Abstract
Trypanosoma cruzi infection drives the expansion of remarkably focused CD8+ T cell responses targeting epitopes encoded by variant trans-sialidase (TS) genes. Infection of C57BL/6 mice with T. cruzi results in up to 40% of all CD8+ T cells committed to recognition of the dominant TSKB20 and subdominant TSKB18 TS epitopes. However, despite this enormous response, these mice fail to clear T. cruzi infection and subsequently develop chronic disease. One possible reason for the failure to cure T. cruzi infection is that immunodomination by these TS-specific T cells may interfere with alternative CD8+ T cell responses more capable of complete parasite elimination. To address this possibility, we created transgenic mice that are centrally tolerant to these immunodominant epitopes. Mice expressing TSKB20, TSKB18, or both epitopes controlled T. cruzi infection and developed effector CD8+ T cells that maintained an activated phenotype. Memory CD8+ T cells from drug-cured TSKB-transgenic mice rapidly responded to secondary T. cruzi infection. In the absence of the response to TSKB20 and TSKB18, immunodominance did not shift to other known subdominant epitopes despite the capacity of these mice to expand epitope-specific T cells specific for the model antigen ovalbumin expressed by engineered parasites. Thus, CD8+ T cell responses tightly and robustly focused on a few epitopes within variant TS antigens appear to neither contribute to, nor detract from, the ability to control T. cruzi infection. These data also indicate that the relative position of an epitope within a CD8+ immunodominance hierarchy does not predict its importance in pathogen control.
INTRODUCTION
Though eukaryotic pathogens potentially express hundreds of thousands of antigenic peptides, in most cases, a reproducible hierarchy of dominant and subdominant T cells recognizing specific peptides expands in response to infection in a given host. Such immunodominance in CD8+ T cell responses is commonly observed in animal models of infection as well as humans infected with viral, bacterial, and protozoal pathogens (1–3). The generation of immunodominance hierarchies can be attributed to numerous factors (4–8), including competition for space and essential resources by dominant T cell clones (immunodomination) (9). Immunodominance likely benefits the host since energy and resources are invested in the most relevant antigen-specific T cells capable of pathogen clearance while eliciting minimal immunopathology. T cell recognition of epitopes located in conserved proteins may place evolutionary pressure on pathogens, selecting for mutants that are less fit and therefore more easily controlled. However, epitope loss mutations that benefit the pathogen by allowing escape of immune recognition may in turn evolve. Immunodominance can also be detrimental to the host because overzealous CD8+ T cell responses can cause severe immunopathology, as is the case for reinfections in hosts with highly focused preexisting immunity or cross-reacting T cell populations (10). Persistently infecting pathogens also pose a problem, since long-term antigen persistence can drive chronic immunopathology (11, 12). Further, it is hypothesized that immunodominance of noncritical antigens may be utilized by pathogens as an immune evasion mechanism.
In contrast to viral and bacterial models, in which immunodominance has been extensively studied (1, 2), less is known concerning immunodominant CD8+ T cells and their importance for control of intracellular protozoan parasites. Having relatively large genomes and stage-regulated proteomes, these eukaryotic pathogens are more complex than viral and bacterial pathogens in terms of the array of antigens expressed by individual stages occurring within the same host. Furthermore, many parasites of medical importance infect humans chronically or can reinfect immune individuals, suggesting that the immunity developed toward these pathogens is “insufficient” (13). Recent studies have described CD8+ T cell immunodominance during infection with Trypanosoma cruzi (14, 15), an obligate intracellular parasite that often persists for the lifetime of its mammalian host (16). Though the genome of T. cruzi contains several large gene families encoding surface proteins (20 to >1,000 annotated genes per family) (17, 18), many of which gain access to major histocompatibility complex class I (MHC-I) presentation (19), the majority of the T. cruzi-specific CD8+ T cells generated in C57BL/6 mice target a highly restricted set of epitopes encoded by the large, strain-variant trans-sialidase (TS) family of genes (14, 15). The dominant TSKB20 (ANYKFTLV)-specific CD8+ T cells expand to represent 20 to 30% of the CD8+ population, and the subdominant TSKB18 (ANYDFTLV)-specific population represents 4 to 10% of CD8+ T cells during the acute peak of the response (14). These parasite-specific CD8+ T cells are maintained for the lifetime of mice at relatively high levels and exhibit robust effector functions during the chronic phase of infection (20). Nevertheless, mice fail to completely clear T. cruzi despite these high-frequency parasite-specific CD8+ T cell populations (20).
We previously analyzed the importance of immunodominant TS-specific CD8+ T cells during T. cruzi infection and observed that mice tolerized against either TSKB20 or TSKB74 (a cross-reactive peptide recognized by TSKB18-specific CD8+ T cells [14]) alone, or simultaneously, exhibited modest increases in parasite load during the peak of acute infection, though ultimately they were similar to control-treated mice with respect to control of the acute infection (21). Since the TS gene family is dramatically and selectively expanded in T. cruzi (22) and TS gene sequences exhibit considerable intra- and interstrain variability (14, 17), it is hypothesized that this gene family is involved in immune evasion (21, 23–27). The observation that immune control is generated independent of CD8+ T cell recognition of the identified immunodominant TS-derived epitopes indicates that the described TS-focused CD8+ responses are not necessary and may even inhibit the generation of alternative CD8+ responses more capable of eliminating the parasite via immunodomination. To determine if diverting the focus of parasite-specific CD8+ T cells away from these TS-encoded epitopes alters the ability of T. cruzi to persist chronically in infected hosts, we have generated transgenic mouse lines expressing the TSKB20 and TSKB18 epitopes as self-antigens, ensuring central tolerance in the context of infection. We find that TSKB peptide transgenic (TSKB-Tg) mice generate effective CD8+ T cell responses that compensate for the absence of the known immunodominant TS-specific responses and mediate long-term control of persisting parasites. As expected, the normally nondominant CD8+ T cells maintained an effector/effector-memory phenotype in lymphoid tissues of TSKB-Tg mice. Furthermore, the normally nondominant CD8+ T cells were retained after drug-induced cure and exhibited an anamnestic response to secondary challenge. Ultimately, T. cruzi established a low-level chronic infection in both wild-type (WT) and TSKB-Tg mice, demonstrating that highly focused immunodominance is not required for, nor is correlated with, effective pathogen control. These results also suggest that the search for effective vaccines may need to extend beyond targets that dominate the normal response to pathogens, particularly those with highly complex genomes.
MATERIALS AND METHODS
Generation of TSKB20 and TSKB18 transgenic mice.
For transgenic expression of the TSKB20 epitope (ANYKFTLV) or TSKB18 epitope (ANYDFTLV), a modified version of chicken ovalbumin (HA-OVA) was inserted into the eukaryotic expression vector pBroad3 (InvivoGen, San Diego, CA), which contains the mouse ROSA26 promoter and the human beta-globin gene 3′ untranslated region (UTR). Briefly, OVA was cloned from pTEX.HA-OVA (19) into pDONR-201 (Invitrogen, Carlsbad, CA). The sequence encoding the SIINFEKL epitope was replaced with a 54-bp oligonucleotide encoding TSKB20 or TSKB18 (including the endogenous residues flanking SIINFEKL) between StuI and AvaII. A 591-bp fragment of modified OVA was amplified from pDONR-201-OVA/TSKB20 or pDONR-201-OVA/TSKB18 with a PCR using primers designed to add 5′ BglII followed by ATGGCC and 3′ TGA followed either by EcoRI or by HindIII-PvuII-XbaI. These fragments were digested and ligated into the pBroad3 multiple cloning site between BglII and EcoRI to generate pBroad-TSKB20 or between BglII and XbaI to generate pBroad-TSKB18. The fidelity of transgenic TSKB20 and TSKB18 epitope expression in the context of the ovalbumin gene fragment was validated by generating transgenic EL4 tumor cells expressing either OVA/TSKB20 or OVA/TSKB18. C57BL/6 mice were injected intraperitoneally (i.p.) with the transgenic EL4 cell lines and monitored for the development of transgene-specific CD8+ T cell responses. Mice challenged with EL4-OVA/TSKB20 generated TSKB20-specific CD8+ T cells, and mice challenged with EL4-OVA/TSKB18 expanded only TSKB18-specific CD8+ T cells. The 2,955-bp transgene expression cassette was liberated from the prokaryotic vector by double digestion with PacI and ApaLI, and the purified TSKB20 and TSKB18 transgene constructs were independently microinjected into fertilized C57BL/6 embryos by the MD Anderson Cancer Center Genetically Engineered Mouse Facility (Houston, TX). Transgene-positive founders were identified by PCR amplification of a 937-bp product using a forward primer located in the ROSA26 promoter (GGGAGAAGGGAGCGGAAAAG) and a reverse primer located in the human beta-globin 3′ UTR (ATTAGGCAGAATCCAGATGC). Founder mice were mated with C57BL/6 mice obtained from the National Cancer Institute at Frederick (Frederick, MD) and kept under specific-pathogen-free conditions at the Coverdell Center animal facility (University of Georgia, Athens, GA). Further identification of TSKB20- and/or TSKB18-specific transgenes was made using the described forward primer located in the ROSA26 promoter and the transgene-specific reverse primer EcoRI-MscI (TGGCCAGAATTCTCAATTGA) for TSKB20 or HindIII-PvuII (CAGCTGAAGCTTTCAATTGA) for TSKB18 transgene identification.
Parasites and mice.
For T. cruzi infections, 8- to 12-week-old mice were infected i.p. with 1 × 103 trypomastigotes of either strain Brazil or strain Brazil-OVA (19). When indicated, some mice were infected with 1 × 104 trypomastigotes i.p. Trypomastigotes were maintained in tissue culture by serial passage through Vero cells (ATCC, Manassas, VA). C57BL/6 mice obtained from the National Cancer Institute at Frederick (Frederick, MD) and RAG1−/− OT-I TCR transgenic mice (CD45.1) were a kind gift of Kimberly Klonowski (University of Georgia, Athens, GA). For adoptive transfer of OT-I cells, the proportion of CD45.1+ CD8+ T cells per spleen was determined by flow cytometry, spleen cell suspensions were adjusted to 500 OT-I cells per ml, and 100 μl of this suspension was injected intravenously (i.v.) per recipient. B6.SJL mice (CD45.1) from Jackson Laboratory (Bar Harbor, ME) were crossed with WT littermates (CD45.2) or TSKB-Tg (CD45.2) mice to generate WT and TSKB20/18 Tg mice expressing both CD45.1 and CD45.2. All mice were kept under specific-pathogen-free conditions at the Coverdell Center animal facility (University of Georgia, Athens, GA). Mice were euthanized by CO2 inhalation. The University of Georgia Institutional Animal Care and Use Committee approved all animal use protocols.
T cell phenotyping.
For ex vivo lymphocyte phenotyping, spleens were removed and dissociated by rubbing between two glass slides in a medium of hypotonic ammonium chloride to lyse red blood cells. Cell numbers were determined on a Z2 Coulter particle count and size analyzer (Beckman Coulter, Fullerton, CA). For staining directly ex vivo, 5 × 106 washed splenocytes were incubated with antibodies in phosphate-buffered saline (PBS) with 1% bovine serum albumin and 0.05% sodium azide (PAB) (both from Sigma). Blood was collected from the retro-orbital sinus into a sodium citrate solution, washed in PAB, and depleted of erythrocytes as described above. TSKB20/Kb and TSKB18/Kb tetramers were synthesized at the Tetramer Core Facility (Emory University, Atlanta, GA) and were labeled with streptavidin-phycoerythrin (SA-PE) or SA-allophycocyanin (SA-APC) (Molecular Probes, Carlsbad, CA). The SIINFEKL/Kb tetramer was prepared as described previously (28) and labeled with PE. Antibodies used were CD8 efluor450, CD4 PE-Cy5, CD44 FITC (where FITC is fluorescein isothiocyanate), KLRG-1 PE-Cy7, CD127 APC-efluor780, CD127 PE-Cy7, CD62L PerCP-Cy5.5 (eBioscience, San Diego, CA), CD11b PE-Cy5, B220 PE-Cy5 (Caltag Laboratories, Burlingame, CA), CD44 APC, CD11a FITC, CD45.1 FITC, and PE (the last two from BD bioscience, San Jose, CA). Cells were stained at 4°C for 30 min, washed with PAB, and fixed in 2% formaldehyde. At least 5 × 105 single lymphocytes were collected for each sample on a Cyan ADP using Summit version 4.3 (Beckman Coulter). FlowJo Flow Cytometry Analysis Software version 9 (Tree Star, Ashland, OR) was used for analyses, and biexponential transformation was applied to the histogram axis.
Benznidazole treatment and adoptive transfer of memory CD8+ T cells.
The trypanocidal drug benznidazole was administered to clear mice of parasites and allow for the generation of T. cruzi-specific memory CD8+ T cell populations. Mice were treated orally with benznidazole (Roche, Basel, Switzerland) as described previously (20). Daily treatments (100 mg/kg body weight) were given for 40 days. For adoptive transfer of memory CD8+ T cells, we used the Dynal mouse CD8 negative isolation kit (Invitrogen) to purify untouched CD8+ T cells from cell suspensions prepared from pooled mouse spleen and lymph nodes. Purity was confirmed as >88% CD8+ T cells prior to cell transfer into recipient naive mice.
Synthetic peptides.
Peptides were synthesized by GenScript (Piscataway, NJ) and solubilized in dimethyl sulfoxide (DMSO) at 5 mM. Peptides were each diluted to 1 μM for stimulation assays. Published peptide sequences are as follows: TSKB20 (ANYKFTLV), TSKB18 (ANYDFTLV), TSKB38 (VNYNFTLV), TSKB60 (LSHSFTLV), TSKB81 (LSHSFTLV), TSKB92 (VGRPTTVV), TSKB388 (ANHRFTLV), ASP-1 (P14) (VNHDFTVV), ASP-2 (P8) (VNHRFTLV), TSA-1 (Pep77.2) (VDYNFTIV), Crz5 (PSVRSSVPL), Crz9 (VPLNKCNRL), Gft16 (SVPIRLLVL), Gft17 (LGFQERNVL), LYT-1p5 (ELTMYKQLL), PAR-1 (PFR-1) (YEIQYVDL), PAR-3 (PFR-3) (RVVSFTQM), and SIINFEKL.
T cell stimulation and intracellular staining.
Splenocytes (1.5 × 106) were stimulated in 96-well round-bottom tissue culture plates (Costar, Corning, NY) at 37°C for 5 h in the presence 1 μM peptide and brefeldin A (Golgi Plug; BD Biosciences). For polyclonal activation, wells were pulsed with 1.5 μg anti-mouse CD3ε (eBioscience) for 1 h at 37°C and excess antibody was removed prior to the addition of cells. For CD107a labeling, 2 μl CD107a FITC (0.5 mg/ml) was added to the well during stimulation at 37°C. Cells were stained with CD8 efluor450 and CD4 on FITC (Caltag) or PE (BD biosciences) followed by intracellular staining with gamma interferon (IFN-γ) APC (BD biosciences) and tumor necrosis factor alpha (TNF-α) PE-Cy7 (eBioscience) according to the Cytofix/Cytoperm kit (BD biosciences). At least 150,000 cells were collected for analysis.
In vivo cytotoxicity assay.
Spleen cells from naive mice were incubated for 1 h at 37°C with 10 μM peptide or medium alone and then labeled with different concentrations of carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes) as described previously (14) to produce CFSE high, medium, and low populations. Equal numbers of CFSE-labeled cells were transferred i.v. into recipients, and after 16 h, splenocytes were isolated and CFSE-labeled cells were detected by flow cytometry. The percentage of specific killing was determined using the following formula: 1 − [(% CFSElo naive/% CFSEmed/hi naive)/(% CFSElo infected/CFSEmed/hi infected)] × 100%.
Real-time PCR.
Mouse hind leg muscles or inguinal fat pads were collected, and lymph nodes were removed prior to DNA extraction as described previously (29). Extracted DNA was analyzed by real-time PCR essentially as described previously (29). PCRs consisted of iQ SYBR green Supermix (Bio-Rad) and primers specific for T. cruzi or mouse genomic DNA (gDNA) (29). An iQ5 Multi-Color real-time PCR detection system was used with iQ5 Standard Edition Optical System Software version 2 (both from Bio-Rad). T. cruzi equivalents were calculated as the quantity of T. cruzi satellite DNA divided by the quantity of mouse TNF-α DNA in each sample.
Statistical analysis.
Statistical significance was calculated between two groups by two-tailed Student's t test or between three or more groups by 1-way analysis of variance (ANOVA) with Bonferroni's multiple-comparison test using Prism 4.0 software (GraphPad Software, San Diego, CA). P values of <0.05 were considered significant.
RESULTS
TSKB20 and TSKB18 peptide transgenic mice exhibit immune tolerance to TS-derived epitopes.
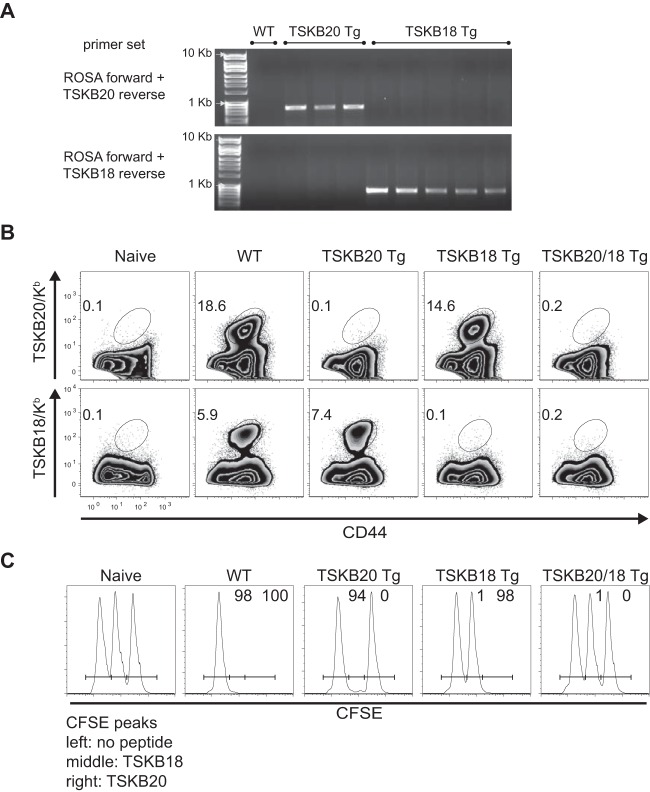
To generate mice expressing the TSKB20 or TSKB18 epitope as neo-self-antigen, we engineered DNA sequences encoding the peptides and cloned each into a transgene vector driven by the ubiquitously expressed ROSA26 promoter. Since the amino acid residues flanking minimal epitopes influence proteolytic cleavage and the processing of antigenic peptides (30) and these sequences vary considerably at the TS genes encoding TSKB20 and TSKB18 (17), we replaced the nucleotides encoding the H-2Kb-restricted SIINFEKL epitope from ovalbumin (OVA) with sequences encoding TSKB20 or TSKB18. The OVA/TSKB20 and OVA/TSKB18 vectors were individually microinjected into fertilized B6 embryos to generate TSKB20 Tg and TSKB18 Tg mouse strains, respectively. Mice expressing both epitopes were obtained by crossing TSKB20 Tg and TSKB18 Tg mice to produce TSKB20/18 Tg mice that can be identified using transgene-specific primers to PCR amplify either transgene sequence (Fig. 1A and data not shown).
FIG 1.
Transgenic expression of TS-derived peptides results in epitope-specific tolerance. (A) PCR products of the transgene construct amplified using TSKB20 or TSKB18 construct-specific primers from gDNA of transgenic founder mice but not from wild-type C57BL/6 gDNA. The primers used were located in the ROSA26 promoter sequence and the unique OVA/TSKB20 or OVA/TSKB18 insert's 3′ restriction sites. (B) Splenocytes from naive or acute-phase T. cruzi-infected WT, TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice were stained for CD44 and TSKB20/Kb or TSKB18/Kb tetramers. Histograms are gated on CD8+ CD4−, and numbers indicate the percentages of CD44hi tetramer-positive CD8+ cells. Data are from infection-matched individuals and are representative of 2 to 4 experiments per acutely infected group. (C) Naive splenocytes were pulsed with 1 μM TSKB20, 1 μM TSKB18, or no peptide and then labeled with high, medium, or low concentrations of CFSE, respectively. At 27 days postinfection, equal numbers of each population were cotransferred i.v. into mice and detected in the spleens after 16 h. Histograms are gated on CFSE+ lymphocytes. Numbers indicate the percentages of specific lysis measured for representative individuals compared with naive mice. Data are from two similar experiments (n = 4 to 7 mice per infected group).
Ubiquitous expression of TSKB20 and TSKB18 should drive clonal deletion of CD8+ T cells recognizing these neo-self-antigens. We assessed epitope-specific central tolerance by infecting TSKB20 Tg, TSKB18 Tg, TSKB20/18 Tg, and WT littermates with T. cruzi strain Brazil and assessing the level of TSKB20/Kb- or TSKB18/Kb-tetramer+ CD8+ T cells at the peak of acute infection (Fig. 1B). Tetramer+ CD8+ T cells specific for the transgene-encoded epitope were not detected in spleens of TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice, though TSKB18/Kb- or TSKB20/Kb-tetramer+ CD8+ T cells readily expanded in TSKB20 Tg and TSKB18 Tg mice, respectively, and activated CD44+ CD8+ T cells expanded normally in all T. cruzi-infected mice (Fig. 1B and data not shown). Furthermore, in vivo cytotoxicity toward the respective transgene-encoded epitope was undetectable during acute T. cruzi infection (Fig. 1C). Thus, transgenic expression of TSKB20 and TSKB18 results in epitope-specific central tolerance and does not prevent the activation and expansion of alternative parasite-specific CD8+ T cells in response to T. cruzi infection.
Long-term control of T. cruzi infection by TSKB20 and TSKB18 transgenic mice.
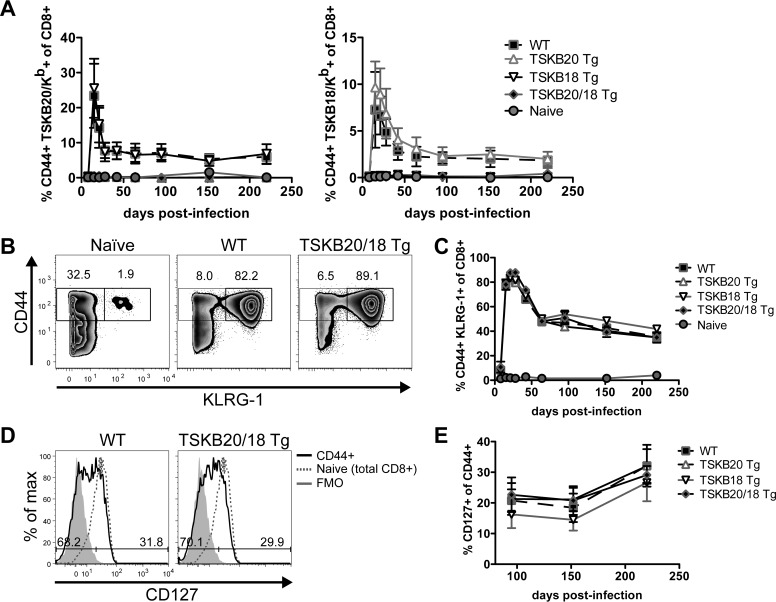
Neither TSKB20- nor TSKB18-specific CD8+ T cells are required for acute control of T. cruzi strain Brazil infection in mice (21). Since T. cruzi infection is persistent lifelong in mice and other hosts, we asked if the TSKB20- or TSKB18-specific CD8+ T cells were essential for continued control of the persistent infection or otherwise contributed to disease development later in infection. We compared the long-term survival of TSKB20 Tg, TSKB18 Tg, TSKB20/18 Tg, and WT littermates infected with T. cruzi and observed no differences in mortality for WT and TSKB-Tg groups up to 1 year postinfection. To determine if infected TSKB-Tg mice maintained T. cruzi-specific immunity, we monitored their peripheral blood for expansion of TSKB20/Kb- or TSKB18/Kb-tetramer+ CD8+ T cells as well as activated CD8+ T cells that had upregulated surface expression of CD44 and KLRG-1, a marker of short-lived effector T cells (31), since we lacked other defined epitopes or MHC tetramers to directly track T. cruzi-specific effector CD8+ T cells other than the TSKB20- and TSKB18-specific CD8+ T cells. Though infected TSKB-Tg mice never expanded tetramer+ CD8+ T cells recognizing their respective transgenic epitope (Fig. 2A), they maintained expanded populations of CD8+ T cells recognizing the alternative nontolerogenic TSKB epitope (Fig. 2A). Furthermore, all infected groups of mice maintained frequencies of CD8+ T cells expressing CD44 and KLRG-1 comparable to that of WT mice (Fig. 2B and C), indicative of the normally robust T cell response throughout infection. During chronic T. cruzi infection, parasite persistence is required for the maintenance of elevated numbers of epitope-specific CD8+ T cells with an effector memory phenotype (20), so these data suggest that all groups of mice remained similarly infected with T. cruzi. In contrast to mice that have fully resolved T. cruzi infection by drug-induced cure (20), the reexpression of surface CD127 on antigen-experienced CD44+ CD8+ T cells occurred slowly in infected TSKB-Tg mice but at a rate similar to that of the chronically infected WT mice (Fig. 2D and E) (20, 32). The accumulation of CD127-expressing CD8+ T cells indicated that a subset of antigen-experienced CD8+ T cells acquired a resting memory phenotype in both WT and TSKB-Tg groups, reflecting decreased levels of parasites due to successful immune control, but not elimination.
FIG 2.
Long-term immunity to T. cruzi by TSKB-peptide Tg mice. (A) TSKB20/Kb+ or TSKB18/Kb+ CD8+ T cells were measured longitudinally in peripheral blood of naive or T. cruzi-infected WT, TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice as described foe Fig. 1B. (B) CD44hi KLRG-1+ CD8+ T cells detected in blood of naive or T. cruzi-infected TSKB20/18 Tg and WT littermates at 21 days postinfection. Histograms are gated on CD8+ CD4− lymphocytes, and numbers indicate the proportions of cells within gates. (C) CD44hi KLRG-1+ CD8+ T cells measured longitudinally in blood of mice analyzed as in panel B. (D) CD127 staining on peripheral blood CD44hi-gated CD8+ T cells (black line) compared with CD127 staining on total CD8+ T cells from a naive individual (dashed line). Filled gray histograms are CD44hi-gated CD8+ T cells not stained for CD127. Numbers indicate the proportions of CD44hi-gated CD8+ T gated cells expressing low (left) or high (right) levels of CD127 at 220 days postinfection. Data in panels B and D are representative of 2 identical experiments (n = 4 to 8 per infected group). (E) Proportion of CD44hi-gated CD8+ T cells stained positive for CD127 measured longitudinally in blood of mice analyzed as in panel D. Data in panels A, C, and E are means ± standard deviations (SD) (n = 3 to 11 per infected group) from one experiment (black squares are WT, up triangles are TSKB20 Tg, down triangles are TSKB18 Tg, diamonds are TSKB20/18 Tg, and circles are uninfected naive mice) and are representative of two similar longitudinal experiments.
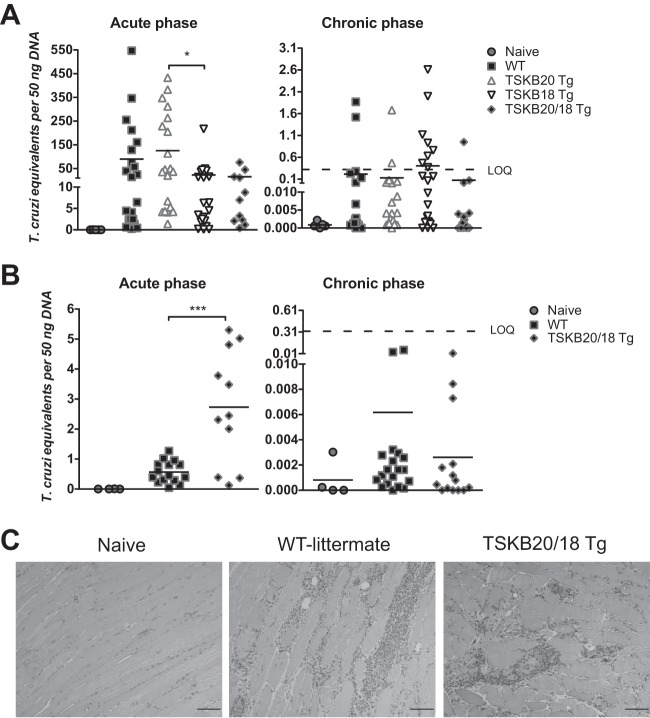
If immunodomination by CD8+ T cells recognizing specific TS-derived epitopes facilitates immune escape by T. cruzi, then mice tolerized against the most dominant TS-specific responses might be expected to develop parasite-specific CD8+ T cell responses that are more capable of controlling infection and preventing disease than responses in mice who have not been tolerized. To determine if parasite control was enhanced in the absence of the normally immunodominant TS-specific CD8+ T cells, we measured parasite loads by quantitative PCR (qPCR) detection of T. cruzi DNA in skeletal muscle and fat (sites of T. cruzi strain Brazil persistence [33, 34]) of acutely infected and chronically infected (>8 months postinfection) TSKB-Tg and WT littermates (Fig. 3A and B). Parasite DNA was more readily detected in skeletal muscle during acute infection (Fig. 3A), and the acutely infected WT mice had a level of parasite DNA similar to that of the TSKB-Tg groups (the TSKB20 Tg had relatively more parasites than did TSKB18 Tg mice [P < 0.05]). Since our previous studies showed that mice tolerized against both TSKB20 and TSKB18 had a slightly higher peak parasite load in muscle than did control mice (21), we also assessed fat tissue for parasite DNA to determine if control in TSKB-Tg was maintained in other tissues known to harbor T. cruzi. Unlike what was seen in muscle tissue (Fig. 3A), we detected an increased level of parasite DNA in fat tissues of TSKB20/18 Tg mice compared with infection-matched WT mice (P < 0.001) (Fig. 3B), though as previously this difference was observed only during the acute phase (Fig. 3B). Similar low levels of parasite DNA were detected in muscle and fat of all groups during the chronic phase (Fig. 3A and B), and members of each group exhibited parasite levels below the limit of quantification (<0.32 parasites per 50 ng DNA). Furthermore, accumulation levels of inflammatory infiltrates as a result of persisting parasites were similar, as assessed by hematoxylin and eosin (H&E) staining of skeletal muscle, in all chronically infected groups (Fig. 3C). Thus, recognition of the immunodominant TSKB20 and TSKB18 epitopes is not required for long-term immune control of T. cruzi, and persistent parasite load was not significantly altered in the absence of these responses.
FIG 3.
Control of persistent T. cruzi infection by TSKB-peptide Tg mice. (A, B) The quantity of T. cruzi DNA in skeletal muscle (A) and fat (B) was detected by real-time PCR in mice during the acute phase (18 to 21 days postinfection) and chronic phase (238 to 337 days postinfection). Data points are individual mice (squares are WT, up triangles are TSKB20 Tg, down triangles are TSKB18 Tg, diamonds are TSKB20/18 Tg, and circles are uninfected naive mice) and bars are means from 4 cumulative acute-phase experiments (n = 11 to 21 per infected group) and 5 cumulative chronic-phase experiments (n = 14 to 23 per infected group). LOQ refers to the limit of detectable quantification based on serially diluted T. cruzi-spiked tissue DNA standards (0.32 parasite equivalents per 50 ng DNA). (C) Representative hematoxylin and eosin-stained skeletal muscle sections from naive or infected WT and TSKB20/18 Tg mice at 337 days postinfection. Scale bar, 200 μM. *, P < 0.05; ***, P < 0.001.
Effector phenotype CD8+ T cells compensate for the absence of TSKB20 and TSKB18 immunodominance.
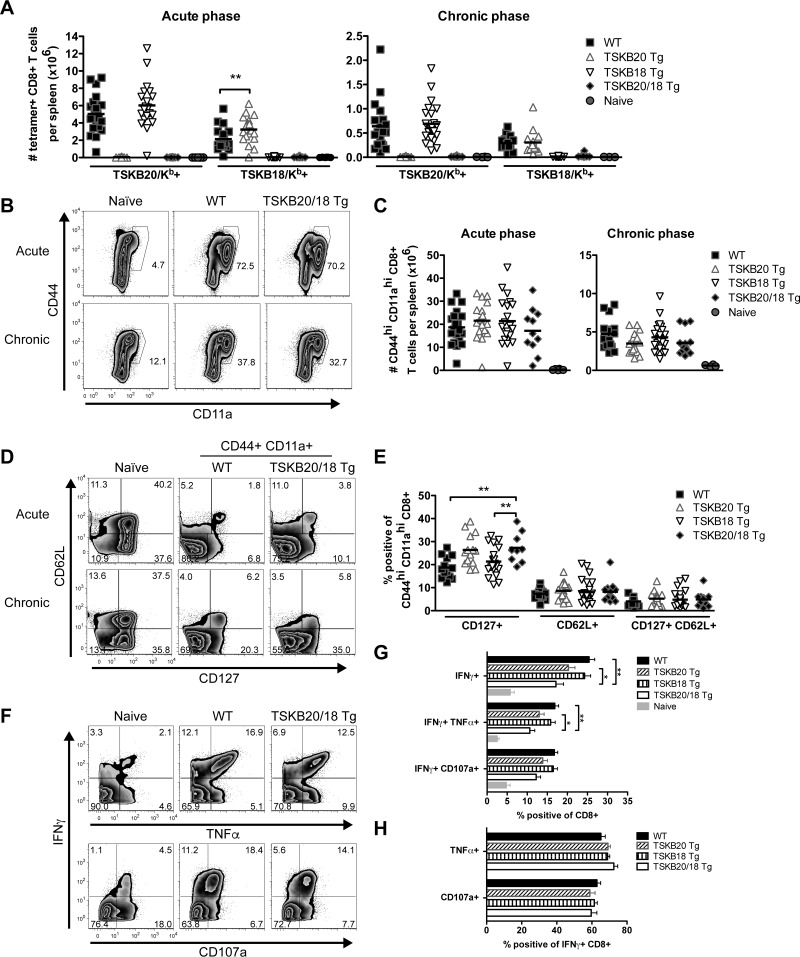
On average, TSKB20 Tg mice had a significantly increased number of the normally subdominant TSKB18-specific CD8+ T cells in their spleens at the peak of infection (1.5-fold more than WT [P < 0.01]) (Fig. 4A), yet the TSKB18 response rarely expanded to the extent of the TSKB20-specific CD8+ T cells in WT or TSKB18 Tg mice (2.4-fold more TSKB20/Kb+ than TSKB18/Kb+ CD8+ T cells in WT spleens) (Fig. 4A). Furthermore, the TSKB18 response was not significantly enhanced in TSKB20 Tg mice during the chronic phase of infection (Fig. 4A, WT versus TSKB20 Tg, P = 0.85). Therefore, normally nondominant CD8+ T cells likely expand and perform critical effector functions that mediate effective parasite control in the absence of the described immunodominant CD8+ T cells.
FIG 4.
Effector CD8+ T cell populations expand and exhibit an effector-memory phenotype in the absence of TSKB20 and TSKB18 immunodominance. (A) The total number of TSKB20/Kb+ or TSKB18/Kb+ CD8+ T cells per spleen of naive or T. cruzi-infected WT, TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice at 18 to 21 days postinfection (left) or 238 to 337 days postinfection (right). (B) Representative staining for CD44 and CD11a expression during acute (day 18) and chronic (day 238) phases of infection. Histograms are gated on CD8+ CD4− events, and numbers represent percentages of gated events. (C) The total number of CD44hi CD11ahi CD8+ T cells per spleen of age-matched naive or T. cruzi-infected WT, TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice. (D) Representative staining for CD127 and CD62L expression during acute (day 18) and chronic (day 238) phases of infection. Histograms are gated on CD8+ CD4− for age-matched naive mice and further gated on CD44hi CD11ahi antigen-experienced cells for infected WT and TSKB20/18 Tg mice. (E) Proportion of CD44hi CD11ahi-gated CD8+ splenocytes that expressed the indicated marker during the chronic phase (238 to 337 days postinfection). Data points in panels A, C, and E are individual mice (squares are WT, up triangles are TSKB20 Tg, down triangles are TSKB18 Tg, diamonds are TSKB20/18 Tg, and circles are uninfected naive mice), and bars are means from 4 cumulative experiments in the acute phase (n = 11 to 21 per infected group) and 5 cumulative experiments in the chronic phase (n = 14 to 23 per infected group). (F) Representative intracellular staining of IFN-γ and TNF-α (top) or surface accumulation of CD107a (bottom) after 5 h of stimulation with plate bound anti-CD3. Histograms are gated on CD8+ CD4− events, and numbers indicate the proportions of events within the gated quadrant. Data are from 338 days postinfection. (G) Cumulative data presented in panel F. Data are from 4 cumulative experiments in the chronic phase (238 to 337 days postinfection) (n = 14 to 24 per infected group). (H) Data are from 3 cumulative experiments in the chronic phase (238 to 337 days postinfection) (n = 11 to 17 per infected group). Filled black bars are WT, forward-slashed bars are TSKB20 Tg, vertical-slashed bars are TSKB18 Tg, open bars are TSKB20/18 Tg, and filled gray bars are uninfected naive mice). Data are means + standard errors of the means (SEM). *, P < 0.05; **, P < 0.01.
If normally nondominant CD8+ T cells indeed compensate for the absence of the TSKB20 and TSKB18 responses, then T. cruzi-infected TSKB-Tg mice should maintain effector CD8+ T cell populations with numbers similar to or greater than those of WT mice during the acute and chronic phases. Compared with infected WT mice, the TSKB-Tg groups had similar proportions of CD8+ T cells expressing the markers of previous antigen encounter, CD44 and CD11a (Fig. 4B), and the total number of antigen-experienced CD8+ T cells per spleen was not different between groups during the acute and chronic phases of infection (Fig. 4C). Interestingly, a greater proportion of CD44hi CD11ahi CD8+ T cells expressed CD127 in spleens of TSKB20/18 Tg mice (Fig. 4E) (P < 0.01 than in WT and TSKB18 Tg groups), though no differences in CD62L expression were observed (Fig. 4E), suggesting that the antigen-experienced CD8+ T cells may encounter antigen slightly less frequently in chronically infected TSKB20/18 Tg mice.
To determine if CD8+ T cells suffer increased evidence of immune exhaustion in the absence of control by the immunodominant TSKB20 and TSKB18 responses, we stimulated T cell receptors (TCRs) of spleen-derived CD8+ T cells from chronically infected TSKB20/18 Tg and WT mice and measured the production of the cytokines IFN-γ and TNF-α (Fig. 4F, top) and the release of cytotoxic granule contents as indicated by surface CD107a staining (Fig. 4F, bottom). Interestingly, despite accumulating numbers of effector CD8+ T cells that were otherwise similar to those of WT mice during the acute phase of infection (see Fig. S1 in the supplemental material), the TSKB20/18 Tg mice had fewer CD8+ T cells producing IFN-γ alone (P < 0.01) or both IFN-γ and TNF-α simultaneously (P < 0.01) than did WT mice during chronic infection (Fig. 4G), possibly due to the contraction of the effector CD8+ T cells in response to antigen clearance. Importantly, chronically infected WT and TSKB-Tg groups maintained similar proportions of multifunctional IFN-γ-producing CD8+ T cells that also produced TNF-α or degranulated in response to in vitro stimulation (Fig. 4H). Collectively, these data demonstrate that the compensating effector CD8+ T cells responding to undefined subdominant epitopes in TSKB20/18 Tg mice appear functionally equivalent to the immunodominant TSKB20- and TSKB18-specifc CD8+ T cells in WT mice.
Memory CD8+ T cells respond to challenge with T. cruzi in the absence of the TSKB20- and TSKB18-specific responses.
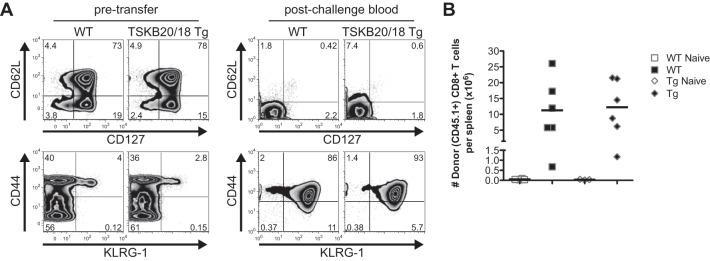
An important property of pathogen-specific T cells is the development of immunological memory after antigen clearance. Pathogen-specific memory cells proliferate and rapidly acquire effector functions that enhance control upon subsequent pathogen reencounter. To test the memory capacity of the normally subdominant CD8+ T cells maintained in TSKB20/18 Tg mice after T. cruzi clearance, we induced immunological memory via an infection and drug treatment/cure protocol (20) and used these mice as a source of memory CD8+ T cells for transfer into naive congenic recipients. As expected, the majority of CD8+ T cells isolated from drug-cured WT and TSKB20/18 Tg mice >200 days posttreatment expressed the memory markers CD127 and CD62L (Fig. 5A, left), and relatively few expressed the short-lived effector marker KLRG-1 (Fig. 5A, left). Following cell transfer and T. cruzi challenge, the majority of these CD8+ T cells rapidly lost expression of both CD127 and CD62L (Fig. 5A, right) and acquired a KLRG-1+ phenotype (Fig. 5A, right) in the peripheral blood. By 21 days postchallenge, spleens of WT and TSKB20/18 Tg mice accumulated expanded numbers of donor-derived CD8+ T cells (Fig. 5B).
FIG 5.
Memory CD8+ T cell responses in the absence of TSKB20- and TSKB18-specific CD8+ T cells. WT littermates or TSKB20/18 Tg mice (both CD45.1/CD45.2) were infected with 1 × 103 T. cruzi organisms, treated orally with benznidazole daily for 40 consecutive days, beginning at 15 days postinfection, and sacrificed >200 days after cessation of treatment. CD8+ T cells (2 × 106) from spleens and lymph nodes of treated WT or TSKB20/18 Tg mice were transferred into uninfected WT or TSKB20/18 Tg (both CD45.2), respectively. Recipients were challenged the following day with 1 × 104 T. cruzi organisms. (A) Surface phenotype of CD45.1+ CD8+ T cells before adoptive transfer (left) and in the peripheral blood of recipients 13 days after challenge (right). Numbers indicate percentages of events in the gate. (B) Total number of donor CD45.1+ CD8+ T cells recovered from spleens of challenged mice (filled symbols) at 21 days postinfection.
Immunodominance does not shift to CD8+ T cells recognizing previously identified parasite-derived epitopes.
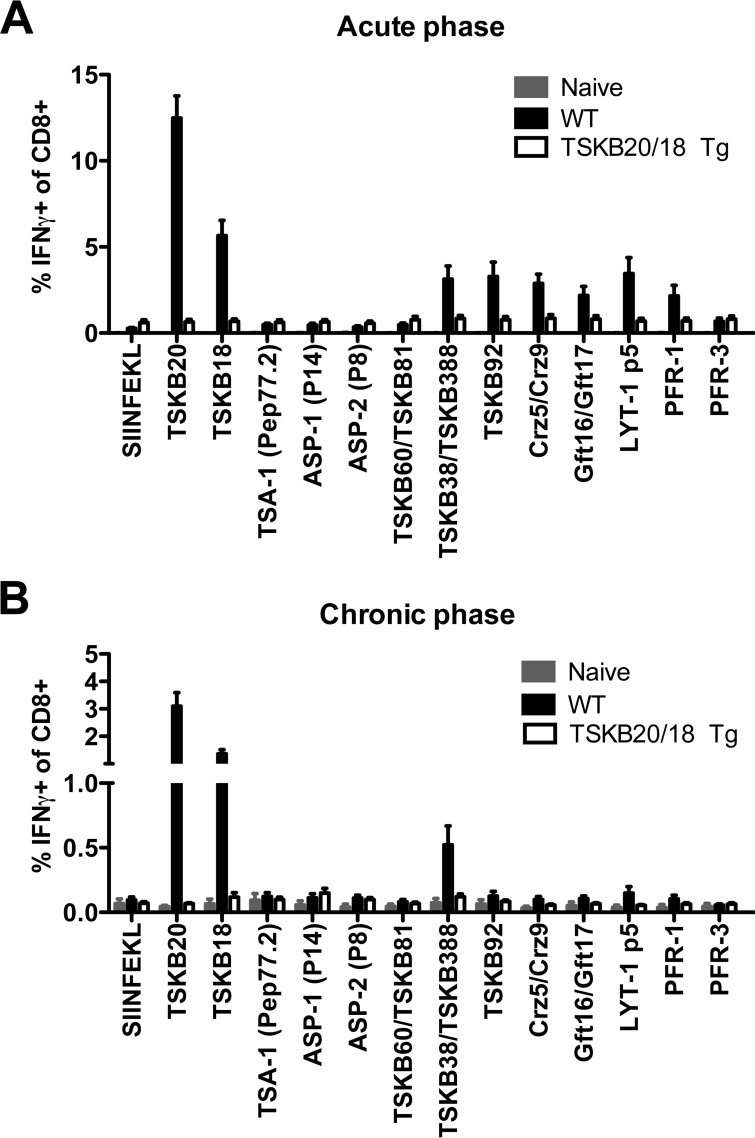
To determine if compensating effector CD8+ T cells that develop in infected TSKB20/18 Tg mice recognize previously identified parasite-derived epitopes, we incubated spleen cells from acutely or chronically infected WT and TSKB20/18 Tg mice with H-2Kb-restricted peptides and assessed specific responsiveness by intracellular staining for IFN-γ. The assay included epitopes encoded by the TS vaccine candidates TSA-1 (Pep77.2) (35, 36), ASP-1 (P14), and ASP-2 (P8) (37, 38) as well as variant TS peptides encoded at homologous (TSKB60, TSKB81, TSKB38, TSKB388) and nonhomologous (TSKB92) positions of TS genes (14). We also assessed responsiveness to non-TS epitopes encoded by cruzipain (Crz5 and Crz9) and β-galactofuranosyl transferase (Gft16 and Gft17) gene family members (14, 32, 39), LYT-1 (LYT-1p5) (40), and T. cruzi's paraflagellar rod proteins (PAR-1-derived PFR-1 and PAR-3-derived PFR-3) (41). The irrelevant H-2Kb-restricted epitope, SIINFEKL, was used as a negative control. Peptide-induced IFN-γ production was observed in CD8+ T cells from infected WT mice but not TSKB20/18 Tg mice at both acute (Fig. 6A) and chronic (Fig. 6B) time points. We also considered that unknown epitopes encoded by PAR-1, PAR-3, LYT-1, or the recently described KMP-11 (42) might be targets of effector CD8+ T cells in TSKB20/18 Tg mice, but in vitro coculture with C57BL/6-derived cell lines transfected with the open reading frames (ORFs) encoding these proteins failed to elicit cytokine production by CD8+ T cells from infected TSKB20/18 Tg mice (see Fig. S2 in the supplemental material). Thus, in the absence of the immunodominant TSKB20 and TSKB18 responses, the main focus of the responding CD8+ T cells cannot be attributed to these tested parasite-derived epitopes.
FIG 6.
Subdominant parasite-derived epitopes are not recognized by CD8+ T cells in infected TSKB20/18 Tg mice. (A, B) Cumulative data from peptide-stimulated splenocytes from naive and acutely (A) or naive and chronically infected (B) WT or TSKB20/18 Tg mice. CD8+ CD4− gated events were assessed for IFN-γ staining after 5 h of incubation with 1 μM indicated peptide alone or indicated combined peptide pool. Data are representative of 5 acute-phase experiments at 18 to 21 days postinfection (n = 11 to 17 mice per infected group) and 4 chronic-phase experiments at 238 to 338 days postinfection (n = 11 to 13 mice per infected group). Filled gray bars are uninfected naive mice, filled black bars are WT, and open bars are TSKB20/18 Tg. Data are means +SEM.
Parasite-specific CD8+ T cells recognize an engineered dominant epitope in the absence of the TSKB20 and TSKB18 response.
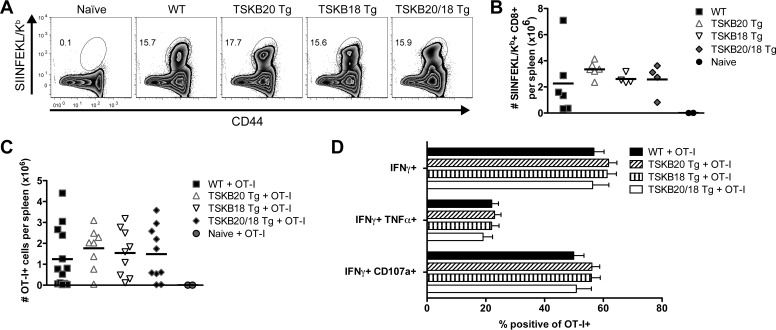
Though our observations supported the hypothesis that effector CD8+ T cells control T. cruzi in the absence of the described immunodominant responses (Fig. 2 to 4), we lacked conclusive evidence of their specificity for bona fide parasite-derived epitopes (Fig. 6). To confirm that the effector CD8+ T cells generated by infected TSKB20/18 Tg mice are specific for parasite antigen, we infected WT and TSKB-Tg mice with T. cruzi strain Brazil parasites stably transfected with the model antigen ovalbumin (Brazil-OVA) (43), allowing us to track parasite-specific CD8+ T cells recognizing the OVA-derived SIINFEKL epitope. Both WT and TSKB-Tg groups expanded SIINFEKL/Kb-tetramer+ CD44hi CD8+ T cells (Fig. 7A), and similar numbers of SIINFEKL-specific CD8+ T cells were observed in spleens of all groups infected with Brazil-OVA (Fig. 7B). Since the T cells that survive thymic selection differ between WT and mice transgenically expressing TSKB20 and TSKB18, we considered that this altered TCR repertoire might impact the ability of the endogenous SIINFEKL-specific CD8+ T cells to respond in the TSKB peptide-tolerant mice. However, transferred TCR transgenic OT-I cells specific for SIINFEKL expanded to the same extent in the Brazil-OVA-infected WT and TSKB-Tg mice (Fig. 7C). Similar proportions of responding OT-I cells produced cytokines and degranulated when incubated with SIINFEKL (Fig. 7D) in the WT and the TSKB-Tg mice. These data demonstrate that in the absence of T cells responding to immunodominant epitopes, alternative parasite-derived epitopes are targeted by effector CD8+ T cells. However, these neoimmunodominant T cells do not necessarily expand more or function better in the absence of TSKB20 and TSKB18 immunodomination.
FIG 7.
Engineered immunodominant SIINFEKL-specific response in the absence of TSKB20 and TSKB18 immunodominance. (A) Representative SIINFEKL/Kb-tetramer staining of splenocytes from naive or WT, TSKB20 Tg, TSKB18 Tg, or TSKB20/18 Tg mice infected with T. cruzi Brazil-OVA at 22 days postinfection. Histograms are gated on CD8+ CD4−, and the numbers indicate percentages of CD44hi SIINFEKL/Kb+ of CD8+ cells. (B) The total number of SIINFEKL/Kb+ CD8+ T cells from spleens of naive or Brazil-OVA infected WT, TSKB20 Tg, TSKB18 Tg, and TSKB20/18 Tg mice at 21 to 22 days postinfection. Data are cumulative results from 2 experiments (n = 4 to 6 per infected group). (C) Total number of CD45.1+ OT-I cells from spleens of naive or Brazil-OVA-infected WT, TSKB20 Tg, TSKB18 Tg, and TSKB20/18 Tg mice at 21 days postinfection. All mice received 50 OT-I cells i.v. prior to infection. Several infected individuals did not have detectable OT-I cells and were omitted from the analysis. Data points in panels B, C, and E represent individual mice (squares are WT, up triangles are TSKB20 Tg, down triangles are TSKB18 Tg, diamonds are TSKB20/18 Tg, and circles are uninfected naive mice), and bars are means. (D) Proportion of OT-I (CD45.1+) CD8+ T cells stained positive for the indicated marker in response to 5 h SIINFEKL peptide stimulation from spleens of Brazil-OVA-infected mice at 21 days postinfection. Filled black bars are results for WT, forward-slashed bars for TSKB20 Tg, vertical-slashed bars for TSKB18 Tg, and open bars for TSKB20/18 Tg mice. Data in panels C and D are cumulative results for 2 experiments (n = 8 to 13 per infected group).
DISCUSSION
How critical for control of infection are CD8+ T cells specific for only a few of the thousands of potential pathogen-derived epitopes? Despite numerous studies on immunodominant CD8+ T cells, surprisingly few have attempted to elucidate the relationship between immunodominance and protective immunity (2, 4, 44, 45). This is especially true for parasitic diseases, for which relatively few studies have been conducted to determine the mechanisms accounting for immunodominance (3, 13–15, 46, 47) and the roles of these parasite-specific CD8+ T cells in controlling infection (21, 48–50). Some suggest that narrow responses favor establishment of chronic infections (4, 51), whereas broadly focused responses are more capable of long-term control (27, 51).
We addressed the importance of CD8+ T cells recognizing the described immunodominant epitopes encoded by T. cruzi's TS gene family by preventing the generation of these responses using tolerizing protocols (reference 21 and this study). We find that immune control of T. cruzi is generated independent of recognition of the dominant TSKB20 and TSKB18 epitopes since TSKB20 and TSKB18 transgenic mice, as well as TSKB20/18 double transgenic mice, produce potent CD8+ T cell responses and survive up to 1 year after T. cruzi infection. Chronic T. cruzi infection drives disease development over long periods; altering the CD8+ T cell immunodominance hierarchies by tolerized mice could result in subtle differences in parasite control that over time may result in altered disease progression. However, chronically infected WT and TSKB Tg mice exhibited similarly low levels of disease in persistently infected tissues. The TSKB20-specific response particularly stands out as one of the most dominant of the CD8+ T cell responses observed in any pathogen infection. Yet this unusually strong immunodominant response is quite dispensable with respect to control of the infection. Thus, the quality, frequency, or phenotype of highly dominant pathogen-specific T cells may not always provide a reliable measure of the overall quality of immune control (as shown in this study and recently reviewed by Tscharke et al. [52]).
Vaccines that boost CD8+ T cells recognizing TS-derived epitopes can enhance control of T. cruzi (21, 38, 53, 54), but only to a limited degree, as TS-based immunizations fail to provide sterilizing protection from infection (55). Given this less-than-stellar protective capacity of current vaccines targeted for TS family molecules, identification of normally subdominant CD8+ T cells that could be expanded by vaccination and that may provide better protection is of significant interest. However, identifying the antigen specificity of these subdominant T cell populations is hampered by their low frequency during infection. The TSKB20/18 double-transgenic mice offered the opportunity to elevate some of these normally subdominant responses, and the demonstration that mice lacking TSKB20 showed modestly increased numbers of the normally subdominant TSKB18-specific CD8+ T cells was encouraging for this approach. However, we ruled out a number of predicted “next-best” subdominant T. cruzi-derived epitopes and in fact found that many of the previously characterized epitope-specific responses are substantially reduced in the infected TSKB20/18 Tg mice, suggesting that these responses might be attributable to cross-reactive recognition by the promiscuous TCRs of TSKB20- and TSKB18-specific CD8+ T cells (14). Alternatively, the TSKB20- and TSKB18-specific effector CD8+ T cells may help the priming and/or maintenance of CD8+ T cells recognizing these otherwise cryptic epitopes by chemokine-dependent recruitment of polyclonal naive T cells to dendritic cells copresenting both dominant and subdominant epitopes (56). Clearly, parasite-specific CD8+ T cells of unknown specificity expand and compensate to provide TSKB20/18 Tg mice with a highly functional effector T cell pool. Future experiments to identify the specificity of T cells that control infection in TSKB-Tg mice should focus on nonvariant antigens that are surface expressed and predicted to be released by the parasite, thus making them available for recognition during infection with diverse strains of T. cruzi (19). A recent study (57) showing that T. cruzi's flagellum is shed early during host cell invasion and is a source of protective T cell epitopes demonstrates the potential utility of assessing the biological relevance of a pathogen's antigenic molecules, rather than simply relying on what the immune system normally focuses on, in the design of antipathogen vaccines.
T. cruzi's TS family exhibits considerable intra- and interstrain variability in sequence (17, 58–60) and expression patterns (25), which impacts the generation of CD8+ immunodominance hierarchies (14, 15). We and others (21, 23–27, 61) hypothesize that variant TS genes function as a means of immune evasion, and it has been suggested that immunodominance by TS epitope-specific CD8+ T cells could limit the generation of potentially more effective parasite-focused responses capable of complete parasite eradication (21, 27, 62). If immunodomination by TSKB20- and TSKB18-specific CD8+ T cells inhibits development of other potentially more protective CD8+ responses, one would predict that TSKB20/TSKB18 Tg mice should exhibit better long-term control of T. cruzi than their WT counterparts. However, this is not what we found. There are a number of reasons why this approach did not yield these expected outcomes. T. cruzi's genome contains thousands of variant TS genes (17, 18), as well as other gene families of surface-expressed proteins that encode variant MHC-I-restricted epitopes (14). Since our manipulations have deleted CD8+ T cells recognizing only a few of these TS-encoded epitopes, it is likely that other TS-encoded epitopes or other large gene family member-encoded epitopes replaced TSKB18 and TSKB20 as the main, but not sufficiently protective, targets of the anti-T. cruzi CD8+ T cells in the TS18/20 Tg mice. Additionally, simply the vast number and the quantity of potentially competing TS may serve as a “smoke screen,” thus obscuring the less abundant nonvariant epitopes that may be better targets for immune elimination among the milieu of evolving antigenic variants. A test of this hypothesis will require ablating the TS genes from T. cruzi's genome, a challenging endeavor given the thousands of TS genes. However, recent developments in the clustered regularly interspaced short palindromic repeat(s) (CRISPR)/Cas9 genome-editing technology may provide us with this opportunity in the near future (63).
Development of T cell-based vaccines has focused on enhancing immunodominant T cells prior to pathogen encounter, though boosting subdominant or cryptic responses has also been suggested as an important goal for vaccines (62, 64). Induction of broadly focused T cell responses might be particularly important for therapeutic vaccines that enhance control of chronically infecting pathogens for which the immunodominant response has already proven ineffective at establishing an immunological cure. In agreement with other studies (65), our data suggest that TS-based vaccination strategies will fail when applied in the field, in part because T. cruzi strains express unique versions of TS genes (14) and also because even under controlled experimental conditions, the recognition of immunodominant TS-derived epitopes is neither necessary nor sufficient for protection.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Angel Padilla for technical assistance and Julie Nelson of the Center for Tropical and Emerging Global Diseases Flow Cytometry Facility at the University of Georgia. Courtney Boehlke and Cecilia Pérez Brandán assisted with the design and construction of the TSKB20 and TSKB18 transgene vectors. Gretchen Cooley prepared the SIINFEKL/Kb tetramer. Kim Klonowski and Stephen Dalton generously shared reagents and gave technical advice. We thank members of the Tarleton Research Group for helpful discussion.
This work was supported by U.S. National Institutes of Health grants R01 AI22070 and R01 AI33106 to R.L.T. C.S.R. was supported by award numbers T32AI060546 and T32AI074492 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00241-16.
REFERENCES
- 1.Wong P, Pamer EG. 2003. CD8 T cell responses to infectious pathogens. Annu Rev Immunol 21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell JW. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Jordan KA, Hunter CA. 2010. Regulation of CD8+ T cell responses to infection with parasitic protozoa. Exp Parasitol 126:318–325. doi: 10.1016/j.exppara.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallimore A, Hengartner H, Zinkernagel R. 1998. Hierarchies of antigen-specific cytotoxic T-cell responses. Immunol Rev 164:29–36. doi: 10.1111/j.1600-065X.1998.tb01205.x. [DOI] [PubMed] [Google Scholar]
- 5.Yewdell JW, Bennink JR. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol 17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. 2006. Memory of mice and men: CD8(+) T-cell cross-reactivity and heterologous immunity. Immunol Rev 211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard N, Shastri N. 2010. Topological journey of parasite-derived antigens for presentation by MHC class I molecules. Trends Immunol 31:414–421. doi: 10.1016/j.it.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Goldszmid RS, Sher A. 2010. Processing and presentation of antigens derived from intracellular protozoan parasites. Curr Opin Immunol 22:118–123. doi: 10.1016/j.coi.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kedl RM, Kappler JW, Marrack P. 2003. Epitope dominance, competition and T cell affinity maturation. Curr Opin Immunol 15:120–127. doi: 10.1016/S0952-7915(02)00009-2. [DOI] [PubMed] [Google Scholar]
- 10.Welsh RM, Fujinami RS. 2007. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol 5:555–563. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin H, Wherry EJ. 2007. CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol 19:408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Padilla AM, Bustamante JM, Tarleton RL. 2009. CD8+ T cells in Trypanosoma cruzi infection. Curr Opin Immunol 21:385–390. doi: 10.1016/j.coi.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarleton RL. 2005. New approaches in vaccine development for parasitic infections. Cell Microbiol 7:1379–1386. doi: 10.1111/j.1462-5822.2005.00589.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin DL, Weatherly DB, Laucella SA, Cabinian MA, Crim MT, Sullivan S, Heiges M, Craven SH, Rosenberg CS, Collins MH, Sette A, Postan M, Tarleton RL. 2006. CD8+ T-cell responses to Trypanosoma cruzi are highly focused on strain-variant trans-sialidase epitopes. PLoS Pathog 2:e77. doi: 10.1371/journal.ppat.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzelepis F, de Alencar BC, Penido ML, Claser C, Machado AV, Bruna-Romero O, Gazzinelli RT, Rodrigues MM. 2008. Infection with Trypanosoma cruzi restricts the repertoire of parasite-specific CD8+ T cells leading to immunodominance. J Immunol 180:1737–1748. doi: 10.4049/jimmunol.180.3.1737. [DOI] [PubMed] [Google Scholar]
- 16.Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. 2008. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest 118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 18.Weatherly DB, Boehlke C, Tarleton RL. 2009. Chromosome level assembly of the hybrid Trypanosoma cruzi genome. BMC Genomics 10:255. doi: 10.1186/1471-2164-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg N, Nunes MP, Tarleton RL. 1997. Delivery by Trypanosoma cruzi of proteins into the MHC class I antigen processing and presentation pathway. J Immunol 158:3293–3302. [PubMed] [Google Scholar]
- 20.Bustamante JM, Bixby LM, Tarleton RL. 2008. Drug-induced cure drives conversion to a stable and protective CD8+ T central memory response in chronic Chagas disease. Nat Med 14:542–550. doi: 10.1038/nm1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg CS, Martin DL, Tarleton RL. 2010. CD8+ T cells specific for immunodominant trans-sialidase epitopes contribute to control of Trypanosoma cruzi infection but are not required for resistance. J Immunol 185:560–568. doi: 10.4049/jimmunol.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, Ghedin E, Peacock C, Bartholomeu DC, Haas BJ, Tran AN, Wortman JR, Alsmark UC, Angiuoli S, Anupama A, Badger J, Bringaud F, Cadag E, Carlton JM, Cerqueira GC, Creasy T, Delcher AL, Djikeng A, Embley TM, Hauser C, Ivens AC, Kummerfeld SK, Pereira-Leal JB, Nilsson D, Peterson J, Salzberg SL, Shallom J, Silva JC, Sundaram J, Westenberger S, White O, Melville SE, Donelson JE, Andersson B, Stuart KD, Hall N. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 23.Frasch AC. 2000. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today 16:282–286. doi: 10.1016/S0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 24.Martin D, Tarleton R. 2004. Generation, specificity, and function of CD8+ T cells in Trypanosoma cruzi infection. Immunol Rev 201:304–317. doi: 10.1111/j.0105-2896.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- 25.Atwood JA III, Weatherly DB, Minning TA, Bundy B, Cavola C, Opperdoes FR, Orlando R, Tarleton RL. 2005. The Trypanosoma cruzi proteome. Science 309:473–476. doi: 10.1126/science.1110289. [DOI] [PubMed] [Google Scholar]
- 26.Hoft DF, Eickhoff CS, Giddings OK, Vasconcelos JR, Rodrigues MM. 2007. Trans-sialidase recombinant protein mixed with CpG motif-containing oligodeoxynucleotide induces protective mucosal and systemic Trypanosoma cruzi immunity involving CD8+ CTL and B cell-mediated cross-priming. J Immunol 179:6889–6900. doi: 10.4049/jimmunol.179.10.6889. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues MM, Alencar BC, Claser C, Tzelepis F. 2009. Immunodominance: a new hypothesis to explain parasite escape and host/parasite equilibrium leading to the chronic phase of Chagas' disease? Braz J Med Biol Res 42:220–223. doi: 10.1590/S0100-879X2009000300001. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi DN, Hung DT, Wiley DC. 1992. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A 89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummings KL, Tarleton RL. 2003. Rapid quantitation of Trypanosoma cruzi in host tissue by real-time PCR. Mol Biochem Parasitol 129:53–59. doi: 10.1016/S0166-6851(03)00093-8. [DOI] [PubMed] [Google Scholar]
- 30.Eisenlohr LC, Yewdell JW, Bennink JR. 1992. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med 175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefrancois L, Obar JJ. 2010. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol Rev 235:206–218. doi: 10.1111/j.0105-2896.2010.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bixby LM, Tarleton RL. 2008. Stable CD8+ T cell memory during persistent Trypanosoma cruzi infection. J Immunol 181:2644–2650. doi: 10.4049/jimmunol.181.4.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Tarleton RL. 1999. Parasite persistence correlates with disease severity and localization in chronic Chagas' disease. J Infect Dis 180:480–486. doi: 10.1086/314889. [DOI] [PubMed] [Google Scholar]
- 34.Combs TP, Nagajyothi Mukherjee S, de Almeida CJG, Jelicks LA, Schubert W, Lin Y, Jayabalan DS, Zhao D, Braunstein VL, Landskroner-Eiger S, Cordero A, Factor SM, Weiss LM, Lisanti MP, Tanowitz HB, Scherer PE. 2005. The adipocyte as an important target cell for Trypanosoma cruzi infection. J Biol Chem 280:24085–24094. doi: 10.1074/jbc.M412802200. [DOI] [PubMed] [Google Scholar]
- 35.Wizel B, Nunes M, Tarleton RL. 1997. Identification of Trypanosoma cruzi trans-sialidase family members as targets of protective CD8+ TC1 responses. J Immunol 159:6120–6130. [PubMed] [Google Scholar]
- 36.Wizel B, Garg N, Tarleton RL. 1998. Vaccination with trypomastigote surface antigen 1-encoding plasmid DNA confers protection against lethal Trypanosoma cruzi infection. Infect Immun 66:5073–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Low HP, Santos MA, Wizel B, Tarleton RL. 1998. Amastigote surface proteins of Trypanosoma cruzi are targets for CD8+ CTL. J Immunol 160:1817–1823. [PubMed] [Google Scholar]
- 38.Garg N, Tarleton RL. 2002. Genetic immunization elicits antigen-specific protective immune responses and decreases disease severity in Trypanosoma cruzi infection. Infect Immun 70:5547–5555. doi: 10.1128/IAI.70.10.5547-5555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotner J, Tarleton R. 2007. Endogenous CD4(+) CD25(+) regulatory T cells have a limited role in the control of Trypanosoma cruzi infection in mice. Infect Immun 75:861–869. doi: 10.1128/IAI.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fralish BH, Tarleton RL. 2003. Genetic immunization with LYT1 or a pool of trans-sialidase genes protects mice from lethal Trypanosoma cruzi infection. Vaccine 21:3070–3080. doi: 10.1016/S0264-410X(03)00121-X. [DOI] [PubMed] [Google Scholar]
- 41.Wrightsman RA, Luhrs KA, Fouts D, Manning JE. 2002. Paraflagellar rod protein-specific CD8+ cytotoxic T lymphocytes target Trypanosoma cruzi-infected host cells. Parasite Immunol 24:401–412. doi: 10.1046/j.1365-3024.2002.00479.x. [DOI] [PubMed] [Google Scholar]
- 42.Lasso P, Mesa D, Cuellar A, Guzman F, Bolanos N, Rosas F, Velasco V, Thomas Mdel C, Lopez MC, Gonzalez JM, Puerta CJ. 2010. Frequency of specific CD8+ T cells for a promiscuous epitope derived from Trypanosoma cruzi KMP-11 protein in chagasic patients. Parasite Immunol 32:494–502. doi: 10.1111/j.1365-3024.2010.01206.x. [DOI] [PubMed] [Google Scholar]
- 43.Padilla AM, Simpson LJ, Tarleton RL. 2009. Insufficient TLR activation contributes to the slow development of CD8+ T cell responses in Trypanosoma cruzi infection. J Immunol 183:1245–1252. doi: 10.4049/jimmunol.0901178. [DOI] [PubMed] [Google Scholar]
- 44.Doherty PC, Christensen JP. 2000. Accessing complexity: the dynamics of virus-specific T cell responses. Annu Rev Immunol 18:561–592. doi: 10.1146/annurev.immunol.18.1.561. [DOI] [PubMed] [Google Scholar]
- 45.Holtappels R, Simon CO, Munks MW, Thomas D, Deegen P, Kuhnapfel B, Daubner T, Emde SF, Podlech J, Grzimek NK, Oehrlein-Karpi SA, Hill AB, Reddehase MJ. 2008. Subdominant CD8 T-cell epitopes account for protection against cytomegalovirus independent of immunodomination. J Virol 82:5781–5796. doi: 10.1128/JVI.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frickel EM, Sahoo N, Hopp J, Gubbels MJ, Craver MP, Knoll LJ, Ploegh HL, Grotenbreg GM. 2008. Parasite stage-specific recognition of endogenous Toxoplasma gondii-derived CD8+ T cell epitopes. J Infect Dis 198:1625–1633. doi: 10.1086/593019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, Shastri N. 2008. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol 9:937–944. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar KA, Sano G, Boscardin S, Nussenzweig RS, Nussenzweig MC, Zavala F, Nussenzweig V. 2006. The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature 444:937–940. doi: 10.1038/nature05361. [DOI] [PubMed] [Google Scholar]
- 49.Mishra S, Rai U, Shiratsuchi T, Li X, Vanloubbeeck Y, Cohen J, Nussenzweig RS, Winzeler EA, Tsuji M, Nussenzweig V. 2011. Identification of non-CSP antigens bearing CD8 epitopes in mice immunized with irradiated sporozoites. Vaccine 29:7335–7342. doi: 10.1016/j.vaccine.2011.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafalla JC, Bauza K, Friesen J, Gonzalez-Aseguinolaza G, Hill AV, Matuschewski K. 2013. Identification of targets of CD8(+) T cell responses to malaria liver stages by genome-wide epitope profiling. PLoS Pathog 9:e1003303. doi: 10.1371/journal.ppat.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posnett DN, Engelhorn ME, Houghton AN. 2005. Antiviral T cell responses: phalanx or multipronged attack? J Exp Med 201:1881–1884. doi: 10.1084/jem.20050928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tscharke DC, Croft NP, Doherty PC, La Gruta NL. 2015. Sizing up the key determinants of the CD8(+) T cell response. Nat Rev Immunol 15:705–716. doi: 10.1038/nri3905. [DOI] [PubMed] [Google Scholar]
- 53.Fujimura AE, Kinoshita SS, Pereira-Chioccola VL, Rodrigues MM. 2001. DNA sequences encoding CD4+ and CD8+ T-cell epitopes are important for efficient protective immunity induced by DNA vaccination with a Trypanosoma cruzi. Gene Infect Immun 69:5477–5486. doi: 10.1128/IAI.69.9.5477-5486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katae M, Miyahira Y, Takeda K, Matsuda H, Yagita H, Okumura K, Takeuchi T, Kamiyama T, Ohwada A, Fukuchi Y, Aoki T. 2002. Coadministration of an interleukin-12 gene and a Trypanosoma cruzi gene improves vaccine efficacy. Infect Immun 70:4833–4840. doi: 10.1128/IAI.70.9.4833-4840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigues MM, de Alencar BC, Claser C, Tzelepis F, Silveira EL, Haolla FA, Dominguez MR, Vasconcelos JR. 2009. Swimming against the current: genetic vaccination against Trypanosoma cruzi infection in mice. Mem Inst Oswaldo Cruz 104(Suppl 1):281–287. [DOI] [PubMed] [Google Scholar]
- 56.Hugues S, Scholer A, Boissonnas A, Nussbaum A, Combadiere C, Amigorena S, Fetler L. 2007. Dynamic imaging of chemokine-dependent CD8+ T cell help for CD8+ T cell responses. Nat Immunol 8:921–930. doi: 10.1038/ni1495. [DOI] [PubMed] [Google Scholar]
- 57.Kurup SP, Tarleton RL. 2014. The Trypanosoma cruzi flagellum is discarded via asymmetric cell division following invasion and provides early targets for protective CD8(+) T cells. Cell Host Microbe 16:439–449. doi: 10.1016/j.chom.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kahn S, Vanvoorhis WC, Eisen H. 1990. The major 85-kD surface antigen of the mammalian form of Trypanosoma cruzi is encoded by a large heterogeneous family of simultaneously expressed genes. J Exp Med 172:589–597. doi: 10.1084/jem.172.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kahn SJ, Wleklinski M. 1997. The surface glycoproteins of Trypanosoma cruzi encode a superfamily of variant T cell epitopes. J Immunol 159:4444–4451. [PubMed] [Google Scholar]
- 60.Millar AE, Kakn SJ. 2000. The SA85-1.1 protein of the Trypanosoma cruzi trans-sialidase superfamily is a dominant T-cell antigen. Infect Immun 68:3574–3580. doi: 10.1128/IAI.68.6.3574-3580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freire-de-Lima L, Alisson-Silva F, Carvalho ST, Takiya CM, Rodrigues MM, DosReis GA, Mendonca-Previato L, Previato JO, Todeschini AR. 2010. Trypanosoma cruzi subverts host cell sialylation and may compromise antigen-specific CD8+ T cell responses. J Biol Chem 285:13388–13396. doi: 10.1074/jbc.M109.096305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez MR, Silveira EL, de Vasconcelos JR, de Alencar BC, Machado AV, Bruna-Romero O, Gazzinelli RT, Rodrigues MM. 2011. Subdominant/cryptic CD8 T cell epitopes contribute to resistance against experimental infection with a human protozoan parasite. PLoS One 6:e22011. doi: 10.1371/journal.pone.0022011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. 2015. CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. mBio 6:e02097-14. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sette A, Peters B. 2007. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Curr Opin Immunol 19:106–110. doi: 10.1016/j.coi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 65.Haolla FA, Claser C, de Alencar BC, Tzelepis F, de Vasconcelos JR, de Oliveira G, Silverio JC, Machado AV, Lannes-Vieira J, Bruna-Romero O, Gazzinelli RT, Dos Santos RR, Soares MB, Rodrigues MM. 2009. Strain-specific protective immunity following vaccination against experimental Trypanosoma cruzi infection. Vaccine 27:5644–5653. doi: 10.1016/j.vaccine.2009.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.