Abstract
Immune modulation is a hallmark of patent filarial infection, including suppression of antigen-presenting cell function and downmodulation of filarial antigen-specific T cell responses. The mammalian target of rapamycin (mTOR) signaling pathway has been implicated in immune regulation, not only by suppressing T cell responses but also by regulating autophagy (through mTOR sensing amino acid availability). Global proteomic analysis (liquid chromatography-tandem mass spectrometry) of microfilaria (mf)-exposed monocyte-derived dendritic cells (DC) indicated that multiple components of the mTOR signaling pathway, including mTOR, eIF4A, and eIF4E, are downregulated by mf, suggesting that mf target this pathway for immune modulation in DC. Utilizing Western blot analysis, we demonstrate that similar to rapamycin (a known mTOR inhibitor), mf downregulate the phosphorylation of mTOR and its regulatory proteins, p70S6K1 and 4E-BP1, a process essential for DC protein synthesis. As active mTOR signaling regulates autophagy, we examined whether mf exposure alters autophagy-associated processes. mf-induced autophagy was reflected in marked upregulation of phosphorylated Beclin 1, known to play an important role in both autophagosome formation and autolysosome fusion, in induction of LC3II, a marker of autophagosome formation, and in induced degradation of p62, a ubiquitin-binding protein that aggregates protein in autophagosomes and is degraded upon autophagy that was reduced significantly by mf exposure and by rapamycin. Together, these results suggest that Brugia malayi mf employ mechanisms of metabolic modulation in DC to influence the regulation of the host immune response by downregulating mTOR signaling, resulting in increased autophagy. Whether this is a result of the parasite-secreted rapamycin homolog is currently under study.
INTRODUCTION
Filarial infection in humans is initiated by a mosquito-derived third-stage larva deposited in the skin, which enters the body, molts to the fourth larval stage, and matures into lymphatic tissue-dwelling adult male and female worms, a process that takes about 3 to 12 months. Adult females, after copulation, release progeny microfilariae (mf), a stage believed to be associated with many of the systemic immunologic defects seen with chronic lymphatic filarial infection. Microfilariae released in the lymphatics can travel through circulation and encounter a variety of antigen-presenting cells (APC), including monocytes and dendritic cells (DC). APC dysfunction has been postulated to be one of the causes of impaired antigen-specific T cell activation commonly seen among patients chronically infected with this parasite (1), a dysfunction that is manifested through a secreted soluble antigen(s) that alters Toll-like receptor (TLR) signaling, induces apoptotic cell death (2, 3), and affects chemokine-mediated trafficking (4). Although these studies highlight some of the important proteins and processes affected, we still do not have a comprehensive understanding of the signaling pathways that are modulated in APCs by these parasites. In this study, using a global shotgun proteomic analysis of human DCs exposed to the mf stage of the filarial parasite Brugia malayi (one of the causative agents of lymphatic filariasis), we have demonstrated that these parasites downregulate some of the important metabolic pathways, including the mammalian target of rapamycin (mTOR).
mTOR, a serine/threonine kinase, is composed of two distinct multiprotein complexes: mTOR complex 1 (mTORC1) and mTORC2 (reviewed in reference 5). The activity of mTORC1 is regulated by intracellular signals as well as signals from the environment, including growth factors and nutrients (6, 7). Moreover, mTOR regulates protein synthesis through the phosphorylation and activation of the ribosomal S6 kinase (p70S6K1) and phosphorylation and inactivation of the repressor of translation, eukaryotic initiation factor 4E-binding protein (4E-BP1; reviewed in reference 8). Phosphorylation of both of these proteins can be blocked by rapamycin, resulting in inhibition of protein translation (9, 10).
Intracellular parasites as well as viruses have developed strategies to manipulate the host translation machinery to benefit their own replication through regulation of mTOR. There is evidence for both the stimulation and inhibition of mTOR depending on the infection. While Toxoplasma gondii maintains mTOR-dependent cellular growth of infected cells (11), Leishmania protease inhibits mTOR in macrophages, leading to enhanced parasite replication (12).
mTOR also plays a key role in regulating the balance between cell growth and autophagy, the major cellular digestion process that removes organelles and other macromolecules. In fact, activation of mTORC1 by nutrients and growth factors leads to inhibition of autophagy through the phosphorylation of several autophagy-related proteins (reviewed in references 13 and 14), including Beclin 1, LC3II, and p62.
In the present study, we show that mf from the extracellular parasite B. malayi inhibit the mTOR pathway in human DC by downregulating the phosphorylation of mTOR itself and also by inhibiting the basal and activation-induced phosphorylation of p70S6K and 4EBP1. In addition, mf induce autophagy in human DC by upregulating the phosphorylation of Beclin 1 (a central regulator of autophagy), by inducing LC3II (an autophagic vacuole-associated protein [15]), and by inducing the degradation of p62, a ubiquitin-binding receptor and marker of conventional autophagy. Together, our data suggest that extracellular helminth parasites employ (presumably through the production of soluble factors) mechanisms of metabolic modulation in DC to influence the regulation of the host immune response by downregulating mTOR signaling, resulting in autophagy.
MATERIALS AND METHODS
Ethics statement.
Human monocytes were isolated from leucopacks from healthy donors by counterflow centrifugal elutriation under Institutional Review Board (IRB)-approved protocols from the Department of Transfusion Medicine (Clinical Center, National Institutes of Health [NIH], Bethesda, MD). All donors provided informed written consent.
mf preparations.
Live B. malayi mf (provided under contract with the University of Georgia, Athens, GA) were collected by peritoneal lavage of infected jirds and separated from peritoneal cells by Ficoll diatrizoate density centrifugation. The mf were then washed repeatedly in RPMI medium with antibiotics and cultured overnight at 37°C in 5% CO2.
In vitro generation of DC.
Human monocytes were cultured at 50 × 106 per 6-well plate in serum-free medium for 2 h, after which the medium was removed and complete medium (RPMI 1640 medium [Lonza, Allendale, NJ] supplemented with 20 mM glutamine [Lonza], 2% heat-inactivated human AB serum [Gemini Bioproducts, Sacramento, CA], 100 IU/ml penicillin, and 100 g/ml streptomycin [Biofluids, Inc., Rockville, MD]) added. Recombinant human interleukin-4 (IL-4) and recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech, Rocky Hill, NJ) were added to the culture at 50 ng/ml on days 1, 4, and 6 of culture.
Stimulation of DC for mass spectrometry.
Live mf were added on day 6 at final concentrations of 50,000 per well (per 1 × 106 to 2 × 106 DCs). This number of mf was chosen to reflect in vivo numbers of mf in individuals with approximately 1,000 mf per ml of blood (containing 0.02 to 0.04 DCs). DCs were exposed to live mf for 48 h and then harvested using Versene-EDTA (Biofluids Inc.). The cells were washed twice with phosphate-buffered saline (PBS) (without Ca2+/Mg2+), counted by trypan blue exclusion, and used for functional studies. DCs harvested at day 8 were repeatedly shown to be 98% pure by flow cytometry (FACSCalibur; Becton Dickinson, Sunnyvale, CA). Cells were harvested, lysed, and prepared for label-free mass spectrometry.
Protein isolation and sample preparation.
mf-exposed and unexposed DC from four donors were lysed, and protein concentrations were quantified by bicinchoninic acid (BCA) assay. Equivalent amounts of proteins from mf-exposed and unexposed DC of each donor were resolved on the same NuPAGE 4 to 12% Bis-Tris precast gel. The set of lanes on the same gel were cut into the same number of gel slices. Gel slices were then destained, digested overnight with trypsin at 37°C, and extracted to be analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
NanoRPLC-MS/MS.
Each set of peptides extracted from mf-exposed and -unexposed DC gel slices was analyzed using Nanobore reverse-phase liquid chromatography-tandem MS (nanoRPLC-MS/MS) with blanks between each run. NanoRPLC was performed using an Agilent 1100 Nanoflow LC system (Agilent Technologies, Palo Alto, CA) coupled online with a linear ion trap (LIT) MS (LTQ; ThermoElectron, San Jose, CA). NanoRPLC columns were slurry packed in-house with 3-μm, 300-Å-pore-size, C18 phase (VYDAC, Hesperia, CA) filters in a 75-μm-inner-diameter, 10-cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a flame-pulled tip. After sample injection, the column was washed for 30 min with 98% mobile phase A (0.1% formic acid in water) at 0.5 μl/min, and peptides were eluted using a linear gradient of 2% mobile phase B (0.1% formic acid in acetonitrile) to 42% mobile phase B in 40 min at 0.25 μl/min and then to 98% mobile phase B for an additional 30 min. The LIT-MS was operated in a data-dependent mode in which each full MS scan was followed by five tandem MS scans where the five most abundant molecular ions were dynamically selected for collision-induced dissociation (CID) using a normalized collision energy of 35%.
Protein identification and quantification.
Proteins were identified by searching the nanoRPLC-MS/MS data using SEQUEST against a UNIPROT-derived human proteome database downloaded from the European Bioinformatics Institue (EBI) and B. malayi RefSeq proteome database (NCBI). Methionine oxidation was included as a dynamic modification in the database search. Only fully tryptic peptides with up to two miscleavages that met certain criteria (delta correlation [ΔCn] of ≥0.1 and charge state-dependent cross correlation [Xcorr] of ≥1.9 for [M + H]1+, ≥2.2 for [M + 2H]2+, and ≥3.5 for [M + 3H]3+) were considered legitimately identified. Protein identifications were accepted if they contained at least 2 identified peptides. The relative abundance of the proteins (number of unique peptides) was normalized by normalized spectral abundance factor (NSAF), where NSAF is represented as
Stimulation of DC for immunoblotting and autophagy.
DC were harvested at day 6 with Versene-EDTA (Biofluids Inc.), washed twice with PBS (without Ca2+/Mg2+), counted by trypan blue exclusion, and cultured at 1 × 106 per ml in serum-free media (RPMI 1640 medium) in 15-ml conical tubes. Cells were either left unexposed or were exposed to 50,000 live mf per tube or to rapamycin (100 ng/ml) (Sigma-Aldrich Co, St. Louis, MO) for 5 to 60 min. For basal expression of pmTOR, pp70S6K, and p4EBP1, cells were processed immediately for immunoblotting. For expression of these proteins following activation, cells were activated with lipopolysaccharide (LPS; 1 μg/ml; Invivogen, San Diego, CA) for an additional 30 min and then processed for immunoblotting.
To measure autophagy, 1 × 106 DC were left unexposed or were exposed to live mf at concentrations of 5,000 (lo), 25,000 (med), or 50,000 (hi) or to rapamycin (100 ng/ml) in 2% heat-inactivated human AB serum (Gemini Bioproducts) supplemented with 100 IU/ml penicillin and 100 g/ml streptomycin (Biofluids, Inc.) overnight.
Immunoblot analysis.
Cell lysates were prepared and boiled for 5 min in Laemmli sample buffer (Bio-Rad Laboratories, Inc., Hercules, CA); 20 μl of protein was then loaded into a 1.5-mm 4 to 20% Tris gel and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). After blocking using 5% nonfat milk for 1 h, the membranes were incubated overnight at 4°C with rabbit anti-pmTOR, rabbit anti-p70S6K, rabbit anti-p4EBP1, rabbit anti-pBeclin 1, or rabbit anti-LC3A/B (all antibodies were from Cell Signaling Technology, Inc., Beverly, MA). After washing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Cell Signaling Technology) at 1:2,000 at room temperature for 2 h. For the β-actin control, membranes were stripped in stripping buffer (Thermo Scientific, Grand Island, NJ) for 10 min and reprobed with anti-β-actin (Cell Signaling Technology) antibody. Proteins were detected by a chemiluminescence detection system (Cell Signaling Technology). β-Actin was used as an internal control because of low background detection and a molecular weight distinct from those of the proteins of interest. LC3II/actin ratios were calculated by quantification of intensity of LC3II and actin bands and graphed. ImageJ (http://rsb.info.nih.gov/ij/) was used to quantify the intensity of bands in the immunoblots. P values were calculated based on protein expression in exposed DC compared to unexposed DC.
Confocal microscopy.
Cells were harvested after overnight incubation with mf or chloroquine (Sigma-Aldrich) and washed with PBS three times, followed by fixation with 2% prewarmed paraformaldehyde for 20 min. The cells were then treated with 1× permeabilizing buffer (BD Biosciences) according to the manufacturer's instructions and then blocked for 60 min at 4°C in blocking buffer with 5% bovine serum albumin (BSA). After washing once with PBS, cells were incubated overnight at 4°C in 1:100 rabbit LC3B/MAP1LC3B antibody (DyLight 488; Novus Biologicals) for detection of LC3II. Following three washes with PBS, cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at a final concentration of 2 μg/ml, followed by an additional three washes. Slide covers were mounted using Mowiol mounting reagent (Calbiochem, San Diego, CA). Confocal images were collected using a Leica DMI 6000 confocal microscope (Leica Microsystems, Exton, PA) enabled with a 63× oil immersion objective (numeric aperture, 1.4). Images were acquired using highly sensitive hybrid detectors to achieve a maximum signal-to-noise ratio for LC3 cluster analysis. Images were deconvolved using Huygens deconvolution software with fixed parameters across the sample (Scientific Volume Imaging B.V., The Netherlands) and further analyzed using Imaris image processing software (Bitplane, South Windsor, CT). Cluster size (dots) was fixed to 0.3 μm across the sample using an Imaris spot analysis process to pick the total number of clusters. The relative difference in intensity was quantified by counting the fluorescent dots in five random microscopic fields with approximately 5 to 10 cells each.
ELISA for p62 analysis.
DC pellets were washed with PBS and lysed with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors. p62 expression was measured with a p62 enzyme-linked immunosorbent assay (ELISA) kit (Enzo Life Sciences, Farmingdale, NJ) according to the manufacturer's instructions. The range of the standard curve was 625 to 40,000 pg/ml. For each assay, all samples were assayed in duplicate. Results were normalized per milligram of protein from the cell lysate.
Statistical analysis.
The statistical significance of regulated proteins (DC that were exposed to live mf of B. malayi [DC/mf] versus DC) was calculated by one-way analysis of variance (ANOVA) (JMP11; SAS). The P values were calculated using the normalized spectral abundance factors (NSAF) for each protein (see Fig. 1 and also Table S1 in the supplemental material). The nonparametric Wilcoxon signed-rank test was used for Fig. 3 to 6. All statistical analyses were performed with GraphPad Prism 6.0 (GraphPad Software, Inc.).
FIG 1.
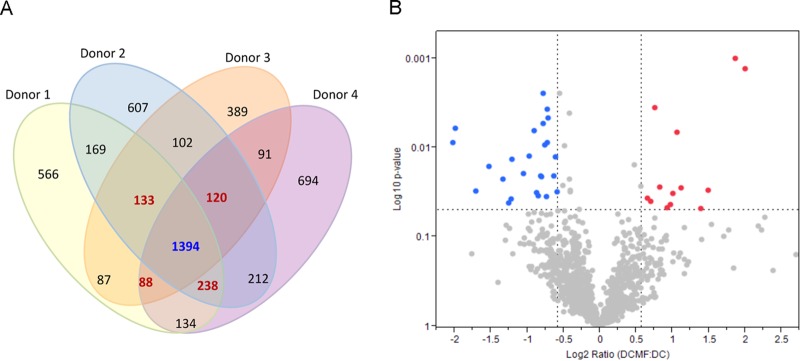
Proteomic analyses of unexposed and mf-exposed human DC. Human DC, unexposed or mf exposed for 48 h, were harvested, lysed, processed, and analyzed by LC-MS/MS. (A) Venn diagram representing the number of proteins identified in human DC (unexposed or mf exposed) from 4 healthy donors. A total of 1,394 proteins (marked in blue) were identified in all four donors in both DC and mf-exposed DC. (B) Volcano plot of significantly regulated proteins by mf in human DC (>1.5-fold) with log10 values (P values) on the x axis and log2 ratios (fold change) on the y axis. Significantly upregulated proteins are shown in red, and significantly downregulated proteins are shown in blue. The P values were calculated using the normalized spectral abundance factors (NSAF) for each protein. See also Fig. S1 and Table S1 in the supplemental material.
FIG 3.
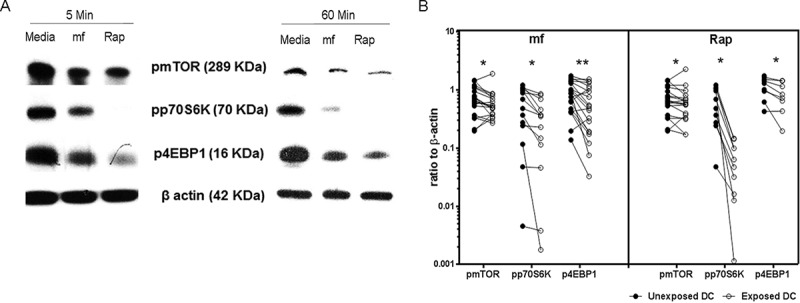
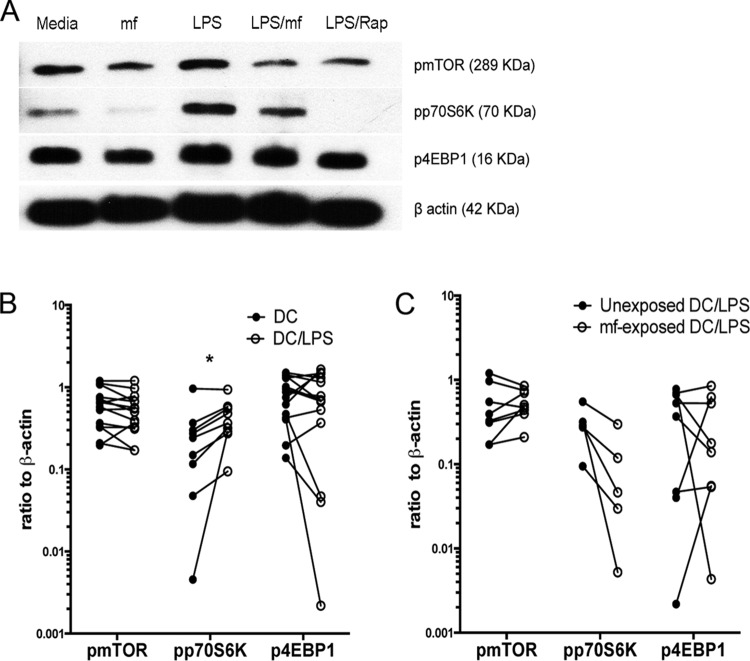
DC expression of pmTOR, pp70S6K, and p4EBP1 is significantly downregulated by mf. Immunoblots of lysates from human DC unexposed (in medium alone) or exposed to mf or rapamycin (Rap; 100 nM) for 5 min or 60 min. (A) One representative image for 5 or 60 min of pmTOR, pp70S6K, p4EBP1, and β-actin expression using Western blot analysis. (B) Line graphs represent the ratio of intensity of protein to β-actin in either unexposed DC (closed circles) or following 60 min of exposure to either mf or rapamycin (open circles) in each independent donor. *, P ≤ 0.05; **, P ≤ 0.001.
FIG 6.
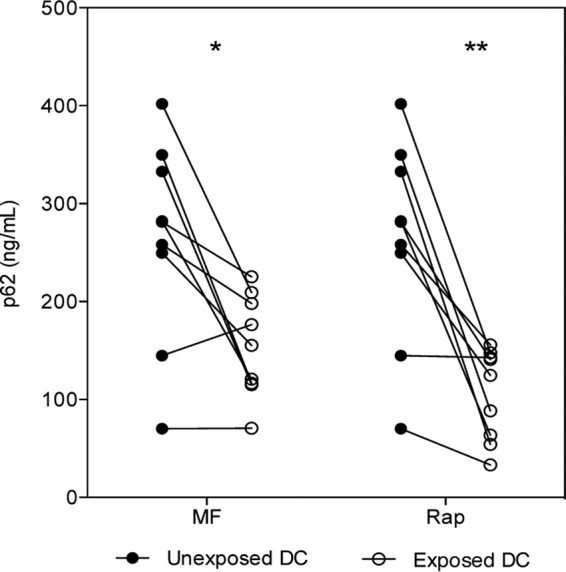
mf reduce p62 levels. DC left unexposed (closed circles) or exposed to mf or rapamycin (Rap; open circles) for 24 h were harvested and lysed, and intracellular concentrations of p62 (ng/ml) levels were measured by ELISA. Each line represents an independent donor. P values were calculated based on levels of p62 in exposed DC compared to unexposed DC. *, P ≤ 0.05; **, P ≤ 0.001.
Accession number.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD004522.
RESULTS
Proteomic analysis of DC and DC/mf.
Previously, we demonstrated that the function and viability of monocyte-derived DC (mDC) are altered by the mf of B. malayi (2, 3). To gain insight into the mechanisms involved in this altered function, we performed global shotgun proteomic analysis of mDC that were either unexposed (DC) or were exposed to live mf of B. malayi (DC/mf) for 48 h. A total of 5,024 human proteins (see Table S1 in the supplemental material) were identified, of which 1,394 were present in the DCs of all four donors (DC and DC/mf) (Fig. 1A); 1,973 proteins were found in at least 3/4 donors (Fig. 1A). Interestingly, there were no proteins that were discretely expressed in parasite-exposed or -unexposed DC. To examine if there was a correlation between a previously studied transcriptomic analysis (16) and the abundance of identified proteins from the current study, normalized fold changes (DC/mf over DC) were assessed as a function of the corresponding fold change in values from the microarrays. A significant correlation was observed between the 2 studies, highlighting the significantly increased expression levels of ICAM-1 and IL-1RA (r = 0.3307, P < 0.0001) (see Fig. S1 in the supplemental material).
Analysis of differentially expressed proteins induced by mf in DC resulted in the identification of 207 proteins that were 1.5-fold up- or downregulated over DC alone. Of these 207 proteins, 13 were significantly upregulated and 25 were significantly downregulated (P < 0.05 by one-way ANOVA) (Fig. 1B; see also Table S1 in the supplemental material). Among these 38 proteins, the 5 that were most significantly upregulated were intercellular cell adhesion molecule 1 (ICAM1; P05362), normal mucosa of esophagus-specific gene 1 protein (NMES1; Q9C002), interleukin-1 receptor antagonist (IL1RA; P18510); antigen peptide transporter 1 (TAP1; Q03518), and tryptophanyl-tRNA synthetase, cytoplasmic (WARS; P23381) (Fig. 1B; see also Table S1). Notable among the significantly downregulated proteins were stabilin-1 (STAB1; Q9NY15), adenosine deaminase CECR1 (CECR1; Q9NZK5), HLA class II histocompatibility antigen gamma chain (CD74; P04233), macrophage mannose receptor 1 (MRC1; CD206; Q5VSK2), and phospholipase D3 (Q8IV08).
In addition, LC-MS/MS analysis of DC/mf samples resulted in the detection of 539 proteins of B. malayi origin and 13 proteins of Wolbachia origin. Of the 539 B. malayi-specific proteins, 53 were detected in DC/mf samples from all 4 donors, and 118 were detected in any three DC/mf samples (see Table S2 in the supplemental material).
mf modulate the expression of proteins associated with mTOR and its related pathways in human DC.
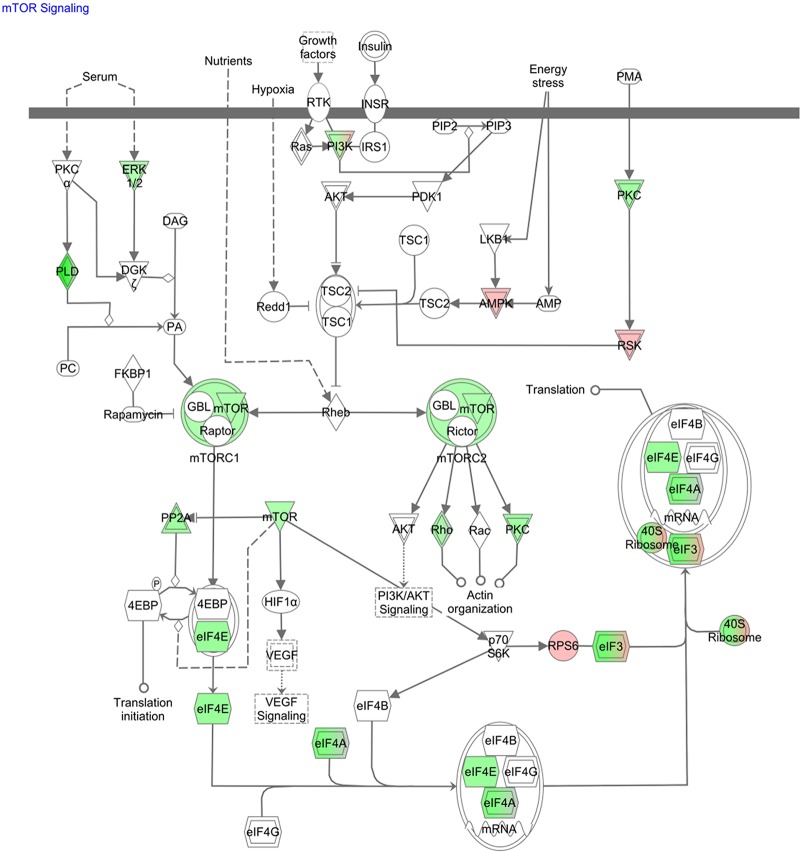
Utilizing Ingenuity pathway analysis (IPA), subcellular localization of the proteins identified in DC and DC/mf revealed that a large proportion were cytosolic (see Table S1 in the supplemental material). Furthermore, DC exposed to mf exhibited significant enrichment of enzymes, including proteases, phosphatases, and kinases (see Table S1 in the supplemental material). Interestingly, mf-exposed DC showed a significant downregulation of important metabolic pathways, including the mTOR pathway, as well as pathways involved in eukaryotic initiation factor (eIF2) signaling, the regulation of eIF4, p70S6K signaling, and protein ubiquitination (Fig. 2). Downmodulation of mTOR by mf was of particular interest, in that this pathway is a master growth regulator and is important in many cellular activities. In fact, through activation of mTOR by environmental signals, phosphorylation of downstream proteins such as p70S6K (also shown to be downmodulated by mf; Fig. 2) can facilitate cellular growth.
FIG 2.
mf regulated mTOR signaling pathway in human DC. Ingenuity pathway analyses of 1,394 proteins identified in all four donors resulted in identification of mTOR and its related pathways (eIF2 and p70S6K) as significantly regulated by mf in human DC; downregulated proteins are illustrated in green and upregulated proteins in red. See also Table S2 in the supplemental material.
Basal expression of pmTOR, pp70S6K, and p4EBP1 in human DC is significantly downregulated by mf.
To further confirm the proteomics data, we investigated the effect of live mf on mTOR pathway protein expression in DC utilizing immunoblot analysis, examining whether mf modulate the activity of mTOR, the translational repressor 4EBP1, and p70S6K. In agreement with the proteomics data, exposure of DC to live mf significantly downregulated the phosphorylation of mTOR, p70S6K, and 4EBP1, similar to that seen for rapamycin, a process seen as early as 5 min (Fig. 3A) and that was sustained for at least 60 min (Fig. 3A and B) following exposure to mf. These data suggest that live mf of B. malayi significantly modulate mTOR in human DC.
Preexposure of human DC to mf downmodulates LPS-induced phosphorylation of pp70S6K.
mTOR signaling can be activated in response to TLR agonists such as LPS (17). We therefore assessed whether mf can inhibit LPS-induced mTOR activation in human DC. Our data indicate that 30 min of exposure to LPS significantly augmented the phosphorylation of p70S6K1 (Fig. 4A and B). Furthermore, preexposure of DC to mf (Fig. 4A and C) or to rapamycin (data not shown) downregulated the phosphorylation of p70S6K1 following LPS activation. Since LPS did not significantly induce the phosphorylation of mTOR or 4E-BP1 (Fig. 4A and B), preexposure to mf did not alter the phosphorylation of these two proteins in DC following activation (Fig. 4C). These data collectively suggest that mf inhibit the mTOR pathway in human DC at the basal level (Fig. 3) and also prevent the activation of this pathway in response to mTOR activators such as LPS (Fig. 4).
FIG 4.
Preexposure of human DC to mf downmodulates LPS-induced phosphorylation of pp70S6K. Human DC were either unexposed (medium alone) or preexposed to mf or rapamycin (Rap) for 60 min and then either left in medium alone or activated with LPS for another 30 min. (A) One representative image of pmTOR, pp70S6K, and p4EBP1 using immunoblotting is shown. (B) Line graphs represent the ratio of intensity of protein to β-actin in either unexposed DC (closed circles) or following 30 min of exposure to LPS (1 μg/ml; open circles). *, P < 0.05. (C) Line graphs represent the ratio of intensity of protein to β-actin in either DC exposed to LPS (1 μg/ml; closed circles) or preexposed to mf for 60 min and following 30 min of exposure to LPS (1 μg/ml; open circles).
mf induce the phosphorylation of Beclin 1 and conversion of LC3II, suggesting the induction of autophagy in human DC.
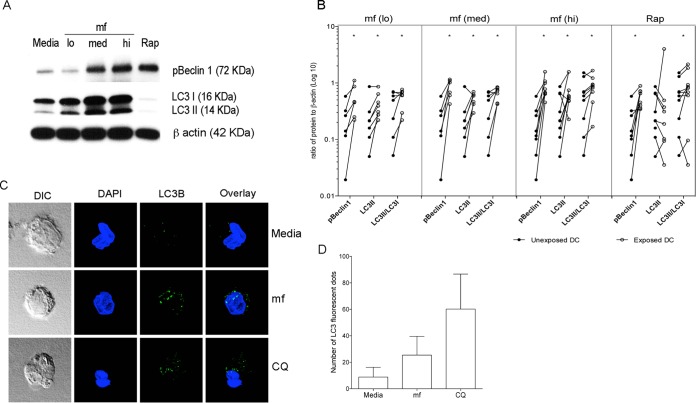
Having established that mf inhibit activation of the mTOR pathway in human DC, both basally and in response to activation signals, we next investigated whether mf are involved in the regulation of autophagy in human DC, a process previously linked to the inhibition of mTOR (14). Following exposure to various numbers of mf or to rapamycin, a known inducer of autophagy, expression of Beclin 1 (a component of the phosphatidylinositol 3-kinase [PI3K] complex) and LC3 (microtubule-associated protein light chain 3), both of which are required for autophagy, were assessed by immunoblotting (Fig. 5). As can be seen, live mf significantly upregulated the phosphorylation of Beclin 1 (Fig. 5A and B) and induced the conversion of LC3II to the same degree as rapamycin. mf also significantly upregulated the ratio of LC3II to LC3I (Fig. 5B), suggesting that mf induce the lipidated form of LC3 that is known to be recruited to autophagosomes and associated with the autophagosome membranes.
FIG 5.
mf induce phosphorylation of Beclin 1 and conversion of LC3II in human DC. DC left unexposed (medium alone) or exposed to mf at different concentrations (5,000 mf/1 × 106 [lo], 25,000 mf/1 × 106 [med], or 50,000 mf/1 × 106 [hi]) or rapamycin (Rap) for 24 h were harvested and lysed. (A) One representative image of phospho-Beclin 1, LC3I, and LC3II expression using Western blot analysis is shown. (B) Line graphs represent the ratio of intensity of protein to β-actin in each independent donor in either unexposed DC (closed circles) or exposed DC (open circles). *, P ≤ 0.05; **, P ≤ 0.001. (C) DC unexposed (medium alone) or exposed to mf or chloroquine (CQ) for 24 h were harvested and stained with anti-LC3B rabbit polyclonal antibody (LC3II). DIC (differential interference contrast) and fluorescent images of DC stained with LC3 (green) and nucleus (blue). The mf-exposed DC show increased LC3II granules compared with the unexposed DC. (D) Cumulative average fluorescent clusters (0.3-μm size), measured in unexposed DC and in DC exposed to mf or CQ from at least 5 to 10 cells, were averaged and plotted. Error bars represent standard deviations (SD) from 5 to 10 cells. DC geometric mean (GM), 6.7; DC/CQ GM, 54.6; DC/mf GM, 22.1.
To further demonstrate mf-induced autophagy in DC, we measured LC3II accumulation using confocal microscopy and staining with anti-LC3B antibody (Fig. 5C). Exposure of human DC to mf induced LC3II accumulation as measured by an increase in fluorescent staining of autophagosomes (Fig. 5C). The data indicate that mf induce the accumulation of LC3II in autophagosomes, as shown by an increase in LC3II fluorescence in DC/mf (geometric mean [GM], 22.1 fluorescent dots) compared to LC3II fluorescence in DC not exposed to mf (GM, 6.7) (Fig. 5D). To further demonstrate LC3II accumulation in autophagosomes, we used chloroquine (CQ) as a control (Fig. 5C). CQ blocks autophagy by inhibiting lysosomal proteases and autophagosome-lysosome fusion, therefore resulting in accumulation of LC3II.
mf induce autophagy in human DC by p62.
The p62 protein, also known as sequestosome 1, serves as a link between LC3 and ubiquitinated proteins and is involved in the autophagic machinery. An alternative method for detecting the autophagic flux is by measurement of p62 degradation (18), as decreased levels of p62 suggest induction of autophagy. Hence, we measured the expression of p62 in human DC after exposure to mf. Both mf (P = 0.02) and rapamycin (P = 0.003) significantly inhibited the levels of p62 in human DC compared with those seen in unstimulated DC (Fig. 6). These data collectively suggest that live mf induce autophagy in human DC, a process that involves the inhibition of the mTOR pathway in these cells.
DISCUSSION
Although helminth parasites appear to affect APC function, the mechanism by which this occurs is largely unknown. Previously we have shown that live mf of B. malayi impair the viability and function of monocyte-derived DCs (2, 3). The current study elucidates the mechanism underlying this impaired function and suggests that cell death operates through the inhibition of the mTOR signaling pathway and the induction of autophagy by this extracellular parasite.
DC are critical components in the development of immunity against a variety of pathogens by interpreting various pathogen-derived signals and by polarizing T-cell responses (19). In the presence of inflammatory stimuli or infectious organisms, immature DCs undergo maturation that is an orchestrated process involving the regulation of a variety of signaling pathways. Global transcriptional and proteomic profiling of different stages of DC have attempted to gain insights into these maturation and activation processes, with most studies utilizing monocyte-derived DC that are differentiated from monocytes in vitro (20–23). To gain an understanding of the mechanisms involved in filarial parasite-induced DC dysfunction, we initially utilized global proteomic profiling, a process that demonstrated 207 intracellular proteins being altered (either up or down). Twenty-five of these proteins, including HLA class II histocompatibility antigen gamma chain (CD74) and DC-SIGN (CD209), were significantly downregulated (see Table S1 in the supplemental material), providing more evidence for the suppressive effect of mf on human DC. In fact, mf-induced downregulation of DC-SIGN has been suggested to play a role in the infection of human DC with Mycobacterium tuberculosis (24).
Proteomic analysis also strongly suggested that mf markedly impaired the mTOR pathway (Fig. 2), a finding corroborated experimentally (Fig. 3). These findings differed from studies using antigens from Schistosoma mansoni, including soluble egg antigens (SEA) and the major immunomodulatory component omega-1 (12, 25), in which the helminth-induced alterations in mTOR function differed from those induced by rapamycin. In marked contrast, extracellular live B. malayi mf behaved more like rapamycin by significantly inhibiting the phosphorylation of the two downstream targets, 4EBP1 and p70S6k, in DC (P < 0.05) (Fig. 3A and B); this inhibition required live parasites in that parasite antigen (or heat-killed mf) failed to alter mTOR-related processes (data not shown). Previous work has demonstrated that DC activation and longevity following exposure to TLR agonists relies on mTOR (26). Interestingly, our data indicate that mf can inhibit mTOR activation (particularly phosphorylation of p70S6K1) following exposure to the TLR agonist LPS (Fig. 4) or to poly(I·C) (data not shown).
Intracellular parasites, viruses, and bacteria are known to be capable of activating or inhibiting the mTOR pathway to ensure their survival and replication (reviewed in references 27 and 28). The dephosphorylation of 4EBP1 by the intracellular parasite Leishmania major has been reported in macrophages, a process that allowed for the parasite's survival (12). Using a different species of Leishmania, Leishmania donovani, IL-12 and IL-10 production was modulated through the mTOR pathway (29). In contrast to Leishmania infection, infection with Toxoplasma gondii maintained mTOR-dependent cellular growth that was independent of S6K and 4EBP1, suggesting a parasite-specific mechanism of mTOR regulation (12). Our results demonstrate that extracellular helminth parasites inhibit the mTOR pathway in DC both at the basal level and in response to TLR activation (Fig. 3 and 4), likely through internalization of actively secreted/excreted molecules.
The mTOR complex also plays an important role in T cell differentiation and activation (30). In CD4+ T cells, mTORC1 selectively regulates Th1 and Th17 differentiation (31, 32), and complete inhibition of mTOR in CD4+ T cells was shown to result in the generation of regulatory T cells (33). How mf-induced mTOR inhibition in DC affects CD4+ and CD8+ differentiation is an important area of research that needs further exploration. Studies from our laboratory suggest that in patent filarial infection, there is an expansion of regulatory T cells (34). Whether this is the direct effect of a downregulation of mTOR in the DCs of infected individuals is an open question. Studies have indicated that the inhibition of mTOR by rapamycin supports the induction of tolerogenic DCs (35–37), a phenomenon that needs to be further investigated in the context of filarial infection.
It has been well documented that inhibition of mTOR by nutrient starvation can induce autophagy (reviewed in references 37–39). Furthermore, reactivation of mTOR leads to the termination of autophagy and the initiation of lysosome formation (40). Therefore, we determined whether mf induce autophagy in human DC through mTOR inhibition. Proteomic analyses resulted in the identification of several autophagy-associated proteins. However, only two, ATG3 and ATG7, were identified in all 4 donors (see Table S1 in the supplemental material). Nevertheless, none of these proteins was shown to be significantly upregulated (>1.5-fold) by mf in DC. It is likely that the limitations of processing by mass spectrometry and/or kinetics of DC exposure to mf (48 h for proteomics, 24 h for other assays) contributed to the lack of identification of autophagy-related proteins using proteomics.
To assess autophagy, we measured the induction of phosphorylation of Beclin 1 (41), since starvation-induced autophagy has been shown to occur through the MK2/MK3-dependent phosphorylation of Beclin 1 (42). In the present study, we were able to demonstrate that live mf (at the three concentrations tested) significantly induced the phosphorylation of Beclin 1 in human DC (Fig. 5A and B). We further examined the impact of mf on the formation of autophagosomes in DC. LC3 is the only known mammalian protein that specifically associates with the autophagosome membrane; LC3I, a diffuse cytosolic form, conjugates with phosphatidylethanolamine upon autophagy induction, resulting in the autophagosome-membrane-associated LC3II form (43, 44). In this study, live mf not only induced the LC3II protein but, similar to rapamycin, also significantly increased the LC3II/LC3I ratio (Fig. 5B). As the amount of LC3II is closely associated with the number of autophagosomes, it can serve as a good indicator of autophagosome formation (45). We further confirmed the induction of LC3II in DC in response to mf utilizing confocal microscopy by measuring LC3II fluorescence (Fig. 5C, phagosomal staining), demonstrating that mf (like chloroquine) blocks the degradation of LC3II (46) and results in LC3II accumulation in DC (Fig. 5C and D).
To confirm that the increased levels of LC3II were not due to inhibition of autophagic degradation and did not represent a blockade in autophagosome maturation, a second method was used for detecting the autophagic flux. We measured ubiquitin-binding receptor p62 degradation, a marker of conventional autophagy whose expression correlates inversely with LC3I-II conversion. p62 is a scaffolding adaptor protein that can interact with both LC3II and polyubiquitinated protein, leading to self-degradation as well as degradation of polyubiquitinated proteins that are in autophagosomes (47–49). mf significantly induced the degradation of p62, providing independent corroboration that mf could activate the autophagy machinery in DC.
Induction of autophagy by intracellular pathogens has been previously reported (reviewed in reference 50). Indeed, autophagy is an important mechanism used by host immune cells to kill intracellular pathogens (e.g., Helicobacter pylori [51], Leishmania amazonensis [52], and Toxoplasma gondii [53]) or to provide some defense against intracellular pathogens (50). Furthermore, various bacteria and viruses regulate autophagy to avoid degradation through this pathway (54, 55). HIV, M. tuberculosis, and T. gondii can prevent autophagic degradation by affecting signaling processes that regulate the autophagy pathway (56, 57). These data collectively suggest that pathogens can act to regulate their own degradation through the autophagy machinery. Many intracellular pathogens, however, have developed strategies to avoid autophagy or inhibit autophagy for their survival (reviewed in reference 50). Listeria organisms, for example, inhibit autophagy as an important mechanism for survival (58). In other organisms, such as T. gondii, the parasites derive nutrients and promote their intracellular growth by the induction of autophagy (53).
How the induction of autophagy might be beneficial to B. malayi mf is still not known; for example, it may provide nutrients for parasite survival or facilitate degradation. Nevertheless, there is mounting evidence that apoptosis and autophagy coexist within the same cell and that these pathways share common upstream signals (59). Whether autophagy and the apoptotic machinery work in parallel as a consequence of exposure of DC to mf (2, 60) is an important question. Our study suggests that B. malayi mf employ mechanisms of metabolic modulation in DC that influence the host immune response by downregulating mTOR signaling, resulting in increased autophagy, a process that may be the result of parasite-secreted rapamycin-like molecules.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Kubofcik for technical advice and help and Cathy Steel for critical reading of the manuscript and for editorial help.
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00174-16.
REFERENCES
- 1.Semnani RT, Keiser PB, Coulibaly YI, Keita F, Diallo AA, Traore D, Diallo DA, Doumbo OK, Traore SF, Kubofcik J, Klion AD, Nutman TB. 2006. Filaria-induced monocyte dysfunction and its reversal following treatment. Infect Immun 74:4409–4417. doi: 10.1128/IAI.01106-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semnani RT, Venugopal PG, Mahapatra L, Skinner JA, Meylan F, Chien D, Dorward DW, Chaussabel D, Siegel RM, Nutman TB. 2008. Induction of TRAIL- and TNF-alpha-dependent apoptosis in human monocyte-derived dendritic cells by microfilariae of Brugia malayi. J Immunol 181:7081–7089. doi: 10.4049/jimmunol.181.10.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semnani RT, Venugopal PG, Leifer CA, Mostbock S, Sabzevari H, Nutman TB. 2008. Inhibition of TLR3 and TLR4 function and expression in human dendritic cells by helminth parasites. Blood 112:1290–1298. doi: 10.1182/blood-2008-04-149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semnani RT, Mahapatra L, Dembele B, Konate S, Metenou S, Dolo H, Coulibaly ME, Soumaoro L, Coulibaly SY, Sanogo D, Seriba Doumbia S, Diallo AA, Traore SF, Klion A, Nutman TB, Mahanty S. 2010. Expanded numbers of circulating myeloid dendritic cells in patent human filarial infection reflect lower CCR1 expression. J Immunol 185:6364–6372. doi: 10.4049/jimmunol.1001605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev 18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 6.Noda T, Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 7.Scott RC, Schuldiner O, Neufeld TP. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunn GJ, Fadden P, Haystead TA, Lawrence JC Jr. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J Biol Chem 272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 10.Beugnet A, Tee AR, Taylor PM, Proud CG. 2003. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J 372:555–566. doi: 10.1042/bj20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Weiss LM, Orlofsky A. 2009. Intracellular parasitism with Toxoplasma gondii stimulates mammalian-target-of-rapamycin-dependent host cell growth despite impaired signalling to S6K1 and 4E-BP1. Cell Microbiol 11:983–1000. doi: 10.1111/j.1462-5822.2009.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaramillo M, Gomez MA, Larsson O, Shio MT, Topisirovic I, Contreras I, Luxenburg R, Rosenfeld A, Colina R, McMaster RW, Olivier M, Costa-Mattioli M, Sonenberg N. 2011. Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe 9:331–341. doi: 10.1016/j.chom.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. 2010. mTOR regulation of autophagy. FEBS Lett 584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YC, Guan KL. 2015. mTOR: a pharmacologic target for autophagy regulation. J Clin Investig 125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA Jr, Behrns KE, Leeuwenburgh C, Kim JS. 2011. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology 141:2188–2199. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaussabel D, Semnani RT, McDowell MA, Sacks D, Sher A, Nutman TB. 2003. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 102:672–681. doi: 10.1182/blood-2002-10-3232. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz F, Heit A, Dreher S, Eisenacher K, Mages J, Haas T, Krug A, Janssen KP, Kirschning CJ, Wagner H. 2008. Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur J Immunol 38:2981–2992. doi: 10.1002/eji.200838761. [DOI] [PubMed] [Google Scholar]
- 18.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonna M, Pulendran B, Iwasaki A. 2006. Dendritic cells at the host-pathogen interface. Nat Immunol 7:117–120. doi: 10.1038/ni0206-117. [DOI] [PubMed] [Google Scholar]
- 20.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, Mann M. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Pereira SR, Faca VM, Gomes GG, Chammas R, Fontes AM, Covas DT, Greene LJ. 2005. Changes in the proteomic profile during differentiation and maturation of human monocyte-derived dendritic cells stimulated with granulocyte macrophage colony stimulating factor/interleukin-4 and lipopolysaccharide. Proteomics 5:1186–1198. doi: 10.1002/pmic.200400988. [DOI] [PubMed] [Google Scholar]
- 22.Richards J, Le Naour F, Hanash S, Beretta L. 2002. Integrated genomic and proteomic analysis of signaling pathways in dendritic cell differentiation and maturation. Ann N Y Acad Sci 975:91–100. doi: 10.1111/j.1749-6632.2002.tb05944.x. [DOI] [PubMed] [Google Scholar]
- 23.Buschow SI, Lasonder E, van Deutekom HW, Oud MM, Beltrame L, Huynen MA, de Vries IJ, Figdor CG, Cavalieri D. 2010. Dominant processes during human dendritic cell maturation revealed by integration of proteome and transcriptome at the pathway level. J Proteome Res 9:1727–1737. doi: 10.1021/pr9008546. [DOI] [PubMed] [Google Scholar]
- 24.Talaat KR, Bonawitz RE, Domenech P, Nutman TB. 2006. Preexposure to live Brugia malayi microfilariae alters the innate response of human dendritic cells to Mycobacterium tuberculosis. J Infect Dis 193:196–204. doi: 10.1086/498912. [DOI] [PubMed] [Google Scholar]
- 25.Hussaarts L, Smits HH, Schramm G, van der Ham AJ, van der Zon GC, Haas H, Guigas B, Yazdanbakhsh M. 2013. Rapamycin and omega-1: mTOR-dependent and -independent Th2 skewing by human dendritic cells. Immunol Cell Biol 91:486–489. doi: 10.1038/icb.2013.31. [DOI] [PubMed] [Google Scholar]
- 26.Amiel E, Everts B, Freitas TC, King IL, Curtis JD, Pearce EL, Pearce EJ. 2012. Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. J Immunol 189:2151–2158. doi: 10.4049/jimmunol.1103741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shertz CA, Cardenas ME. 2011. Exploiting and subverting Tor signaling in the pathogenesis of fungi, parasites, and viruses. PLoS Pathog 7:e1002269. doi: 10.1371/journal.ppat.1002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsalikis J, Croitoru DO, Philpott DJ, Girardin SE. 2013. Nutrient sensing and metabolic stress pathways in innate immunity. Cell Microbiol 15:1632–1641. [DOI] [PubMed] [Google Scholar]
- 29.Cheekatla SS, Aggarwal A, Naik S. 2012. mTOR signaling pathway regulates the IL-12/IL-10 axis in Leishmania donovani infection. Med Microbiol Immunol 201:37–46. doi: 10.1007/s00430-011-0202-5. [DOI] [PubMed] [Google Scholar]
- 30.Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. 2015. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J Clin Investig 125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol 12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. 2010. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity 32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metenou S, Dembele B, Konate S, Dolo H, Coulibaly SY, Coulibaly YI, Diallo AA, Soumaoro L, Coulibaly ME, Sanogo D, Doumbia SS, Traore SF, Mahanty S, Klion A, Nutman TB. 2010. At homeostasis filarial infections have expanded adaptive T regulatory but not classical Th2 cells. J Immunol 184:5375–5382. doi: 10.4049/jimmunol.0904067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taner T, Hackstein H, Wang Z, Morelli AE, Thomson AW. 2005. Rapamycin-treated, alloantigen-pulsed host dendritic cells induce ag-specific T cell regulation and prolong graft survival. Am J Transplant 5:228–236. doi: 10.1046/j.1600-6143.2004.00673.x. [DOI] [PubMed] [Google Scholar]
- 36.Turnquist HR, Sumpter TL, Tsung A, Zahorchak AF, Nakao A, Nau GJ, Liew FY, Geller DA, Thomson AW. 2008. IL-1beta-driven ST2L expression promotes maturation resistance in rapamycin-conditioned dendritic cells. J Immunol 181:62–72. doi: 10.4049/jimmunol.181.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Itakura E, Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberstein A, Jeffrey PD, Shi Y. 2007. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 42.Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. 2015. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife 4:e05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, Cuervo AM, Seglen PO. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 44.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. 2007. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res 67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 45.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng Y, Kohli L, Klocke BJ, Roth KA. 2010. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neurol Oncol 12:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauvy C, Meijer AJ, Codogno P. 2009. Assaying of autophagic protein degradation. Methods Enzymol 452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
- 48.Mizushima N, Yoshimori T. 2007. How to interpret LC3 immunoblotting. Autophagy 3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 49.Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. 2010. Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Investig Ophthalmol Vis Sci 51:6030–6037. doi: 10.1167/iovs.10-5278. [DOI] [PubMed] [Google Scholar]
- 50.Skendros P, Mitroulis I. 2012. Host cell autophagy in immune response to zoonotic infections. Clin Dev Immunol 2012:910525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YH, Wu JJ, Lei HY. 2009. The autophagic induction in Helicobacter pylori-infected macrophage. Exp Biol Med (Maywood, NJ) 234:171–180. doi: 10.3181/0808-RM-252. [DOI] [PubMed] [Google Scholar]
- 52.Cyrino LT, Araujo AP, Joazeiro PP, Vicente CP, Giorgio S. 2012. In vivo and in vitro Leishmania amazonensis infection induces autophagy in macrophages. Tissue Cell 44:401–408. doi: 10.1016/j.tice.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Weiss LM, Orlofsky A. 2009. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J Biol Chem 284:1694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. 2005. Escape of intracellular Shigella from autophagy. Science 307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 55.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, Schwartz O, Deretic V, Piguet V. 2010. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. 2010. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 59.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. 2009. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 60.Semnani RT, Liu AY, Sabzevari H, Kubofcik J, Zhou J, Gilden JK, Nutman TB. 2003. Brugia malayi microfilariae induce cell death in human dendritic cells, inhibit their ability to make IL-12 and IL-10, and reduce their capacity to activate CD4+ T cells. J Immunol 171:1950–1960. doi: 10.4049/jimmunol.171.4.1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.