Abstract
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is a foodborne pathogen that causes bloody diarrhea and hemolytic uremic syndrome throughout the world. A defining feature of EHEC pathogenesis is the formation of attaching and effacing (AE) lesions on colonic epithelial cells. Most of the genes that code for AE lesion formation, including a type three secretion system (T3SS) and effectors, are carried within a chromosomal pathogenicity island called the locus of enterocyte effacement (LEE). In this study, we report that a putative regulator, which is encoded in the cryptic E. coli type three secretion system 2 (ETT2) locus and herein renamed EtrB, plays an important role in EHEC pathogenesis. The etrB gene is expressed as a monocistronic transcript, and EtrB autoregulates expression. We provide evidence that EtrB directly interacts with the ler regulatory region to activate LEE expression and promote AE lesion formation. Additionally, we mapped the EtrB regulatory circuit in EHEC to determine a global role for EtrB. EtrB is regulated by the transcription factor QseA, suggesting that these proteins comprise a regulatory circuit important for EHEC colonization of the gastrointestinal tract.
INTRODUCTION
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is a foodborne pathogen that causes severe bloody diarrhea, which may be associated with complications, including hemolytic uremic syndrome (1). A defining feature of EHEC infection is the formation of attaching and effacing (AE) lesions on colonic enterocytes. AE lesions are characterized by intimate attachment of EHEC to epithelial cells, rearrangement of the actin cytoskeleton, and effacement of the microvilli (2–4). Most of the genes that mediate AE lesion formation are carried within the locus of enterocyte effacement (LEE) pathogenicity island (5). The LEE is comprised of five major operons, LEE1 to LEE5 (6–8), which encode a type three secretion system (T3SS) (E. coli type three secretion system-1 [ETT1]) (2), an adhesin (intimin) (3) and its receptor (Tir) (4), and secreted effectors (9–13). The LEE also encodes regulatory proteins, including the LEE1-encoded Ler that activates expression of all of the LEE genes (8) as well as GrlA and GrlR, which positively and negatively influence LEE expression, respectively (14–16). Additionally, the LEE-encoded T3SS translocates effector proteins encoded outside the LEE that are also important for virulence (15, 17–23).
In addition to the LEE-encoded T3SS, EHEC carries another locus that encodes a nonfunctional T3SS, named E. coli T3SS-2 (ETT2) (24, 25), which shares homology to the Salmonella T3SS-1 (26). The ETT2 pathogenicity island encodes five predicted or characterized transcription factors. A study by Zhang et al. showed that two of these, EtrA and EivF, repress LEE expression and adherence to epithelial cells, whereas YgeH displayed no regulatory phenotype (27). The other ETT2-encoded putative transcription factors, YqeI and YgeK, herein renamed EtrB, have not been characterized.
EHEC controls expression of virulence traits through complex regulatory circuits that are responsive to metabolites, host hormones, and bacterial signaling molecules, in addition to other environmental cues (28–37). For example, expression of the transcription factor QseA is induced through bacterial cell signaling as well as by ethanolamine, an abundant metabolite in the intestine (33, 38). QseA is an LysR-type regulator that activates LEE expression by directly binding the ler promoter as well as promoting grlA transcription (16, 38–40). Additionally, QseA controls expression of genes located in several O-islands (which are regions of the genome not carried in E. coli K-12 strains [24, 25]), and importantly, QseA positively regulates etrB expression (39). Because ETT2-encoded proteins have been shown to influence virulence and because genetic data indicate that etrB is part of the QseA regulatory cascade, we hypothesized that EtrB plays a role in modulating EHEC virulence gene expression. Our findings indicate that EtrB activates LEE expression, not only through direct regulation but also by repressing expression of eivF and etrA. Moreover, we mapped the EtrB regulon and report that EtrB also modulates expression of genes encoding distinct functions, including the non-LEE-encoded effector NleA, a fimbrial adhesin, a small RNA (sRNA), and maltose and tryptophan metabolism.
MATERIALS AND METHODS
Strains, plasmids, growth conditions, and recombinant DNA techniques.
Strains and plasmids used in this study are listed in Table 1. Standard methods were used to perform plasmid purification, PCR, ligation, restriction digests, transformations, and gel electrophoresis. Luria-Bertani (LB) broth (Invitrogen) or Dulbecco's modified Eagle's medium (DMEM; Invitrogen) was used to grow bacteria. For experiments with the qseA deletion strain (VS145), bacteria were grown overnight in LB broth, diluted 1:100 in low-glucose DMEM, and then grown at 37°C, aerobically to late exponential growth phase (optical density at 600 nm [OD600] of 1.0). For all other experiments, bacteria were grown overnight in LB broth, diluted 1:100 in low-glucose DMEM, and grown for 6 h statically at 37°C under a 5% CO2 atmosphere. Streptomycin was added to overnight cultures of EHEC at a final concentration of 50 μg/ml. Overnight cultures of strains carrying pGEN, pDL01, or pDL03 contained ampicillin to a final concentration of 100 μg/ml. The nonpolar EHEC 86-24 etrB mutant (DL01) was constructed using lambda red mutagenesis (41). The mutant was complemented with etrB under the endogenous promoter cloned into pGEN-MCS (42) (Addgene MTA) using the restriction enzymes HindIII and NcoI (NEB). When complement data are shown, the wild-type (WT) and ΔetrB strains contain empty vector controls. Strains and plasmids were confirmed by DNA sequencing. Primers used to generate the etrB deletion and complemented strains are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 86-24 | Wild-type EHEC (serotype O157:H7) | 51 |
| DL01 | 86-24 etrB mutant | This study |
| DL02 | etrB mutant containing pGEN-MCS | This study |
| DL03 | DL01 containing plasmid pDL01 (etrB complement) | This study |
| DL04 | 86-24 containing pGEN-MSC | This study |
| DL05 | 86-24 containing pDL03 (etrB::lux fusion) | This study |
| DL06 | etrB mutant containing pDL03 (etrB::lux fusion) | This study |
| VS145 | 86-24 qseA mutant | 38 |
| VS151 | VS145 with plasmid pVS150 (qseA complement) | 38 |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| Plasmids | ||
| pKD3 | pANTSλ derivative containing FRT-flanked kanamycin resistance | 41 |
| pKD46 | Lambda red recombinase expression plasmid | 41 |
| pCP20 | Temperature-sensitive replication and thermal induction of FLP synthesis | 41 |
| pGEN-MCS | Cloning vector | 42 |
| pGEN-luxCDABE | Pem7-lux; synthetic promoter, constitutive lux expression (Ampr) | 42 |
| pMAL-c5x | Cloning vector | NEB |
| pVS150 | EHEC 86-24 qseA in pACYC177 | 38 |
| pMK08 | EHEC 86-24 qseA in pET28 | 39 |
| pDL01 | EHEC 86-24 etrB in pGEN | This study |
| pDL02 | EHEC 86-24 etrB in pMAL-C5x | This study |
| pDL03 | EHEC 86-24 etrB in pGEN-luxCDABE | This study |
TABLE 2.
Primers used in this study
| Primer name | Sequence | Primer use |
|---|---|---|
| etrB_GenComp_F | AAGCTTGTATTCTTTGGATTTTGCTTA | etrB complement |
| etrB_GenComp_R | CCATGGACTAGGCTTAATGAACTAGA | etrB complement |
| etrB_MalComp_F | CCTGCAGGTATAGTGCACACACCCATAC | etrB::MBP fusion |
| etrB_MalComp_R | CCATGGGAATGATGGGGGCCGAACTC | etrB::MBP fusion |
| etrB_Lux_F | GTTTAAACTAATTATATTTCATTATTATTTCATC | etrB::lux fusion |
| etrB_Lux_R | TACGTATTAACCCATTTTACGAGTTC | etrB::lux fusion |
| etrB_LR_F | TATTTTAGGAGAATTTGCAGGTGGAATGATGGGGGCCGAACTCGTAAAATTGTGTAGGCTGGAGCTGCTTC | etrB mutant |
| etrB_LR_R | CAAAATGAGCCTAAAGCCTCTTTTTTTTATATAGTGCACACACCCATACGTCATATGAATATCCTCCTTAG | etrB mutant |
| etrB_RT_F1 | GGGCCGAACTCGTAAAATGG | qRT-PCR |
| etrB_RT_R1 | ATACGCATCCTTTCGCACCT | qRT-PCR |
| ler_RT_F1 | CGACCAGGTCTGCCC | qRT-PCR |
| ler_RT_R1 | GCGCGGAACTCATC | qRT-PCR |
| grlA_RT_F1 | CCGGTTGTTCCAGGACTTTC | qRT-PCR |
| grlA_RT_R1 | TAAGCGCCTTGAGATTTTCATTT | qRT-PCR |
| escC_RT_F1 | GCGTAAACTGGTCCGGTACGT | qRT-PCR |
| escC_RT_R1 | TGCGGGTAGAGCTTTAAAGGCAAT | qRT-PCR |
| escV_RT_F1 | TCGCCCCGTCCATTGA | qRT-PCR |
| escV_RT_R1 | CGCTCCCGAGTGCAAAA | qRT-PCR |
| espA_RT_F1 | TCAGAATCGCAGCCTGAAAA | qRT-PCR |
| espA_RT_R1 | CGAAGGATGAGGTGGTTAAGCT | qRT-PCR |
| eae_RT_F1 | GCTGGCCCTTGGTTTGATCA | qRT-PCR |
| eae_RT_R1 | GCGGAGATGACTTCAGCACTT | qRT-PCR |
| rpoA_RT_F1 | GCGCTCATCTTCTTCCGAAT | qRT-PCR |
| rpoA_RT_R1 | CGCGGTCGTGGTTATCTG | qRT-PCR |
| ygeI_RT_F1 | TAGCGAATGCAACGGGTGAT | qRT-PCR |
| ygeI_RT_R1 | GACGCCATCCATGTTGAAACT | qRT-PCR |
| yqeK_RT_F1 | ATGGACATTGAGTTTTCGCAGA | qRT-PCR |
| yqeK_RT_R1 | CCCATGATGTTGTTTGCGTGA | qRT-PCR |
| ryeA_RT_F1 | AGATGACGACGCCAGGTTTT | qRT-PCR |
| ryeA_RT_R1 | ACCAGAACGGGCGGTTTTTA | qRT-PCR |
| tnaA_RT_F1 | TGTACACCGAGTGCAGAACC | qRT-PCR |
| tnaA_RT_R1 | CCGTCATACAGACCTACGGC | qRT-PCR |
| eivF_RT_F1 | TTGTTTGCTGATGCCTTGCC | qRT-PCR |
| eivF_RT_R1 | CGCTGCTCAGATAAGTGGCT | qRT-PCR |
| etrA_RT_F1 | TGCAAGTCTTTTCCAGTGATGTC | qRT-PCR |
| etrA_RT_R1 | CCAACGCAACTAAATCGCTGT | qRT-PCR |
| malK_RT_F1 | CGCAATCGATCAAGTGCAGG | qRT-PCR |
| malK_RT_R1 | GTCAGCGATATCACTCGGCA | qRT-PCR |
| nleA_RT_F1 | TGTTGAAGGCTGGAAGTTTGTTT | qRT-PCR |
| nleA_RT_R1 | CCGCTACAGGGCGATATGTT | qRT-PCR |
| Z4498_RT_F1 | CTTGGCAAAAGTGGGCTCTT | qRT-PCR |
| Z4498_RT_R1 | CCGCATCGTCAATACGGATA | qRT-PCR |
| stx2a _RT_F1 | ACCCCACCGGGCAGTT | qRT-PCR |
| stx2a _RT_R1 | GGTCAAAACGCGCCTGATA | qRT-PCR |
| etrB_prom_F1 | TATTATTTCATCAATGTATTCTTT | EMSA |
| etrB_prom_R1 | CGATTTAACCCATTTTACGA | EMSA |
| kan_EMSA_F1 | CCGGAATTGCCAGCTGGGGCG | EMSA |
| kan_EMSA_R1 | TCTTGTTCAATCATGCGAAACGATCC | EMSA |
| ler_emsaF | ATGCAATGAGATCTATCTTA | EMSA |
| ler_emsaR | AATATTTTAAGCTATTAGCG | EMSA |
| amp_emsaF | GGAATTCGAAAGGGCCTCGTGATACGC | EMSA |
| amp_emsaR | CGGGATCCGGTGAGCAAAAACAGGAAGG | EMSA |
| Z4175_R | GATAGCATAGGGAAGAACAG | RT-PCR |
| Z4177_F | TCACTGGCTCAGGTTTAATG | RT-PCR |
| Z4178_F | GGAATGTCCATATTGCATATC | RT-PCR |
| Z4176_F | TTACAACTCATAGCTGATGG | RT-PCR |
| Z4176_R | TTCCATACGCATCCTTTCGC | RT-PCR |
| etrB_PE_R | CGACTGTTTTCCTGCTTAAC | RT-PCR |
Measurement of etrB expression.
The plasmid pGEN-luxCDABE (42) was used to create the etrB expression plasmid named pDL03. For this, approximately 332 bp of the etrB promoter region was inserted upstream of luxCDABE using the restriction enzymes PmeI and SnaBI (NEB). Luminescence was measured using a Victor Wallac luminometer (Perkin-Elmer). Luminescence was corrected for the OD600 value for each condition. Statistical significance was determined by Student's t test.
RNA extraction and qRT-PCR.
RNA purification and quantification of RNA transcription were performed as described previously (43). RNA was extracted from three biological replicate cultures of each strain/condition using a RiboPure Bacteria RNA isolation kit (Ambion). The amplification efficiency and template specificity of each of the primer pairs (listed in Table 2) were validated, and reaction mixtures were prepared as previously described (44). Quantitative real-time PCR (qRT-PCR) was performed using a one-step reaction with an ABI 7500-Fast sequence detection system (Applied Biosystems). Data were collected using ABI Sequence Detection software, version 1.2 (Applied Biosystems). All data were normalized to levels of rpoA and analyzed using the comparative cycle threshold (CT) method (45). Target gene expression levels were compared by the relative quantification method (45). Statistical significance was determined by Student's t test.
Reverse transcriptase PCR (RT-PCR).
SuperScript II reverse transcriptase (Invitrogen) and random primers were used to create cDNA from RNA samples. The cDNA was used for PCR with gene-specific primers (Table 2). Genomic DNA was used as a positive control, and a reaction without reverse transcriptase was used as a negative control.
FAS assay.
Fluorescent actin staining (FAS) assays were performed as described previously (46). Briefly, overnight bacterial cultures were grown in LB broth at 37°C and then diluted 1:100 to infect HeLa cells. Infected HeLa cells were grown on coverslips for 6 h at 37°C with 5% CO2. Subsequently, the coverslips were washed and fixed with formaldehyde, and then the membranes were permeabilized with 0.2% Triton-X and stained with fluorescein isothiocyanate (FITC)-labeled phalloidin to visualize actin. Bacteria and HeLa cell nuclei were stained with propidium iodide. AE lesions formed by each strain were enumerated for at least 400 HeLa cells in each experiment. Two independent experiments with three biological replicates of each condition were performed. Statistical significance was determined by Student's t test.
Secreted protein immunoblotting.
Secreted proteins were collected as previously described (2). Secreted proteins from culture supernatants were separated from bacterial cells using centrifugation and filtration. SDS-PAGE and immunoblotting were performed as previously described (47). Samples were subjected to immunoblotting with rabbit polyclonal antiserum to EspA and visualized with enhanced chemiluminescence (Bio-Rad). Coomassie blue staining was used to visualize bovine serum albumin (BSA) loading controls. Expression of EspA was quantified from three replicate samples using ImageJ and normalized to BSA levels. Expression levels are shown relative to those of the WT. Three independent experiments were performed.
Purification of EtrB and QseA.
The EtrB protein was fused with the maltose-binding protein (MBP) using a pMAL-C5x vector (NEB) with the restriction enzymes NcoI and SbfI (NEB) to create pDL02. E. coli strain BL21(DE3) containing pDL02 was grown at 37°C in LB broth with glucose (0.2% final concentration) and ampicillin (100 μg/ml) to an OD600 of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 M, and protein expression was induced overnight at 16°C. Cells were then pelleted by centrifugation at 4,000 × g for 10 min and resuspended in column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA). The cells were lysed with an EmulsiFlex instrument. The lysed cells were centrifuged at 4°C, and the supernatant was loaded onto a gravity column (Qiagen) with amylose resin. The column was washed with column buffer, and protein was eluted from the column using column buffer containing 10 mM maltose. Fractions containing purified EtrB were confirmed by SDS-PAGE and Western analysis. The His-tagged QseA protein from plasmid pMK08 was purified as previously described (39). Briefly, the E. coli strain BL21(DE3) containing pMK08 was grown to an OD600 of 0.5 and then induced with 0.4 M IPTG for 3 h. Cells were lysed as described above, and purification was performed using gravity columns (Qiagen) with nickel beads. The column was washed with nickel wash buffer (50 mM NaPO4, 300 mM NaCl, 20 mM imidazole), and protein was eluted using nickel wash buffer containing 250 mM imidazole.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed using the purified EtrB-MBP, QseA-His, and PCR-amplified DNA probes (Table 2), as previously described (48). DNA probes were end labeled using T4 polynucleotide kinase (NEB) with [32P]ATP (Perkin-Elmer) (47). End-labeled probes were purified using Invitrogen NucAway spin columns. EMSAs were performed by adding increasing amounts of purified QseA or EtrB protein to end-labeled probe in binding solution [500 μg/ml BSA, 50 ng of poly(dI-dC), 60 mM HEPES, pH 7.5, 5 mM EDTA, 3 mM dithiothreitol (DTT), 300 mM KCl, and 25 mM MgCl2] and incubated at room temperature for 25 min. A 1% Ficoll solution was added to the reaction mixtures immediately before the samples were loaded on the gel. The reaction mixtures were electrophoresed on a 6% polyacrylamide gel for approximately 6 h at 150 V, dried, and imaged with a PhosphorImager (Molecular Dynamics). Bands were quantified using ImageQuant software.
Primer extension.
Primer extension analysis was performed as previously described (39). Briefly, primer etrB_PE_R (Table 2) was end labeled as described above. A total of 40 μg of RNA, isolated from strain 86-24, was used to generate cDNA using a Primer Extension System with an avian myeloblastosis virus (AMV) reverse transcriptase kit (Promega). The resultant cDNA was precipitated, electrophoresed on a 6% polyacrylamide-urea gel next to a sequencing reaction (Affymetrix). Amplified genomic DNA from strain 86-24 was used to generate a sequencing ladder using primers etrB_prom_F1 and etrB_PE_R for the etrB promoter.
Microarray.
Affymetrix E. coli Genome 2.0 gene arrays were used to compare gene expression of strain 86-24 to that of DL01 (ΔetrB) as previously described (33). The RNA processing, labeling, hybridization, and slide-scanning procedures were performed as described in the Affymetrix GeneChip Expression Analysis Technical Manual (49). Data analyses from the array were performed as previously described (50). The Affymetrix GeneChip Command Console (AGCC) software was used to obtain the output from scanning a single replicate of the Affymetrix GeneChip E. coli Genome 2.0 array for each of the biological conditions, according to the manufacturer's instructions. Data were normalized using robust multiarray analyses (RMA), and the resulting data were compared to determine genes whose expression was increased or decreased in response to the presence of etrB.
RESULTS
Identification and characterization of etrB.
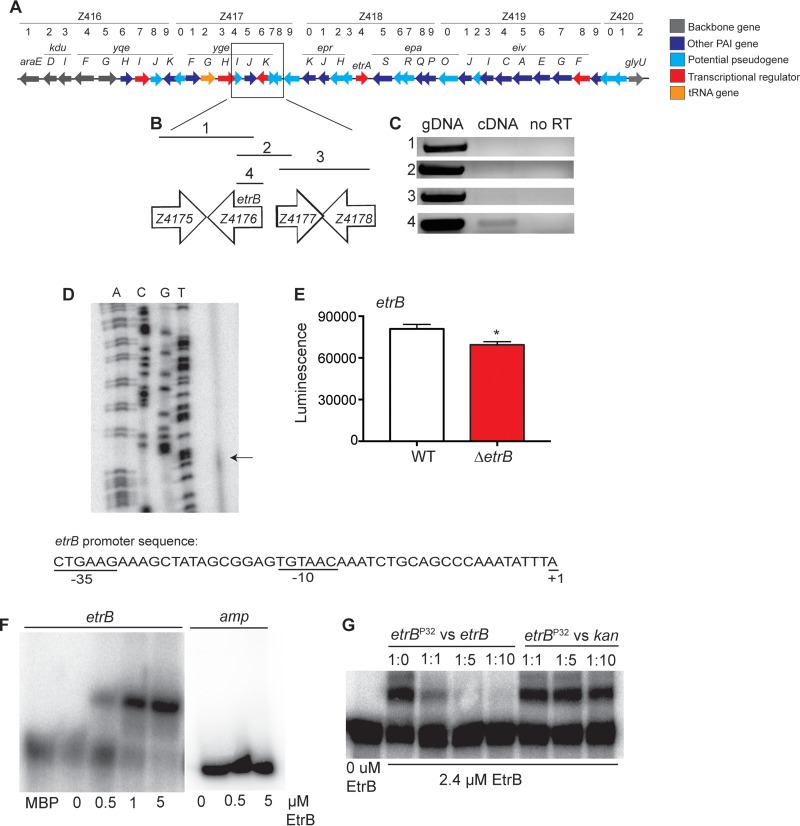
The gene etrB (ygeK, MG1655 genome; Z4176, EDL933 genome; ECs712, Sakai genome [24, 25]) is carried in the ETT2 pathogenicity island (Fig. 1A) and is predicted to encode a 16-kDa protein that shares homology to NarL-type transcription factors, including the Salmonella regulatory protein SsrB (26). The etrB open reading frame (ORF) is present in several E. coli strains; however, bioinformatics analyses indicate that etrB is a pseudogene in nonpathogenic E. coli K-12 strains (26). In pathogenic EHEC strains, the potential functionality of EtrB is less clear. The Sakai and EDL933 annotated genome sequences predict different ORFs, with etrB encoding a 663-bp gene in Sakai or a 447-bp gene in EDL933 (24, 25). We studied EHEC pathogenesis using the strain 86-24, which was isolated from a patient suffering hemorrhagic colitis (51) and has been used in several EHEC animal models (52–59). To begin to characterize etrB in strain 86-24, we performed RT-PCR to determine whether etrB was expressed as part of a transcript with the adjacent up- or downstream genes (depicted in Fig. 1B). For this, we used cDNA synthesized from RNA that was purified from WT EHEC grown statically for 6 h in DMEM. No PCR products were obtained in reactions that included primers specific for the flanking genes (Fig. 1B and C); however, a PCR product was visible when primers specific for etrB were used in the reaction (Fig. 1C). These findings indicate that etrB is expressed in EHEC strain 86-24 but that it is not cotranscribed with the immediate up- or downstream genes.
FIG 1.
etrB expression and autoregulation. (A) Schematic representation of the ETT2 genetic locus in EHEC. PAI, pathogenicity island. (B) Inset of etrB and adjacent genes. Lines with numbers indicate regions amplified and correspond to PCRs shown in panel C. (C) RT-PCR of etrB and adjacent genes. Genomic DNA (gDNA) was used as a positive control, and a reaction without RT was used as a negative control. (D) Primer extension assay of etrB. The first four lanes show the etrB sequencing ladder. The arrow represents the transcription start site of etrB. The promoter sequence of etrB is shown with the predicted transcription start site and −10 and −35 regions. (E) Expression of etrB in WT 86-24 or the ΔetrB strain transformed with the etrB::lux reporter (n = 3; error bars represent the geometric means ± standard deviations). *, P ≤ 0.05. (F) EMSA of the etrB promoter and amp negative-control promoter region with MBP or EtrB::MBP. (G) Competition assays with EtrB. The assay was performed with increasing amounts of the unlabeled etrB probe or the unlabeled kan probe as a negative control.
To further characterize etrB expression and map the promoter region, we performed primer extension analyses. For this, we designed a primer approximately 50 bp downstream from the etrB translational start codon. Then, primer extension analysis was performed using cDNA synthesized from RNA. The primer extension results revealed one transcriptional start site in the etrB promoter (Fig. 1D), which we mapped to approximately 44 bp upstream of the translational start site. These data are consistent with the annotation of the EDL933 genome. These data suggest a −10 sequence, TGTAAC, and a −35 sequence, CTGAAG, which contain two and three mismatches from the σ70 consensus sequences (TATAAT and TTGACA), respectively, and which are separated by 15 nucleotides (Fig. 1D).
To begin to characterize EtrB expression, we examined whether EtrB autoregulates transcription. For this, we generated a deletion of etrB. The deletion of etrB did not impact the EHEC growth rate under our experimental conditions, as the wild-type (WT) and ΔetrB strains reached similar ODs after 6 h of static growth in DMEM (WT OD600, 0.841 ± 0.008; ΔetrB strain OD600, 0.842 ± 0.004). Then, we transformed the WT and the ΔetrB strains with a plasmid containing the etrB promoter fused to the luxCDABE gene cluster that encodes bacterial luciferase (60). Expression of etrB::lux was significantly decreased in the ΔetrB strain compared to that in the WT (Fig. 1E), indicating that EtrB positively autoregulates expression.
Proteins belonging to the NarL family bind DNA to regulate transcription (61). Therefore, we performed EMSAs to investigate whether EtrB binds its own promoter to regulate transcription. For this, we constructed a plasmid that expresses a fusion protein in which the C terminus of MBP was fused to the N terminus of EtrB. EMSAs indicated that EtrB directly binds its promoter to regulate expression (Fig. 1F). To confirm specificity of binding, we performed EMSAs with purified MBP alone as well as competition EMSAs. Purified MBP did not bind the etrB promoter (Fig. 1F). Moreover, EtrB binding was outcompeted by the addition of unlabeled etrB probe, whereas no competition was observed when increasing amounts of cold kan probe were added (Fig. 1G). Altogether, these findings indicate that etrB is expressed and encodes a functional protein that controls its own expression. Accordingly, we renamed the ygeK gene etrB (ETT2 transcriptional regulator B), according to the nomenclature of Zhang et al. (27).
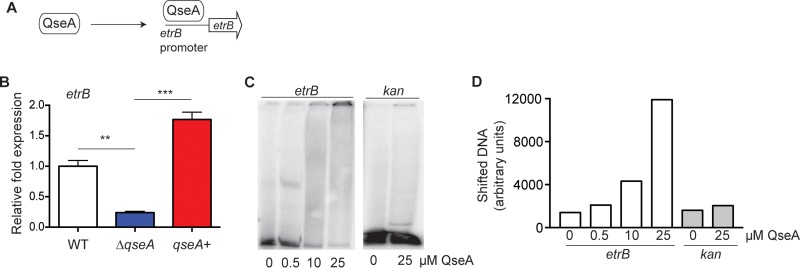
We originally identified etrB in a microarray study as being activated by QseA (39). Here, we confirmed that QseA influences etrB transcription and tested whether this was through direct interaction (Fig. 2A). First, we performed qRT-PCR analyses using RNA extracted from WT, the ΔqseA, or the qseA (qseA+) complemented strain. Transcription of etrB was significantly decreased in the ΔqseA strain compared to the level in the WT, and complementation of the ΔqseA strain restored etrB expression to WT levels (Fig. 2B). To determine whether QseA influenced etrB expression directly, we performed EMSAs. We observed a shift with the addition of purified QseA to radiolabeled etrB promoter DNA; however, no shift was observed using the negative-control kan DNA (Fig. 2C). Quantification of radiolabeled DNA confirmed that the amount of shifted etrB DNA correlated with increasing amounts of QseA added to the reaction mixture (Fig. 2D). Collectively, these data indicate that etrB is a direct target of QseA regulation.
FIG 2.
QseA activates etrB expression. (A) Schematic of model being tested. (B) qRT-PCR of etrB expression in WT 86-24, the ΔqseA strain, and the qseA complemented strain (qseA+) (n = 3; error bars represent the geometric means ± standard deviations). **, P ≤ 0.005; ***, P ≤ 0.0005. (C) EMSA of the etrB promoter and the kan negative-control promoter regions with QseA protein. (D) Quantification of shifted etrB or kan DNA depicted in panel C.
EtrB activates LEE transcription.
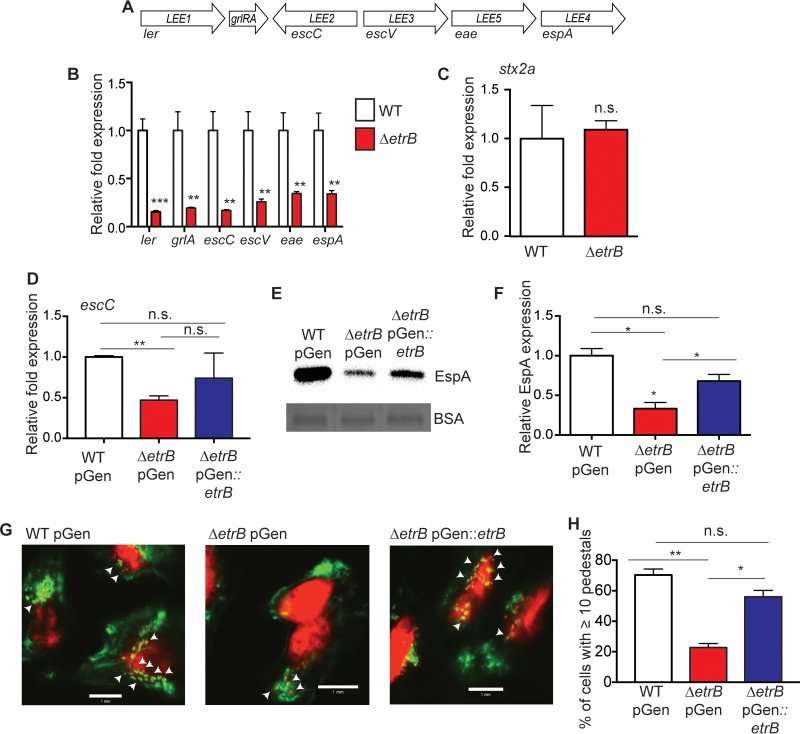
To determine whether EtrB plays a role in directing virulence gene expression in EHEC, we performed qRT-PCR and measured expression of one gene in each LEE operon (Fig. 3A), as well as expression of the stx2a gene, which encodes Shiga toxin. Transcription of ler, located in LEE1, as well as transcription of grlA and operons LEE2 to LEE5, was significantly decreased in the ΔetrB strain compared to that in the WT (Fig. 3B); however, no differences in stx2a expression levels were measured (Fig. 3C). Additionally, trans complementation with etrB on a low-copy-number plasmid nearly restored LEE mRNA and protein to WT levels (Fig. 3D to F).
FIG 3.
EtrB activates expression of the LEE. (A) Schematic of the LEE pathogenicity island. (B) qRT-PCR of ler, grlA, escC, escV, eae, and espA in WT 86-24 and the ΔetrB strains. (C) Expression of stx2a by qRT-PCR in WT 86-24 and the ΔetrB strains. (D) Expression of escC by as determined by qRT-PCR in the WT 86-24 strain transformed with an empty vector control, the ΔetrB strain transformed with an empty vector control, and the ΔetrB strain complemented with etrB. (E) Representative Western blot of the LEE-encoded EspA secreted protein in WT 86-24 transformed with an empty vector control, the ΔetrB strain transformed with an empty vector control, and the ΔetrB strain complemented with etrB. Bovine serum albumin (BSA) is shown as a loading control. (F) Quantification of EspA expression from three independent assays from EHEC strains as described for panel E. (G) FAS assay with WT 86-24 transformed with an empty vector control, the ΔetrB strain transformed with an empty vector control, and the ΔetrB strain complemented with etrB. HeLa nuclei and bacteria were stained red with propidium iodide, and HeLa cell actin cytoskeleton was stained green with FITC-phalloidin. AE lesions are observed as punctate green structures associated with bacterial cells and are indicated by arrowheads. Scale bar, 1 mm. (H) Percentage of HeLa cells with greater than 10 AE lesions. Statistical significance is shown relative to results with the WT 86-24 strain unless otherwise indicated. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005; ns, not significant.
To functionally test the impact of EtrB on LEE expression, we assessed AE lesion formation. For this, we performed an FAS assay (46) and determined the number of pedestals formed on epithelial cells by WT EHEC, the ΔetrB strain, and the etrB complemented strain. In agreement with the LEE expression data, we measured significantly fewer pedestals when HeLa cells were infected with the ΔetrB strain than when cells were infected with WT EHEC, and the etrB plasmid was able to complement the ΔetrB strain (Fig. 3G and H).
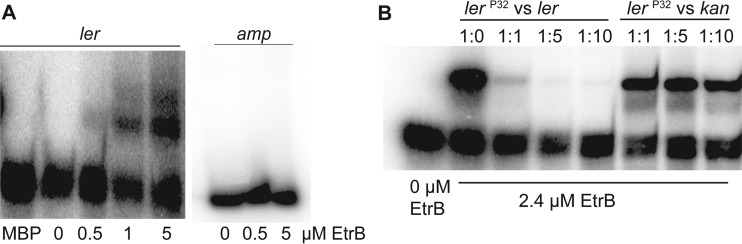
To determine whether EtrB directly regulates LEE expression, we performed EMSAs. For these experiments, we generated a probe containing the entire ler regulatory region. EtrB shifted the radiolabeled ler DNA but not the negative-control amp promoter (Fig. 4A). To ensure specificity of binding, we performed EMSAs using MBP alone as well as competition EMSAs, as described for the etrB promoter. In these assays, no shift was observed with MBP alone (Fig. 4A). Additionally, the unlabeled ler probe competed for EtrB binding at a ratio of 1:1 (labeled probe to unlabeled probe) (Fig. 4B); however, the negative-control kan probe exhibited no competition for binding (Fig. 4B). These findings indicate a specific and direct interaction between EtrB and the ler (LEE1) regulatory region.
FIG 4.
EtrB directly binds to the ler promoter region to activate LEE expression. (A) EMSA of the ler promoter and amp negative-control promoter region with MBP or EtrB::MBP. (B) Competition assays with EtrB. The assay was performed with increasing amounts of the unlabeled ler probe or the unlabeled kan probe as a negative control.
EtrB transcriptome analyses.
To investigate the global role of EtrB in EHEC gene regulation, we performed transcriptome analyses using the Affymetrix E. coli Genome 2.0 gene microarrays. These data revealed that EtrB functions to both positively and negatively influence gene expression in EHEC. For example, 46 gene probe sets were decreased and 70 probe sets were increased in the ΔetrB strain compared to levels in the WT (using a ≥2-fold change in expression as the cutoff for differentially regulated probes) (see Table S1 in the supplemental material). Genes regulated by EtrB included virulence factors, such as the LEE genes and non-LEE-encoded effectors, adhesins and ETT2-located genes, genes important for metabolism, and the noncoding RNA, RyeA/SraC.
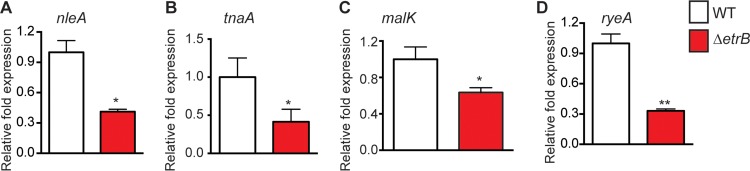
We confirmed a subset of transcripts that were differentially regulated in the array. NleA is an effector that is encoded outside the LEE but which is secreted through the LEE-encoded T3SS. In EHEC, NleA has diverse functions and has been shown to disrupt tight junctions of epithelial cells, inhibit protein secretion, and modulate the host immune response (21, 62, 63). Moreover, an nleA deletion strain is attenuated during murine infection, highlighting its importance to EHEC pathogenesis (21). EtrB positively influences nleA expression as nleA transcription was decreased in the ΔetrB strain compared to that in the WT (Fig. 5A). NleA expression and secretion are at least partly dependent on Ler (64); thus, the decrease in NleA in the ΔetrB strain may be an indirect result of EtrB-dependent regulation of LEE expression.
FIG 5.
EtrB-positive targets of regulation. qRT-PCR of indicated genes (described in the main text) was performed in WT 86-24 and the ΔetrB strain (n = 3; error bars represent the geometric means ± standard deviations). Statistical significance is shown relative to results with the WT 86-24 strain. *, P ≤ 0.05; **, P ≤ 0.005.
In addition to activating expression of virulence factors, EtrB positively influences transcription of genes involved in metabolism and posttranscriptional gene expression. For example, genes involved in tryptophan and maltose utilization were decreased in the ΔetrB strain (Fig. 5B and C). Additionally, transcript levels of the sRNA RyeA/SraC were decreased in the ΔetrB strain compared to the level in the WT (Fig. 5D). RyeA/SraC is present as a 270-bp RNA during exponential growth and is processed to a shorter 150-bp RNA during stationary phase (65–67). These findings suggest that RyeA/SraC plays a role during the stress response; however, the biological role of this sRNA is not known.
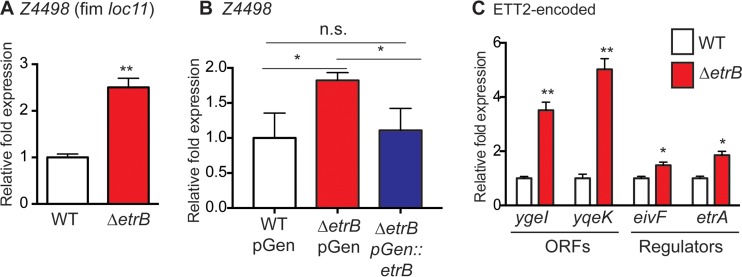
EHEC encodes 14 to 16 fimbrial loci which mediate attachment to epithelial cells. The fimbrial locus 11 (loc11) belongs to the chaperone-usher family of adhesins. These types of fimbriae typically are composed of a chaperone, an usher, and a major fimbrial subunit and may also include additional minor subunits (68). The microarray data indicated that EtrB represses expression of loc11. This locus contains seven ORFs, all of which have been shown to be expressed and cotranscribed (69); therefore, we confirmed the microarray data by measuring expression of the first gene in this operon, Z4498, which is predicted to encode the major fimbrial subunit. Expression of Z4498 was significantly increased in the ΔetrB strain compared to the level in WT EHEC (Fig. 6A), and complementation restored expression to WT levels (Fig. 6B). Collectively, these data suggest that EtrB is important for coordinating expression of fimbrial- and LEE-mediated adherence.
FIG 6.
EtrB negatively regulates transcription of fimbrial locus 11 and genes located in the ETT2. (A) qRT-PCR of Z4498 in WT 86-24 and the ΔetrB strain. (B) qRT-PCR of Z4498 in the WT 86-24 strain transformed with an empty vector control, the ΔetrB strain transformed with an empty vector control, and the ΔetrB strain complemented with etrB (Δn = 3; error bars represent the geometric means ± standard deviations). (C) qRT-PCR of indicated genes in WT 86-24 and the ΔetrB strain. Statistical significance is shown relative to results with WT 86-24 unless otherwise indicated. *, P ≤ 0.05; **, P ≤ 0.005; ns, not significant.
The ETT2 pathogenicity island carries 35 ORFs that are predicted to encode effectors, components of a T3SS, and transcription factors and to harbor pseudogenes (24–26). The microarray data indicated that EtrB represses expression of ORFs located in the ETT2 pathogenicity island. For example, the expression levels of the ORFs ygeI and yqeK were significantly increased in the ΔetrB strain compared to expression in the WT (Fig. 6C). Additionally, expression of the ETT2-located regulators eivF and etrA was increased in the ΔetrB strain (Fig. 6C).
DISCUSSION
The ETT2 locus is present in the majority of E. coli strains; however, many of the ETT2 gene clusters carry mutations and deletions, suggesting that the ETT2 T3SS is not functional (26). Despite this fact, deletions of ETT2-encoded regulatory or structural proteins impact virulence in EHEC as well as in meningitis-causing E. coli strains (27, 70, 71). However, no biochemical evidence of how ETT2-located genes affect pathogenesis has been reported. EtrB belongs to the NarL family of transcription factors, which can function independently or function as part of a two-component system (TCS) (72, 73). The etrB gene is not located adjacent to a putative histidine kinase, which is typical for a cognate TCS (74). A previous study predicted that EtrB might function as an orphan response regulator and demonstrated that purified EtrB could be phosphorylated in vitro (75); however, the physiological relevance of this was not further examined. Our studies indicate that EtrB is able to bind target DNA in the absence of phosphorylation (Fig. 1 and 4), suggesting that EtrB acts as an independent transcription factor.
We mapped the EtrB regulon, and our data revealed that EtrB plays a broad role in EHEC gene expression, affecting expression of genes important for virulence, metabolism, and posttranscriptional gene expression (Fig. 3 to 6 and summarized in 7). Specifically, EtrB activates LEE expression through direct interaction with the LEE1 regulatory region (Fig. 4). This is distinct from the regulatory influence of EtrA and EivF, which repress LEE expression (27). Interestingly, etrA and eivF expression was increased in the ΔetrB strain (Fig. 6C), suggesting that EtrB functions to promote AE lesion formation not only by direct interaction with the LEE1 regulatory region but also by repressing expression of negative regulators of the LEE.
FIG 7.
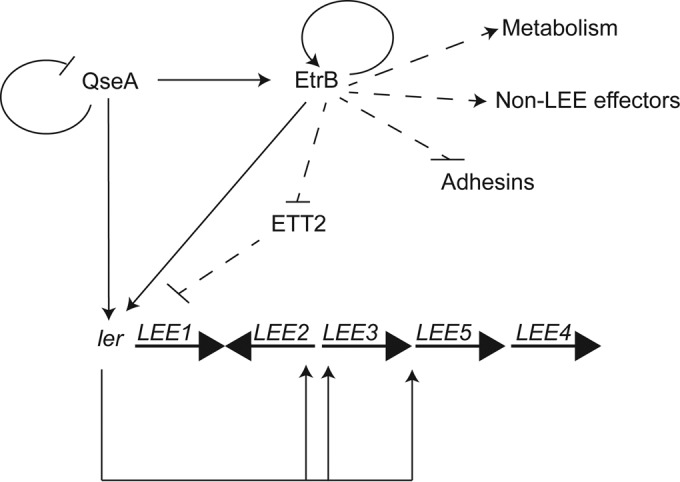
Model of the EtrB regulatory circuit. Lines with arrows indicate positive regulation; lines with bars indicate negative regulation. Solid lines represent direct interactions that have been biochemically defined; dashed lines represent interactions that occur indirectly or that have not been shown to bind biochemically to the target.
Finally, we provide biochemical evidence showing that etrB is a direct regulatory target of QseA. Therefore, we propose that QseA and EtrB function in a coherent feed-forward loop (FFL) (76). This FFL has important implications for the regulatory dynamics of LEE expression. For example, the coordination of multiple regulators might act to amplify environmental cues and promote AE lesion formation. Additionally, a previous study demonstrated that QseA negatively regulates its own transcription (77). In this model, as QseA levels decrease due to autoregulation, EtrB could still act to promote LEE expression, thereby prolonging expression to ensure efficient colonization of the gastrointestinal tract. Overall, this study has identified EtrB as an important regulator of gene expression in EHEC and provides a mechanistic understanding as to how ETT2-encoded regulators influence bacterial pathogenesis.
Supplementary Material
ACKNOWLEDGMENT
We thank members of the Kendall lab for insightful comments and suggestions for this project.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00407-16.
REFERENCES
- 1.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299–1300. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci U S A 92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 5.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott SJ, Hutcheson SW, Dubois MS, Mellies JL, Wainwright L, Batchelor M, Frankel G, Knutton S, Kaper JB. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol Microbiol 33:1176–1189. [DOI] [PubMed] [Google Scholar]
- 7.Elliott SJ, Wainwright L, McDaniel TK, Jarvis KG, Deng YK, Lai L-C, McNamara BP, Donnenberg MS, Kaper JB. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 28:1–4. [DOI] [PubMed] [Google Scholar]
- 8.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol 33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 9.Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect Immun 69:4027–4033. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. 2005. SepZ/EspZ is secreted and translocated into HeLa cells by the enteropathogenic Escherichia coli type III secretion system. Infect Immun 73:4327–4337. doi: 10.1128/IAI.73.7.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny B, Jepson M. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol 2:579–590. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 12.McNamara BP, Donnenberg MS. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol Lett 166:71–78. doi: 10.1111/j.1574-6968.1998.tb13185.x. [DOI] [PubMed] [Google Scholar]
- 13.Tu X, Nisan I, Yona C, Hanski E, Rosenshine I. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol Microbiol 47:595–606. doi: 10.1046/j.1365-2958.2003.03329.x. [DOI] [PubMed] [Google Scholar]
- 14.Barba J, Bustamante VH, Flores-Valdez MA, Deng W, Finlay BB, Puente JL. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol 187:7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell R, Sharp F, Rasko D, Sperandio AV. 2007. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J Bacteriol 189:5387–5392. doi: 10.1128/JB.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campellone KG, Robbins D, Leong JM. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell 7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Dahan S, Wiles S, LaRagione RM, Best A, Woodward MJ, Stevens MP, Shaw RK, Chong Y, Knutton S, Phillips AD, Frankel G. 2005. EspJ is a prophage-carried type III effector protein of attaching and effacing pathogens that modulates infection dynamics. Infect Immun 73:679–686. doi: 10.1128/IAI.73.2.679-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Angulo VA, Deng W, Thomas NA, Finlay BB, Puente JL. 2008. Regulation of expression and secretion of NleH, a new non-locus of enterocyte effacement-encoded effector in Citrobacter rodentium. J Bacteriol 190:2388–2399. doi: 10.1128/JB.01602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garmendia J, Phillips AD, Carlier MF, Chong Y, Schüller S, Marches O, Dahan S, Oswald E, Shaw RK, Knutton S, Frankel G. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 21.Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 22.Mundy R, Petrovska L, Smollett K, Simpson N, Wilson RK, Yu J, Tu X, Rosenshine I, Clare S, Dougan G, Frankel G. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun 72:2288–2302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw RK, Smollett K, Cleary J, Garmendia J, Straatman-Iwanowska A, Frankel G, Knutton S. 2005. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 disrupt the microtubule network of intestinal epithelial cells. Infect Immun 73:4385–4390. doi: 10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 25.Perna NT, Plunkett Gr Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 26.Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, Pallen MJ. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol 186:3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Chaudhuri RR, Constantinidou C, Hobman JL, Patel MD, Jones AC, Sarti D, Roe AJ, Vlisidou I, Shaw RK, Falciani F, Stevens MP, Gally DL, Knutton S, Frankel G, Penn CW, Pallen MJ. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect Immun 72:7282–7293. doi: 10.1128/IAI.72.12.7282-7293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abe H, Tatsuno I, Tobe T, Okutani A, Sasakawa C. 2002. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 70:3500–3509. doi: 10.1128/IAI.70.7.3500-3509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connolly JP, Goldstone RJ, Burgess K, Cogdell RJ, Beatson SA, Vollmer W, Smith DG, Roe AJ. 2015. The host metabolite d-serine contributes to bacterial niche specificity through gene selection. ISME J 9:1039–1051. doi: 10.1038/ismej.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. 2008. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect Immun 76:5247–5256. doi: 10.1128/IAI.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes DT, Clarke MB, Yamamoto K, Rasko D, Sperandio AV. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. doi: 10.1128/mBio.00172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. 2009. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 35.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. 2012. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio 3:e00280-12. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang B, Feng L, Wang F, Wang L. 2015. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat Commun 6:6592. doi: 10.1038/ncomms7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperandio V, Li CC, Kaper JB. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect Immun 70:3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kendall MM, Rasko D, Sperandio AV. 2010. The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 76:1306–1321. doi: 10.1111/j.1365-2958.2010.07174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp FC, Sperandio V. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect Immun 75:2432–2440. doi: 10.1128/IAI.02003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson CJ, Clark DE, Adli M, Kendall MM. 2015. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog 11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun 74:5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Applied Biosystems. 1997. Applied Biosystems Prism 7700 Sequence Detection System: user bulletin 2. Perkin-Elmer Corp., Norwalk, CT. [Google Scholar]
- 46.Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 57:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2 ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Affymetrix. 2009. GeneChip expression analysis technical manual. Affymetrix, Santa Clara, CA. [Google Scholar]
- 50.Willsey GG, Wargo MJ. 2016. Sarcosine catabolism in Pseudomonas aeruginosa is transcriptionally regulated by SouR. J Bacteriol 198:301–310. doi: 10.1128/JB.00739-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7. Ann Intern Med 109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 52.Dean-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun 66:4560–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest 92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O'Brien AD. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun 63:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohawk KL, Melton-Celsa AR, Robinson CM, O'Brien AD. 2010. Neutralizing antibodies to Shiga toxin type 2 (Stx2) reduce colonization of mice by Stx2-expressing Escherichia coli O157:H7. Vaccine 28:4777–4785. doi: 10.1016/j.vaccine.2010.04.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohawk KL, Melton-Celsa AR, Zangari T, Carroll EE, O'Brien AD. 2010. Pathogenesis of Escherichia coli O157:H7 strain 86-24 following oral infection of BALB/c mice with an intact commensal flora. Microbial Pathog 48:131–142. doi: 10.1016/j.micpath.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasko D, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez-Juarez RC, Kuriakose JA, Rasko D, Ritchie AJM, Kendall MM, Slater TM, Sinha M, Luxon BA, Popov VL, Waldor MK, Sperandio V, Torres AG. 2008. CadA negatively regulates Escherichia coli O157:H7 adherence and intestinal colonization. Infect Immun 76:5072–5081. doi: 10.1128/IAI.00677-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woods JB, Schmitt CK, Darnell SC, Meysick KC, O'Brien AD. 2002. Ferrets as a model system for renal disease secondary to intestinal infection with Escherichia coli O157:H7 and other Shiga toxin-producing E. coli. J Infect Dis 185:550–554. doi: 10.1086/338633. [DOI] [PubMed] [Google Scholar]
- 60.Lane MC, Simms AN, Mobley HLT. 2007. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol 189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 62.Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, Metalnikov P, Rizg K, Thomas NA, Lee SF, Hartland EL, Hardwidge PR, Pawson T, Strynadka NC, Finlay BB, Schekman R, Gruenheid S. 2007. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe 2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Yen H, Sugimoto N, Tobe T. 2015. Enteropathogenic Escherichia coli uses NleA to inhibit NLRP3 inflammasome activation. PLoS Pathog 11:e1005121. doi: 10.1371/journal.ppat.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roe AJ, Tysall L, Dransfield TA, Wang D, Fraser-Pitt D, Mahajan A, Constantinidou C, Inglis N, Downing A, Talbot R, Smith DG, Gally DL. 2007. Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157:H7. Microbiology 153:1350–1360. doi: 10.1099/mic.0.2006/003707-0. [DOI] [PubMed] [Google Scholar]
- 65.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11:941–950. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 66.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acid Res 31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nuccio S-P, Bäumler AJ. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol. Rev 71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gonyar LA, Kendall MM. 2014. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ideses D, Gophna U, Paitan Y, Chaudhuri RR, Pallen MJ, Ron EZ. 2005. A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J Bacteriol 187:8164–8171. doi: 10.1128/JB.187.23.8164-8171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao Y, Xie Y, Perace D, Zhong Y, Lu J, Tao J, Guo X, Kim KS. 2009. The type III secretion system is involved in the invasion and intracellular survival of Escherichia coli K1 in human brain microvascular endothelial cells. FEMS Microbiol Lett 300:18–24. doi: 10.1111/j.1574-6968.2009.01763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ishihama A. 2012. Prokaryotic genome regulation: a revolutionary paradigm. Proc Jpn Acad Ser B Phys Biol Sci 88:485–508. doi: 10.2183/pjab.88.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem 280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 76.Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 77.Sircili MP, Walters M, Trabulsi LR, Sperandio V. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun 72:2329–2337. doi: 10.1128/IAI.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.