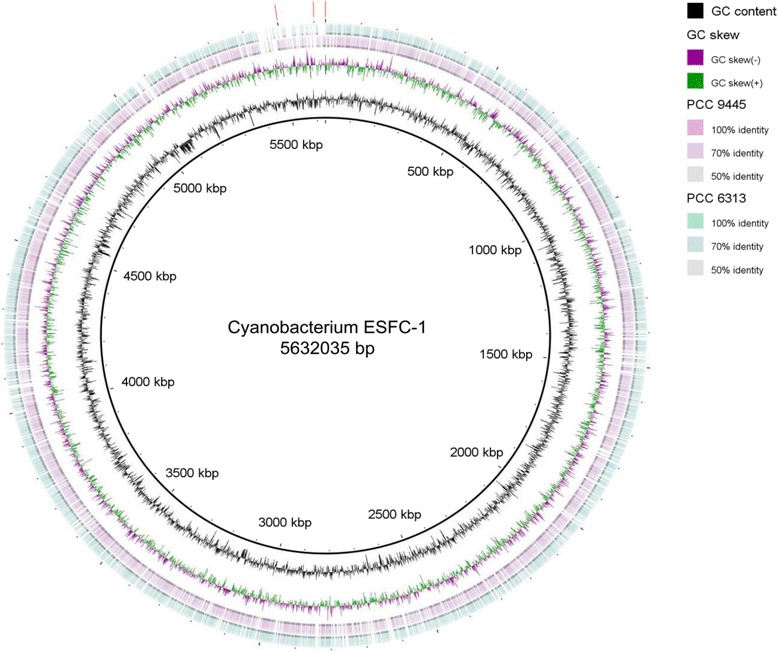
Circularized representation of the three contigs of the genome of strain ESFC-1, with comparisons to Spirulina spp. PCC 9445 and PCC 6313, the closest relatives to ESFC-1 with genomes available. From center to outside: GC content, GC skew, genes on forward strand, genes on reverse strand. Red lines denote the location of the three contigs in this map. The identified genomic island begins at approximately 5050 kbp. Map was generated by BRIG [46]
Table 5.
Near full-length 16S rRNA gene similarity matrix for ESFC-1, the two most-closely related cyanobacteria Spirulina spp. PCC 6313 and PCC 9445 (based on the phylogeny in Fig. 2), and the highest-scoring BLAST hit from the NCBI database (Aphanocapsa sp. HBC6, accession EU249123)
Insights from the genome sequence
ESFC-1 is a nitrogen-fixing cyanobacterium [3]; all structural genes for nitrogenase were detected (nifHDK operon; A3MYDRAFT_2398-2400), as were genes required for uptake of nitrate and reduction to ammonia (narB and nirA; A3MYDRAFT_3316 and A3MYDRAFT_3311, respectively). However, ESFC-1’s sister taxa, SpirulinaPCC 6313 and PCC 9445, as determined by the phylogenomic analysis presented in Fig. 2, both lack the nif operon. Of the cyanobacterial taxa found in the phylogenomic tree, the highest scoring BLAST hit to the translated nifH sequence of ESFC-1 belongs to Halothece sp. PCC 7418 (second best score overall, 87 % amino acid identity, E-value 0.0). ESFC-1 has homologs required for assimilatory sulfur reduction, and a homolog for a sulfide:quinine oxidoreductase, suggesting it can utilize hydrogen sulfide as an electron donor to photosystem I; a useful trait in mat environments that may periodically become anoxic/sulfidic [23, 24].
ESFC-1 has a full complement of homologs for photosystems I and II, the cytochrome b6f complex, photosynthetic electron transport and ATP synthesis. Detected phycobilisome gene homologs indicate a phycocyanin-rich genotype, with phycocyanin, allophycocyanin core, and linker peptide homologs present. ESFC-1 appears to lack the ability to chromatically adapt; phycoerythrin and phycoerythrocyanin genes appear to be absent. A single set of phycocyanin homologs are present (cpcBA; A3MYDRAFT_2965-2964).
Complete sets of genes were detected for the Calvin-Benson cycle, the pentose phosphate pathway, Entner-Doudoroff pathway and glycolysis/gluconeogenesis. TCA cycle and the carbon dioxide concentrating mechanism genes were detected, with 18 hat/hatR homologs found. One locus, for malate dehydrogenase (EC 1.1.1.37) was not detected, however, a putative low-identity alternative protein homolog was found. Strain ESFC-1 contains the necessary gene homologs for most of the lactate and mixed acid fermentation pathways.
Extended insights
Cyanobacterium ESFC-1 appears to lack either a functional uptake or bi-directional hydrogenase. Neither the JGI nor RAST annotations detected these sequences. Extensive manual searches for the hox cluster genes (hoxEFUYH), encoding the bi-directional hydrogenase, and the hupSL genes encoding the uptake hydrogenase commonly found in N-fixing cyanobacteria, were unsuccessful. Similarly, the hydrogenase-maturation enzymes hypFCDE were not found, although the hypAB locus was detected (A3MYDRAFT_0781-0782). Comparative analyses of strain ESFC-1 with the two closely-related Spirulina strains revealed they both lack nif and hupL, but unlike ESFC-1, both possess hupS homologs and the hox operon. The hypFCDEAB homologs were found dispersed throughout their genomes; a comparative blastp analysis between the SpirulinaPCC 6313hox operon and best hits within strain ESFC-1 did not reveal any evidence of synteny for nearby loci.
The genome of ESFC-1 contains an approximately 56 kbp region of low GC content, with several putatively horizontally transferred ORFs (Fig. 3). Based on the IMG annotation, 51 ORFs were identified (A3MYDRAFT_4511 – A3MYDRAFT_4561), with a global GC content of 39.67 % and individual GC content for loci ranging from 29 to 47 %, compared with the global genome GC content of 46.47 %. At the phylum level and higher, IMG designated 112 ORFs as putatively horizontally transferred within the entire genome; of these 16, or 14.2 %, were found within this island (11 from Proteobacteria, and one each from Bacteroidetes, Chloroflexi, Deferribacteres, Firmicutes and Nitrospirae). Additional blastp analysis against the non-redundant protein database (excluding environmental sequences) for these ORFs revealed 19 have best hits to non-cyanobacterial sequences. Several additional sequences in this island did have best hits to cyanobacterial sequences, but these cyanobacterial homologs appeared to be restricted to ESFC-1 and a few closely-related Cyanobacteria. The remaining high-scoring hits for these genes belonged to other bacterial phyla, suggesting that a gene transfer event for these loci into the Cyanobacteria occurred in a common ancestor shared by ESFC-1 and the other closely-related Cyanobacteria. In total, as many as 32 loci in the island region may have been horizontally transferred, either recently or into an ancestor of ESFC-1. Predicted gene functions for this region are primarily involved with lipopolysaccharide and outer membrane synthesis, including several methyltransferase-like and glycosyltransferase-like enzymes, such as homologs to the rfaB, rfaG, rfaS, loci and to the rfaL and rfbX loci related to O-antigen synthesis. Seven of these proteins have been detected via proteomics of an ESFC-1 culture, four of which were extracellular [6], suggesting genes in this island may play a role in extracellular polysaccharide and cell wall synthesis and maintenance. Many Cyanobacteria secrete exopolysaccharides with distinct structures and roles, both in protection from stress (ultraviolet radiation, osmotic and metals) and possibly for carbon storage [25]. This genomic island may provide an advantage to ESFC-1 in stress protection, as has been shown with other cyanobacterial genomic islands [26].
Conclusions
Despite representing a genus-level divergence within the Cyanobacteria, based on both 16S rRNA and phylogenomic analyses, the genome of ESFC-1 appears to belong to a typical filamentous cyanobacterium. However, the ESFC-1 genome is striking in its apparent lack of uptake or bi-directional hydrogenases expected within a diazotrophic cyanobacterium.
Although the uptake hydrogenase hupSL is found dispersed through the cyanobacterial radiation, to our knowledge, strain ESFC-1 is one of very few N-fixing cyanobacteria to lack this gene [27]. The uptake hydrogenase is generally considered an integral part of the energetically expensive process of N-fixation, allowing the cyanobacterium to recapture hydrogen produced by nitrogenase activity [28]. However, a deficient mutant of Anabaena sp. PCC 7120, lacking the large subunit of the uptake hydrogenase, hupL, demonstrated similar growth and N-fixation rates compared to the wildtype, but with enhanced hydrogen production under N-fixation conditions [29]. Given this, and the fact that ESFC-1 is known to be an active nitrogen-fixer in situ in the Elkhorn Slough intertidal mat community, it appears that some Cyanobacteria do not require a classical uptake hydrogenase, yet still perform this critical ecological role.
The bi-directional hydrogenase hox is also common within Cyanobacteria. It is thought to play roles in fermentation and potentially as an electron valve during photosynthesis to maintain proper redox conditions [30–32]. However, as suggested by Tamagnini et al. [33], the physiological role of the bi-directional hydrogenase in Cyanobacteria is unsettled. A recent analysis of 36 cyanobacterial strains indicated that a bi-directional hydrogenase was necessary for hydrogen production via fermentation in these cyanobacteria [34]. Consistent with its apparent lack of a bi-directional hydrogenase, strain ESFC-1 has been shown to produce hydrogen under N-fixation, but not fermentation conditions under laboratory conditions (data not shown). One possible explanation is that strain ESFC-1 ferments under anoxic conditions via lactate or homolactate fermentation pathways, as found within the genome. Such fermentation is known from filamentous Cyanobacteria, and allows for maintenance of redox without concomitant production of hydrogen gas [30]. Both Spirulina spp. PCC 6313 and PCC 9445 possess homologues for the hox genes, so this absence in strain ESFC-1 is best explained by loss, consistent with the uneven distribution of the hup and hox genes in the cyanobacterial radiation [33].
Finally, since the ESFC-1 genome is not closed, the absent hox, hup and hyp genes are possibly in missing regions, or simply were not detected in the automated annotation process. However, extensive manual searches of the genome failed to find any putative hydrogenases. A search of the draft genome for the 107 marker genes commonly used to estimate completeness in metagenomic analyses found all 107 [35], suggesting the absence of these three gene groups is genuine.
Despite the apparent lack of a functional hydrogenase, strain ESFC-1 has been shown to be a dominant and active member of the Elkhorn Slough community. Further, it appears to be globally distributed. Although this distribution appears more limited compared to the cosmopolitan C. chthonoplastes, both nifH and 16S rRNA gene environmental sequences similar to ESFC-1 (>95 %) have been observed in the intertidal mats at Guerrero Negro, Mexico [36], and in lake sediments in Daqing, China (unpublished data, accession KJ176902). An isolate, Leptolynbya sp. LEGE 07176, from the intertidal zone in Portugal [37] may represent a second isolate of this lineage. As one of the only known N-fixing cyanobacteria natively lacking an uptake hydrogenase, this organism may be a suitable target for hydrogen production research. Future studies of ESFC-1 should experimentally confirm the lack of functioning hydrogenase proteins, and explore the nature and energetics of fermentation and N-fixation, and the ecological consequences for an organism that lacks these key enzymes.
Acknowledgements
Funding was provided by the US. DOE Genomic Science Program under contract SCW1039. Sequencing and support was provided by Community Sequencing Project #701 'Microbial Interactions in Extremophilic Mat Communities' at the DOE JGI. Work conducted by the U.S. DOE-JGI was supported by the Office of Science of the U.S. DOE Under Contract No. DE-AC02-05CH11231. Work at LLNL was performed under the auspices of the US DOE at Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. We thank Jeff Cann, Associate Wildlife Biologist, Central Region, California Department of Fish and Wildlife, for coordinating our access to the Moss Landing Wildlife Area.
Authors’ contributions
RCE drafted the manuscript, conceived the study, conducted the phylogenetics and light microscopy, and genome analysis. RKS performed the electron and fluorescence microscopy characterizations, genome analysis, and helped to draft the manuscript. LPB performed the cyanobacterial isolation, conceived the study, and participated in the study design. AMD and DW performed sample preparation and laboratory experiments, and participated in the study design. JZL participated in the study design and genome analysis. BMB and JPR conceived the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- CLIP-PE
Cre-LoxP Inverse PCR Paired-End
- EPS
Extracellular polymeric matrix
- ESFC
Elkhorn Slough filamentous cyanobacterium
References
- 1.Bebout BM, Paerl HW, Crocker KM, Prufert LE. Diel interactions of oxygenic photosynthesis and N2 fixation (acetylene reduction) in a marine microbial mat community. Appl Eviron Microbiol. 1987;53:2353–62. doi: 10.1128/aem.53.10.2353-2362.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stal LJ. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 1995;131:1–32. doi: 10.1111/j.1469-8137.1995.tb03051.x. [DOI] [PubMed] [Google Scholar]
- 3.Woebken D, Burow LC, Prufert-Bebout L, Bebout BM, Hoehler TM, Pett-Ridge J, et al. Identification of a novel cyanobacterial group as active diazotrophs in a coastal microbial mat using NanoSIMS analysis. ISME J. 2012;6:1427–39. doi: 10.1038/ismej.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burow LC, Woebken D, Marshall IPG, Lindquist EA, Bebout BM, Prufert-Bebout L, et al. Anoxic carbon flux in photosynthetic microbial mats as revealed by metatranscriptomics. ISME J. 2013;7:817–29. doi: 10.1038/ismej.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burow LC, Woebken D, Bebout BM, McMurdie PJ, Singer SW, Pett-Ridge J, et al. Hydrogen production in photosynthetic microbial mats in the Elkhorn Slough estuary, Monterey Bay. ISME J. 2012;6:863–74. doi: 10.1038/ismej.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart RK, Mayali X, Lee JZ, Everroad RC, Hwang M, Bebout B, et al. Cyanobacterial reuse of extracellular organic carbon in microbial mats. ISME J. 2016;10:1240–51. doi: 10.1038/ismej.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster JS, Green SJ, Ahrendt SR, Golubic S, Reid RP, Hetherington KL, et al. Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J. 2009;3:573–87. doi: 10.1038/ismej.2008.129. [DOI] [PubMed] [Google Scholar]
- 8.Everroad RC, Woebken D, Singer SW, Burow LC, Kyrpides N, Woyke T, et al. Draft genome sequence of an oscillatorian cyanobacterium, strain ESFC-1. Genome Announc. 2013;1:e00527–13. doi: 10.1128/genomeA.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 10.Castenholz RW, Rippka R, Herdman M, Wilmotte M. Subsection III. (Formerly Oscillatoriales Elenkin 1934). In: Boone DR, Castenholz RW, editors. Bergey's manual of systematic bacteriology, vol. 539–562. 2nd ed. New York: Springer; 2001.
- 11.Protocols - DOE Joint Genome Institute. http://jgi.doe.gov/user-program-info/pmo-overview/protocols-sample-preparation-information/. Accessed 25 May 2016.
- 12.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–8. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 13.Peng Z, Zhao Z, Nath N, Froula JL, Clum A, Zhang T, et al. Generation of long insert pairs using a Cre-LoxP inverse PCR approach. PLoS One. 2012;7:e29437. doi: 10.1371/journal.pone.0029437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler J, MacCallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, et al. ALLPATHS: de novo assembly of wholegenome shotgun microreads. Genome Res. 2008;18:810–20. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–94. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 18.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 19.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:344–8. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen Y, Jørgensen BB, Padan E, Shilo M. Sulfide-dependent anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. Nature. 1975;257:489–92. doi: 10.1038/257489a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jørgensen BB, Revsbech NP, Blackburn TH, Cohen Y. Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl Environ Microbiol. 1979;38:46–58. doi: 10.1128/aem.38.1.46-58.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev. 2009;33:917–41. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- 26.Stuart RK, Dupont CL, Johnson DA, Paulsen IT, Palenik B. Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response relative to those of open-ocean strains. Appl Environ Microbiol. 2009;75:5047–57. doi: 10.1128/AEM.00271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig M, Schulz-Friedrich R, Appel J. Occurrence of hydrogenases in cyanobacteria and anoxygenic photosynthetic bacteria: Implications for the phylogenetic origin of cyanobacterial and algal hydrogenases. J Mol Evol. 2006;63:758–68. doi: 10.1007/s00239-006-0001-6. [DOI] [PubMed] [Google Scholar]
- 28.Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wünschiers R, Lindblad P. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindbald P, Christensson K, Lindberg P. Photoproduction of H2 by wildtype Anabaena PCC 7120 and a hydrogen uptake deficient mutant: from laboratory to outdoor culture. Int J Hydrogen Energ. 2002;27:1271–81. doi: 10.1016/S0360-3199(02)00111-8. [DOI] [Google Scholar]
- 30.Stal LJ, Moezelaar R. Fermentation in cyanobacteria. FEMS Microbiol Rev. 1997;21:179–211. doi: 10.1016/S0168-6445(97)00056-9. [DOI] [Google Scholar]
- 31.Appel J, Schulz R. Hydrogen metabolism in organisms with oxygenic photosynthesis: hydrogenases as important regulatory devices for a proper redox poising? J Photoch Photobiol B. 1998;47:1–11. doi: 10.1016/S1011-1344(98)00179-1. [DOI] [Google Scholar]
- 32.Appel J, Phunpruch S, Steinmüller K, Schulz R. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch Microbiol. 2000;173:333–8. doi: 10.1007/s002030000139. [DOI] [PubMed] [Google Scholar]
- 33.Tamagnini P, Leitão E, Oliveira P, Ferreira D, Pinto F, Harris DJ, et al. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol Rev. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- 34.Kothari A, Potrafka R, Garcia-Pichel F. Diversity in hydrogen evolution from bidirectional hydrogenases in cyanobacteria from terrestrial, freshwater and marine intertidal environments. J Biotechnol. 2012;162:105–14. doi: 10.1016/j.jbiotec.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 35.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woebken D, Burow LC, Behnam F, Mayali X, Schintlmeister A, Fleming ED, et al. Revisiting N2 fixation in Guerrero Negro intertidal microbial mats with a functional single-cell approach. ISME J. 2015;9:485–96. doi: 10.1038/ismej.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brito Â, Ramos V, Seabra R, Santos A, Santos CL, Lopo M, et al. Culture-dependent characterization of cyanobacterial diversity in the intertidal zones of the Portuguese coast: a polyphasic study. Syst Appl Microbiol. 2012;35:110–9. doi: 10.1016/j.syapm.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87(12):4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castenholz RW. General characteristics of the cyanobacteria. In: Boone DR, Castenholz RW, editors. Bergey's manual of systematic bacteriology. 2. New York: Springer; 2001. pp. 474–87. [Google Scholar]
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A. 2013;110:1053–8. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–20. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 46.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]


