Abstract
Vibrio cholerae O1 El Tor strains have been responsible for pandemic cholera since 1961. These strains have evolved over time, spreading globally in three separate waves. Wave 3 is caused by altered El Tor (AET) variant strains, which include the strain with the signature ctxB7 allele that was introduced in 2010 into Haiti, where it caused a devastating epidemic. In this study, we used phenotypic analysis to compare an early isolate from the Haiti epidemic to wave 1 El Tor isolates commonly used for research. It is demonstrated that the Haiti isolate has increased production of cholera toxin (CT) and hemolysin, increased motility, and a reduced ability to form biofilms. This strain also outcompetes common wave 1 El Tor isolates for colonization of infant mice, indicating that it has increased virulence. Monitoring of CT production and motility in additional wave 3 isolates revealed that this phenotypic variation likely evolved over time rather than in a single genetic event. Analysis of available whole-genome sequences and phylogenetic analyses suggested that increased virulence arose from positive selection for mutations found in known and putative regulatory genes, including hns and vieA, diguanylate cyclase genes, and genes belonging to the lysR and gntR regulatory families. Overall, the studies presented here revealed that V. cholerae virulence potential can evolve and that the currently prevalent wave 3 AET strains are both phenotypically distinct from and more virulent than many El Tor isolates.
INTRODUCTION
The pathogen Vibrio cholerae induces the severe diarrheal disease cholera. The current seventh global pandemic of cholera has been ongoing for more than 50 years and is due to El Tor O1 strains that originally emerged in Indonesia in 1961 (1). These strains have spread globally in three separate waves (2–4). The wave 1 “typical” El Tor (ET) strains caused disease from the early 1960s through the middle 1990s. Since then, originating from within the El Tor, several successful strain lineages have arisen. The wave 2 strains included the O139 variant that acquired novel O-antigen genes (5) and hybrid variants that acquired a second prophage (2, 3). Since the late 1990s, a lineage known as the wave 3 altered El Tor (AET) V. cholerae has come to predominate as the major cause of human cholera disease. These strains are identified in part by a ctxB gene in which two nucleotide changes converted the cholera toxin B subunit (CT-B) sequence from that of ET (ctxB2 allele) to a sequence identical to that of classical strains (ctxB1 allele) that circulated during pandemics of the 18th and 19th centuries (2). The AET ctxB1 allele was acquired in two stages. A wave 1 ET strain acquired a classical CTX-2 prophage that then recombined with the El Tor CTX-1 prophage, giving rise to the unique CTX-3 prophage. It is postulated that CTX-3 then naturally transduced into a new genetic background that lacked any CTX prophages, ultimately giving rise to the AET (6).
Although they share some common polymorphisms, the wave 2 hybrid and wave 3 AET strains have diverged, with the AET acquiring numerous unique genetic changes (3). The AET strains have a gyrA mutation for nalidixic acid resistance and integrating conjugative elements (ICE) that confer multiple drug resistance, including to streptomycin and chloramphenicol (3, 7, 8). The ICEVchIn5 present in ∼77% of strains has recently been shown to also encode a repressor of V. cholerae natural competence (9, 10). Thereby, the AET strains are highly stable in their genome sequences, as they are suppressed in their ability to acquire genetic content by horizontal gene acquisition. Further, spontaneous mutation rates are low in V. cholerae (estimated at only 3.3 to 6.8 high-quality single nucleotide polymorphisms [hqSNPs]/year [3, 11, 12]), such that the global spread of these strains has been easily tracked by monitoring synapomorphic hqSNP accumulation in whole-genome sequences (3, 10, 11, 13–16).
Over the course of the spread of the AET, signature point mutations in known virulence genes have been acquired. The tcpA gene for the major colonization factor toxin-coregulated pilus (TCP) gained the polymorphism N89S, although the functional consequence of this change is unknown (7, 17). The rtxA gene for the multifunctional, autoprocessing repeats-in-toxin (MARTX) toxin acquired a stop codon (rtxA°) and the toxin became nonfunctional (18). Within the tcpAN89S rtxA° strain, the CTX-6b prophage polymorphisms arose, including a 3-nucleotide (nt) deletion in the gene rstB and an H20N mutation in ctxB (the ctxB7 allele) (7, 18). The functional consequence of the CtxB H20N change is unknown, but its proximity to the leader peptidase processing site suggests that it could affect toxin maturation. This clonal variant of V. cholerae with tcpAN89S, rtxA°, and ctxB7 alleles was first detected in Kolkata, India, in 2004 (7). It was also identified as the cause of outbreaks in Orissa, India, in 2007 (19), in Cameroon, Zimbabwe, and South Africa in 2009, and in Nepal in 2010 (8, 13, 14, 18). The strain has been the predominant cause of cholera in India and Bangladesh at least to 2012 (7, 20, 21). This is also the strain that was introduced into Haiti in 2010 (10, 14–16).
Clinical reports have revealed that the AET strains induce a more dehydrating diarrhea (22), and isolation of AET strains has been specifically associated with patients with severe diarrheal symptoms (17). Human studies have revealed a proinflammatory phase of cholera disease (23), a deviation from the traditional definition of ET cholera as “noninflammatory,” although no comparative studies have been conducted. During epidemics, clinical observation of increased diarrheal severity and the apparent explosive spread of these strains when introduced has led to the AET being referred to as “hypervirulent” (17, 24).
Despite extensive genomic research tracking the global spread of V. cholerae strains, very little research has examined if and how these strains have changed. We have undertaken a phenotypic analysis of an AET strain isolated during the Haiti epidemic in comparison to well-studied laboratory wave 1 isolates to understand whether AET strains are distinct, accounting for the rapid onset of the epidemic. The selected AET strain 2010EL-1786 was isolated in 2010 at the start of the Haiti cholera epidemic and is advantageous to characterize, as the entire genome sequence has been determined (13). We found that the strain has indicators of hypervirulence, with increased production of cholera toxin and hemolysin, increased motility, reduced production of cyclic-di-guanylate (c-di-GMP), and a reduced ability to form biofilms. This increased pathogenic potential was directly demonstrated as the strain outcompeted other strains in cocolonization studies. Analysis of strains representing the accumulation of mutations during the spread of the AET revealed that increased toxin and motility phenotypes likely arose over time, suggesting positive selection for increased virulence.
MATERIALS AND METHODS
Bacterial strains.
2010EL-1786 (Haiti, 2010) was obtained from the ATCC (BAA-2163). ET strains N16961 (Bangladesh, 1971), P27459 (Bangladesh, 1976), E7946 (Bahrain, 1978), C6709 (Peru, 1991), and MO10 (India, 1992) and classical strain O395 (India, 1965) are clinical isolates modified to have spontaneous streptomycin resistance and are part of the Satchell strain collection originally obtained from J. Mekalanos (Harvard Medical School, Boston, MA). A1552 (Peru, 1992) is a clinical isolate modified to have rifampin resistance from the Yildiz strain collection originally obtained from G. Schoolnik (Stanford University, Palo Alto, CA). AET isolates CP1041, CP1042, CP1038, and CP1048, sequenced by Hasan et al. (11), were acquired from the BEI Resources repository (Manassas, VA). All AET strains were confirmed to be streptomycin and nalidixic acid resistant, and strains CP1038 and CP1048 were confirmed to lack the HeLa cell rounding indicative of inactivation of rtxA. Unless otherwise noted, bacteria were grown in Luria-Bertani (LB) broth and agar supplemented with streptomycin (100 μg/ml) or rifampin (100 μg/ml). Deletion of hlyA and lacZ was done as previously described (25, 26).
Phenotypic assays.
Hemolysis was determined from bacteria washed with 190 μl of phosphate-buffered saline (PBS) at 5 × 108 CFU per ml mixed in a microtiter dish with 10 μl of PBS-washed sheep red blood cells (RBCs) (7249007; Lampire Biological Products, Pipersville, PA). Plates were incubated (30°C for 5 h with shaking) and then centrifuged (4,100 rpm for 10 min at 4°C). One hundred microliters of supernatant fluid was transferred to a new microtiter dish, and absorbance at 540 nm was read. Percent hemolysis was corrected for background lysis based on a PBS-only control and 100% lysis of RBCs suspended in distilled water.
V. cholerae treatment of THP-1 monocytic cells and quantification of interleukin 1β (IL-1β) secreted to supernatant fluids were conducted by enzyme-linked immunosorbent assay (ELISA) as previously described (26).
The production of cholera toxin (CT) was determined by CT-ganglioside assay as previously described (27) from cultures inoculated with 10 μl of an overnight culture of V. cholerae into 10 ml of AKI+NaHCO3 medium, incubated statically for 4 h, and then shaken overnight at 37°C (28). Assay plates were prepared using GM1-ganglioside sodium salt (AG-CN2-9000-M005; Adipogen Life Sciences, San Diego, CA) and rabbit polyclonal anti-CT antibody (3062; Sigma). Due to unexpected large variability between assays, particularly among AET strains, all assays were normalized to fold increase compared to the value for the least active strain in the assay.
To monitor motility, V. cholerae cells were inoculated to 20 ml of 0.3% LB agar in a 100-mm dish. Plates were incubated (37°C for 24 h) and then digitally imaged. The area of the colony was calculated using Caliper Living Image software.
Flow cell biofilms were generated as previously described (29). Confocal images were obtained with a 40× dry objective on a Zeiss LSM 5 PASCAL laser scanning confocal microscope and were processed using Imaris (Bitplane, Zurich, Switzerland). Quantitative analyses were performed using the COMSTAT software package (http://www.comstat.dk).
For surface attachment, 100 μl of V. cholerae grown to an optical density at 600 nm (OD600) of 0.05 in 2% LB was applied to polyvinyl chloride microtiter dishes and then incubated at 25°C for 6 h. Plates were washed by immersion in water, and adherent biomass was stained with 1% crystal violet for 15 min, followed by washing in water. The remaining crystal violet was solubilized in 100 μl of 95% ethanol for 15 min and transferred to a fresh microtiter plate, and absorbance was determined at 595 nm.
c-di-GMP extraction and quantification were performed with strains grown in LB as described previously (30). The c-di-GMP concentration was calculated based on a standard curve from 50 nM to 10 μM generated from pure c-di-GMP suspended in 184 mM NaCl (Biolog Life Science Institute, Bremen, Germany). The assay is linear from 50 nM to 10 μM, with an R2 of 0.999. c-di-GMP levels were normalized to total protein per milliliter of culture. Protein concentration was determined using a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL) from cells lysed in 2% sodium dodecyl sulfate.
Infection studies.
Animal protocols were approved by the Northwestern University IACUC. Infant mouse colonization studies were conducted with log-phase bacterial cultures mixed 1:1 inoculated into 5- to 6-day-old ICR mice as previously described (25). The colonization index at 22 h postinfection was calculated from homogenized intestine diluted and plated to LB with antibiotics and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to distinguish inoculated strains.
Statistical analysis.
Statistical methods as noted in figure legends were conducting using GraphPad Prism 6.0 for MacIntosh software (San Diego, CA).
RESULTS
Overproduction of secreted toxins by an AET isolate from Haiti.
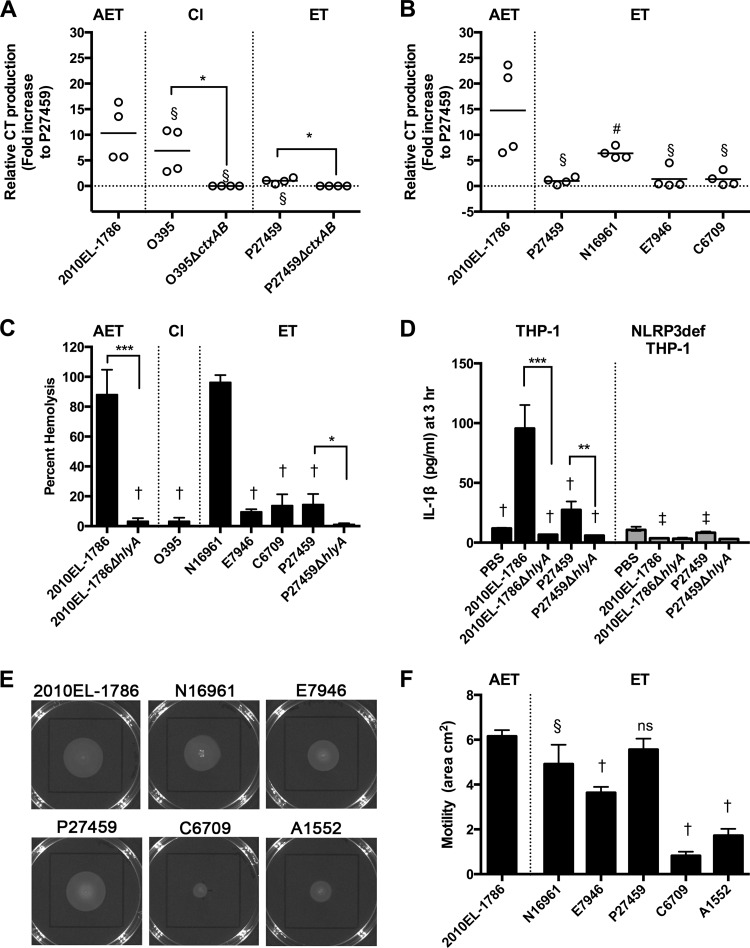
CT is the primary virulence factor of V. cholerae and is responsible for the massive diarrhea linked to this disease (1). To examine if the strain introduced into Haiti produces CT, CT secretion into AKI+NaHCO3 medium was monitored. ET strains are known to produce less CT than classical strains, and consistent with this, the ET strain P27459 produced less CT than classical strain O395, with both showing detectable CT compared to isogenic ΔctxAB negative-control strains (Fig. 1A). We found that the Haiti isolate 2010EL-1786 produced 5- to 16-fold more CT than P27459, matching closely the amount produced by O395. Isolate 2010EL-1786 produced 7- to 24-fold-greater amounts of CT than commonly studied ET strains (E7946, C6709, and P27459) (Fig. 1B). Another common ET isolate, N16961, produced more CT than other ET strains, which was expected, as this strain has previously been found to produce more CT than other ET strains and, as observed in this study for 2010EL-1786 (Fig. 1B), at levels that overlapped the amount expressed by AET strains (31). These results confirm results by others that 2010EL-1786 expresses ∼2-fold more CT than N16961 and ∼10-fold more than C6706 (a strain closely related to C6709) (17) and that AET strains in general produce more CT and induce secretion of more fluid in rabbit ileal loops (31).
FIG 1.
Toxin production and motility by a variety of V. cholerae isolates. (A and B) Cholera toxin production in AKI+NaHCO3 medium determined by ELISA. Individual data points for two assays performed in duplicate are shown to demonstrate range. To account for differences in raw values across two assays, merged data were normalized to the mean CT production (nanograms per milliliter per OD unit) observed for P27459, the lowest value in each assay. Lines indicate the means. (C) Hemolysis of sheep RBCs normalized to 100% lysis by 1% Triton X-100. (D) Secretion of IL-1β to medium from THP-1 monocytic-like cells or NLRP3-deficient THP-1 cells. Hemolysis and IL-1β assay results are shown as means ± standard deviations (SDs) of merged data from biological triplicates for at least two independent experiments. (E and F) Soft-agar motility of V. cholerae inoculated in the center of the plate and (E) photographed or (F) quantified as area using the Caliper Lumina LTE IVIS imager, shown as means ± SDs from biological triplicates. Symbols represent statistically significant difference by one-way analysis of variance (ANOVA) followed by Dunnett's multiple-comparison test compared to 2010EL-1786 in the same panel (#, P < 0.05; §, P < 0.01; †, P < 0.001) or the same sample between THP-1 or NLRP3-deficient THP-1 panels (‡, P < 0.001). Asterisks indicate statistically significant difference of indicated samples by Student's two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). ns, not significant.
The Haiti isolate and other AET strains have a null mutation in the rtxA gene that encodes another V. cholerae toxin known as the MARTX toxin (18). The existence of this mutation is curious, as studies of rtxA+ ET strains showed that the MARTX toxin positively contributes to V. cholerae intestinal colonization (32). However, its contribution to virulence is redundant with that of another toxin, the hlyA-encoded hemolysin/cytolysin HlyA (32). Thus, we hypothesized that rtxA° strains hyperproduce not only CT but also hemolysin, compensating for the loss of rtxA.
The 2010EL-1786 strain induced lysis of RBCs and displayed significantly greater hemolytic activity than did ET strains E7946, C6709, and P27459. The hemolytic activity was similar to that with N16961. Consistent with its natural deletion of hlyA, the classical strain O395 was not hemolytic and served as the negative control (Fig. 1C).
The ability of V. cholerae to produce hemolysin has recently been linked to rapid induction of the NLRP3 inflammasome, resulting in release of IL-1β from human THP-1 monocytic-like cells (26). Consistent with an increase in hemolysis, 2010EL-1786 also stimulated a significant increase in IL-1β release from THP-1 cells, and this release was dependent upon NLRP3 and on production of hemolysin (Fig. 1D).
Thus, 2010EL-1786 produces more CT and more hemolysin than most El Tor strains and toxin activity similar to or greater than that of N16961. These changes can account for the increased severity of disease and inflammation noted for these strains (22, 23).
An AET isolate from Haiti is more motile than most El Tor strains.
The heightened production of two toxins suggests a global change in the regulatory control of virulence in the Haiti isolate. Regulation of hlyA has been linked to the flagellar regulatory network that controls bacterial motility (33). When tested, 2010EL-1786 was found to be hypermotile, spreading across a greater area from the center of a motility agar plate than most of the tested ET strains (Fig. 1E and F). As seen with toxin assays, N16961 was only slightly less motile than 2010EL-1786. Surprisingly, P27459 also showed increased motility and was not different from 2010EL-1786, suggesting that this strain has a unique mutation(s) linked to motility independent of CT and hemolysin control. The increased motility of 2010EL-1786 was most striking compared to that of ET strains isolated in Peru, including C6709 and A1552.
Low production of c-di-GMP in the Haiti AET isolate.
Flagellar motility phenotypes of V. cholerae are in part linked to the intracellular concentrations of the secondary messenger molecule cyclic-di-guanylate (c-di-GMP), with low c-di-GMP promoting a virulence state with high motility and increased toxin production and high c-di-GMP promoting the environmental state, including formation of biofilms (34).
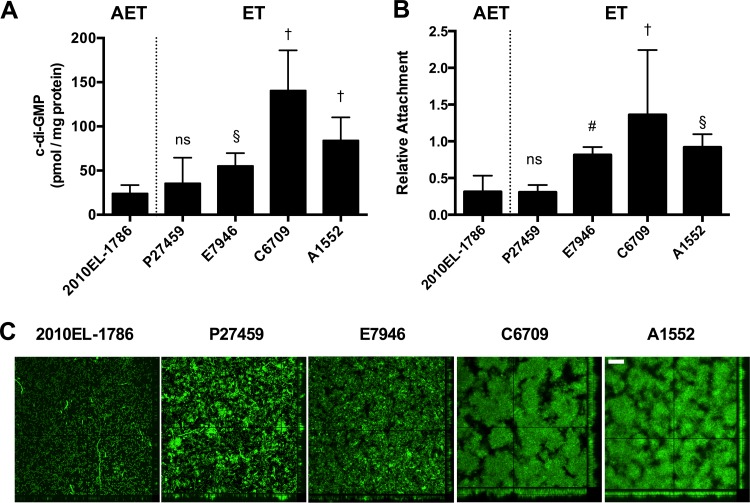
Indeed, 2010EL-1786 produced less c-di-GMP than other ET strains, particularly C6709 (Fig. 2A). Strain A1552 was added as a comparison strain, as it is commonly studied for environmental persistence and known to form robust biofilms (29, 30). It also showed much greater production of c-di-GMP than 2010EL-1786 (Fig. 2A). N16961 was not tested, as its naturally occurring hapR null mutation is expected to alter c-di-GMP production through its control of multiple diguanylate cyclase and phosphodiesterase genes and N16961 was not considered a relevant comparison strain (35).
FIG 2.
c-di-GMP and biofilm formation by a variety of V. cholerae isolates. (A) Intracellular concentrations of c-di-GMP from bacteria grown in LB. Data are pooled from four biological replicates in three independent assays. (B) Initial attachment of V. cholerae to 96-well polyvinyl chloride plates normalized to that of A1552. Data are pooled from 6 to 14 biological replicates from three independent assays, with outliers eliminated using Grubb's algorithm. For both assays, bars indicate means ± SDs. Statistical significance is marked as described for Fig. 1. (C) Biofilm of V. cholerae expressing green fluorescent protein in LB after 24 h of growth in a flow cell. Images show the top view of the biofilm and z-stack series of a lateral slice to show density and channels. Two biological replicates were performed in triplicate. Images presented are from one representative experiment. Bar, 50 µm.
When assessed for the ability to adhere and form structured biofilms in LB (Fig. 2B and C), strains A1552 and C6709 engineered to express gfp adhered well to the surface and produced dense structured biofilms in the flow cell assay. ET strains P27459 and E7946 formed biofilms that were less dense but retained channels indicative of a well-formed three-dimensional biofilms. By comparison, 2010EL-1786 showed weak initial adherence of bacteria to the surface but did not ultimately form a structured biofilm, and bacteria remained predominantly as a single layer (Fig. 2C). This suggests that this strain is severely hampered in biofilm formation.
2010EL-1786 has greater fitness for infant mouse colonization.
Overall, the AET Haiti isolate has an altered phenotypic profile typified by loss of MARTX toxin activity and competence (9, 10, 18) but also increased toxin production, increased motility, reduced biofilm production, and reduced c-di-GMP, all phenotypes associated with increased virulence (36). Therefore, we speculated that this isolate might have greater fitness for infant mouse colonization.
In a competitive colonization assay, 2010EL-1786 outcompeted typical ET strains (Fig. 3A), ranging from a minimum of a 3.3-fold-increased colonization (lowest value for N16961) to a maximum of 1,260 (highest value for C6709). ET strains N16961, P27459, and E7946 actually showed a slightly higher growth rate in rich broth, indicating that the competitive fitness of 2010EL-1786 is not due to its faster growth (Fig. 3B). In fact, when the competitive indices are corrected for the lower growth rate of 2010EL-1786, the increased fitness of the strain becomes even more evident (Fig. 3C).
FIG 3.
Increased fitness of 2010EL-1786 during intestinal infection. Strains as indicated were each competed with 2010EL-1786 ΔlacZ for identification as white colonies on plates containing X-Gal. Shown are infant mouse intestinal colonization (A) and in vitro growth (B) in LB, with competitive index calculated as the ratio of CFU of 2010EL-1786 ΔlacZ/strain indicated recovered (output) divided by the ratio that was inoculated (input). Values above 1 indicate greater fitness of 2010EL-1786, with the horizontal lines indicating observed variation. (C) Values obtained in panel A were divided by the mean in vitro competitive colonization index from panel B to correct for differences in growth rates of some competed strains. Asterisks indicate statistically significant difference of indicated samples by Student's two-tailed t test for each competition compared to 2010EL-1786 competed against itself (*, P < 0.05; **, P < 0.01). ns, not significant.
Isolates C6709 and A1552 from the 1990s outbreak in Peru were even less competitive in the experiment, with a 10- to 1,000-fold competitive index favoring colonization by 2010EL-1786. This reduced fitness is in part due to in vitro growth defects of a factor of 5 to 10. However, growth rate differences do not account for all the competitive advantages of 2010EL-1786, because the strain still shows a significant increase in competitive fitness when corrected for the difference in growth rate (Fig. 3C). These data show that 2010EL-1786 has increased fitness for colonization compared to all isolates selected from those commonly used for laboratory research.
Increased cholera toxin production and motility arise independently.
To further investigate the emergence of hypervirulence, the SNP data set published by Hasan et al. (11) was analyzed for synapomorphic, nonsynonymous SNPs (nSNPs). Additional data sets of Mutreja et al. (3), Hendriksen et al. (14), Lazinski and Camilli (37), and the Genome Bioinformatics Group of University of California at Santa Cruz (http://archaea.ucsc.edu/genomes/bacteria) were compared, and the sequential acquisition of these nSNPs was confirmed against the whole-genome phylogenetic tree published by Eppinger et al. (15). From these analyses, 68 nSNPs that arose within the wave 2 strains (28 mutations) and wave 3 AET strains (32 mutations) or that were found solely in at least 10 sequenced Haiti isolates (8 mutations) were identified (see Table S1 in the supplemental material). Of these, 17 were identified as potentially significant, as they occur in known or putative regulatory, virulence, or missense repair genes (Table 1).
TABLE 1.
Selected synapomorphic hqSNPs in sequenced V. cholerae strainsa

Data drawn primarily from supplemental information published by Hasan et al. (11). E7946 and A1552 SNP data are from comparisons to N16961 published by Lazinski and Camilli (37) and online at http://archaea.ucsc.edu/, respectively. The gene designation number from the V. cholerae N16961 complete genome (VC no.) is indicated. fs, frameshift; aa, amino acids; HTH, helix-turn-helix; Nal, nalidixic acid. Gray shading in SNP columns indicates that the nucleotide is the same as that found in N16961, while white indicates polymorphism. A dash indicates that the residue is deleted.
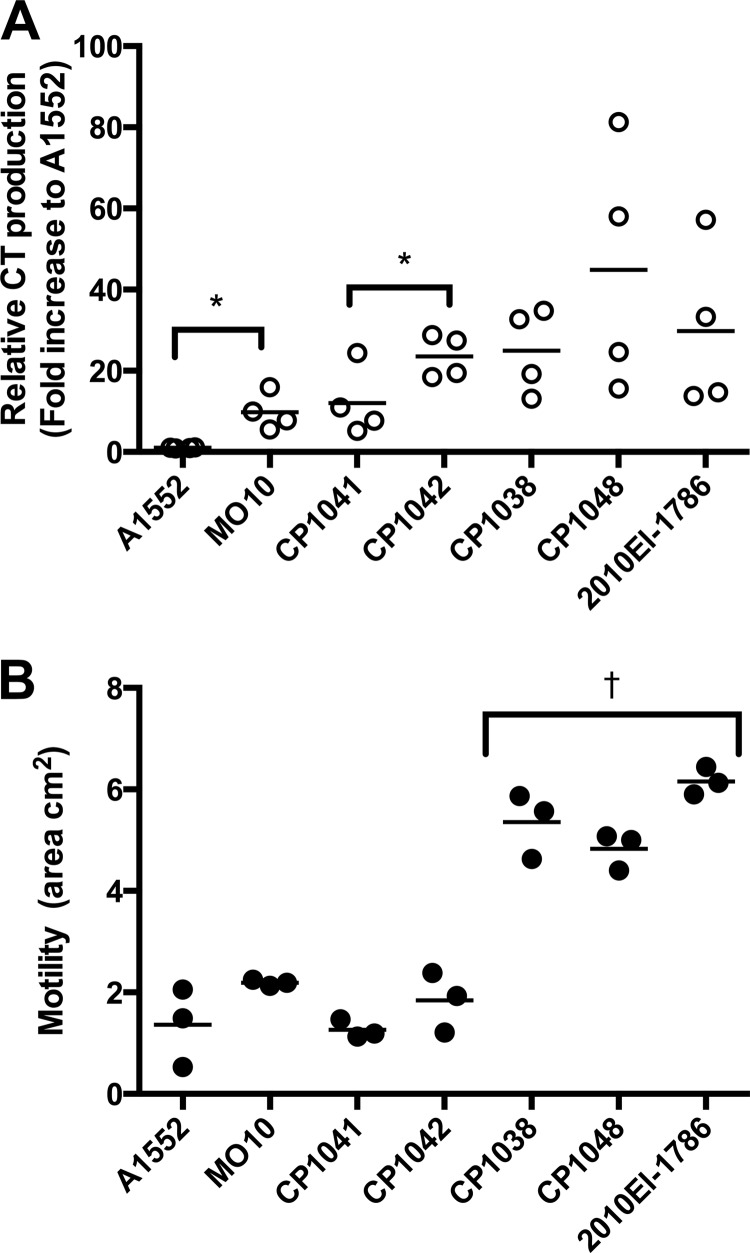
As an initial step to understanding hypervirulence, we sought to distinguish if CT production and motility increased slowly over time or if the increases possibly occurred as a single genetic event. Strains A1552, M010, CP1041, CP1042, CP1038, and CP1048 are known to represent sequential stages in the evolution of the Haiti isolate (15) and thus were selected as strains to initially assess CT and motility (Fig. 4).
FIG 4.
Cholera toxin and motility during evolution of AET. Cholera toxin production in AKI+NaHCO3 medium was determined by ELISA. Individual data points for two assays performed in duplicate are shown to demonstrate range. To account for differences in raw values across two assays, merged data were normalized to the mean CT production (nanograms per milliliter per OD unit) observed for A1552. Lines indicate the means. Asterisks indicate statistically significant difference of indicated samples by Student's two-tailed t test comparing each successive evoultionary step; *, P < 0.05. (B) Soft-agar motility of V. cholerae inoculated in the center of the plate quantified as area using the Caliper Lumina LTE IVIS imager. Lines indicate the means of biological triplicates. One-way ANOVA followed by Turkey's multiple-comparison test of all pairwise samples showed that indicated strains are statistically different from all other samples (†, P < 0.001).
CT production was low for A1552 (ranging from 82 to 158 ng/ml/OD unit) (Fig. 4A). The production of CT then shows stepwise increases first in the wave 2 strain MO10 (Fig. 4A), which shares point mutations with other wave 2 strains, including hybrid strains B33 and MJ-1236, and the later wave 3 AET strains (Table 1; see also Table S1 in the supplemental material). These include frameshift mutations in two diguanylate cyclase genes, cdgM (vc1376) and cdgL (vc2285), and an A189T point mutation in the ligand binding domain of a putative gluconate utilization system gnt-I transcriptional regulator linked to intestinal colonization by genome-wide transposon sequencing (Tn-Seq) analysis (38). The increased production of CT compared to that of A1552 suggests that the root of hypervirulence may reach into the wave 2 strains prior to the emergence of the AET wave 3 strains.
Within the AET wave 3 strains (marked by the presence of nalidixic acid resistance and the ctxB1 allele), CT production shows a trend of increased production as the strains evolve (Fig. 4A). Although raw values for the five AET wave 3 strains tested were all higher than that of A1552 (ranging from 700 to 5,000 ng/ml/OD unit), the values also became more variable across replicates, which could not be resolved through repeated assays or attributed to technical issues but rather seems to represent a natural heterogeneity. Notable mutations across the sequential strains first include a G107S mutation in the histone-like protein H-NS, which is in the DNA binding domain of this repressor that controls transcription of genes that encode hemolysin, MARTX, and CT (39). A recent study showed that deletion of hns uncouples virulence gene expression from its normal control by ToxR, which could account in part for the increased toxin production (40). Also accumulating over time are synapomorphic mutations in two lysR family transcriptional regulatory genes, vc2383 and vc1617; notably, deletion of vc1617 resulted in increased colonization fitness in infant mice for strain C6706 (41).
Another notable mutation is found in vieA, a regulatory gene that is normally repressed by H-NS and is part of a signal system controlling c-di-GMP levels and virulence (39, 42). The L79F mutation in the receiver domain could dysregulate the EAL phosphodiesterase domain of this protein to reduce c-di-GMP levels in the bacterium and thereby variably increase CT production as observed.
The aggregate accumulation of mutants affecting CT production is starkly contrasted with results for motility (Fig. 4B). Compared to A1552, one of the least motile strains in the initial screen (Fig. 1E), MO10, CP1041, and CP1042 do not show a hypermotile phenotype. However, CP1038 and CP1048 showed hypermotility that was not significantly different from that of 2010EL-1786 (Fig. 4B). This indicates that hypermotility arose as a single discrete step in evolution. It is notable that this step includes a large bottleneck of 17 nSNPs (see Table S1 in the supplemental material) that includes the mutation in vieA and null mutations in rtxA and acfC, a gene linked to intestinal colonization (43) (Table 1). The impact of these 17 mutations could be at least split in half by analysis of the sequenced strain CIRS201 (see Table S1), but a source of this strain could not be identified. CP1048 is very similar to CP1038, differing only by three additional nSNP point mutations. These include the appearance of the ctxB7 allele, which identifies the Haiti-like strains worldwide and shows that the ctxB H20N mutation apparently arose in an already hyper-CT-producing, hypermotile background. This strain also has a truncation in chemotaxis gene cheA-3 that removes the C-terminal CheW-like regulatory domain, but this mutation did not affect motility, consistent with complete deletion of cheA-3 having minimal effect on motility compared to the result with cheA-2 (44).
Overall, based thus far on assessment of only CT production and motility, it seems that hypervirulence accumulated in sequential stages for CT production compared to a possible bottleneck selection for hypermotility alongside other selected phenotypes. These results suggest that since the AET first emerged in the late 1990s, ongoing positive selection for increased pathogenetic traits led to emergence of a hypervirulent clone that was ultimately introduced into Haiti.
DISCUSSION
In recent decades, cholera incidence has increased worldwide, with an emerging profile of epidemics arising after founding events. Data from Mutreja et al. (3) support the idea that all cholera disease is currently due to a single clonal lineage that has spread globally since 1961 in three distinct waves. With the wave 3 AET V. cholerae, a particularly successful clone emerged around 2004 that is defined by signature gene alleles tcpAN89S, rtxA°, and ctxB7 (7). This lineage has been a dominant cause of disease in recent years. Indeed, at least through 2012, this strain has accounted for nearly all cholera monitored in India and Bangladesh (7, 20, 21). The strain has spread across all major continents since its first detection in 2004 (7), including isolation in Asia and Africa and accounting for recent outbreaks in the Caribbean (7) and Mexico (45).
This study reveals that this successful lineage is unique in parameters that indicate that it is hypervirulent. The strain produces excess hemolysin and CT and is more motile than commonly studied wave 1 El Tor strains. A previous study found that 2010EL-1786 (also known as BAA-2613) showed increased lethality compared to El Tor strains during single-strain infection, with 100% of infant mice inoculated with 2010EL-1786 succumbing to infection, while 50% of mice inoculated with C6706 and 30% of mice inoculated with N16961 survived (17). In our study, we found that 2010EL-1786 outcompeted all tested wave 1 strains during competitive intestinal colonization, even after adjusting for growth rate. This finding suggests that the successful AET could have emerged as the predominant global clone due to positive selection of increased virulence, ultimately outcompeting other strains through repeated cycles of human transmission. Such a selection could be enhanced by an ability to significantly increase and to more successfully defend against V. cholerae-stimulated inflammation in the mouse intestine (26, 46–48).
On the positive side, this strain was also found to have low c-di-GMP production and to form poor biofilms, which can be associated with poor environmental persistence (34). Notably, during the Haiti epidemic, this strain isolated from patients was not sampled from the environment during the early stages of the epidemic (11). However, environmental sampling conducted more recently suggests that this strain has accumulated new positively selected mutations since its introduction into Haiti. It is now sampled from the environment in Haiti and may be endemic (12). These data suggest that this clone has a high capacity to evolve, transitioning rapidly from a hypervirulent to an environmental persistence state. Poor environmental survival may also explain why this strain may be abating in Bangladesh since 2012 (20). The ability of this strain to evolve is possibly accelerated due to uncharacterized point mutations in DNA mismatch repair proteins MutH and MutL (Table 1), which may compensate for the loss of genetic variation due to horizontal gene transfer when competence is inhibited by the presence of ICEVchIn5 (9, 10).
Extensive hqSNP analyses have been done tracking synapomorphic point mutations that arose over time as part of studies to track the source of the Haiti epidemic (3, 10, 11, 13–16). 2010EL-1786 has 68 point mutations resulting in protein mutations that differentiate it from wave 1 El Tor strains. The sequential acquisition and estimated timing of appearance of these mutations have been analyzed by whole-genome sequence phylogeny (15). Through examination of CT and motility of strains representative of the sequential accumulation of mutations (see Table S1 in the supplemental material), it seems that changes over time account for the increased toxicity of these strains, possibly due to point mutations and null mutations found in several regulatory genes, including the diguanylate cyclase genes cdgL and cdgM, the gntR-like gene vc0289, the hns gene vc1130, the lysR-like genes vc2383 and vc1617, and the vieA gene vc1652. The mutation in vieA might also be linked to hypermotility, as three different strains with this mutation showed the hypermotile phenotype. Additional changes in regulation might also occur due to nucleotide variations in intergenic regions that could include changes in promoters or changes in small RNAs (sRNAs), although these were not specifically analyzed for this study.
Future studies should look beyond phenotypic assays to changes in global transcriptional profiles, particularly using paired genetically modified strains different by a single nucleotide. Further parsing of the contribution of each mutation would also require detailed quantitative assessment of virulence in mice or by a global Tn-Seq-type analysis of colonization in an AET wave 3 strain.
Yet until these extensive studies are conducted, evidence presented here that wave 3 AET clones are phenotypically distinct from strains previously studied in laboratory settings highlights the importance for pathogenesis researchers to consider a wider range of strains in their investigations. In addition, investigation of outbreaks could be modified to include not only identification of surface O antigen but also at least some sequencing analysis to search for the signature markers of this strain. If it is established that an outbreak is due to a hypervirulent clone, containment and treatment measures should be modified to account for the potentially dramatic differences in likelihood of human-to-human spread, disease severity, and environmental survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Satchell and Yildiz laboratories for discussions. Jazel Dolores is thanked for technical assistance, and Qiangli Zhang is thanked for aid with high-performance liquid chromatography-tandem mass spectrometry.
We declare no conflicts of interest.
The funders had no role in study design, data collection, or the decision to submit the work for publication.
c-di-GMP quantification was performed at the UCSC Mass Spectrometry Facility, which is funded by NIH grant S10-RR20939.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00189-16.
REFERENCES
- 1.Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 2.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore S, Thomson N, Mutreja A, Piarroux R. 2014. Widespread epidemic cholera caused by a restricted subset of Vibrio cholerae clones. Clin Microbiol Infect 20:373–379. doi: 10.1111/1469-0691.12610. [DOI] [PubMed] [Google Scholar]
- 5.Comstock LE, Maneval D Jr, Panigrahi P, Joseph A, Levine MM, Kaper JB, Morris JG Jr, Johnson JA. 1995. The capsule and O antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect Immun 63:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim EJ, Lee D, Moon SH, Lee CH, Kim SJ, Lee JH, Kim JO, Song M, Das B, Clemens JD, Pape JW, Nair GB, Kim DW. 2014. Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog 10:e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. 2014. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS One 9:e112973. doi: 10.1371/journal.pone.0112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutar BM, Rajpara N, Upadhyay H, Ramamurthy T, Bhardwaj AK. 2013. Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS One 8:e56477. doi: 10.1371/journal.pone.0056477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalia AB, Seed KD, Calderwood SB, Camilli A. 2015. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc Natl Acad Sci U S A 112:10485–10490. doi: 10.1073/pnas.1509097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azarian T, Ali A, Johnson JA, Mohr D, Prosperi M, Veras NM, Jubair M, Strickland SL, Rashid MH, Alam MT, Weppelmann TA, Katz LS, Tarr CL, Colwell RR, Morris JG Jr, Salemi M. 2014. Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from Haiti reveals diversification driven by positive selection. mBio 5:e01824-14. doi: 10.1128/mBio.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P. 2011. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis 17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2:e00157-11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppinger M, Pearson T, Koenig SS, Pearson O, Hicks N, Agrawal S, Sanjar F, Galens K, Daugherty S, Crabtree J, Hendriksen RS, Price LB, Upadhyay BP, Shakya G, Fraser CM, Ravel J, Keim PS. 2014. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. mBio 5:e01721-14. doi: 10.1128/mBio.01721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. 2011. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J Clin Microbiol 49:3739–3749. doi: 10.1128/JCM.01286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolores J, Satchell KJ. 2013. Analysis of Vibrio cholerae genome sequences reveals unique rtxA variants in environmental strains and an rtxA-null mutation in recent altered El Tor isolates. mBio 4:e00624-12. doi: 10.1128/mBio.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Jain M, Goel AK, Bhadauria S, Sharma SK, Kamboj DV, Singh L, Ramamurthy T, Nair GB. 2009. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J Med Microbiol 58:234–238. doi: 10.1099/jmm.0.002089-0. [DOI] [PubMed] [Google Scholar]
- 20.Rashid MU, Rashed SM, Islam T, Johura FT, Watanabe H, Ohnishi M, Alam M. 19 October 2015. CtxB1 outcompetes CtxB7 in Vibrio cholerae O1, Bangladesh. J Med Microbiol doi: 10.1099/jmm.0.000190. [DOI] [PubMed] [Google Scholar]
- 21.Kumar P, Mishra DK, Deshmukh DG, Jain M, Zade AM, Ingole KV, Goel AK, Yadava PK. 2014. Vibrio cholerae O1 Ogawa El Tor strains with the ctxB7 allele driving cholera outbreaks in south-western India in 2012. Infect Genet Evol 25:93–96. doi: 10.1016/j.meegid.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Qadri F, Bhuiyan TR, Dutta KK, Raqib R, Alam MS, Alam NH, Svennerholm AM, Mathan MM. 2004. Acute dehydrating disease caused by Vibrio cholerae serogroups O1 and O139 induce increases in innate cells and inflammatory mediators at the mucosal surface of the gut. Gut 53:62–69. doi: 10.1136/gut.53.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qadri F, Raqib R, Ahmed F, Rahman T, Wenneras C, Das SK, Alam NH, Mathan MM, Svennerholm AM. 2002. Increased levels of inflammatory mediators in children and adults infected with Vibrio cholerae O1 and O139. Clin Diagn Lab Immunol 9:221–229. doi: 10.1128/CDLI.9.2.221-229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam M, Islam A, Bhuiyan NA, Rahim N, Hossain A, Khan GY, Ahmed D, Watanabe H, Izumiya H, Faruque AS, Akanda AS, Islam S, Sack RB, Huq A, Colwell RR, Cravioto A. 2011. Clonal transmission, dual peak, and off-season cholera in Bangladesh. Infect Ecol Epidemiol 1:7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fullner KJ, Mekalanos JJ. 1999. Genetic characterization of a new type IV pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun 67:1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queen J, Agarwal S, Dolores JS, Stehlik C, Satchell KJ. 2015. Mechanisms of inflammasome activation by Vibrio cholerae secreted toxins vary with strain biotype. Infect Immun 83:2496–2506. doi: 10.1128/IAI.02461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardel CL, Mekalanos JJ. 1994. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol 235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 28.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol 30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 29.Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GC, Yildiz FH. 2015. c-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog 11:e1005068. doi: 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. 2010. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol 192:4541–4552. doi: 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh-Banerjee J, Senoh M, Takahashi T, Hamabata T, Barman S, Koley H, Mukhopadhyay AK, Ramamurthy T, Chatterjee S, Asakura M, Yamasaki S, Nair GB, Takeda Y. 2010. Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J Clin Microbiol 48:4283–4286. doi: 10.1128/JCM.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivier V, Queen J, Satchell KJ. 2009. Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PLoS One 4:e7352. doi: 10.1371/journal.pone.0007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F, Klose KE. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol 191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. 2015. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun 73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54:25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Ayala JC, Benitez JA, Silva AJ. 2015. RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One 10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazi MI, Conrado AR, Mey AR, Payne SM, Davies BW. 2016. ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog 12:e1005570. doi: 10.1371/journal.ppat.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogard RW, Davies BW, Mekalanos JJ. 2012. MetR-regulated Vibrio cholerae metabolism is required for virulence. mBio 3:e00236-12. doi: 10.1128/mBio.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun 56:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gosink KK, Kobayashi R, Kawagishi I, Hase CC. 2002. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J Bacteriol 184:1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Díaz-Quiñonez JA, Hernandez-Monroy I, Montes-Colima NA, Moreno-Perez MA, Galicia-Nicolas AG, Lopez-Martinez I, Ruiz-Matus C, Kuri-Morales P, Ortiz-Alcantara JM, Garces-Ayala F, Ramirez-Gonzalez JE. 2016. Biochemical and full genome sequence analyses of clinical Vibrio cholerae isolates in Mexico reveals the presence of novel V. cholerae strains. Microbes Infect 18:322–328. doi: 10.1016/j.micinf.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Bishop AL, Patimalla B, Camilli A. 2014. Vibrio cholerae-induced inflammation in the neonatal mouse cholera model. Infect Immun 82:2434–2447. doi: 10.1128/IAI.00054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olivier V, Salzman NH, Satchell KJ. 2007. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect Immun 75:5043–5051. doi: 10.1128/IAI.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Queen J, Satchell KJ. 2013. Promotion of colonization and virulence by cholera toxin is dependent on neutrophils. Infect Immun 81:3338–3345. doi: 10.1128/IAI.00422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.