Abstract
Helicobacter pylori secretes a pore-forming VacA toxin that has structural features and activities substantially different from those of other known bacterial toxins. VacA can assemble into multiple types of water-soluble flower-shaped oligomeric structures, and most VacA activities are dependent on its capacity to oligomerize. The 88-kDa secreted VacA protein can undergo limited proteolysis to yield two domains, designated p33 and p55. The p33 domain is required for membrane channel formation and intracellular toxic activities, and the p55 domain has an important role in mediating VacA binding to cells. Previous studies showed that the p55 domain has a predominantly β-helical structure, but no structural data are available for the p33 domain. We report here the purification and analysis of a nonoligomerizing mutant form of VacA secreted by H. pylori. The nonoligomerizing 88-kDa mutant protein retains the capacity to enter host cells but lacks detectable toxic activity. Analysis of crystals formed by the monomeric protein reveals that the β-helical structure of the p55 domain extends into the C-terminal portion of p33. Fitting the p88 structural model into an electron microscopy map of hexamers formed by wild-type VacA (predicted to be structurally similar to VacA membrane channels) reveals that p55 and the β-helical segment of p33 localize to peripheral arms but do not occupy the central region of the hexamers. We propose that the amino-terminal portion of p33 is unstructured when VacA is in a monomeric form and that it undergoes a conformational change during oligomer assembly.
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that persistently colonizes the stomach in about half of the human population worldwide (1–3). Most people tolerate the presence of this organism for long periods without any adverse consequences, but a small subset of H. pylori-infected individuals develop gastric adenocarcinoma or peptic ulcer disease.
H. pylori is a relatively noninvasive organism that lives in the gastric mucus layer overlying gastric epithelial cells, sometimes adhering to the gastric epithelium. Many H. pylori-induced alterations in the gastric mucosa are attributable to the actions of secreted bacterial proteins on host cells (4–6). One such protein is the secreted vacuolating toxin, VacA (4, 7–10). VacA causes multiple alterations in gastric epithelial cells, including swelling of endosomes (vacuolation) (11, 12), permeabilization of mitochondrial membranes, and activation of mitogen-activated protein kinases (4, 7–9). In addition to its effects on gastric epithelial cells, VacA can disrupt the functions of many types of immune cells (T cells, B cells, neutrophils, eosinophils, and monocytes) (4, 7–9, 13–16). The cellular activities of VacA are attributed mainly to its ability to form anion-selective channels in host cells (17–21). VacA lacks sequence similarity to any other known bacterial toxins.
H. pylori strains isolated from unrelated individuals are genetically heterogeneous, and the risk of developing gastric cancer or peptic ulcer disease is determined in part by characteristics of the H. pylori strains with which persons are infected (22). All H. pylori strains contain a vacA gene, but there is a high level of sequence variation among vacA alleles from different strains (23). Several families of vacA alleles (designated s1 or s2, i1 or i2, and m1 or m2) have been described based on variation in specific regions of the gene (22, 24, 25). Individuals infected with H. pylori strains harboring type s1, i1, or m1 forms of vacA have an increased risk of gastric cancer or peptic ulcer disease compared to individuals infected with strains harboring type s2, i2, or m2 forms of vacA (22).
The 88-kDa VacA protein secreted by H. pylori can assemble into multiple types of water-soluble flower-shaped oligomeric structures, including both double-layered forms (dodecamers and tetradecamers) and single-layered forms (hexamers and heptamers) (26–31). The single-layered water-soluble oligomers are proposed to be structurally related to VacA membrane channels (17, 19, 31). By using single-particle electron microscopy and the random conical tilt approach, the three-dimensional structures of several VacA oligomeric conformations have been determined at ∼15-Å resolution (31). Mutant forms of VacA that fail to form water-soluble oligomeric structures lack toxin activity (20, 32, 33), which supports the view that VacA oligomerization is required for toxin activity.
Secretion of VacA is believed to proceed via a type V or autotransporter mechanism (6, 34–38). The secreted 88-kDa VacA protein can undergo limited proteolysis to yield two domains, designated p33 and p55, which remain together after proteolysis (27, 36, 39–42). The p55 domain has an important role in mediating VacA binding to cells (41, 43). Studies of VacA proteins expressed in transiently transfected HeLa cells have shown that the entire p33 domain plus the amino-terminal part of the p55 domain are required for intracellular toxin activity (39, 44, 45). The p33 domain has an important role in targeting VacA to mitochondria (46–48). Within the p33 domain, a predicted hydrophobic region about 30 amino acids in length is present near the amino terminus (20, 49). This region contains multiple GXXXG motifs, sequences that are predicted to mediate transmembrane dimerization (21, 50). Site-directed mutagenesis of several amino acids within the amino-terminal hydrophobic region, including the GXXXG motifs, abolishes membrane channel-forming activity and vacuolating toxin activity (21, 49, 51).
Analysis of crystals formed by a recombinant p55 protein revealed that this domain has a predominantly β-helical structure (52, 53). Similar to the VacA p55 domain, several other bacterial proteins secreted through a type V or autotransporter pathway (such as pertactin) are also known to have a predominantly β-helical structure (54). Despite considerable effort by multiple labs, there has been no progress thus far in experimentally defining the structure of the VacA p33 domain. Efforts to elucidate the structure of the p33 domain have been hindered by its insolubility (42), and efforts to determine the crystal structure of p88 VacA secreted by H. pylori have been hindered by its assembly into a heterogeneous assortment of oligomeric forms. Several studies have proposed structural models for regions of the p33 domain (47, 52, 55, 56), particularly the predicted transmembrane domain that contains the GXXXG repeats (55), but at present there are no experimental data to either validate or refute the proposed models. In this study, we report crystallization and preliminary structural analysis of a nonoligomerizing VacA protein, which provides the first insight into structural properties of the p33 domain.
MATERIALS AND METHODS
Bacterial growth conditions.
Wild-type (WT) H. pylori 60190 (ATCC 49503) was used as the parent strain for all of the experiments described here. H. pylori strains were routinely grown on Trypticase soy agar plates containing 5% sheep blood or in sulfite-free brucella broth (57) supplemented with cholesterol lipid concentrate (Gibco) (BB-cholesterol) (58). Mutant strains were cultured initially on brucella agar plates containing 10% fetal bovine serum (FBS) supplemented with chloramphenicol (5 μg/ml) or metronidazole (3.75 μg/ml) and subsequently grown on medium without antibiotics. H. pylori strains were grown at 37°C in ambient air containing 5% CO2.
Generation of H. pylori mutant strains.
H. pylori strains producing VacA proteins containing a strep tag (Strep-tag II) were constructed using a previously described negative selection approach involving use of a cat::rdxA cassette (59). One strain was engineered to produce VacA containing a strep tag introduced at amino acid 312 (VacA-Str312), and another strain produced VacA with a deletion of amino acids 346 and 347 and containing the strep tag [VacA-Δ(346-347)-Str312]. The presence of nucleotide sequences encoding the strep tag and the Δ346-347 mutation was confirmed by PCR and nucleotide sequence analysis of PCR products amplified from mutant strains. Strains producing VacA proteins containing strep tags introduced at alternate sites within the p55 domain (Str427, Str480, Str524, or Str808) were generated using the same approach.
Purification of VacA.
H. pylori strains engineered to produce strep-tagged VacA were grown in BB-cholesterol broth for 48 h. After centrifugation of the cultures, VacA and other proteins in the culture supernatant were precipitated by addition of ammonium sulfate (50% saturated solution). The precipitated proteins were pelleted by centrifugation and resuspended in phosphate-buffered saline (PBS) containing 1 mM EDTA and 0.02% sodium azide. Strep-tagged VacA proteins were purified using a protocol modified from Schmidt et al. (60). In brief, protein preparations were incubated with Strep-Tactin resin (IBA), and the resin was then loaded into a gravity column. After washing of the column with wash buffer (50 mM Tris, 150 mM NaCl [pH 8.0]), VacA was eluted with elution buffer (50 mM Tris, 150 mM NaCl, and 5 mM d-desthiobiotin [pH 8.0]). WT oligomeric VacA (without a strep tag) was purified from culture supernatants of WT H. pylori 60190 by gel filtration using a Superose 6 column, as described previously (12, 27, 31).
Analysis of cell-vacuolating activity.
HeLa cells were grown in minimal essential medium (modified Eagle medium containing Earle's salts) supplemented with 10% FBS in a 5% CO2 atmosphere at 37°C, and AGS cells were grown in RPMI 1640 medium supplemented with 10% FBS, 10 mM HEPES, penicillin, and streptomycin. Purified oligomeric forms of VacA (wild type, VacA-Str312, or VacA-Str808) were acid activated by the slow addition of 200 mM HCl until a pH of 3.0 was reached (61, 62). The acidified VacA preparations were added to the tissue culture medium overlying cells (supplemented with 5 mM ammonium chloride), followed by incubation at 37°C for the times specified for individual experiments. To quantify cell vacuolation, HeLa cells were seeded at a density of 1.2 × 104 cells/well into 96-well plates 1 day prior to the addition of VacA proteins. VacA-induced vacuolation of HeLa cells was detected by inverted light microscopy and quantified by a neutral red uptake assay (12, 63). The neutral red uptake assay provided a quantitative measurement of HeLa cell vacuolation but was not useful for studies of AGS gastric epithelial cells. Therefore, VacA-induced vacuolation of AGS cells was monitored by inverted light microscopy and differential interference contrast (DIC).
Fluorescence microscopy.
Purified WT VacA and VacA-Δ(346-347)-Str312 were labeled with Alexa Fluor 488 (Molecular Probes) according to the manufacturer's instructions (42, 64, 65). AGS cells were then incubated with the acid-activated, fluorescently labeled proteins (5 μg/ml). The cells were washed 1× with PBS, fixed with 4% paraformaldehyde, and stained with Hoechst 33342 to label the DNA. The cells were imaged with a DeltaVision Elite microscope equipped with a 60× 1.4 NA lens (Olympus) and a Cool SnapHQ2 CCD camera (Photometrics). Z-sections spaced 200 nm apart were acquired and deconvolved with SoftWorx (GE Healthcare). Images are a maximum intensity projection of three Z-sections. ImageJ was used for image processing.
Electron microscopy.
VacA proteins [3 μl of 10-μg/ml protein solutions of VacA-Δ(346-347)-Str312 or acidified WT VacA; 3 μl of a 40-μg/ml protein solution for nonacidified WT VacA or VacA-Str312] were spotted onto glow-discharged copper-mesh grids (EMS). The grids were then washed two times and stained in 0.7% uranyl formate (31). Images were collected on an FEI Morgagni run at 100 kV at a magnification of ×28,000 and then recorded on an AMT 1Kx1K charge-coupled device camera.
Crystallization and structure determination.
Purified VacA-Δ(346-347)-Str312 in elution buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 5 mM d-desthiobiotin [pH 8.0]) was concentrated to at least 5 mg/ml. Initial crystal trials were performed using a Mosquito Nanoliter-Dispenser (TTP Labtech) and a Rock Imager (Formulatrix) for automated analysis of sitting drops. A hanging-drop method was used to further optimize crystallization of proteins. VacA and precipitants were mixed in a 1:1 ratio at 21°C. Cubic crystals grew in reservoirs containing 0.8 to 1.8 M sodium chloride and 0.1 M sodium acetate at pH 4.6 to 5. X-ray data were collected from single crystals on an LS-CAT beamline 21 ID-D at the Advanced Photon Source (Argonne, IL) at 100 K. Diffraction data were indexed, integrated, and scaled using the HKL2000 software package (66) (Table 1). Phases were determined by molecular replacement using the p55 structure (52) and the program MOLREP (67). The electron density map was visualized using Coot (68). Although the maps were not of sufficient resolution to allow assignment of the sequence register, a continuous chain of 165 residues could be modeled as a polyalanine β-helix.
TABLE 1.
X-ray data collection statistics
| Parameter | Value(s)a |
|---|---|
| Unit cell dimensions | |
| a, b, c (Å) | 330.7, 330.7, 330.7 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution, outer shell (Å) | 50.0–4.5 (4.58–4.50) |
| Rmerge (%) | 10.9 (52.1) |
| Mean I/σ〈I〉 | 10.4 (2.75) |
| Completeness (%) | 99.7 (97.8) |
| Redundancy | 7.4 (6.6) |
| No. of unique observations | 35,776 (1,747) |
Outer-resolution bin statistics are given in parentheses.
Rmerge = Σhkl(Σi|Ihkl,i − 〈Ihkl〉)/Σhkl,i〈Ihkl,i〉, where Ihkl,i, is the intensity of an individual measurement of the reflection with the Miller indices h, k, and l, and 〈Ihkl〉 is the mean intensity of that reflection.
RESULTS
Purification of a nonoligomerizing VacA protein.
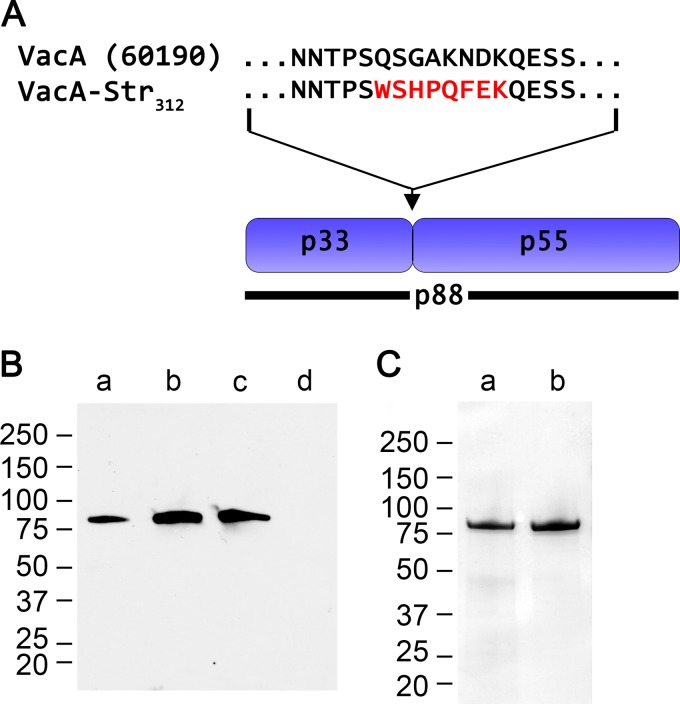
Previous studies have shown that mutagenesis of VacA residues 346 and 347, located within the p55 domain of VacA, abolishes the capacity of VacA to cause vacuolation of host cells as well as its capacity to assemble into water-soluble oligomeric structures (33, 39). The nonoligomerizing Δ346-347 mutant VacA protein cannot be purified using the gel filtration methods developed for purification of WT VacA oligomers (33). To overcome this problem, we engineered an H. pylori strain that produced a VacA Δ346-347 mutant protein with a strep tag introduced at the junction between p33 and p55 domains, designated VacA-Δ(346-347)-Str312 (Fig. 1A). The VacA-Δ(346-347)-Str312 protein was secreted into the extracellular space without any defects in secretion compared to WT untagged VacA (Fig. 1B), and the strep-tagged protein could be purified from culture supernatant using streptactin resin. When analyzed by SDS-PAGE under denaturing conditions, the purified strep-tagged protein had a mass of ∼88 kDa, similar to WT VacA (Fig. 1C) (12, 69). Also similar to WT VacA, the VacA-Δ(346-347)-Str312 protein spontaneously underwent limited proteolytic degradation into 55- and 33-kDa fragments during prolonged storage (data not shown).
FIG 1.
Introduction of a strep tag into VacA. The p88 VacA protein secreted by H. pylori strain 60190 (GenBank U05676) contains two domains, designated p33 and p55. H. pylori 60190 was engineered as described in Materials and Methods to allow production of VacA proteins with a strep tag introduced at the junction between p33 and p55 domains. (A) The figure compares the amino acid sequence of the p33-p55 junction in WT VacA with the corresponding sequence in a modified VacA protein containing the strep tag (WSHPQFEK) introduced at amino acid 312. (B) H. pylori strains were grown in broth culture, and unconcentrated broth culture supernatants were analyzed by immunoblotting using polyclonal anti-VacA serum. Lanes: a, strain 60190, producing WT VacA; b, strain producing VacA with a strep tag introduced at position 312 (VacA-Str312); c, strain producing VacA-Δ(346-347)-Str312; d, vacA-null mutant. (C) SDS-PAGE and Coomassie blue staining of strep-tagged VacA proteins, purified as described in Materials and Methods. Lanes: a, VacA-Str312; b, VacA-Δ(346-347)-Str312.
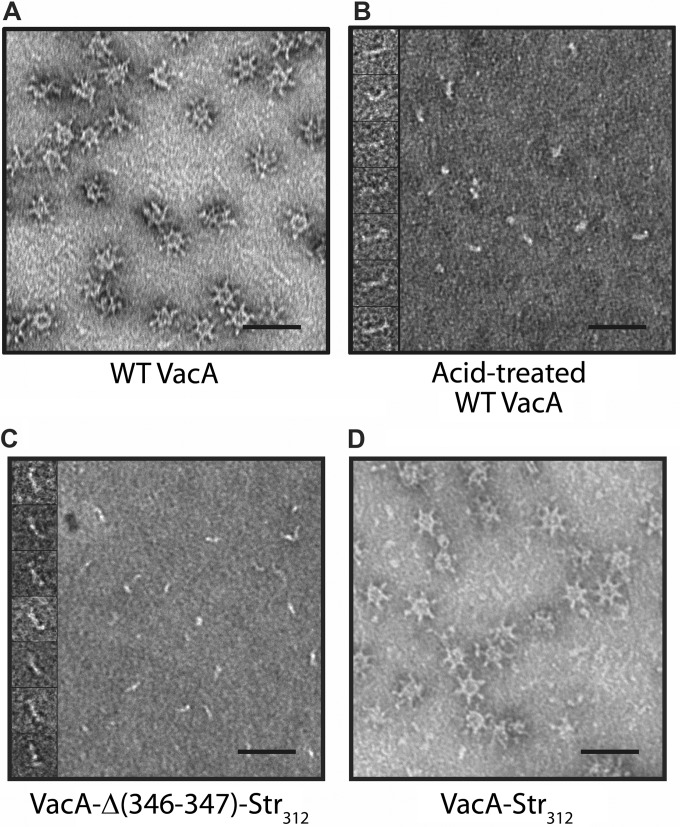
Under nondenaturing conditions at neutral pH, WT VacA assembles into water-soluble flower-shaped oligomeric structures (26–31) (Fig. 2A); however, when exposed to low pH, VacA disassembles into monomers (Fig. 2B). Electron microscopy (EM) analysis of purified VacA-Δ(346-347)-Str312 at neutral pH showed that, as expected, it failed to assemble into oligomeric structures (Fig. 2C). Thus, purified VacA-Δ(346-347)-Str312 at neutral pH appears similar to acid activated WT VacA. VacA-Str312 (containing the strep tag but not mutated at residues 346 to 347) retained the capacity to assemble into oligomeric structures, indicating that the presence of the strep tag does not block oligomerization (Fig. 2D). These EM results, combined with the SDS-PAGE analysis of protein purity (Fig. 1), provided evidence that the purified VacA-Δ(346-347)-Str312 protein was a homogeneous and uniform preparation of VacA monomers resembling acidified WT VacA.
FIG 2.
EM analysis of purified wild-type VacA and VacA-Δ(346-347)-Str312. VacA proteins were purified from H. pylori broth culture supernatant as described in Materials and Methods. (A to D) EM analysis of purified WT VacA at neutral pH (A), WT VacA exposed to low-pH conditions as described in Materials and Methods (B), VacA-Δ(346-347)-Str312 at neutral pH (C), and VacA-Str312 at neutral pH (D). The insets show high-magnification images of representative particles. Scale bar, 50 nm for all panels.
Functional analysis of a nonoligomerizing VacA protein.
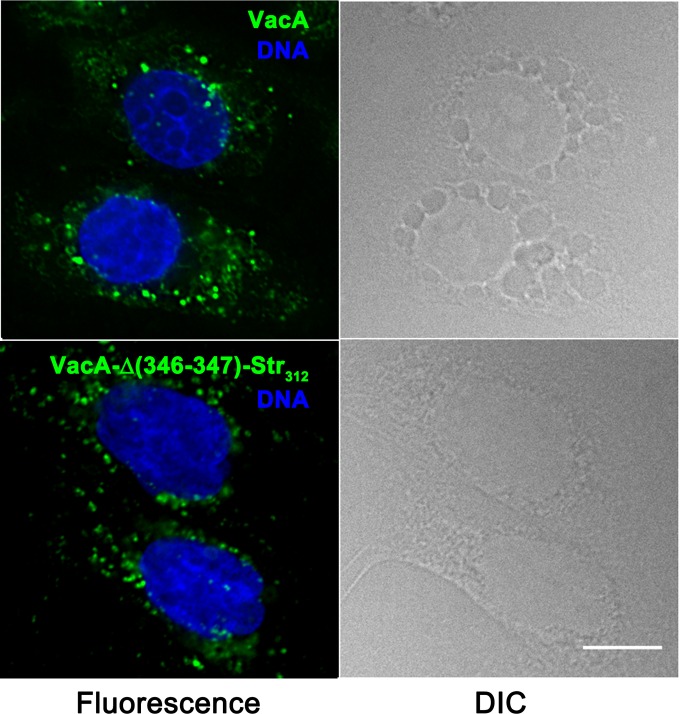
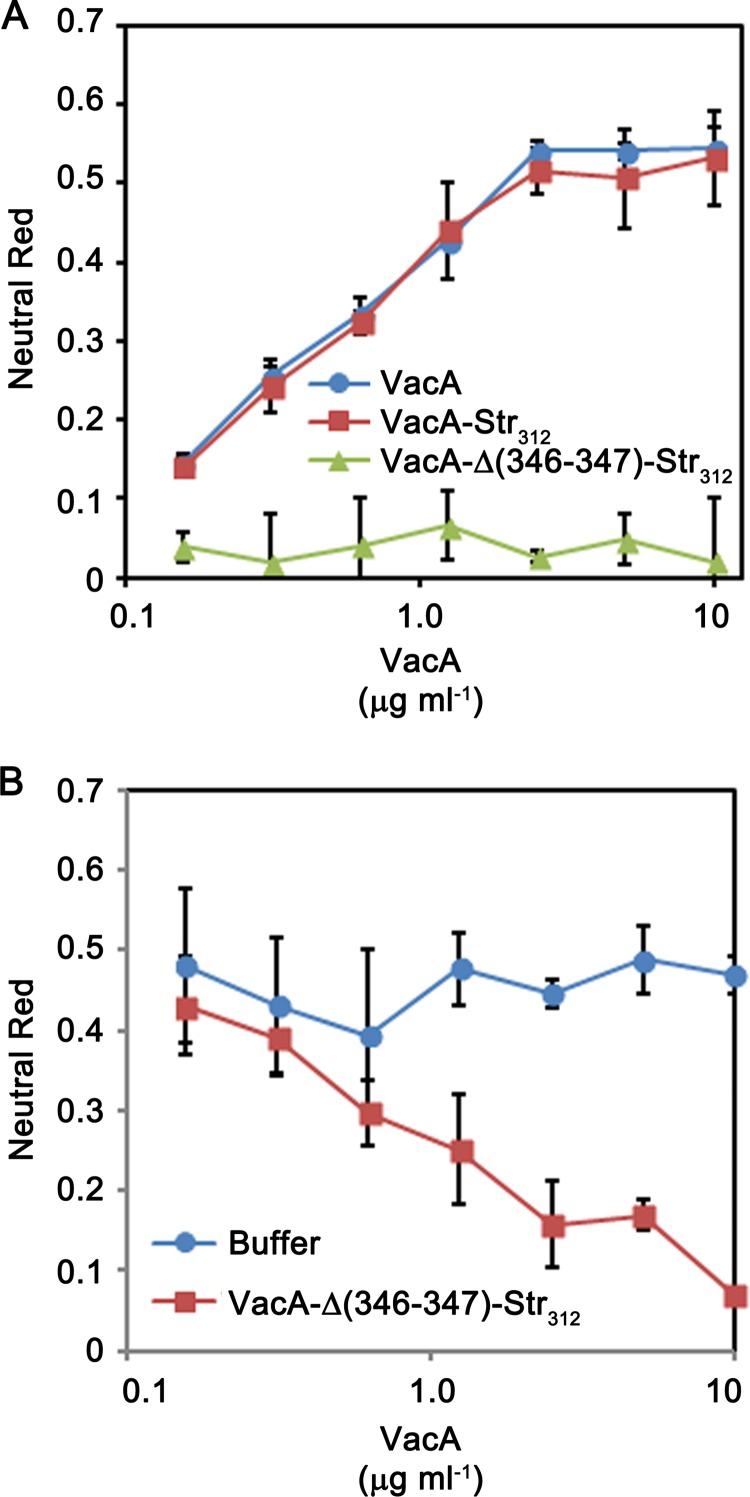
When added to cultured AGS gastric epithelial cells, WT VacA is internalized into the cells and induces the formation of large intracellular vacuoles (Fig. 3). VacA-Δ(346-347)-Str312 was internalized by AGS cells, similar to wild-type VacA, but lacked the capacity to cause vacuolation of these cells (Fig. 3). Similarly, VacA-Δ(346-347)-Str312 lacked the capacity to cause vacuolation of HeLa cells, based on both microscopic visualization of cells (data not shown) and a quantitative neutral red uptake assay (Fig. 4A). VacA-Str312 (containing the strep tag but not mutated at residues 346 to 347) retained the capacity to cause cell vacuolation (Fig. 4A). Acid-activated VacA-Str312 exhibited increased vacuolating activity compared to nonactivated VacA-Str312, similar to what is observed with WT VacA (data not shown). These results indicate that the Δ346-347 mutation (and not the strep tag) was responsible for the nonvacuolating phenotype of VacA-Δ(346-347)-Str312. Consistent with previous studies of a VacA Δ346-347 mutant protein (33), the purified VacA-Δ(346-347)-Str312 protein inhibited the activity of WT VacA when HeLa cells were treated with a mixture of both proteins (Fig. 4B). Thus, despite lack of vacuolating activity, the nonoligomeric VacA-Δ(346-347)-Str312 protein retained the capacity to be secreted by H. pylori, enter host cells, and block the activity of WT VacA, suggesting that the strep-tagged monomer is folded.
FIG 3.
Internalization of VacA-Δ(346-347)-Str312 by gastric epithelial cells. AGS cells were treated for 4 h in the presence of 5 mM ammonium chloride with 5 μg/ml of either acid-activated WT VacA (top panels) or VacA-Δ(346-347)-Str312 (bottom panels), each labeled with Alexa Fluor 488. The cells were then washed with PBS, fixed with 4% paraformaldehyde, and stained with Hoechst to label DNA. Fluorescence and DIC images are shown. Scale bar, 10 μm.
FIG 4.
Analysis of vacuolating toxic activity. H. pylori 60190 (producing WT VacA), a strain producing VacA-Str312, and a strain producing VacA-Δ(346-347)-Str312 were grown in broth cultures, and VacA proteins were purified as described in Materials and Methods. (A) HeLa cells were incubated with the indicated final concentrations of purified VacA proteins, and cell vacuolation was quantified by neutral red uptake assay (as determined by the optical density at 600 nm [OD600]). WT VacA and VacA-Str312 exhibited cell-vacuolating activity (corresponding to high neutral red uptake values), whereas VacA-Δ(346-347)-Str312 lacked vacuolating activity. (B) WT VacA (10 μg/ml) was incubated with the indicated concentrations of VacA-Δ(346-347)-Str312 or buffer only, and the mixtures were then added to HeLa cells in the presence of 5 mM ammonium chloride. Cell vacuolation was quantified by neutral red uptake assay (i.e., based on the OD600). VacA-Δ(346-347)-Str312 inhibited the activity of WT VacA.
Crystallization of VacA-Δ(346-347)-Str312.
WT VacA protein assembles into a heterogeneous assortment of oligomeric structures (26–31), which is suboptimal for crystal formation. We reasoned that the purified VacA-Δ(346-347)-Str312 protein might be more suitable for crystallization than WT VacA. In initial crystallization trials, we tested more than 1,000 crystallization conditions, as described in Materials and Methods, and observed that VacA-Δ(346-347)-Str312 formed cubic crystals.
Conditions for crystal formation were optimized by the hanging-drop method, and crystals were analyzed as described in Materials and Methods. Diffraction data were indexed, integrated, scaled, and merged with HKL2000. The crystals were space group I23 with a 330.7-Å cell edge. The highest resolution obtained was about 4.2 Å. Efforts to improve the resolution by forming crystals under alternate conditions or by dehydrating crystals were not successful. Molecular replacement using the 2.4-Å p55 domain of VacA as the search model revealed a solution where each asymmetric unit has two monomers. A packing arrangement of two monomers per asymmetric unit in a 330-Å3 cubic I23 space group indicates that the crystals are 88% solvent. Although this is high, we observed a full array of crystal packing contacts that allow for the formation of the cubic lattice.
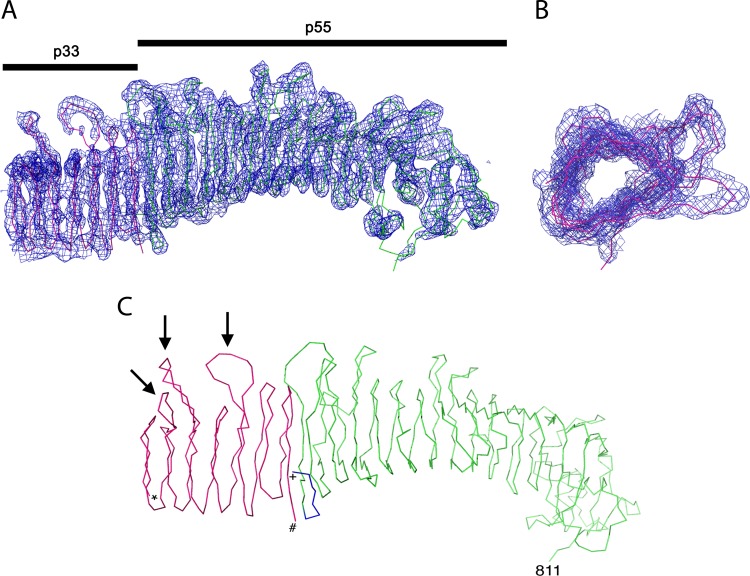
Analysis of the electron density map of VacA-Δ(346-347)-Str312 crystals revealed that the previously determined structure of the p55 domain (residues 355 to 811) was present (colored green in Fig. 5) and also allowed us to map amino acids at the N terminus of p55 (residues 348 to 354) that were not part of the previously determined p55 structure (Fig. 5C, colored blue). Notably, residue 348 (located at the N-terminal end of the p55 structure) is directly adjacent to the two amino acids that were deleted in the nonoligomerizing VacA-Δ(346-347)-Str312 protein crystallized in the present study. The electron density map contained not only the p55 structure (containing predominantly a β-helical fold) but also an additional β-helix segment consisting of 7 rungs and about 165 amino acids (Fig. 5, colored red). The additional β-helix segment is characterized by three long loops that project on one side of the β-helix (Fig. 5C). The loops are located on four rungs such that one loop is separated from the other two by a rung with a simple tight turn. The space and length of these loops are unique relative to the loop structures observed in p55. Therefore, the additional β-helical structure detected in the electron density map is not simply an additional copy of the p55 β-helix. The most plausible interpretation is that the additional β-helical structure represents a portion of the p33 domain. Consistent with this interpretation, analysis of dissolved crystals by mass spectrometry analysis revealed the presence of peptides assigned to both p33 and p55 (representing about 83% coverage of the 88-kDa protein).
FIG 5.
Density map of VacA-Δ(346-347)-Str312. VacA-Δ(346-347)-Str312 was crystallized, and the crystals were analyzed as described in Materials and Methods. (A) The structure of the p55 domain is colored green, and the structure of a portion of the p33 domain is colored red. A 2Fo−Fc map contoured at 1.5 sigma reveals the presence of an extended β-helix structure in the p33 domain. Although the junction of p33 with the p55 fragment is not resolved, the density map suggests the presence of a β-helix segment in p33 consisting of 7 rungs and about 165 amino acids (red coloration). (B) Rotated view of the β-helix segment within the p33 domain. (C) Line representation of the VacA-Δ(346-347)-Str312 structure. The portion of p55 corresponding to the previously determined structure (residues 355 to 811) (52) is shown in green, and additional p55 residues mapped in the present study are shown are shown in blue. A β-helical portion of the p33 domain is shown in red. *, N-terminal end of p33 β-helix; #, C-terminal end of p33 β-helix; +, N-terminal end of p55 domain. Arrows designate distinctive loops in the p33 domain.
As shown in Fig. 5C, we were unable to resolve the junction between the p55 and p33 domains within the electron density map. Previous studies showed that the p55-p33 junction is susceptible to proteolysis (27, 36, 40), and the sequences of VacA proteins produced by different H. pylori strains vary considerably in this region, exhibiting differences in length (due to insertions or sequence duplications), as well as differences in amino acid sequence. Therefore, it has been predicted that the p55-p33 junction is a flexible region (36). The failure to resolve this region in the present study is consistent with the presence of an unstructured region. Although the electron density maps were not of sufficient resolution to allow assignment of the sequence register of the p33 β-helix, we can generate hypotheses about the p33 sequence region that forms this β-helix based on use of structure prediction programs, as considered further in the Discussion.
We also generated H. pylori strains that produced VacA-Δ(346-347) proteins with strep tags inserted at several other sites within the p55 domain (Str427, Str480, Str524, and Str808). Each of these proteins was secreted by H. pylori, and each formed cubic crystals similar to those formed by VacA-Δ(346-347)-Str312, suggesting that the strep tag could be placed at multiple locations without altering the protein structure. To further test this hypothesis, we engineered a strain that produced a VacA-Str808 protein (without the Δ346-347 mutation) and found that the VacA-Str808 protein had vacuolating toxin activity similar to that of wild-type VacA (data not shown). Analysis of crystals formed by the VacA-Δ(346-347) proteins with strep tags at alternate sites did not improve the resolution and did not allow resolution of the structure of the p33-p55 junction.
Localization of p55 and p33 domains within the VacA oligomeric structure.
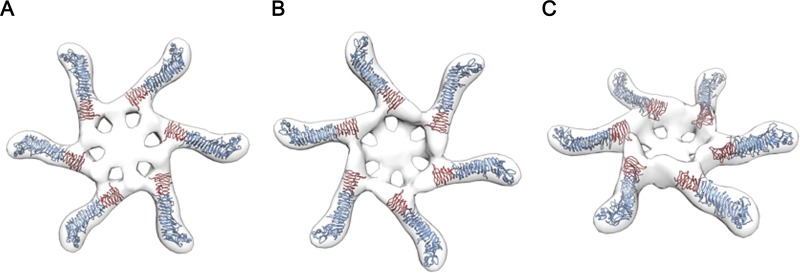
The three-dimensional structure of water-soluble VacA oligomers has been analyzed previously using negative stain EM and random conical tilt methods, yielding a 15-Å structure (31). Water-soluble hexameric structures are predicted to be structurally related to the membrane-inserted VacA channel (17, 19, 31). Therefore, we sought to fit the VacA-Δ(346-347)-Str312 model into an EM map of a VacA hexamer. As expected, p55 domains localized to peripheral arms of the hexamer and p33 domains localized closer to the center of the hexamer (Fig. 6). Notably, a large region within the center of the oligomer was not occupied by the density map of the VacA-Δ(346-347)-Str312 crystal structure. As considered further in the Discussion, we propose that the central hub of the oligomer is composed of an N-terminal portion of VacA (for example, amino acids 1 to 120) that was not part of the p33 β-helical domain of the current crystal structure.
FIG 6.
Model depicting assembly of VacA monomers into hexameric structures. The density map of VacA-Δ(346-347)-Str312 was fitted into an EM map of a VacA hexamer (31). The figure illustrates bottom, top, and side views of the hexamer. Blue indicates p55 domains, and red indicates an ∼165-amino-acid β-helical segment within the p33 domain.
DISCUSSION
In a previous study, the crystal structure of the VacA p55 domain was determined by performing experiments with a recombinant p55 protein (52). Efforts to determine the structure of the p33 domain using the same approach have not been successful, probably due to the relative insolubility of the recombinant p33 domain (42), and efforts to determine the structure of p88 proteins produced by either H. pylori or E. coli also have been unsuccessful. In the present study, we analyzed the structure of a p88 VacA protein harboring a mutation that blocked oligomer formation. The VacA-Δ(346-347)-Str312 protein appeared to be similar to monomeric forms of WT VacA when analyzed by EM, and it retained several functional properties of the WT protein, including the capacity to be secreted by H. pylori and the capacity to enter gastric epithelial cells.
The VacA-Δ(346-347)-Str312 protein and VacA proteins with strep tags at alternate sites formed cubic crystals under multiple conditions, but none of the density maps were at a resolution that allowed assignment of the sequence register. We used multiple approaches in an effort to improve the level of diffraction, but none were successful. We speculate that the suboptimal diffraction is attributable to the high solvent content of the crystals. Despite this limitation, we were able to obtain molecular replacement phases for a 4.2-Å data set and thereby obtain the first experimental data pertaining to the structure of the p33 domain.
The present study reveals a region of the p33 domain about 165 amino acids in length that has a predominantly β-helical structure. This result compares favorably to structure predictions that were made using the program BetaWrap Pro (54). Specifically, the BetaWrap Pro program predicts the existence of a stretch of amino acids between residues 120 and 249 as a five-coil β-helix, with a potential for additional coils extending out to residue 278 (52). Based on the current data combined with structure predictions, we propose that the C-terminal region of p33 (approximately residues 120 to 285) has a predominantly β-helical structure.
Relatively little is known about functional properties of the β-helical region of p33. Random mutagenesis experiments revealed only two mutations in this region that abrogated VacA toxin activity (G121R and S246L) (51). Interestingly, comparative sequence analysis of VacA proteins produced by different H. pylori strains reveals heterogeneity in a region (approximately residues 144 to 244, known as the “intermediate region” or the “i-region”), which lies within the putative β-helical portion of p33 (23, 25). H. pylori strains containing i1 forms of VacA have been linked to an elevated risk of gastric cancer, compared to strains containing i2 forms of VacA (25). The i-region is predicted to be at least partially contained within the p33 β-helical structure, but the exact sites of diversity could not be mapped due to the limited resolution of the current structure, and thus far our efforts to crystallize i2 forms of VacA have not been successful. It will be important in future studies to define precisely the structural variation within the i-region and elucidate how structural variation in this region impacts VacA activity.
VacA assembles into multiple types of water-soluble oligomeric structures, including single-layered structures containing 6 to 9 components, as well as double-layered components containing 12 or 14 components (26–31). The single-layered structures are proposed to be structurally related to the pores that VacA forms in membranes. Protein-protein interactions between sequences in p33 and sequences in p55 are predicted to be required for the formation of oligomeric structures (20, 32, 33, 52, 70). Specifically, VacA residues 49 to 57 (within the p33 domain) and residues 346 to 347 (within p55) are required for VacA oligomerization (32, 33). These sites were not included in the current crystal structure, since residues 346 to 347 were intentionally deleted and residues 49 to 57 are predicted to be localized outside the p33 β-helix. Fitting of the VacA-Δ(346-347)-Str312 structural model into an EM map of hexamers formed by WT VacA reveals that p55 and the β-helical segment of p33 localize to the arms but do not extend into the central region of the hexamer.
The amino-terminal region of VacA is predicted to be highly hydrophobic and contains multiple GXXXG motifs, which are often involved in transmembrane dimerization (12, 20, 21, 50). Mutagenesis studies have shown that this region of VacA is required for membrane channel formation (20, 21). One study proposed a model for the structure of this portion of VacA and suggested that it resembles the structure of an unrelated anion-selective channel, MscS (55). Previous EM studies have shown that WT VacA oligomers have structurally organized spoke-like densities in the central region, whereas this central density is absent in oligomers formed by VacA Δ6-27 mutant proteins (30, 31). Therefore, it has been presumed that VacA amino acids 6 to 27 localize to the center of oligomers and are required for the structural integrity of this region. The current data provide further support for a model in which the central portion of VacA oligomers is formed by the amino-terminal region of VacA. We predict that the central region is formed by an amino-terminal portion of p33 that is unstructured when VacA is in a monomeric form and that it undergoes a conformational change when monomers assemble into oligomeric structures.
In summary, these data constitute the first experimental insights into structural properties of the VacA p33 domain. In future studies, it will be important to define the structure of the p33 domain at a higher level of resolution. In addition, it will be important to define the structure of the amino-terminal portion of p33 and clarify the structure-function relationships important for membrane channel formation.
ACKNOWLEDGMENTS
We thank members of the Cover, Lacy, Ohi, and Spiller labs for helpful discussions.
Funding Statement
Cell Imaging Core Services were supported by the Vanderbilt University Medical Center Digestive Disease Research Center (NIH grant P30DK058404) and the Vanderbilt-Ingram Cancer Center (P30 CA068485), and the Vanderbilt robotic crystallization facility was supported by NIH S10 RR026915.
REFERENCES
- 1.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest 119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR. 2011. Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol Mol Biol Rev 75:84–132. doi: 10.1128/MMBR.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieder G, Fischer W, Haas R. 2005. Interaction of Helicobacter pylori with host cells: function of secreted and translocated molecules. Curr Opin Microbiol 8:67–73. doi: 10.1016/j.mib.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Zanotti G, Cendron L. 2014. Structural and functional aspects of the Helicobacter pylori secretome. World J Gastroenterol 20:1402–1423. doi: 10.3748/wjg.v20.i6.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snider CA, Voss BJ, McDonald WH, Cover TL. 2016. Growth phase-dependent composition of the Helicobacter pylori exoproteome. J Proteomics 130:94–107. doi: 10.1016/j.jprot.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cover TL, Blanke SR. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol 3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 8.Boquet P, Ricci V. 2012. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol 20:165–174. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Kim IJ, Blanke SR. 2012. Remodeling the host environment: modulation of the gastric epithelium by the Helicobacter pylori vacuolating toxin (VacA). Front Cell Infect Microbiol 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. 2016. An overview of Helicobacter pylori VacA toxin biology. Toxins 8:173. doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol 26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 12.Cover TL, Blaser MJ. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem 267:10570–10575. [PubMed] [Google Scholar]
- 13.Gebert B, Fischer W, Weiss E, Hoffman R, Haas R. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 14.Boncristiano M, Paccani SR, Barone S, Ulivieri C, Patrussi L, Ilver D, Amedei A, D'Elios MM, Telford JL, Baldari CT. 2003. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J Exp Med 198:1887–1897. doi: 10.1084/jem.20030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. 2004. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci U S A 101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres VJ, Van Compernolle SE, Sundrud MS, Unutmaz D, Cover TL. 2007. Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J Immunol 179:5433–5440. doi: 10.4049/jimmunol.179.8.5433. [DOI] [PubMed] [Google Scholar]
- 17.Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. 1999. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc Natl Acad Sci U S A 96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, Rappuoli R, Montecucco C, Papini E, Zoratti M. 1999. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J 18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. 1999. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS Lett 450:101–104. doi: 10.1016/S0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 20.Vinion-Dubiel AD, McClain MS, Czajkowsky DM, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke SR, Shao Z, Cover TL. 1999. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J Biol Chem 274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 21.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. 2003. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J Biol Chem 278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 22.Cover TL. 2016. Helicobacter pylori diversity and gastric cancer risk. mBio 7:e01869-15. doi: 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, Bordenstein SR. 2010. Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. J Bacteriol 192:6126–6135. doi: 10.1128/JB.01081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem 270:17771–17777. [DOI] [PubMed] [Google Scholar]
- 25.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 26.Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. 1996. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol 133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cover TL, Hanson PI, Heuser JE. 1997. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol 138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzavecchia S, Bellon PL, Lupetti P, Dallai R, Rappuoli R, Telford JL. 1998. Three-dimensional reconstruction of metal replicas of the Helicobacter pylori vacuolating cytotoxin. J Struct Biol 121:9–18. doi: 10.1006/jsbi.1997.3941. [DOI] [PubMed] [Google Scholar]
- 29.Adrian M, Cover TL, Dubochet J, Heuser JE. 2002. Multiple oligomeric states of the Helicobacter pylori vacuolating toxin demonstrated by cryo-electron microscopy. J Mol Biol 318:121–133. doi: 10.1016/S0022-2836(02)00047-5. [DOI] [PubMed] [Google Scholar]
- 30.El-Bez C, Adrian M, Dubochet J, Cover TL. 2005. High-resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. J Struct Biol 151:215–228. doi: 10.1016/j.jsb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Chambers MG, Pyburn TM, Gonzalez-Rivera C, Collier SE, Eli I, Yip CK, Takizawa Y, Lacy DB, Cover TL, Ohi MD. 2013. Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin. J Mol Biol 425:524–535. doi: 10.1016/j.jmb.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genisset C, Galeotti CL, Lupetti P, Mercati D, Skibinski DA, Barone S, Battistutta R, de Bernard M, Telford JL. 2006. A Helicobacter pylori vacuolating toxin mutant that fails to oligomerize has a dominant negative phenotype. Infect Immun 74:1786–1794. doi: 10.1128/IAI.74.3.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivie SE, McClain MS, Torres VJ, Algood HM, Lacy DB, Yang R, Blanke SR, Cover TL. 2008. Helicobacter pylori VacA subdomain required for intracellular toxin activity and assembly of functional oligomeric complexes. Infect Immun 76:2843–2851. doi: 10.1128/IAI.01664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cover TL, Tummuru MKR, Cao P, Thompson SA, Blaser MJ. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem 269:10566–10573. [PubMed] [Google Scholar]
- 35.Schmitt W, Haas R. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol 12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 36.Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med 179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer W, Buhrdorf R, Gerland E, Haas R. 2001. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori. Infect Immun 69:6769–6775. doi: 10.1128/IAI.69.11.6769-6775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss BJ, Gaddy JA, McDonald WH, Cover TL. 2014. Analysis of surface-exposed outer membrane proteins in Helicobacter pylori. J Bacteriol 196:2455–2471. doi: 10.1128/JB.01768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye D, Blanke SR. 2002. Functional complementation reveals the importance of intermolecular monomer interactions for Helicobacter pylori VacA vacuolating activity. Mol Microbiol 43:1243–1253. doi: 10.1046/j.1365-2958.2002.02818.x. [DOI] [PubMed] [Google Scholar]
- 40.Torres VJ, McClain MS, Cover TL. 2004. Interactions between p-33 and p-55 domains of the Helicobacter pylori vacuolating cytotoxin (VacA). J Biol Chem 279:2324–2331. doi: 10.1074/jbc.M310159200. [DOI] [PubMed] [Google Scholar]
- 41.Torres VJ, Ivie SE, McClain MS, Cover TL. 2005. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J Biol Chem 280:21107–21114. doi: 10.1074/jbc.M501042200. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Rivera C, Gangwer KA, McClain MS, Eli IM, Chambers MG, Ohi MD, Lacy DB, Cover TL. 2010. Reconstitution of Helicobacter pylori VacA toxin from purified components. Biochemistry 49:5743–5752. doi: 10.1021/bi100618g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyrat JM, Lanzavecchia S, Lupetti P, de Bernard M, Pagliaccia C, Pelicic V, Charrel M, Ulivieri C, Norais N, Ji X, Cabiaux V, Papini E, Rappuoli R, Telford JL. 1999. 3D imaging of the 58-kDa cell binding subunit of the Helicobacter pylori cytotoxin. J Mol Biol 290:459–470. doi: 10.1006/jmbi.1999.2877. [DOI] [PubMed] [Google Scholar]
- 44.de Bernard M, Burroni D, Papini E, Rappuoli R, Telford J, Montecucco C. 1998. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect Immun 66:6014–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye D, Willhite DC, Blanke SR. 1999. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J Biol Chem 274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]
- 46.Galmiche A, Rassow J, Doye A, Cagnol S, Chambard JC, Contamin S, de Thillot V, Just I, Ricci V, Solcia E, Van Obberghen E, Boquet P. 2000. The N-terminal 34-kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J 19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domanska G, Motz C, Meinecke M, Harsman A, Papatheodorou P, Reljic B, Dian-Lothrop EA, Galmiche A, Kepp O, Becker L, Gunnewig K, Wagner R, Rassow J. 2010. Helicobacter pylori VacA toxin/subunit p34: targeting of an anion channel to the inner mitochondrial membrane. PLoS Pathog 6:e1000878. doi: 10.1371/journal.ppat.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foo JH, Culvenor JG, Ferrero RL, Kwok T, Lithgow T, Gabriel K. 2010. Both the p33 and p55 subunits of the Helicobacter pylori VacA toxin are targeted to mammalian mitochondria. J Mol Biol 401:792–798. doi: 10.1016/j.jmb.2010.06.065. [DOI] [PubMed] [Google Scholar]
- 49.Ye D, Blanke SR. 2000. Mutational analysis of the Helicobacter pylori vacuolating toxin amino terminus: identification of amino acids essential for cellular vacuolation. Infect Immun 68:4354–4357. doi: 10.1128/IAI.68.7.4354-4357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClain MS, Cao P, Cover TL. 2001. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect Immun 69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClain MS, Czajkowsky DM, Torres VJ, Szabo G, Shao Z, Cover TL. 2006. Random mutagenesis of Helicobacter pylori vacA to identify amino acids essential for vacuolating cytotoxic activity. Infect Immun 74:6188–6195. doi: 10.1128/IAI.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. 2007. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci U S A 104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivie SE, McClain MS, Algood HM, Lacy DB, Cover TL. 2010. Analysis of a beta-helical region in the p55 domain of Helicobacter pylori vacuolating toxin. BMC Microbiol 10:60. doi: 10.1186/1471-2180-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junker M, Schuster CC, McDonnell AV, Sorg KA, Finn MC, Berger B, Clark PL. 2006. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci U S A 103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Chamberlain AK, Bowie JU. 2004. Membrane channel structure of Helicobacter pylori vacuolating toxin: role of multiple GXXXG motifs in cylindrical channels. Proc Natl Acad Sci U S A 101:5988–5991. doi: 10.1073/pnas.0308694101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaidya M, Panchal H. 2012. In silico investigation and structural characterization of virulent factor and a metallo peptidase present in Helicobacter pylori strain J99. Interdiscip Sci 4:302–309. doi: 10.1007/s12539-012-0145-6. [DOI] [PubMed] [Google Scholar]
- 57.Hawrylik SJ, Wasilko DJ, Haskell SL, Gootz TD, Lee SE. 1994. Bisulfite or sulfite inhibits growth of Helicobacter pylori. J Clin Microbiol 32:790–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jimenez-Soto LF, Rohrer S, Jain U, Ertl C, Sewald X, Haas R. 2012. Effects of cholesterol on Helicobacter pylori growth and virulence properties in vitro. Helicobacter 17:133–139. doi: 10.1111/j.1523-5378.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 59.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacterium-host cell interface. PLoS Pathog 7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt TG, Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 61.de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, Fontana A, Rappuoli R, Montecucco C. 1995. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J Biol Chem 270:23937–23940. doi: 10.1074/jbc.270.41.23937. [DOI] [PubMed] [Google Scholar]
- 62.McClain MS, Schraw W, Ricci V, Boquet P, Cover TL. 2000. Acid-activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol Microbiol 37:433–442. doi: 10.1046/j.1365-2958.2000.02013.x. [DOI] [PubMed] [Google Scholar]
- 63.Cover TL, Puryear W, Pérez-Pérez GI, Blaser MJ. 1991. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect Immun 59:1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. 2008. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. PLoS Pathog 4:e1000073. doi: 10.1371/journal.ppat.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radin JN, Gonzalez-Rivera C, Frick-Cheng AE, Sheng J, Gaddy JA, Rubin DH, Algood HM, McClain MS, Cover TL. 2014. Role of connexin 43 in Helicobacter pylori VacA-induced cell death. Infect Immun 82:423–432. doi: 10.1128/IAI.00827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otwinowski Z, Minor W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 67.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 68.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Nguyen VQ, Caprioli RM, Cover TL. 2001. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun 69:543–546. doi: 10.1128/IAI.69.1.543-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torres VJ, McClain MS, Cover TL. 2006. Mapping of a domain required for protein-protein interactions and inhibitory activity of a Helicobacter pylori dominant-negative VacA mutant protein. Infect Immun 74:2093–2101. doi: 10.1128/IAI.74.4.2093-2101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]