Significance
The transcription factor FAR-RED ELONGATED HYPOCOTYL3 (FHY3) is known to play multiple roles at the vegetative stage in Arabidopsis, but its functions in reproductive stage are unclear. We find that FHY3 is required for floral meristem determinacy and shoot apical meristem maintenance by mainly acting as a transcriptional repressor. FHY3 mediates light-regulated CLAVATA3 expression to regulate WUSCHEL expression in shoot apical meristem and directly represses CLAVATA3, but activates SEPALLATA2, to promote floral meristem determinacy. Furthermore, FHY3 may coregulate flower development with three flower-specific MADS-domain transcription factors and four basic helix–loop–helix transcription factors that are involved in photomorphogenesis, and thus may act as a bridge molecule in the cross-talk between external signals and endogenous cues to coordinate plant development.
Keywords: meristem maintenance, meristem determinacy, FHY3, CLV3, SEP2
Abstract
Plant meristems are responsible for the generation of all plant tissues and organs. Here we show that the transcription factor (TF) FAR-RED ELONGATED HYPOCOTYL3 (FHY3) plays an important role in both floral meristem (FM) determinacy and shoot apical meristem maintenance in Arabidopsis, in addition to its well-known multifaceted roles in plant growth and development during the vegetative stage. Through genetic analyses, we show that WUSCHEL (WUS) and CLAVATA3 (CLV3), two central players in the establishment and maintenance of meristems, are epistatic to FHY3. Using genome-wide ChIP-seq and RNA-seq data, we identify hundreds of FHY3 target genes in flowers and find that FHY3 mainly acts as a transcriptional repressor in flower development, in contrast to its transcriptional activator role in seedlings. Binding motif-enrichment analyses indicate that FHY3 may coregulate flower development with three flower-specific MADS-domain TFs and four basic helix–loop–helix TFs that are involved in photomorphogenesis. We further demonstrate that CLV3, SEPALLATA1 (SEP1), and SEP2 are FHY3 target genes. In shoot apical meristem, FHY3 directly represses CLV3, which consequently regulates WUS to maintain the stem cell pool. Intriguingly, CLV3 expression did not change significantly in fhy3 and phytochrome B mutants before and after light treatment, indicating that FHY3 and phytochrome B are involved in light-regulated meristem activity. In FM, FHY3 directly represses CLV3, but activates SEP2, to ultimately promote FM determinacy. Taken together, our results reveal insights into the mechanisms of meristem maintenance and determinacy, and illustrate how the roles of a single TF may vary in different organs and developmental stages.
Plant meristems are responsible for the generation of all plant tissues and organs. Unlike the shoot apical meristem (SAM), whose activity is maintained throughout the life of plants, the floral meristem (FM) is precisely programmed to terminate in a process known as FM determinacy (1). WUSCHEL (WUS) plays a central role in the establishment and maintenance of SAM, inflorescence meristem, and FM, as well as in FM determinacy (2–4). WUS is expressed in the organizing center located beneath the stem cells in the meristem to promote cell proliferation by maintaining stem cell potential (2). The WUS/CLAVATA3 (CLV3) signaling pathway maintains the stabilization of meristem size and the stem cell pool (3, 5). Consistent with WUS overactivation, clv3 mutants have an enlarged SAM and increased numbers of floral organs and whorls (5). In addition to the WUS/CLV3 loop, several other pathways are known to regulate FM determinacy (4). AGAMOUS (AG) encodes a MADS-box transcription factor (TF) and is the lynchpin of the FM determinacy network (4, 6, 7). In the null ag-1 mutant, FM determinacy is severely impaired, resulting in a flower-in-flower phenotype (6). AG inhibits WUS expression through both indirect and direct means (8, 9). A number of other genes have been shown to regulate FM determinacy through the AG pathway or in parallel pathways, and additional players in this critical developmental process await characterization (4).
Floral organs are produced by the FM based on the classic ABC model in Arabidopsis (10, 11). However, the ABC genes were found to be necessary but not sufficient for the determination of floral organ identity (12, 13), and the E class genes—SEPALLATA1 (SEP1), SEP2, SEP3, and SEP4—were subsequently incorporated into the model (14). Although single or double sep mutants produce flowers indistinguishable from those of the wild-type, the sep1sep2sep3sep4 quadruple mutant develops flowers with leaf-like organs in all floral whorls and indeterminate FM activity, thereby demonstrating the functional redundancy of SEP genes and their fundamental roles in floral organ identity and FM determinacy (14, 15). On the other hand, the differing spatiotemporal expression patterns of individual SEP genes are suggestive of their distinct functions in flower development. Once the FM is produced, SEP1 and SEP2 are expressed in all four whorls. However, the mechanisms underlying the regulation of SEP1 and SEP2 expression remain unclear. Moreover, although the precise timing of the early events in floral organ production mediated by SEP genes has been well studied (16, 17), the molecular mechanisms mediated by SEP genes in the context of FM determinacy are largely unknown.
Light is one of the most important environmental signals for plant growth and development. Light could regulate stem cell activity through auxin and cytokinin (18). The detailed mechanisms underlying this process are still waiting dissection. The transposase-derived TF FAR-RED ELONGATED HYPOCOTYL3 (FHY3) was isolated as a key positive regulator of the phytochrome A (phyA) signaling pathway in Arabidopsis (19). FHY3 regulates the expression of target genes by directly binding to FHY3-binding sites (FBS, CACGCGC) (20, 21). ChIP-sequencing (ChIP-seq) analysis and genome-wide gene-expression profiling have identified hundreds of FHY3 target genes predicted to function in diverse environmental, hormonal, and developmental contexts. Under far-red (FR) conditions, FHY3 mainly acts as a transcriptional activator (21). In addition to phytochrome and circadian signaling, FHY3 has been found to function in diverse plant developmental and physiological processes, including UV-B signaling, chloroplast biogenesis, chlorophyll biosynthesis, programmed cell death, ABA signaling, and branching (22). It is important to note that although the functions of FHY3 during the plant vegetative stage are well studied, its roles in flower development remain poorly understood.
In this study, we isolated several fhy3 mutations that dramatically enhanced the FM indeterminacy phenotype of ag-10, a weak ag allele. Through genetic analyses we show that FHY3 is required for FM determinacy and SAM maintenance and that wus is epistatic to fhy3 in FM determinacy. Through ChIP-seq and RNA-seq analyses, we identify hundreds of FHY3 binding sites and FHY3 target genes in floral organs and find that FHY3 mainly acts as a transcriptional repressor during flower development. Further analyses show that FHY3 functions in meristem determinacy and maintenance by directly binding the promoters of CLV3, SEP1, and SEP2, resulting in direct CLV3 repression, direct SEP2 activation, and downstream regulation of WUS expression.
Results
FHY3 Is Required for FM Determinacy and SAM Maintenance.
To identify new players involved in FM determinacy, an ethyl methanesulfonate (EMS) mutagenesis screen was performed in the ag-10 background, as previously reported (9). In contrast to ag-1 null mutants (6), ag-10 exhibits only a weak FM determinacy defect showing a few curved and bulged siliques with additional floral tissue inside (Fig. 1 A and B and Fig. S1A) (9). For screening, we focused on mutants with more bulged siliques throughout the entire plant as an indicator of prolonged FM activity.
Fig. 1.
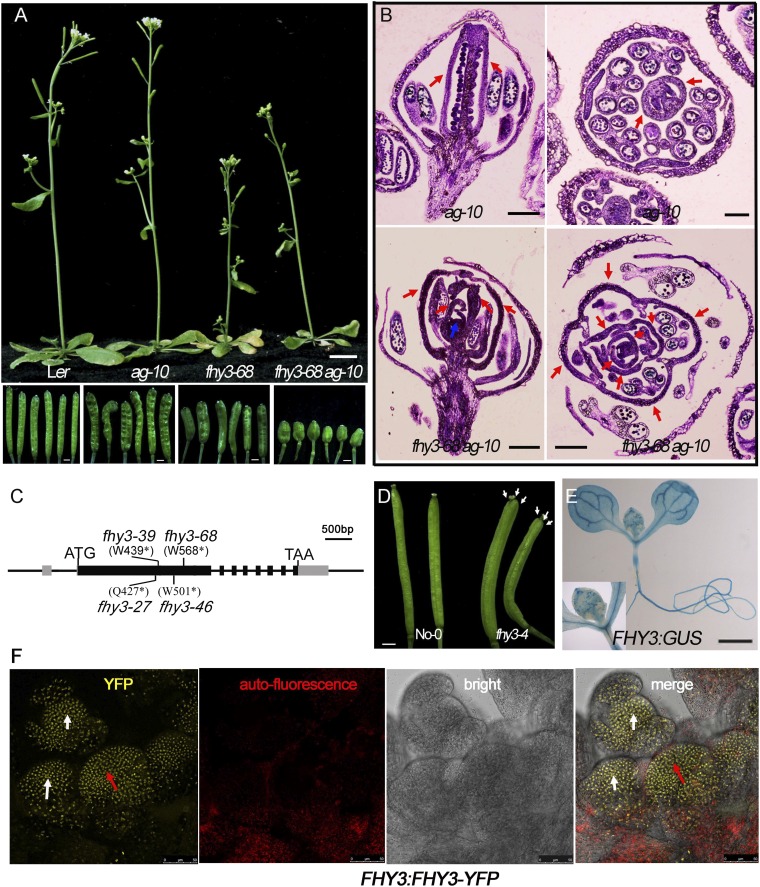
FHY3 is required for FM determinacy and SAM maintenance. (A–C, E, and F) Siliques of Ler (A), ag-10 (B), fhy3-68 ag-10 (C), 35S:FHY3-FLAG fhy3-68 ag-10 (E), and fhy3-68 (F). Carpels marked by red arrows in F. (D) Flowers of fhy3-68 ag-10. Sterile anthers are marked by a white arrow in D. (G) Quantification of inflorescence size (mm) of Ler (n = 15) and fhy3-68 (n = 15). **P < 0.01. (H and I) Inflorescences of Ler (H) and fhy3-68 (I). Dashed lines mark the width used to measure inflorescence size in G. (J and K) SAM (marked by a red arrow) of Ler (J) and fhy3-68 (K). Red lines mark the width of SAM. (Scale bars: 1 mm in A–C, E, and F; 250 µm in D; 500 µm in H and I; 60 µm in J and K.)
Fig. S1.
Various FHY3 mutants and their phenotypes in the wild-type and ag-10 backgrounds. (A) Phenotype of indicated plants (Upper) and siliques (Lower). All siliques of fhy3-68 ag-10 plant are bulged and short. (B) Longitudinal (Left) and transversal (Right) sections of indicated plants siliques. Although ag-10 produced two fused carpels (red arrows), the indeterminate floral meristem (blue arrow) of fhy3-68 ag-10 continued to generate additional organs inside the carpels (red arrows). (C) Gene diagram of FHY3 and the locations of mutations in the various fhy3 mutants used in this work. ATG and TAG correspond to the start and stop codons, respectively. The gray and black rectangles represent the 5′ or 3′ UTRs and coding regions, respectively. The black lines represent introns or intergenic regions. All mutations are nonsense mutations. (Scale bar, 500 bp.) (D) Siliques of No-0 (Left) and fhy3-4 (Right) plants. fhy3-4 siliques had three fused carpels (marked by white arrows). (E) Tissue-specific expression of FHY3 using FHY3:GUS transgenic plant. FHY3 is universally expressed in the seedling and SAM (Inset). (F) FHY3-YFP signal in the SAM and FM of FHY3:FHY3-YFP. Red arrows indicate YFP signal in SAM and white arrows indicate YFP signal in FM. [Scale bars: 1 cm in A (Upper) and 1 mm (Lower); 200 µm in B; 1 mm in D and E; and 50 µm in F.]
Several such mutants were isolated with similar phenotype, showing very short and bulged siliques in whole plants with a mean carpel number of 4.3 ± 0.4 (n = 50) (Fig. 1C) and additional floral organs growing inside (Fig. S1 A and B). The mutants also produced very small petals and sterile anthers (Fig. 1D). Longitudinal and transversal silique sections revealed plenty of floral organs growing inside the pistils from the indeterminate FM (Fig. S1B), resulting in infertile siliques in the mutant in a Landsberg erecta (Ler) background (Fig. S1A). Through genetic mapping, all mutation sites were found in FHY3 and the mutations were named fhy3-27, fhy3-39, fhy3-46, and fhy3-68 (Fig. S1C). fhy3-68 was used for subsequent analysis. Introducing a 35S:FHY3-FLAG transgene into fhy3-68 ag-10 rescued the mutant phenotype, confirming that fhy3-68 was responsible for the enhanced FM determinacy defects (Fig. 1E).
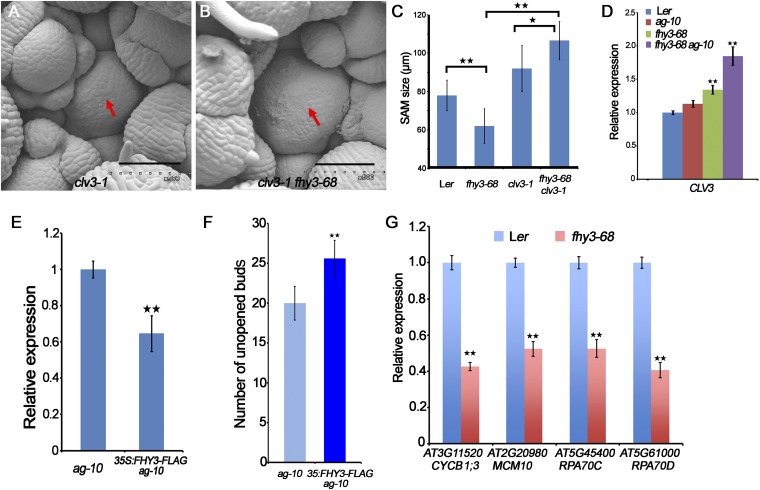
Through outcrossing of fhy3-68 ag-10 with Ler, we obtained the fhy3-68 single mutant. The number of carpels of fhy3-68 was 2.61 ± 0.6 (n = 50), which was similar to that of fhy3-4 plants in the No-0 background (3.2 ± 0.5, n = 50), indicated enhanced or prolonged FM activity of fhy3 (Fig. 1F and Fig. S1D). Taken together, these findings show that FHY3 is required for FM determinacy in Arabidopsis. In addition, we noticed that compared with the wild-type, the inflorescences of fhy3-68 were smaller (Fig. 1 G–I) and the SAM size of fhy3-68 was dramatically reduced (Fig. 1 J and K), indicating that FHY3 is also required for proper SAM maintenance, consistent with the high expression level of FHY3 in SAM and FM (Fig. S1 E and F).
wus Is Epistatic to fhy3 in FM Determinacy.
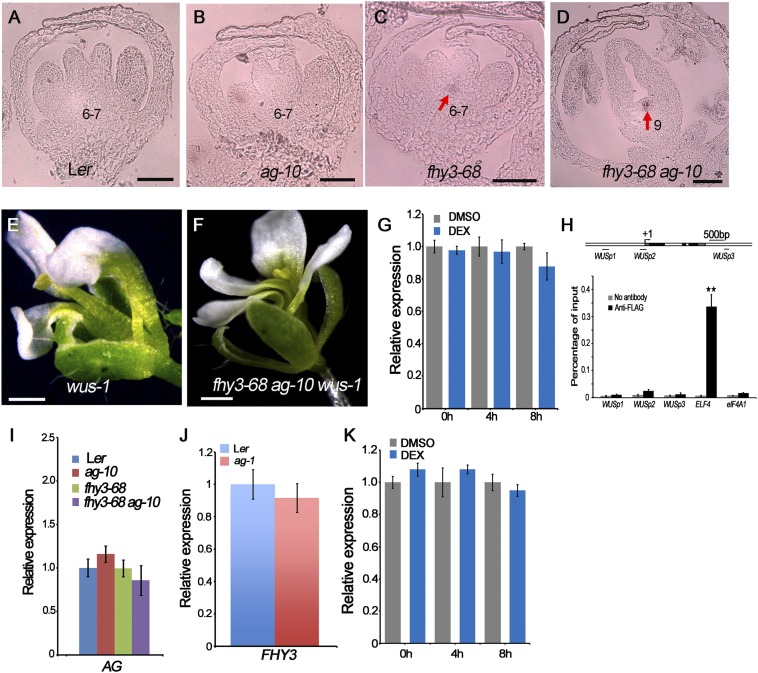
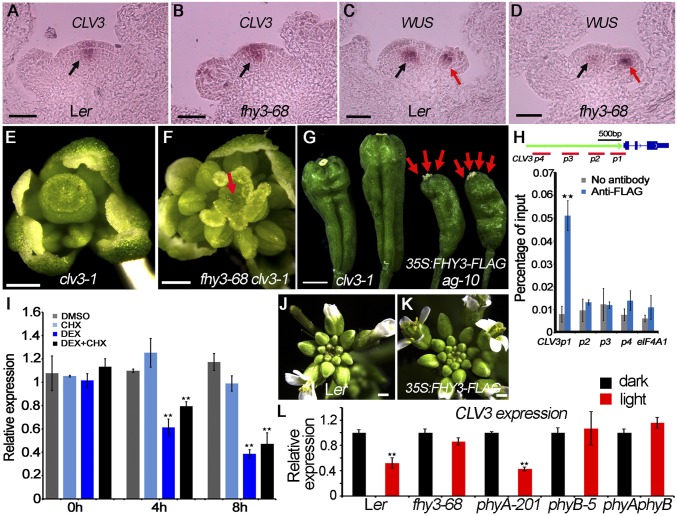
WUS plays a pivotal role in FM initiation, maintenance, and determinacy (2), and several genes have been characterized as FM determinacy factors through their regulation of WUS expression (4). To dissect the interaction between FHY3 and WUS in FM determinacy, we performed in situ hybridization to assess the temporal-spatial expression pattern of WUS. In wild-type, WUS expression is shut off at stage 6 of flower development (23). In ag-10 plants, most flowers have normal WUS expression patterns, whereas a few flowers exhibit WUS expression until stage 7 (Fig. S2 A and B) (9). For fhy3-68, the tested flowers (n = 9) showed slight WUS expression at the end of stage 6, indicating slightly prolonged WUS expression (Fig. S2C). All of the fhy3-68 ag-10 stage 9 flowers examined (n = 9) had obviously prolonged WUS expression (Fig. S2D), showing that FHY3 is required for the temporally precise repression of WUS. To determine the genetic relationship of FHY3 and WUS, we crossed fhy3-68 ag-10 with the wus-1 loss-of-function mutant (2). wus-1 flowers exhibited premature FM termination with normal sepals and petals and one or two stamens (Fig. S2E). The precocious termination of fhy3-68 ag-10 wus-1 flowers resembled that of wus-1 flowers (Fig. S2F), demonstrating that wus-1 was epistatic to fhy3-68 ag-10.
Fig. S2.
Molecular and genetic analysis of WUS and AG and FHY3 in FM determinacy. (A–D) In situ hybridization examining the WUS expression pattern (marked by a red arrow) in Ler (A), ag-10 (B), fhy3-68 (C), and fhy3-68 ag-10 (D). Floral development stage was marked. (Scale bars, 100 µm.) (E and F) Flower phenotypes of wus-1 (E) and fhy3-68 ag-10 wus-1 (F). (Scale bars, 500 µm.) (G) The WUS transcript abundance in FHY3:FHY3-GR fhy3-4 inflorescences measured by real-time PCR. Inflorescences containing stage 8 and younger flowers were treated with DEX or DMSO and then harvested at the indicated time point. UBQ5 served as the internal control. Three biological replicates were performed. (H) ChIP with anti-FLAG antibody to examine FHY3 binding at WUS in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences. The regions examined are diagrammed in the upper row, with “+1” indicating the TSS. The gray, black, and white rectangles represent the 5′ or 3′ UTR, coding regions, and introns or intergenic regions, respectively. (Scale bar, 500 bp). ELF4, a FHY3 target gene, served as a positive control, and eIF4A1 (EU.K.ARYOTIC TRANSLATION INITIATION FACTOR 4A1) served as a negative control. Error bars represent SD from three biological repeats. No significant FHY3 occupancy was detected at WUS. **P < 0.01. (I and J) Real-time RT-PCR to measure AG (I) and FHY3 (J) transcript levels in the indicated plants. UBQ5 served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. (K) Real-time RT-PCR to measure AG transcript levels in FHY3:FHY3-GR fhy3-4 inflorescences. Inflorescences containing stage 8 and younger flowers were treated with DEX or DMSO (control) then harvested at the indicated time point. UBQ5 served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. No significant change in AG transcript levels in the DEX-treated sample compared with the DMSO control indicated that FHY3 does not directly regulate AG expression.
To investigate whether FHY3 is a direct transcriptional regulator of WUS, we used an FHY3:FHY3-GR fhy3-4 transgenic line (20). After 4- and 8-h treatment with dexamethasone (DEX) or dimethyl sulfoxide (DMSO, as a negative control), WUS expression was similar in both treatments (Fig. S2G). Additionally, the ChIP-quantitative PCR (qPCR) data using the 35S:3FLAG-FHY3-3HA fhy3-4 transgenic line (21) revealed no significant enrichment of FHY3 binding activity at the WUS locus, indicating that WUS is not a direct target gene of FHY3 (Fig. S2H).
AG is the major FM terminator and acts via WUS repression (7). Thus, AG expression was analyzed to assess the relationship between FHY3 and AG in FM determinacy. AG transcript levels were normal in fhy3-68 and fhy3-68 ag-10 compared with Ler and ag-10, respectively (Fig. S2I). In addition, the expression of FHY3 was not affected in ag-1 compared with Ler (Fig. S2J), indicating that FHY3 and AG do not regulate each other. RT-qPCR assays using DEX- and DMSO-treated FHY3:FHY3-GR fhy3-4 transgenic plants revealed no significant change in AG transcript levels after DEX treatment, further indicating that FHY3 does not regulate AG expression (Fig. S2K). These results suggest that FHY3 may act independently of the AG pathway in FM determinacy.
Genome-Wide Identification of FHY3 Binding Sites.
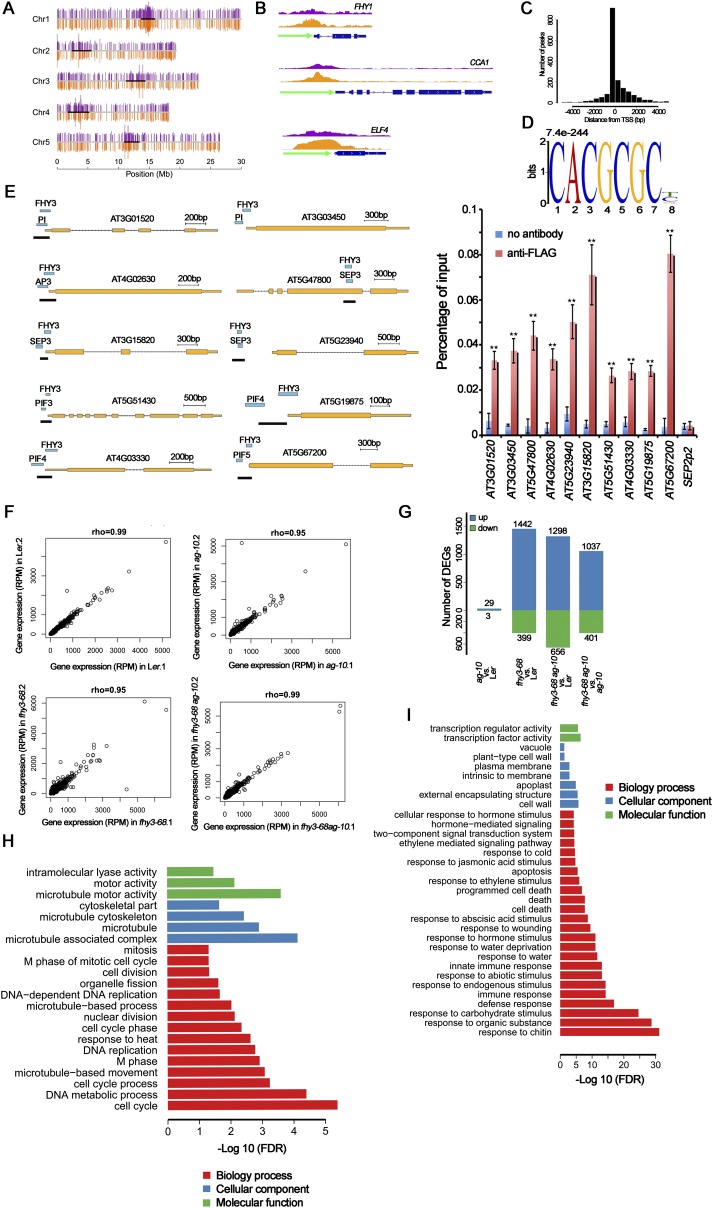
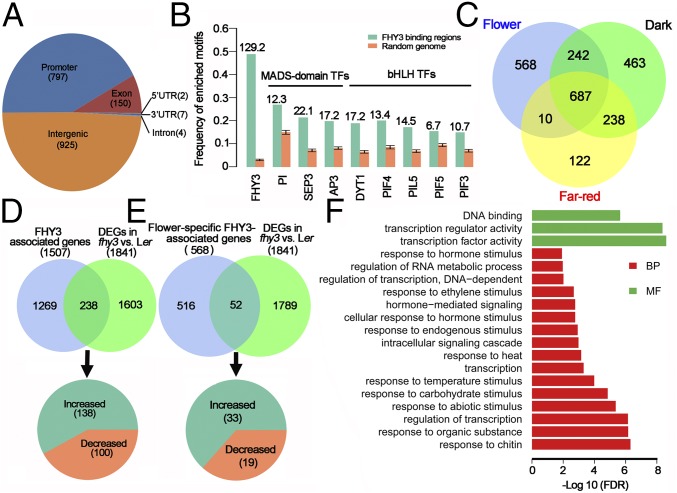
To investigate the molecular mechanisms underlying the FHY3 functions in flower development, we performed ChIP-seq analysis using a 35S:3FLAG-FHY3-3HA fhy3-4 transgenic line (21) to identify the binding sites of FHY3 in floral organs. Inflorescences containing stage 8 and younger flowers were harvested for ChIP-seq analysis. We identified 1,885 FHY3 binding sites (FBSs, P < 5e-3) distributed across the five chromosomes (Fig. S3A and Dataset S1), of which 51% (960) were subsequently assigned to genic regions [from −2,000 bp of the transcription start site (TSS) to the 3′UTR] and grouped into 1,507 genes (Dataset S1), which were referred to as FHY3-associated genes. The remaining 49% (925) FBSs were localized in intergenic regions. Consistent with a previous report (21), 74% (688) of the intergenic FBSs resided in centromeric regions (Fig. 2A). FHY1, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and EARLY FLOWERING4 (ELF4) are well-characterized FHY3 target genes (21). Specific enrichment of FHY3 was detected in the promoter regions rather than the transcribed regions of these genes, confirming the reliability of our ChIP-seq data (Fig. S3B).
Fig. S3.
ChIP-seq and RNA-seq analysis of FHY3 binding sites. (A) Distribution of FHY3 binding sites on the five Arabidopsis chromosomes. The top purple bars and bottom orange bars on each chromosome represent the positions of the FHY3 binding sites from two biological replicates. The scale at the bottom indicates chromosome positions. (B) FHY3 binding sites were found in the promoter region of three known FHY3 target genes FHY1, CCA1, and ELF4. FLAG-FHY3 peaks (purple and orange) from two biological replicates and gene structure are shown in the top, middle, and bottom rows, respectively. (C) FHY3 binding sites are highly enriched in the regions around the TSS. (D) The typical FBS motif (CACGCGC, E-value = 7.4e-244) was identified as a statistically significant motif in the FHY3 binding regions in flower. (E) ChIP-PCR to verify the colocalization of FHY3 and indicated TFs. FLAG-FHY3 and TFs peaks, gene structures and the regions examined by ChIP (marked by black lines) are shown (Left). ChIP to measure FHY3 occupancy at loci in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences (Right). SEP2P2 served as a negative control. Error bars represent SD from three biological replicates. **P < 0.01 compared with no antibody (negative control). (F) Scatterplots of gene expression data from two replicates for each sample. Expression level was normalized to reads per million (RPM). Spearman correlation coefficients were calculated as an indicator of reproducibility between replicates. (G) Number of DEGs identified in pairwise comparisons of RNA-seq data. (H and I) Significantly enriched GO terms in the down-regulated genes (H) and up-regulated genes (I) in fhy3-68 vs. Ler (genes from G).
Fig. 2.
Genome-wide identification of FHY3 binding sites and target genes. (A) Classification of FHY3 binding sites in the Arabidopsis genome. The numbers of binding sites are indicated in parentheses. (B) The binding motifs of several TFs were significantly enriched around the FHY3 binding peaks compared with randomly selected genomic regions. The numbers on the top of columns are z-scores computed from the permutation test. A z-score of 2 or above is considered statistically significant. (C) Venn diagram showing the number and overlap of FHY3-associated genes in flower and seedling under D and FR conditions. (D and E) The FHY3 ChIP-seq data and RNA-seq data were compared to identify FHY3 target genes (D) and flower-specific FHY3 target genes (E). (F) Enrichment of GO terms among flower-specific FHY3 target genes. BP, biological process; MF, molecular function.
Further analysis revealed that more than 42% of the FBSs occurred in promoter and TSS regions of annotated Arabidopsis genes, with the peak regions located at −2,000 to +200 bp from the TSS, confirming the role of FHY3 as a TF (Fig. S3C). We next searched for significantly enriched motifs in FBSs using the Multiple Em for Motif Elicitation (MEME) program, and the FBS motif (CACGCGC) (E-value = 7.4e-244) was identified (Fig. S3D). One or more FBS motifs were found in 47% of the identified FBSs, indicating that FHY3 may bind other motifs or coordinately regulate target genes with other factors (24). To find potential FHY3 cofactors, we investigated whether the binding motifs of other known plant TFs were present in FBSs. Interestingly, the binding motifs of three flower-specific MADS-domain TFs, PISTILLATA (PI), SEP3, and APETALA3 (AP3), were significantly enriched in FBSs (Fig. 2B). Moreover, the binding motifs of PHYTOCHROME INTERACTING FACTOR 3, -4, and -5 (PIF3, -4, and -5) and PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 (PIL5), a group of basic helix–loop–helix (bHLH) TFs that mediate light signal in photomorphogenesis, were also enriched around the FHY3 binding peaks (Fig. 2B). ChIP-qPCR assays showed that the promoters of 10 randomly chosen target genes were indeed bound by FHY3 and one of the aforementioned TFs (Fig. S3E), confirming that these TFs are cofactors of FHY3. These results suggest that FHY3 functions synergistically with flower-specific MADS-domain TFs and bHLH TFs to regulate flower development. Using ChIP-seq data from a previous study (21), we identified 1,630 and 1,057 FHY3-associated genes in seedlings under dark (D) and FR conditions, respectively. A comparison of FHY3-associated genes under three tissues/conditions (D, FR, and flower) uncovered 687 genes that were commonly bound by FHY3 under three tissues/conditions and 568 genes (38% of total genes) that were bound by FHY3 specifically in floral organs (Fig. 2C).
Identification of FHY3 Target Genes in Floral Organs.
To identify FHY3 target genes in floral organs, we compared the gene-expression profiles in the inflorescences containing stage 8 and younger flowers from Ler, ag-10, fhy3-68, and fhy3-68 ag-10 using RNA-seq. For each genotype, the expression profiles of two biological replicates were highly correlated with each other (Fig. S3F), indicating that our RNA-seq data were highly reproducible. Using edgeR, we identified differentially expressed genes (DEGs) for each pairwise comparison using false-discovery rate (FDR) < 0.05 as significance cut-off (Fig. S3G and Dataset S2). Compared with Ler, a very small number of DEGs (29 up-regulated and 3 down-regulated genes) were found in the ag-10 sample, consistent with its weak FM determinacy defects. Among the DEGs in fhy3-68 vs. Ler, 78% (1,442 of 1,841 DEGs) were up-regulated (Fig. S3G and Dataset S2), indicating that FHY3 plays a repressive role in flower development.
Comparing the results of the ChIP-seq and RNA-seq analyses defined 238 FHY3 target genes that were bound directly and transcriptionally regulated by FHY3 in flower development (Fig. 2D and Dataset S3), of which 58% (138 genes) were up-regulated and 42% (100 genes) were down-regulated in fhy3-68 (Fig. 2D). Based on the specific binding sites in flower (Fig. 2C), we identified 52 flower-specific FHY3 target genes, of which 63% (33 genes) were up-regulated in fhy3-68, once again suggesting a repressive role of FHY3 in flower development (Fig. 2E and Dataset S3). Gene Ontology (GO) analysis of the DEGs between fhy3-68 vs. Ler revealed that GO terms related to cell cycle, DNA metabolism, and cell division were enriched in FHY3 up-regulated genes, and GO terms related to defense or immune response, cell death, and hormone signaling were enriched in FHY3 down-regulated genes (Fig. S3 H and I), indicating that FHY3 functions in cell proliferation and environmental and hormone response. Notably, among the flower-specific FHY3 target genes, genes assigned to the terms “transcription factor activity,” “transcription regulator activity,” and “DNA binding” in the “molecular function” category were highly enriched (Fig. 2F), suggesting that these genes may function as early target genes of FHY3 in flower development.
SEP1 and SEP2 Are FHY3 Target Genes.
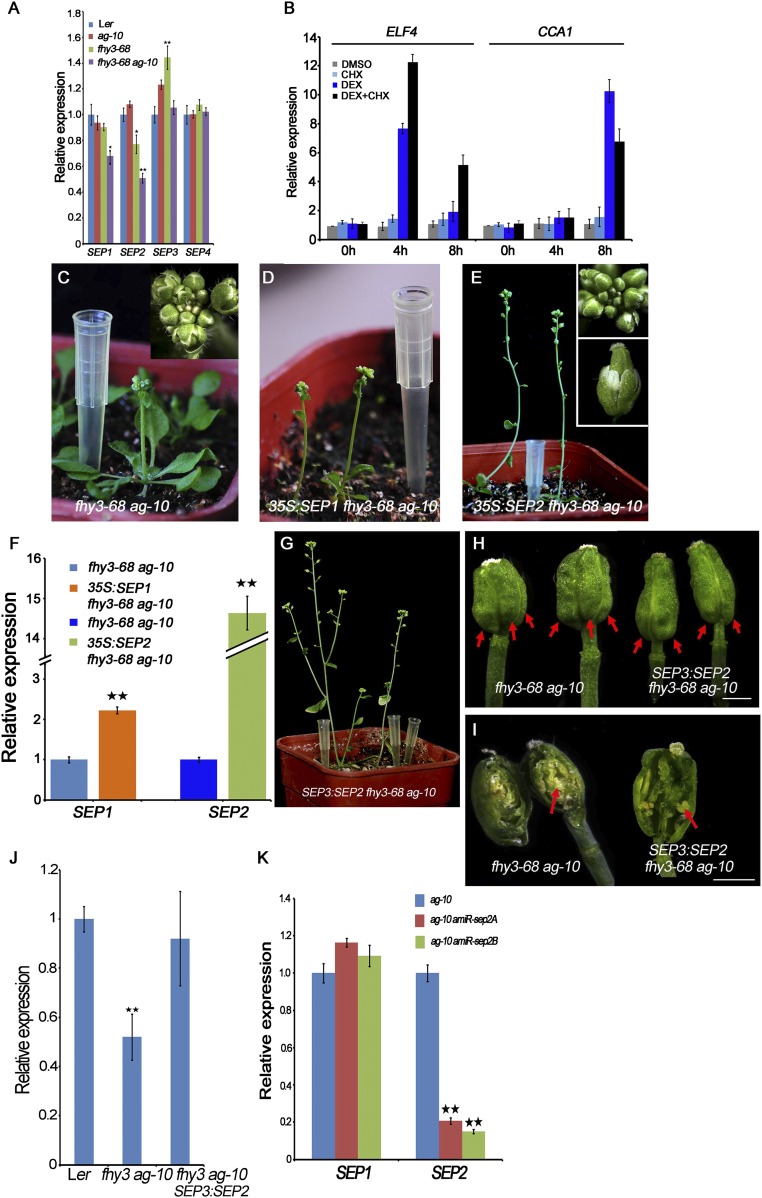
SEP genes have fundamental roles in floral organ identity and FM determinacy. Our genome-wide analyses revealed that SEP1 and SEP2 were putative target genes of FHY3 (Dataset S3). Real-time RT-PCR analysis showed that SEP genes are differentially regulated by FHY3. SEP1 and SEP2 were down-regulated in fhy3-68 ag-10 inflorescences and SEP3 transcripts increased in fhy3-68 (Fig. S4A). The Integrative Genomics Viewer (IGV) showed that FHY3-FLAG peaks were located at the promoter regions of SEP1 and SEP2, respectively (Fig. 3A). We conducted ChIP-qPCR to confirm the occupancy of FHY3 at SEP1 and SEP2 in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences (21). Significant enrichment of FHY3 at the TSS regions but not the intragenic regions of SEP1 and SEP2 (Fig. 3B) demonstrated the direct and specific binding of FHY3 at the promoters of SEP1 and SEP2. We also asked whether FHY3 directly regulates SEP1 and SEP2. In FHY3:FHY3-GR fhy3-4 transgenic plants treated with DEX or DMSO, SEP1 and SEP2 were induced after DEX or DEX/cycloheximide (CHX; a protein synthesis inhibitor) treatment but not DMSO or CHX (Fig. 3C), indicating that FHY3 induces SEP1 and SEP2 expression independent of new protein synthesis (Fig. 3C and Fig. S4B). Collectively, these findings show that SEP1 and SEP2 are FHY3 direct target genes in floral organs.
Fig. S4.
SEP2 mediates the function of FHY3 in FM determinacy. (A) The transcript levels of SEPs in the indicated plants measured by RT-PCR. Inflorescences containing stage 8 and younger flowers were used. (B) The transcript levels of ELF4 and CCA1 in FHY3:FHY3-GR fhy3-4 inflorescences measured by RT-qPCR served as a positive control of Fig. 3C. (C–E) Plants and inflorescences (Inset) of fhy3-68 ag-10 (C), 35S:SEP1 fhy3-68 ag-10 (D), and 35S:SEP2 fhy3-68 ag-10 (E). (F) The SEP1 and SEP2 transcript levels in transgenic plants measured by real-time RT-PCR. (G) Plant of SEP3:SEP2 fhy3-68 ag-10 transgenic plant. The plant developed normally at the vegetative stage. SEP3:SEP2 transgene mainly rescued the FM determinacy defects of fhy3-68 ag-10. However, the smaller inflorescence, shorter petal and sterile anther phenotypes were not rescued. (H) Siliques from plants of the indicated genotypes. The siliques of SEP3:SEP2 fhy3-68 ag-10 were composed of two carpels and were thinner than those of fhy3-68 ag-10. Carpels were indicated by red arrows. (I) Representative sliced-open siliques of indicated plants. Removing the primary carpels from fhy3-68 ag-10 siliques revealed additional floral organs growing inside (red arrow). No more layered carpeliod organs except of stamenoid organs (red arrow) grew inside of the silique of SEP3:SEP2 fhy3-68 ag-10 as in fhy3-68 ag-10, indicating that SEP3:SEP2 transgene mainly rescued the FM indeterminacy of fhy3-68 ag-10. (J) The transcript levels of SEP2 in indicated plants measured by real-time RT-PCR. (K) The transcript levels of SEP1 and SEP2 in amiR-sep2 transgenic plants measured by real-time RT_PCR. In A, B, F, G, and K UBQ5 served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. *P < 0.05 and **P < 0.01. (Scale bars in H and I, 1 mm.)
Fig. 3.
SEP2 mediates the function of FHY3 in FM determinacy. (A) The FHY3-FLAG ChIP-seq peaks (two biological replicates) at SEP1 and SEP2 revealed in IGV. FLAG-FHY3 peaks (purple and orange), gene structure, and the regions examined by ChIP are shown in the top, middle, and bottom rows, respectively. (Scale bars, 500 bp.) (B) ChIP to measure FHY3 occupancy at SEP1 and SEP2 in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences. The regions examined are shown in A. eIF4A1 served as a negative control. Error bars represent SD from three biological replicates. **P < 0.01 compared with no antibody (negative control). (C) The transcript levels of SEP1 and SEP2 in FHY3:FHY3-GR fhy3-4 inflorescences measured by RT-qPCR. Ubiquitin 5 (UBQ5) served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. **P < 0.01. (D and E) Siliques from plants of the indicated genotypes. Carpels were indicated by red arrows; Sliced open siliques were indicated by white arrows. (Scale bars, 1 mm.)
SEP2-Mediated FHY3 Functions in FM Determinacy.
To investigate whether the FHY3 involvement in FM determinacy is mediated by SEP1 and SEP2, SEP1 and SEP2 overexpression constructs under the CaMV35S promoter were generated as previously described (14) and then transformed into an ag-10 fhy3-68/+ population. Compared with fhy3-68 ag-10 plants, all of the 35S:SEP1 fhy3-68 ag-10 and 35S:SEP2 fhy3-68 ag-10 transgenic plants exhibited an early flowering phenotype (Fig. S4 C–E). Real-time RT-PCR confirmed that SEP1 and SEP2 were overexpressed in the transgenic plants (Fig. S4F). Although SEP1 overexpression in 35S:SEP1 fhy3-68 ag-10 plants (n = 12) failed to rescue the FM determinacy defects (Fig. S4D), the siliques of 35S:SEP2 fhy3-68 ag-10 transgenic plants (n = 8) were composed of two carpels with normal gynophores (Fig. 3D and Fig. S4E). Moreover, sliced open siliques showed no more layered carpeloid organs growing inside (compare the Inset in Fig. 3D to Fig. S1B), indicating that SEP2 overexpression rescued the FM determinacy defects of fhy3-68 ag-10. However, it failed to rescue the small inflorescence, short petal, and sterile anther phenotypes of fhy3-68 ag-10 (Fig. S4E). These findings indicate that SEP2 only mediates the function of FHY3 in FM determinacy.
To avoid the pleiotropic phenotypes resulted from constitutive overexpression of SEP2, we generated a SEP3:SEP2 fhy3-68 ag-10 transgene because SEP3 expression was flower-specific and unchanged in fhy3-68 ag-10 (Fig. S4A). Although they showed normal development at the vegetative stage, the transgenic plants also produced short siliques because of the sterile anther (Fig. S4 G and H). The siliques were composed of 2.2 ± 0.3 carpels (n = 30) (Fig. S4H) and sliced-open siliques contained fewer additional organs growing inside than those of fhy3-68 ag-10 (Fig. S4I), indicating that the SEP3:SEP2 transgene could mainly rescue the FM indeterminacy of fhy3-68 ag-10. Real-time PCR analysis showed that the expression of SEP2 reached normal level in the transgenic plants as in Ler (Fig. S4J), indicating that other factors may also mediate the function of FHY3 in FM determinacy besides SEP2.
To further confirm the role of SEP2 in FM determinacy, two artificial miRNAs (amiRNAs) targeting SEP2 (amiR-sep2A and amiR-sep2B) were generated and introduced into the ag-10. All amiR-sep2A ag-10 plants (n = 15) and a small percentage of amiR-sep2B ag-10 plants (8 of 52) produced more bulged siliques with additional organs growing inside, similar to the phenotype of fhy3-68 ag-10 (Fig. 3E). Thus, reduced SEP2 expression enhanced the FM indeterminacy of ag-10. qPCR analysis confirmed the reduced expression of SEP2 but not SEP1 in the amiR-sep2 ag-10 lines (Fig. S4K). Taken together, these findings show that SEP2 mediates the functions of FHY3 in FM determinacy.
FHY3 Contributes to SAM and FM Regulation by Directly Repressing CLV3.
Besides the FM determinacy defects, we found the SAM maintenance was impaired in the fhy3-68 mutant (Fig. 1 J and K). It is well established that CLV3 encodes a secreted peptide that restricts the domain of WUS expression and the size of the stem cell domain (5). We therefore examined the genetic relationship between FHY3 and CLV3. Like clv3-1, fhy3-68 clv3-1 produced a larger SAM than Ler and fhy3-68 (Fig. S5 A–C), indicating that clv3 is epistatic to fhy3 in SAM maintenance. Consistent with these observations, qPCR revealed increased CLV3 transcript levels in fhy3-68 and fhy3-68 ag-10, compared with Ler and ag-10, respectively (Fig. S5D). In situ hybridization was then used to detect CLV3 and WUS expression in Ler and fhy3-68. Whereas CLV3 expression increased and expanded in the SAM of fhy3-68 (Fig. 4 A and B), WUS expression decreased in the SAM of fhy3-68 compared with Ler but remained unchanged in the early stage of FM (Fig. 4 C and D). These findings indicate that CLV3 mediates FHY3 function in the SAM by regulating WUS expression to balance the stem cell pool and SAM size.
Fig. S5.
Genetic and expression analysis of CLV3 and FHY3. (A and B) SAM (marked by a red arrow) of clv3-1 (A) and fhy3-68 clv3-1 (B). (Scale bars, 60 µm.) (C) SAM size of the indicated plants. The inflorescences of 21-d-old plants were measured for Ler (n = 20), fhy3-68 (n = 20), clv3-1 (n = 20), and fhy3-68 ag-10 (n = 12). *P < 0.5 and **P < 0.01. (D) The CLV3 expression in the indicated plants measured by real-time RT-PCR. (E) The CLV3 transcript level examined by RT-qPCR. (F) Number of unopened buds of the indicated plant inflorescences (n = 22). **P < 0.01. (G) Gene expression of cell cycle gene (CYCB1;3) and DNA replication genes (MCM10, RPA70C, and RPA70D) in Ler and fhy3-68. In D, E, and G UBQ5 served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. **P < 0.01.
Fig. 4.
CLV3 mediates FHY3 functions in regulating the stem cell pool in the SAM and FM meristem activity. (A–D) In situ hybridization to examine the expression of CLV3 (A and B) and WUS (C and D) in Ler (A and C) and fhy3-68 (B and D). CLV3 signals are marked by a black arrow in A and B. WUS signals are marked by a black arrow in SAM and a red arrow in FM in C and D. (E and F) Flowers of clv3-1 (E) and fhy3-68 clv3-1 (F). Dome-shaped meristem is marked by a red arrow. (G) Representative siliques of clv3-1 (Left) and 35S:FHY3-FLAG ag-10 (Right) plants. Carpels are marked by red arrows. (H) ChIP to measure FHY3 occupancy at CLV3 in 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences. The regions examined are shown on the Upper panel. CLV3 gene structure was shown. (Scale bar, 500 bp.) eIF4A1 served as a negative control. Error bars represent SD from three biological replicates. **P < 0.01 compared with no antibody (negative control). (I) The CLV3 transcript levels in FHY3:FHY3-GR fhy3-4 inflorescences measured by RT-qPCR. (J and K) Inflorescence of Ler (J) and 35S:FHY3-FLAG (K). 35S:FHY3-FLAG developed a larger inflorescence containing more unopened buds than Ler. (L) The CLV3 transcript levels in seedlings of indicated plants after light treatment measured by RT-qPCR. In I and L, UBQ5 served as the internal control. Three biological replicates were performed. Error bars represent SD from three biological repeats. **P < 0.01. (Scale bars: 50 µm in A–D and 500 µm in E–G, J, and K.)
We next examined the genetic relationship of FHY3 and CLV3 in FM determinacy. In the clv3-1 mutant, the number of floral organs, particularly the stamen number (7.7 ± 0.5; n = 50) and carpel number (4.8 ± 0.6; n = 50), increased (Fig. 4E). The FM indeterminacy of fhy3-68 clv3-1 was more severe than that of clv3-1, with increased stamen number (8.5 ± 0.5; n = 20) and unfused carpels with a dome-shaped meristem growing inside (Fig. 4F). The enhanced FM indeterminacy of fhy3-68 clv3-1 was consistent with the finding that FHY3 acts in CLV3 pathway besides the SEP2 pathway to regulate WUS expression in FM determinacy. Accordingly, we found that 16% (n = 50) siliques of 35S:FHY3-FLAG ag-10 plants were composed of three or more fused carpels, similar to those of weak clv3 mutants (Fig. 4G). qPCR revealed decreased CLV3 transcript levels in the inflorescence of 35S:FHY3-FLAG ag-10 compared with ag-10 (Fig. S5E).
We performed additional ChIP-qPCR to investigate whether CLV3 is an FHY3 direct target gene. The high occupancy of FHY3 in the upstream region of the CLV3 TSS instead of other tested regions suggested that FHY3 binds specifically to the CLV3 promoter (Fig. 4H). We then examined the CLV3 expression in FHY3:FHY3-GR fhy3-4 plants treated with DEX and DEX/CHX or DMSO and CHX (control). RT-qPCR revealed severely attenuated CLV3 transcript levels in the DEX- or DEX/CHX-treated plants within 4 h of treatment (Fig. 4I), indicating that FHY3 directly represses CLV3. Correspondingly, the 35S:FHY3-FLAG plants developed larger inflorescences that contained more unopened buds than Ler (Fig. 4 J and K and Fig. S5F). Taken together, these results suggest that FHY3 functions in FM determinacy and SAM maintenance by directly repressing CLV3.
FHY3 Mediates the Light-Repressed CLV3 Expression.
A recent report showed that light regulates meristem activity by activating cytokinin signaling and repressing CLV3 expression (18). To investigate whether FHY3 is involved in light-regulated meristem activity, we grew seedlings 4 d in the dark after germination, followed by 12-h light treatment and examined the CLV3 expression in diverse genotypes. As expected, CLV3 expression was repressed by light in the wild-type plants (Fig. 4L). However, CLV3 expression did not change significantly in fhy3-68 before and after light treatment (Fig. 4L), indicating that FHY3 is essential for light-regulated expression of CLV3 in SAM. Unexpectedly and interestingly, light-repressed CLV3 expression was observed in the phyA mutant, but not in phyB and phyAphyB mutants (Fig. 4L), indicating that phyB, but not phyA, may be involved in the light-regulated meristem activity. The interaction of FHY3 and phyB in light-regulated meristem activity is an open and interesting question.
Discussion
Plant meristem maintenance and determinacy are critical for plant growth, life cycle, and crop yield. Most studies have been focused on the cross-talk between phytohormones, such as auxin and cytokinin, and gene expression in these processes (25, 26). Little is known whether, and if so how, external signals like light and temperature contribute to meristem regulation. FHY3 is known to play pivotal roles in the phyA signaling and the circadian clock pathways, and other developmental and physiological processes at the vegetative stage (22), but its roles in the reproductive stage remain unclear. Our findings describe the previously unknown functions of FHY3 in SAM maintenance, FM determinacy, as well as petal and stamen development during flower development (Figs. 1 and 3F). Previous studies have shown that most of FHY3 target genes (99% in FR or 78% in D) were activated, indicating that FHY3 mainly acts as a transcriptional activator in seedlings (21). In this study, we identified hundreds flower-specific FHY3-associated genes (Fig. 2C), suggesting that FHY3 has distinctive roles in flower development. Of the DEGs identified in fhy3-68 vs. Ler, 78% were up-regulated, indicating that FHY3 mainly functions as a transcriptional repressor in flower development (Fig. S3G). Notably, the expressions of MYB77 in ethylene signaling, BTB AND TAZ DOMAIN PROTEIN4 (BT4) in gibberellin signaling, and MYBR1 in ABA, auxin, and ethylene signaling were activated in seedlings but repressed in flowers by FHY3 (Dataset S3) (21). The enrichment of genes related to cell cycle, DNA metabolic process, and DNA replication genes among the FHY3-activated genes (Fig. S3H), and the clustering of FHY3 intergenic binding sites in centromeric regions (Fig. S3A), raise the possibility that FHY3 may also function in cell division and epigenetic regulation. These findings underscore how the same TF may have vastly different roles in different organs and developmental stages.
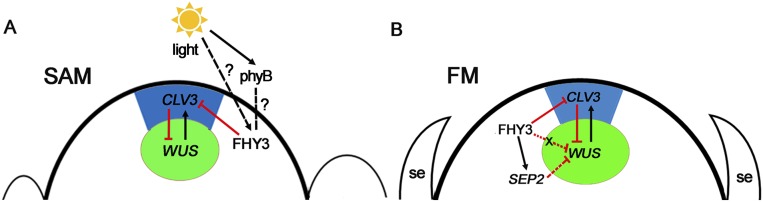
Certain cis-elements, such as the G-box and GCC-box that are bound by bHLH TFs (like PIL5 and PIF3) and bZIP TFs, are enriched around the FHY3 binding sites in the vegetative stage (21). Here, we show that the binding motifs of bHLH TFs PIF3, -4, -5, and PIL5, which mediate light signal in photomorphogenesis, were significantly enriched around the FHY3 binding sites (Fig. 2B and Fig. S3E), indicating that FHY3- and PIFs-mediated light signaling may function in flower development. Furthermore, the binding motifs of AP3, PI, and SEP3 were also enriched in the FHY3 binding regions (Fig. 2B and Fig. S3E). Given that AP3 and PI, B-class MADS-domain TFs, form a protein complex with SEP3 to function in sepal and stamen development (27), our results suggest that FHY3 may coregulate sepal and stamen development with AP3, PI, and SEP3, consistent with the sepal and stamen developmental defects of fhy3 ag-10 (Fig. 1D). Remarkably, we find that FHY3 and phyB are involved in light-repressed CLV3 expression (Fig. 4L). The relationship between FHY3 and pyhB in this process awaits further investigation (Fig. S6A). Combined with recent reports that light could regulate stem cell activity (18), and that two key meristem regulatory genes, BASIC PENTACYSTEINE 3 (BPC3) and AG were uniquely bound by light signal transducer FHY1 and phyA, respectively (28), our results highlight the possibility that FHY3 may act as a bridge molecule in the cross-talk between endogenous cues and external signals to coordinate plant development.
Fig. S6.
Proposed model of FHY3 functions in meristem activity regulation. (A) FHY3 directly represses CLV3 and subsequently alters the WUS/CLV3 regulatory loop to maintain the stem cell pool in the SAM. Both FHY3 and phyB are involved in the light-regulated expression of CLV3, but the exact mechanisms need further investigation. (B) In FM, FHY3 directly represses CLV3 but also directly activates SEP2 to regulate WUS expression to promote FM determinacy. Besides SEP2, other factors (X) may also mediate the function of FHY3 in FM determinacy. se: sepal.
The CLV3/WUS feedback loop was well characterized in meristem maintenance (5). However, more detailed mechanisms in this process are unclear. It was recently reported that HECATE1 (HEC1) is repressed by WUS and in turn represses CLV3 to fine-tune the balance of stem cell proliferation (29). The present findings add FHY3 as a new player in this regulation system. We propose that FHY3 indirectly regulates WUS expression and stem cell pool maintenance through a direct target gene, CLV3 (Fig. S6A). How FHY3 coordinates with HEC1 in terms of stem cell pool maintenance is an open question. Meanwhile, we noticed that the SAM size of fhy3-68 was small but the SAM size of fhy3-68 clv3-1 was larger than that of clv3-1 (Fig. S5 A and B). Therefore, the possibility that FHY3 directly promotes cell proliferation in the SAM besides the CLV3 pathway cannot be ruled out because RNA-seq and GO analysis showed that FHY3-induced genes were enriched for “cell cycle” and “DNA replication” genes (Figs. S3H and S5G).
The small SAM size of fhy3 as a result of the de-repressed CLV3 expression and the reduced WUS expression was paradoxical to the severe FM indeterminacy of fhy3 ag-10 with prolonged WUS expression, which indicates that FHY3 may act through a genetic pathway parallel to the CLV3 pathway in the FM determinacy because the temporal expression of WUS is more important than its relative expression level for FM activity regulation (30). Subsequently, this hypothesis was reinforced by the identification of SEP1 and SEP2 as FHY3 target genes. Overexpression of SEP2 but not SEP1 rescued the FM indeterminacy of fhy3-68 ag-10, and reduced SEP2 expression in ag-10 resulted in FM indeterminacy similar to those of fhy3-68 ag-10 (Fig. 3E and Fig. S4E). It nevertheless remains possible that SEP1 coordinates SEP2 in promoting FM determinacy because the SEP3:SEP2 transgene could only mainly rescue FM indeterminacy of fhy3-68 ag-10. Therefore, it is highly possible that other factors may mediate the function of FHY3 in FM determinacy (Fig. S6B). Collectively, the present findings reveal the dual roles of FHY3 in regulating meristem activity (Fig. S6), and therefore give insights into the mechanisms of SAM maintenance and FM determinacy.
Materials and Methods
All plants were grown in soil and maintained in a greenhouse at 23 °C under LD conditions (16 h of light/8 h of dark). Standard genetic and molecular biology techniques were used for crossing and for the construction of plasmids. Quantitative real-time PCR, in situ hybridization, and ChIP were performed as previously described (9). The primers used for genotyping and construction are listed in Table S1. Details are provided in SI Materials and Methods.
Table S1.
List of primers used in this study
| Primer name | Primer sequence |
| Primers for ChIP | |
| SEP1p1F | 5′-GAGAGCGTGGGAATGAAAGTAAA-3′ |
| SEP1p1R | 5′-AAGAAATATGTGAGGGTGAGGTTAGG-3′ |
| SEP1p2F | 5′-CCTCCTCCCATATGATGACTTCTC-3′ |
| SEP1p2R | 5′-CATCTCTAGTTTTCTTGTTTCTGATTATAGTCT-3′ |
| SEP2p1F | 5′-GGGAGAGAAGGTGTGAGAATGTTT-3′ |
| SEP2p1R | 5′-AGAGATCATTAGGGTTTGGTGATACA-3′ |
| SEP2p2F | 5′-GCCGATCATATCTTCCAGCTAAA-3′ |
| SEP2p2R | 5′-TTCACATTCTTCAACTGCTACATGTT-3′ |
| ELF4F | 5′-CGGACACCGAGGCGAGTA-3′ |
| ELF4R | 5′-ACGCGCGTGTTCTTTGATATC-3′ |
| WUSp1F | 5′-AGATCCGATCACTGAAGTTGCTT-3′ |
| WUSp1R | 5′-TTTGCACGTTGTGTTGAATCC-3′ |
| WUSp2F | 5′-GCACATTTTTCAATAGGGTTT-3′ |
| WUSp2R | 5′-AGGTTTTGTGTGAGAGAGAAGA-3′ |
| WUSp3F | 5′-TTGTAATGGTTTGCTATTGTAC-3′ |
| WUSp3R | 5′-GTTGATTTTGGTCGTTATCC-3′ |
| CLV3p1F | 5′-CCCCTTCTCATTTCATTACCA-3′ |
| CLV3p1R | 5′-GCAGAAAACTCTTCGAATCCA-3′ |
| CLV3p2F | 5′-ATTAGAGTATGTGCCGGTGCC-3′ |
| CLV3p2R | 5′-TGCTGTGGAGGTTCACAACTAA-3′ |
| CLV3p3F | 5′-CAAGGCTCATATAATCCATTCA-3′ |
| CLV3p3R | 5′-CAATATGGATGATACCTTAATCGG-3′ |
| CLV3p4F | 5′-AAGACCTAATAAATTCGTGGACTTG-3′ |
| CLV3p4R | 5′-GAACAATTTTAAAGACATTCGGACC-3′ |
| AT3G01520 F | 5′-GCGAAGGGTTCTTTGTCTTCT-3′ |
| AT3G01520 R | 5′-CACCATCACTTTTGTTGGCTC-3′ |
| AT3G03450 F | 5′-CAAACAATAGTGGCATGCTCTG-3′ |
| AT3G03450 R | 5′-AAAGGAGATTCTGAGAGGAGCA-3′ |
| AT5G47800 F | 5′-CATCGACCCACGTTGACTACT-3′ |
| AT5G47800 R | 5′-TGTTTATCCCCACGAATCTCA-3′ |
| AT4G02630 F | 5′-ATGATGAGTGCAAGTCCCATC-3′ |
| AT4G02630 R | 5′-TTGCTTTTGTTCTGCTTCACA-3′ |
| AT5G23940 F | 5′-GGGAAGTAGAGTCCTTGAAATCC-3′ |
| AT5G23940 R | 5′-GGATTTGATGAAGTTGTTGGG-3′ |
| AT3G15820 F | 5′-AAAGGTAAAGCCCACCAAAGA-3′ |
| AT3G15820 R | 5′-ATACAAGGCGGAGATTTCCAA-3′ |
| AT5G51430 F | 5′-ACTGCGTGGTCTCCAAGTAAA-3′ |
| AT5G51430 R | 5′-AACCAAACCAAAGTAAACGGAA-3′ |
| AT4G03330 F | 5′-CATGGTACCAACCAAATCACA-3′ |
| AT4G03330 R | 5′-TTAAGAGCCGTCTGAAACCAA-3′ |
| AT5G19875 F | 5′-AACACCCATTCAAAGCATACTC-3′ |
| AT5G19875 R | 5′-CCTTCTTCTTGGTCTTCTCTGG-3′ |
| AT5G67200 F | 5′-TCACAGCTGCTTCCCATATTT-3′ |
| AT5G67200 R | 5′-GGAGAATGAAGAAGAAGAAGAAGG-3′ |
| eIF4A1F | 5′-GCTGAGTTGGGAGATCGAAGTT-3′ |
| eIF4A1R | 5′–TCTGGTTCCTCCCGTGTTCT-3′ |
| Primers for real-time PCR | |
| SEP1qPCRF | 5′-CAAGCAAGTTCGGTCCATCA-3′ |
| SEP1qPCRR | 5′-TGCCAAAGCTCTATTGGTTTCA-3′ |
| SEP2qPCRF | 5′-TGCAAGGTCAGTCCCAACAA-3′ |
| SEP2qPCRR | 5′-CCCCACCAGTACTTGCTTAAGATC-3′ |
| SEP3qPCRF | 5′-TGACCAGCTCAACGATCTTCA-3′ |
| SEP3qPCRR | 5′-GCTGGAGTGGCATCTGATACC-3′ |
| SEP4qPCRF | 5′-TGTCCTTTGTGATGCTGAGATTG-3′ |
| SEP4qPCRR | 5′-ATCAACCGTCCTCGCCATAC-3′ |
| FHY3qPCRF | 5′-TTTTGAAAACAACCAAGACCCAATGGTAACAT-3′ |
| FHY3qPCRR | 5′-GCGAGGGGATGGAAGAAAGATGAC-3′ |
| CLV3qPCRF | 5′-ATGGAGAAGCAGAGAAGGCAAA-3′ |
| CLV3qPCRR | 5′-GGGTTCACATGATGGTGCAA-3′ |
| FHY1qPCRF | 5′-GATGAAAGAGGAATCATCTGGA-3′ |
| FHY1qPCRR | 5′-AATCCTCTAAGTTCTGAGTCCCA-3′ |
| AGqPCRF | 5′-CTCAGGAACTTGGAAGGCAGAT-3′ |
| AGqPCRR | 5′-CTCTTTTCTGCATGTAGTCGATTTCA-3′ |
| WUSqPCRF | 5′-GCATCGCCACCACATTCTT-3′ |
| WUSqPCRR | 5′-TGGGCAAACATGGATCATCA-3′ |
| CYCB1.3 F | 5′-TAGCTGAGTTGGGTTTGATGC-3′ |
| CYCB1.3 R | 5′-GCCTGTGTGGAATTTCAGTGT-3′ |
| MCM10 F | 5′-CCAGACAAAAGGCATCAAAGA-3′ |
| MCM10 R | 5′-GGAGTTTCCTTCCTGTGCTCT-3′ |
| RPA70C F | 5′-GATTACCTGCAGCAAACCAAG-3′ |
| RPA70C R | 5′-ACTCATAAGGGTTGGGCAGTT-3′ |
| RPA70D F | 5′-TGGATTTCGTCATTCAACGAT-3′ |
| RPA70D R | 5′-TAACCCGGAAGACATGAGATG-3′ |
| N_UBQ5 | 5′-GGTGCTAAGAAGAGGAAGAAG-3′ |
| C_UBQ5 | 5′-CTCCTTCTTTCTGGTAAACGT-3′ |
| Primers for plasmid construction | |
| FHY3cdsF | 5′-CACCATGGATATAGATCTTCGACTAC-3′ |
| FHY3cdsR | 5′-CGAGTGTCTAGACGCGTCCTCATGC-3′ |
| SEP2cdsF | 5′-CACCATGGGAAGAGGAAGAGTAGAGCTC-3′ |
| SEP2cdsR | 5′-TCACAGCATCCAGCCAGGGATGTAG-3′ |
| SEP1cdsF | 5′-CACCATGGGAAGAGGAAGAGTAGAGCTG-3′ |
| SEP1cdsR | 5′-TCAGAGCATCCACCCCGGGATGT-3′ |
| SEP2cds2F | 5′-CGGATCCATGGGAAGAGGAAGAGTAGAGCTC-3′ |
| SEP2cds2R | 5′-CCGCTCGAGTCACAGCATCCAGCCAGGGATGTAG-3′ |
| SEP3proF | 5′-GCACATCATCTCGATCAATTGTC-3′ |
| SEP3proR | 5′-CCTCTCCAGTGTCCGAAGCAT-3′ |
| amiR-sep2AI | 5′-gaTTCTAGTTGACGCTCAAGCTAtctctcttttgtattcc-3′ |
| amiR-sep2AII | 5′-gaTAGCTTGAGCGTCAACTAGAAtcaaagagaatcaatga-3′ |
| amiR-sep2AIII | 5′-gaTAACTTGAGCGTCTACTAGATtcacaggtcgtgatatg-3′ |
| amiR-sep2AIV | 5′-gaATCTAGTAGACGCTCAAGTTAtctacatatatattcct-3′ |
| amiR-sep2BI | 5′-gaTTATACTGTGTCTTGATGCAGtctctcttttgtattcc-3′ |
| amiR-sep2BII | 5′-gaCTGCATCAAGACACAGTATAAtcaaagagaatcaatga-3′ |
| amiR-sep2BIII | 5′-gaCTACATCAAGACAGAGTATATtcacaggtcgtgatatg-3′ |
| amiR-sep2BIV | 5′-gaATATACTCTGTCTTGATGTAGtctacatatatattcct-3′ |
| Primers for mutant genotyping | |
| ag-10gtF | 5′-CAATGTCTCCCAAAAGAGCCCAGGAACTT-3′ |
| ag-10gtF | 5′-GCAACAAGGCATATAGATTTAATTTG-3′ |
| fhy3-68F | 5′-TTTTGAAAACAACCAAGACCCAATGGTAACAT-3′ |
| fhy3-68R | 5′-CCGCAGCAGCCTAGTCTGCAACTG-3′ |
| wus-1gtF | 5′-AGTAGCCATGTCTATGGATCCATG-3′ |
| wus-1gtR | 5′-CCTCCACCTACGTTGTTGTAATTC-3′ |
| clv3-1gtF | 5′-TGATGAAAATGGAAAGTGAA-3′ |
| clv3-1gtR | 5′-AAAACCGATAAAGCAGAAAG-3′ |
| fhy3-4gtF | 5′-GAAGCCAAAGCTGATCCAGAAATGT-3′ |
| fhy3-4gtR | 5′-GTGGCATCTCGGTTCTCC-3′ |
SI Materials and Methods
Plant Materials and Growth Conditions.
The Arabidopsis thaliana ecotype Landsberg erecta (Ler) was used as wild-type in this study. The ag-10 (9), fhy3-4 (19), wus-1 (2), and clv3-1 (31) mutants and the FHY3p:FHY3-GR fhy3-4 and 35S:3FLAG-FHY3-3HA fhy3-4 transgenic lines were described previously (5). fhy3-68 was isolated in this study from a genetic screen in the ag-10 background. For fhy3-68 genotyping, PCR was performed with primers fhy3-68F and fhy3-68R, and XcmI digestion of the amplified fragment cut wild-type but not fhy3-68 DNA. The double- or triple-mutant combinations in this study were created by cross-pollination of the relevant mutants, and the F2 populations were genotyped to identify plants with the correct genotypes.
All plants were grown in soil and maintained in a greenhouse at 23 °C under LD conditions (16 h of light/8 h of dark). For DEX treatment, the FHY3:FHY3-GR fhy3-4 plants were treated with DMSO or DEX (10 µM) in 0.015% Silwet L-77 by applying the solution onto inflorescences. Total RNA was isolated from the inflorescences at 0, 4, and 8 h after chemical treatment.
Meristem size was measured as previously described (29). SEM imaging was performed with a Hitachi S-3000N Scanning Electron Microscope according to the manufacturer’s instructions.
EMS Mutagenesis and Mapping.
The EMS mutagenesis of ag-10 was previously reported. For map-based cloning, ag-10 fhy3-68 was crossed into ag-10Col to create the mapping population. In the F2 generation, plants with floral determinacy defects were selected for rough mapping, which revealed linkage to the marker K1G2 on chromosome 3. For fine mapping, new simple sequence length polymorphism (SSLP) and cleaved amplified polymorphic sequences (CAPS) markers were designed based on the polymorphisms between Col and Ler (www.arabidopsis.org/Cereon). The mutation was mapped to a region between BAC clones MKA23 and MMP21, and FHY3 was selected as a candidate gene for sequencing.
Plasmid Construction.
To create the 35S:FHY3-FLAG construct, FHY3 cDNA was amplified by PCR using the FHY3cdsF/FHY3cdsR primer pair. The PCR product was cloned into pENTR/D-TOPO (Invitrogen), and the resulting plasmid was sequenced to ensure the integrity of the gene. The plasmid was linearized, and the insert was recombined into modified pEarleyGate100, in which the nucleotides corresponding to the 3xFLAG epitopes (DYKDDDDKDYKDDDDKDYKDDDDK) were inserted into the AvrII and XbaI sites downstream of the attR2 recombination site, using the Gateway LR Clonase kit (Invitrogen). To create 35S:SEP1 and 35S:SEP2, SEP1 and SEP2 cDNA was amplified by PCR using the primer pairs SEP1cdsF/SEP1cdsR and SEP2cdsF/SEP2cdsR, respectively. The PCR products were cloned into pENTR/D-TOPO (Invitrogen) and recombined into pEarleyGate100, as described above. To create 35S:amiR-sep2, two independent suitable target sites for the microRNA, referred to as amiR-sep2A and amiR-sep2B, were identified according to the instructions on wmd3.weigelworld.org. The constructs were cloned following a previously described protocol (32) and recombined into pEarleyGate100.
To generate pSEP3:SEP2, SEP2 cDNA was amplified by RT-PCR using primers SEP2cds2F and SEP2cds2R. The PCR product was cloned into pENTR 1A (Invitrogen) at BamHI and XhoI sites, and then the SEP3 promoter fragment, which was PCR-amplified by SEP3proF and SEP3proR, was inserted into above vector at XmnI site. The resulting plasmid was recombined into pEarleyGate301. The primers used for genotyping and construction are listed in Table S1.
Total RNA Isolation and RT-qPCR Analysis.
Total RNA was isolated from Arabidopsis inflorescences using the RNeasy plant mini kit (Qiagen) and treated with DNaseI (Roche) to eliminate contaminating DNA. M-MLV Reverse Transcriptase (Promega) was used for reverse transcription. Quantitative real-time RT-PCR was performed in triplicate on the Biorad CFX-96 Real-Time PCR system (Bio-Rad) using the SYBR Green RT-PCR kit (DBI Bioscience).
ChIP and in Situ Hybridization.
ChIP was performed as previously described (9). 35S:3FLAG-FHY3-3HA fhy3-4 inflorescences were completely ground in liquid nitrogen and cross-linked with 1% formaldehyde (Sigma-Aldrich) for 10 min on ice. The chromatin complexes were isolated and sonicated into DNA fragments of ∼500 bp. The lysate was precleared then incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich, A2220). The bound chromatin was purified using columns from the Qiagen plasmid extraction kit. Real-time PCR was conducted on the input, no antibody control and antibody-bound DNA in triplicate. For ChIP-seq, the ChIP DNA and TruSeq ChIP Sample Preparation Kit (Illumina) were used to generate Illumina sequencing libraries according to the manufacturer’s instructions.
In situ hybridization was conducted as previously described (9). The synthesis of WUS and CLV3 antisense probes were also as previously described (9). All inflorescences came from 21-d-old plants grown under LD conditions at 23 °C.
ChIP-Seq and RNA-Seq Analysis.
The reference genome sequences and gene annotations of Ler-0 and No-0 were downloaded from the 19 genomes of the Arabidopsis thaliana project website (33) (mus.well.ox.ac.uk/19genomes). For ChIP-seq analysis, paired-end reads of FHY3 ChIP and input libraries were mapped to the No-0 genome using Bowtie2 with default parameters. A total of 21, 24, and 37 million raw reads were obtained from the two replicate ChIP and input library, respectively, 89–92% of which were uniquely mapped to the Arabidopsis genome. The ChIP-seq read depths were visualized in IGV (34). We identified the peaks by MACS (35) (P < 0.005) and merged the overlapped peaks from two replicates. The peaks were classified according to the following criteria: (i) peaks occurring within 2,000-bp upstream and 200-bp downstream of the TSS of a gene were classified as promoter region binding sites; (ii) peaks localized within a gene body were further categorized as CDS, 5′-UTR, 3′-UTR, ncRNA, or intron region binding sites; and (iii) peaks not selected by the two preceding criteria were classified as intergenic region binding sites.
For RNA-seq analysis, single-end reads of the RNA-seq libraries were mapped to the Ler-0 genome using TopHat (36). The read counts of the genes were calculated by htseq-count (37), and edgeR (38) was used to identify DEGs with an FDR < 0.05.
Motif Search and GO Analysis.
The 1-kb sequences (500-bp upstream and 500-bp downstream) surrounding the summits of the peaks were extracted and subjected to motif search by MEME (39). GO analysis was performed using DAVID (40).
To identify potential cofactors of FHY3, the 1-kb sequences (500-bp upstream and 500-bp downstream) surrounding the summits of the FHY3 binding peaks were searched for plant TF binding motifs in JASPAR database (41). We matched the motifs using MOODS (v1.0.2) (42) with the matching score threshold set to 9, and calculated the frequencies of each motif in the FHY3 binding regions. To evaluate the statistical significance of the binding motif enrichment, we used permutation test to calculate the z-score for each motif frequency. In a permutation test, the same number of regions were randomly extracted from the whole genome and scanned by MOODS. This process was repeated 1,000 times to generate the background distribution for each motif. The z-score is calculated as the difference between the motif frequency and the mean divided by the SD. A z-score of 2 or above is considered statistically significant.
Accession Numbers.
Sequence data for the genes in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AG, AT4G18960; CCA1, AT2G46830; CLV3, AT2G27250; CYCB1;3, AT3G11520; eIF4A1, AT3G13920; ELF4, AT2G40080; FHY1, AT1G79790; FHY3, AT3G22170; MCM10, AT2G20980; PHYA: AT1G09570; RPA70C, AT5G45400; RPA70D, AT5G61000; SEP1, AT5G15800; SEP2, AT3G02310; SEP3, AT1G24260; SEP4, AT2G03710; UBQ5, AT3G62250; and WUS, AT2G17950. All raw ChIP-seq data and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession numbers GSE69422, GSE69424, and GSE69425.
Supplementary Material
Acknowledgments
We thank Rae Eden Yumul for valuable advice and language editing. This work was supported by National Science Foundation of China (NSFC) Project 31500985 (to M.Z.); National Basic Research Program of China Grant 2014CB138100; NSFC Project 31471168 (to X.L.); CAS Pioneer Hundred Talents Program (to X.L. and R.L.); and NSFC Grants 31401039 (to L.G.) and 91317312 (to L.X.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE69422, GSE69424, and GSE69425).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602960113/-/DCSupplemental.
References
- 1.Sablowski R. Flowering and determinacy in Arabidopsis. J Exp Bot. 2007;58(5):899–907. doi: 10.1093/jxb/erm002. [DOI] [PubMed] [Google Scholar]
- 2.Laux T, Mayer KFX, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122(1):87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Mayer KFX, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, He Z, Guo L, Liu X. Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol. 2015;168(4):1189–1196. doi: 10.1104/pp.15.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 6.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1(1):37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105(6):805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 8.Sun B, et al. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science. 2014;343(6170):1248559. doi: 10.1126/science.1248559. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, et al. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell. 2011;23(10):3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112(1):1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Buylla ER, et al. Flower development. Arabidopsis Book. 2010;8:e0127. doi: 10.1199/tab.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409(6819):525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 13.Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF. Conversion of leaves into petals in Arabidopsis. Curr Biol. 2001;11(3):182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- 14.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14(21):1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405(6783):200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 16.Irish VF. The flowering of Arabidopsis flower development. Plant J. 2010;61(6):1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013;24(6):612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida S, Mandel T, Kuhlemeier C. Stem cell activation by light guides plant organogenesis. Genes Dev. 2011;25(13):1439–1450. doi: 10.1101/gad.631211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Deng XW. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002;21(6):1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318(5854):1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang X, et al. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell. 2011;23(7):2514–2535. doi: 10.1105/tpc.111.085126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Wang H. Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 2015;20(7):453–461. doi: 10.1016/j.tplants.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 2011;13(5):616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 25.Galinha C, Bilsborough G, Tsiantis M. Hormonal input in plant meristems: A balancing act. Semin Cell Dev Biol. 2009;20(9):1149–1156. doi: 10.1016/j.semcdb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Yadav RK, Tavakkoli M, Xie M, Girke T, Reddy GV. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development. 2014;141(13):2735–2744. doi: 10.1242/dev.106104. [DOI] [PubMed] [Google Scholar]
- 27.Immink RG, et al. SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 2009;10(2):R24. doi: 10.1186/gb-2009-10-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F, et al. Photoreceptor partner FHY1 has an independent role in gene modulation and plant development under far-red light. Proc Natl Acad Sci USA. 2014;111(32):11888–11893. doi: 10.1073/pnas.1412528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster C, et al. A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev Cell. 2014;28(4):438–449. doi: 10.1016/j.devcel.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Prunet N, Morel P, Negrutiu I, Trehin C. Time to stop: Flower meristem termination. Plant Physiol. 2009;150(4):1764–1772. doi: 10.1104/pp.109.141812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121(7):2057–2067. [Google Scholar]
- 32.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue) suppl 2:W369-73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2013;42(Database issue):D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korhonen J, Martinmäki P, Pizzi C, Rastas P, Ukkonen E. MOODS: Fast search for position weight matrix matches in DNA sequences. Bioinformatics. 2009;25(23):3181–3182. doi: 10.1093/bioinformatics/btp554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.