Significance
Within an evolutionary framework, aging and reproduction are intrinsically linked. Although both laboratory and epidemiological studies have observed associations between the timing of reproductive senescence and longevity, it is not yet known whether differences in the age of menopause are reflected in biomarkers of aging. Using our recently developed biomarker of aging, the “epigenetic clock,” we examined whether age at menopause is associated with epigenetic age of blood, saliva, and buccal epithelium. This is a definitive study that shows an association between age of menopause and biological aging (measured using the epigenetic clock). Our results also indicate menopause may accelerate the epigenetic aging process in blood and that age at menopause and epigenetic age acceleration share a common genetic signature.
Keywords: menopause, DNA methylation, aging, WHI, epigenetic clock
Abstract
Although epigenetic processes have been linked to aging and disease in other systems, it is not yet known whether they relate to reproductive aging. Recently, we developed a highly accurate epigenetic biomarker of age (known as the “epigenetic clock”), which is based on DNA methylation levels. Here we carry out an epigenetic clock analysis of blood, saliva, and buccal epithelium using data from four large studies: the Women's Health Initiative (n = 1,864); Invecchiare nel Chianti (n = 200); Parkinson's disease, Environment, and Genes (n = 256); and the United Kingdom Medical Research Council National Survey of Health and Development (n = 790). We find that increased epigenetic age acceleration in blood is significantly associated with earlier menopause (P = 0.00091), bilateral oophorectomy (P = 0.0018), and a longer time since menopause (P = 0.017). Conversely, epigenetic age acceleration in buccal epithelium and saliva do not relate to age at menopause; however, a higher epigenetic age in saliva is exhibited in women who undergo bilateral oophorectomy (P = 0.0079), while a lower epigenetic age in buccal epithelium was found for women who underwent menopausal hormone therapy (P = 0.00078). Using genetic data, we find evidence of coheritability between age at menopause and epigenetic age acceleration in blood. Using Mendelian randomization analysis, we find that two SNPs that are highly associated with age at menopause exhibit a significant association with epigenetic age acceleration. Overall, our Mendelian randomization approach and other lines of evidence suggest that menopause accelerates epigenetic aging of blood, but mechanistic studies will be needed to dissect cause-and-effect relationships further.
Reproductive senescence, concluding in menopause, is a feature of all female mammals (1), but humans are unique in that they experience exceptionally long postreproductive lifespans. Within human populations, the timing of menopause onset has been linked to susceptibility for age-related morbidity and mortality outcomes (1). For instance, observational studies have uncovered associations between a woman’s age at menopause and her subsequent risk of mortality. Results based on 12,134 Dutch women showed that for every 1-y increase in the age of menopause, the age-adjusted mortality rate was decreased by 2% (2).
Although social/behavioral and developmental factors, such as smoking, lifetime socioeconomic circumstances, infant growth, breastfeeding, and childhood cognitive ability have been shown to influence reproductive aging, age at menopause is also considered to be highly heritable, with estimates from twin and sibling studies ranging from about 0.40–0.70 (3–10). A recent large-scale genome-wide association study identified 44 genomic loci with common variants that significantly related to age at menopause (11), and a case-control study comparing centenarian women and those with average lifespans found that individuals from families with a history of longevity also tend to exhibit delayed reproductive aging (12).
Although these and other studies suggest that there might be a relationship between age at menopause and the biological aging rate, it has been difficult to test this hypothesis because of the dearth of molecular biomarkers of aging. Several recent articles describe epigenetic biomarkers of aging based on methylation levels (13–16), drawing on the profound effect of chronological age on DNA methylation (DNAm) levels (17–26). Although previous articles describe epigenetic age measures that apply only to a single tissue, i.e., saliva (13) or blood (14), our recently developed “epigenetic clock” method (based on 353 CpGs) applies to the majority of human tissue and cell types that contain DNA, with the exception of sperm (15). Age acceleration (AgeAccel) effects can be estimated by contrasting DNAm age with an individual’s chronological age. For instance, a woman whose blood has a higher DNAm age than expected based on her chronological age can be said to exhibit positive AgeAccel, i.e., to be aging faster than expected. AgeAccel also has been shown to have a strong genetic basis, with heritability estimates of 40% for older subjects (15, 27). Although the epigenetic clock has been shown to relate to a number of aging-related outcomes (27–30), it is not yet known whether it relates to reproductive aging.
Using data from four large observational studies (described in Table S1)—the Women's Health Initiative (WHI), Invecchiare nel Chianti (InCHIANTI), Parkinson's disease, Environment, and Genes (PEG), and the United Kingdom Medical Research Council National Survey of Health and Development (NSHD)—we examined the associations between epigenetic age and three menopause-related phenotypes: age at menopause, bilateral oophorectomy, and the use of menopausal hormone therapies (MHT). Given the strong heritability of both age at menopause and epigenetic age, we also estimated the genetic correlation between age at menopause and epigenetic aging and carried out a Mendelian randomization analysis to examine causality using the top two SNPs shown previously to be strongly associated with age at menopause (31).
Table S1.
Study population characteristics
| Characteristic | Study | |||
| WHI, n = 1,864 | InCHIANTI, n = 200 | PEG, n = 256 | NSHD, n = 790 | |
| Sample tested | Blood | Blood | Blood/saliva | Buccal |
| Chronological age in years, mean (SD) | 65.3 (7.1) | 70.6 (7.6) | 67.9 (12.0) | 53 (0) |
| Non-Hispanic black, frequency | 0.32 | 0 | 0.01 | 0 |
| Hispanic, frequency | 0.21 | 0 | 0.09 | 0 |
| Current smoker, frequency | 0.102 | 0.125 | 0.04 | 0.21 |
| Former smoker, frequency | 0.364 | 0.120 | 0.38 | 0.30 |
| Age at menopause in years, mean (SD) | 47.4 (6.9) | 49.1 (5.9) | 46.4 (9.0) | 50.3 (3.9) |
| MHT use ever, frequency | 0.39 | 0.12 | 0.71 | 0.57 |
Results
AgeAccel of Blood Versus Age at Menopause.
In our primary analysis, we considered only women whose menopause occurred after age 30 y. As shown in Table 1, based on results from Pearson correlations in each of our three blood datasets (WHI, InCHIANTI, and PEG), metaanalysis showed that age at menopause was significantly associated with epigenetic AgeAccel (P = 0.00091). Similar meta-analytic P values were obtained in our secondary analysis, which excluded women with surgical menopause (P = 0.0083), and in our tertiary analysis, which used all women regardless of age or type (natural/surgical) of menopause (P = 0.0061). Pearson correlation results from individual studies and stratified by race/ethnicity can be found in Fig. S1.
Table 1.
Unadjusted metaanalysis of AgeAccel in blood versus age at menopause
| Exclusion criteria | Metaanalysis Z statistics (P values) |
| Excluded because age at menopause ≤30 y | 3.3 (P = 0.00091) |
| Excluded because of surgical menopause | 2.6 (P = 0.0083) |
| None | 2.7 (P = 0.0061) |
The first and second rows correspond to analysis after excluding women who were 30 y or younger at the age of menopause and women with surgical menopause, respectively. The third row reports findings with no exclusions applied.
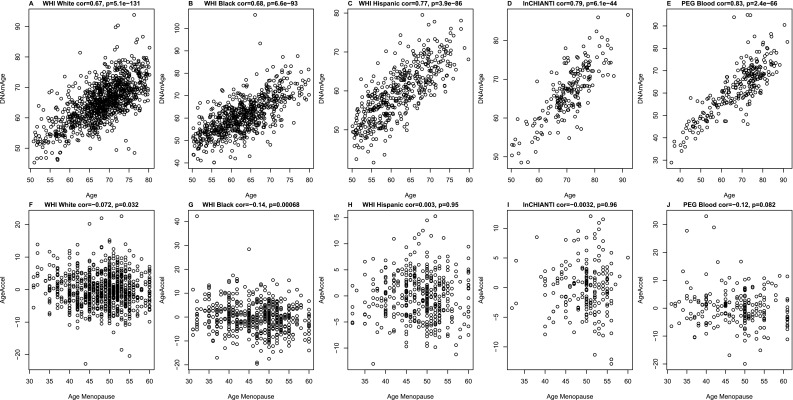
Fig. S1.
Epigenetic age versus age at menopause. (A–E) DNA methylation age (y axis) versus chronological age (x axis) in the WHI (white) (A), WHI (black) (B), WHI (Hispanic) (C), InCHIANTI (D), and PEG (E) studies. (F–J) Correlations between epigenetic AgeAccel (y axis) and age at menopause (x axis) in the WHI (white) (F), WHI (black) (G), WHI (Hispanic) (H), InCHIANTI (I), and PEG (J) studies. Each panel reports a Pearson correlation coefficient and a corresponding P value. The analysis was restricted to women whose age at menopause was >30 y. For all five samples, DNAm age and chronological age are highly correlated. We also find significant inverse associations between epigenetic AgeAccel and age of menopause in the WHI white (F) and WHI black (G) cohorts and a marginally significant inverse association in the PEG cohort (J). Metaanalysis for these and other strata can be found in Table S2.
Multivariate Linear Models Linking AgeAccel with Age at Menopause.
Using blood methylation data from the WHI, InCHIANTI, and PEG studies, we conducted multivariate regression for women who experienced menopause after age 30 y and combined results using metaanalysis to determine whether covariates accounted for the association between epigenetic AgeAccel and age at menopause (Table 2). Models were adjusted for age, race/ethnicity (in the WHI and PEG cohorts), smoking status, age at menarche, and MHT use. Additionally, models run using PEG data were also adjusted for Parkinson’s disease (PD). To retain the moderate number of women for whom data on MHT use were missing in the PEG study, those who were missing data were coded as “never,” and a dummy variable for missing was added to the model. After adjusting for possible confounders, we found that higher epigenetic AgeAccel is associated with a younger age at menopause (MetaP = 8.32 × 10−4).
Table 2.
Multivariate metaanalysis of AgeAccel in blood versus age at menopause
| AgeAccel | β-coefficient (P value) | Meta P value | ||
| WHI | InCHIANTI | PEG | ||
| Age at menopause | −0.063 (0.001) | −0.012 (0.772) | −0.060 (0.350) | 8.32 × 10−4 |
| Chronological age | 0.012 (0.530) | −0.010 (0.825) | −0.051 (0.305) | 0.837 |
| Non-Hispanic black | 0.008 (0.980) | — | −5.113 (0.201) | 0.714 |
| Hispanic | −0.918 (0.008) | — | −1.905 (0. 321) | 0.005 |
| Former smoker | −0.312 (0.234) | 0.446 (0.648) | −1.177 (0.238) | 0.198 |
| Current smoker | −0.189 (0.666) | −0.870 (0.394) | −1.378 (0.613) | 0.427 |
| MHT | 0.041 (0.871) | 0.940 (0.368) | 2.864 (0.018) | 0.269 |
| Age at menarche | −0.055 (0.501) | 0.280 (0.179) | −0.020 (0.950) | 0.821 |
| PD status | — | — | 1.075 (0.282) | — |
For all models, women with age at menopause <30 y were excluded.
Time Since Menopause and Surgical Menopause Are Associated with Epigenetic AgeAccel.
To examine whether menopause may contribute to accelerated aging, we tested the association between epigenetic aging and time since menopause using multivariate models that adjust for race/ethnicity (WHI and PEG studies) and smoking (Table S2). Results showed that the variable “time since menopause” was associated with AgeAccel (β = 0.038, P = 0.007) in the WHI and in our metaanalysis (AgeAccel, P = 0.017).
Table S2.
Multivariate metaanalysis of AgeAccel in blood versus years since menopause
| AgeAccel | β-coefficient (P value) | Meta P value | ||
| WHI | InCHIANTI | PEG | ||
| Years since menopause | 0.038 (0.007) | −0.001 (0.994) | −0.013 (0.766) | 0.017 |
| Non-Hispanic black | 0.185 (0.519) | — | −4.731 (0.237) | |
| Hispanic | −0.771 (0.022) | — | −1.417 (0. 456) | |
| Former smoker | −0.303 (0.249) | 0.409 (0.674) | −0.952 (0.335) | |
| Current smoker | −0.066 (0.879) | −0.793 (0.424) | −1.120 (0.681) | |
| MHT | 0.111 (0.660) | 1.019 (0.316) | 3.080 (0.010) | |
| Age at menarche | −0.068 (0.408) | −0.278 (0.182) | −0.006 (0.989) | |
| Parkinson’s disease status | — | — | 0.751 (0.443) | |
The menopause of a substantial number of women resulted from bilateral oophorectomy (i.e., n = 127 women in the WHI white, 112 in the WHI black, and 50 women in the WHI Hispanic cohorts and 48 women in the PEG study). We evaluated the effect of surgical menopause on epigenetic AgeAccel among women whose bilateral oophorectomy took place before age 50 y (Fig. 1). The association was highly consistent across the blood datasets (Fig. 1), leading to a significant metaanalysis P value for all three measures of AgeAccel: Stouffer's Z = 3.2; P = 0.0014.
Fig. 1.
Epigenetic AgeAccel versus surgical menopause status. The x axis of each plot reports surgical menopause status before age 50 y; i.e., “yes” denotes the group of women who experienced surgical menopause before age 50 y, and “no” corresponds to the group of women who did not undergo bilateral oophorectomy at any age before the blood draw. The bar plots report mean values of AgeAccel (± 1 SE) and the P value from a Student t test. The six panels show data from WHI (white) (A), WHI (black) (B), WHI (Hispanic) (C), InCHIANTI (D), and PEG (E) samples and all samples combined (F). A metaanalysis based on Stouffer's Z method indicates that AgeAccel is significantly positively associated with surgical menopause status (P = 0.0018).
Genetic Correlation and Mendelian Randomization Between Epigenetic AgeAccel and Age at Menopause.
Using the WHI data (n = 1,940), we conducted a bivariate restricted maximum likelihood (REML) analysis to examine the overlap in genetic variants that accounted for the heritability of both age at menopause and epigenetic AgeAccel. As shown in Table 3, the 10,769,392 autosomal SNPs or indel markers included in the analysis accounted for about 38% of the variance in age at menopause and 65% of the variance in AgeAccel. The genetic correlation (or pleiotropy) between AgeAccel and age of menopause was marginally significant with rG = −0.256 (one-sided P = 0.054).
Table 3.
GCTA bivariate REML results
| Statistic | Value |
| Heritability of AgeAccel (SE) | 0.651 (0.13) |
| Heritability of Age at menopause (SE) | 0.384 (0.14) |
| Genetic correlation with age at menopause (P value) | −0.256 (0.054) |
| Genetic variance (SE) | 18.71 (3.90) |
| Genetic covariance) (SE) | −4.70 (3.69) |
| Residual variance (SE) | 10.03 (3.61) |
| Residual covariance (SE) | 1.69 (3.47) |
| Log likelihood | −9017.31 |
Although we clearly demonstrate that age at menopause relates to epigenetic AgeAccel, our cross-sectional data make it difficult to dissect causal relationships. Mendelian randomization (reviewed in ref. 32)—the random assortment of genes from parents to offspring that occurs during gamete formation and conception—provides one method for assessing the causal nature of associations. We tested the hypothesis that menopause leads to an acceleration of epigenetic aging by leveraging the two most highly significant SNPs from a genome-wide association study for age at menopause, rs11668344 (replication P value = 2.65 × 10−18) and rs16991615 (replication P value = 7.90 × 10−21). If menopause accelerates epigenetic aging, then one would expect these two SNPs also to relate to epigenetic AgeAccel, as was found for rs11668344 (P = 0.031) (Table 4). Additionally, the directionality of the associations is consistent with the causal model SNP → age at menopause → AgeAccel, i.e., the minor allele is associated with earlier age at menopause and higher epigenetic AgeAccel. On the other hand, rs16991615 was not associated with epigenetic AgeAccel (P = 0.763).
Table 4.
Mendelian randomization
| Chromosome | SNP | Base pair position | Minor/major alleles | β-coefficient (P value) |
| 19 | rs11668344 | 55833664 | G/A | 0.506 (0.031) |
| 20 | rs16991615 | 5948227 | A/G | 0.151 (0.763) |
Analysis of Buccal Epithelium and Saliva.
We are not aware of any biological reason suggesting that blood tissue stands out when it comes to studying menopausal effects. Several previous datasets demonstrate that the epigenetic clock method also applies to buccal epithelium (33). Therefore we also examined epigenetic age measured in 790 buccal epithelium samples from women participating in the NSHD study from the UK Medical Research Council, a birth cohort of men and women born in Britain in March 1946.
Unlike our results in blood, MHT was associated with a significantly lower epigenetic AgeAccel of buccal epithelium (P = 0.00078) (Fig. 2A), whereas age at natural or surgical menopause did not correlate with epigenetic AgeAccel across the postmenopausal buccal samples (n = 419) (Fig. 2 B and D), and no significant association was found between AgeAccel and menopausal status at age 53 y (n = 469 postmenopausal and 321 premenopausal samples) (Fig. 2C).
Fig. 2.
Epigenetic age analysis of buccal samples from the NSHD. The measure of AgeAccel was defined as the difference between DNAm age and the mean DNAm age in this birth cohort. The scatter plot (B) reports Pearson correlation coefficients and corresponding P values, and the bar plots (A, C, and D) report the P value (±1 SE) from a nonparametric group comparison test (Kruskal–Wallis test). Epigenetic AgeAccel (y axis) is associated with MHT (P = 0.00078) (A) but not with age at menopause (B), menopausal status (C), or surgical menopause (D).
We also analyzed saliva samples from 113 women from the PEG study, 16 of whom had undergone surgical menopause. As in buccal epithelium, age at menopause and time since menopause were not associated with epigenetic AgeAccel in saliva. However, the directionality and size of the effect were consistent with results in blood (r = −0.11, P = 0.3), suggesting that the null association could reflect the low sample size. However, surgical menopause was associated with increased AgeAccel of saliva (P = 0.0079) (Fig. 3).
Fig. 3.
Epigenetic age analysis of saliva samples from PEG. The measure of AgeAccel in saliva was defined in the same way as in blood. The scatter plots (A and B) report biweight midcorrelation coefficients and corresponding P values, and the bar plots (C and D) report the P value (±1 SE) from a nonparametric group comparison test (Kruskal–Wallis test). (A) There is a strong correlation between epigenetic age (y axis) and chronological age. (B) Although the correlation between epigenetic AgeAccel and age at menopause in saliva is about twice that of the association in blood, the finding is not significant. (C) However, we do observe an association between epigenetic AgeAccel in saliva and surgical menopause (P = 0.0079). (D) No association was observed between epigenetic AgeAccel and MHT (P = 0.4).
Discussion
To the best of our knowledge, ours is the first study showing that (i) the epigenetic age of blood has a negative correlation with age at menopause; (ii) surgical menopause is associated with an increased epigenetic age of blood and saliva; (iii) MHT is inversely associated with epigenetic AgeAccel of buccal epithelium; and (iv) an SNP that relates to age at menopause also relates to epigenetic AgeAccel. Although our study demonstrates that postmenopausal women with a late onset of menopause are epigenetically younger than women with an early onset of menopause, dissecting cause-and-effect relationships is challenging. Here we discuss several causal scenarios that could explain the reported findings (Fig. S2).
Fig. S2.
Potential causal relationships between age at menopause and epigenetic age. (A) One causal model assumes that both age at menopause and epigenetic AgeAccel reflect biological aging, which is a latent construct. (B) Another potential causal model assumes that menopause leads to an increase of epigenetic age via an acceleration of the biological aging process.
The first causal model, age at menopause ← biological age → epigenetic AgeAccel, assumes that both age at menopause and epigenetic AgeAccel are indicator variables of a latent variable, which can be thought of as true biological age. There is evidence that risk factors for heart disease, which arguably accelerate biological age, likely contribute to an earlier age of menopause, rather than the commonly held notion that early menopause is a risk factor for heart disease (34). Consistent with evolutionary theories of aging, a genetic predisposition to later menopause may coincide with an innate protection against early mortality (35). In this study, we observe a suggestive genetic correlation between age at menopause and epigenetic AgeAccel which may suggest that increased age at menopause is genetically linked to decreased epigenetic AgeAccel. This observation advocates for a common genetic etiology and is consistent with both variables being indirect measures of biological age; genetic variants of these variables influence biological aging therefore may lead to both slower reproductive aging and lower epigenetic age.
Another causal model assumes that menopause leads to an increase of epigenetic age. This model is supported by the following lines of evidence. First, we find that longer time since menopause (irrespective of age at menopause) is associated with increased epigenetic AgeAccel. Second, bilateral oophorectomy is associated with increased epigenetic AgeAccel in both blood and saliva. This finding is congruent with a growing body of evidence suggesting that the premature loss of ovarian function caused by bilateral oophorectomy performed before natural menopause contributes to increased susceptibility to premature death, cardiovascular disease, dementia, parkinsonism, osteoporosis, and bone fractures (33). This model is also consistent with findings that transplantation of young ovaries into old mice significantly increases lifespan (36). Third, our study demonstrated that MHT, which arguably counters some of the effects of menopause, is associated with decreased epigenetic AgeAccel of buccal epithelium (but not of blood). Fourth, our Mendelian randomization analysis provided evidence of a causal pathway in which menopause accelerates epigenetic aging in blood: One of the most significant SNPs for age of menopause (rs11668344) also relates to epigenetic AgeAccel in blood. Although we acknowledge that each of these arguments in support of the causal model has pitfalls, in aggregate our results strongly support the causal model: menopause → epigenetic AgeAccel.
In moving forward, it will be important to examine longitudinal change in epigenetic age of blood before and after women transition from pre- to postmenopausal status. Additionally, it will be important to examine how long it takes accelerated epigenetic aging effects to become apparent following surgical menopause and whether they were apparent before surgical menopause. For instance, if accelerated aging is triggered by menopause, then one would expect AgeAccel to increase as a function of time since bilateral oophorectomy. However, if the condition that led to a surgical removal of both ovaries (e.g., fibroids, menstrual disorders, or endometriosis) is driving AgeAccel, one would not see a linear increase in epigenetic AgeAccel over time. In the NSHD study, it was reported that women who underwent bilateral oophorectomy tended to do so as a result of reported fibroids, menstrual disorders, or endometriosis (or some combination of these) (Table S3).
Table S3.
Reported causes of hysterectomy for women in the NSHD
| Reason for hysterectomy, from hospital records and self-reports | Hysterectomy and oophorectomy status, prior to 2009 | |||
| No operation | Oophorectomy, bilateral or unilateral | Hysterectomy only | Total | |
| Fibroids | 35 | 29 | 64 | |
| Endometriosis | 5 | 4 | 9 | |
| Fibroids and endometriosis | 3 | 0 | 3 | |
| Menstrual disorders | 24 | 35 | 59 | |
| Disorders of the uterus | 0 | 2 | 2 | |
| Prolapse | 4 | 12 | 16 | |
| Cancer | 7 | 7 | 14 | |
| Neoplasm unspecified | 1 | 1 | 2 | |
| Benign neoplasms | 1 | 0 | 1 | |
| Inflammatory disease | 2 | 0 | 2 | |
| Noninflammatory disorders | 4 | 2 | 6 | |
| Other | 2 | 0 | 2 | |
| Had operation but reason unknown, pre-2009 | 3 | 5 | 8 | |
| Subtotal | 91 | 97 | ||
| Unknown (operation after 2009) | 24 | 7 | 609 | |
| Total | 578 | 115 | 104 | 797 |
Although our study detected an association between epigenetic age in blood and age at menopause, similar associations were not found for epigenetic age estimates from buccal epithelium. One potential explanation is that aging measures in the various tissues capture different phenomena. Although we found a robust correlation between AgeAccel in blood and saliva (r = 0.70, P = 1.4E-12) (Fig. S3), when comparing the measures of AgeAccel in women with data from both blood and buccal epithelium, we found that the correlation is relatively weak (r = 0.20, P = 0.013) (Fig. S4). At first sight, the low correlation of AgeAccel between tissues is surprising. However, we interpret it as follows. The epigenetic clock is a strong predictor of age in multiple tissues in comparisons across individuals but is not necessarily correlated across tissues within a single individual. Stress factors and other perturbations act in a largely tissue-specific manner (37). Therefore one would not expect all tissue within an individual to age at the same rate, and these differences in rate potentially account for the low/moderate correlation between epigenetic AgeAccel in blood and buccal epithelium.
Fig. S3.
Correlation between epigenetic AgeAccel in blood and saliva. Using data from women in the PEG study for whom blood and saliva measures were available, we examined the association between epigenetic AgeAccel in the two tissues. We found that epigenetic AgeAccel in saliva may be a good proxy for measures in blood, given that the two are correlated at r = 0.70 (P = 1.4E-12).
Fig. S4.
Correlation between epigenetic AgeAccel in blood and buccal cells. Using data from women in the NSHD study for whom data from both blood and buccal epithelium were available, we found that the correlation of epigenetic AgeAccel in these two tissues is relatively weak (r = 0.20, P = 0.013), suggesting they may capture distinct phenomena.
To the best of our knowledge, our study is the first to demonstrate that reproductive age, bilateral oophorectomy, and MHT relate to measures of epigenetic AgeAccel, but the reported associations are specific to blood, buccal epithelium, or saliva. Future studies are warranted to examine these effects in other tissues. Because epigenetic age captures aspects of biological age (27–30), our study strongly suggests that the hormonal changes that accompany menopause accelerate biological aging in women.
Methods
Information on sample characteristics and recruitment for the WHI, PEG, InCHIANTI, and NSHD studies can be found in SI Methods.
DNAm Data.
All DNAm datasets used the Illumina Infinium 450K platform. These data were generated by following the standard protocol of Illumina methylation assays, which quantifies methylation levels by the β value using the ratio of intensities between methylated and unmethylated alleles. Specifically, the β value is calculated from the intensity of the methylated (M, corresponding to signal A) and unmethylated (U, corresponding to signal B) alleles as the ratio of fluorescent signals: β = Max(M,0)/[Max(M,0)+Max(U,0)+100]. Thus, β values range from 0 (completely unmethylated) to 1 (completely methylated). For our blood datasets (the WHI and PEG cohorts) we used background-corrected β values. Buccal cells were normalized with the same method from the original publications (38, 39). The correlation between DNAm age and chronological age is highly robust with respect to different normalization methods because (i) the epigenetic clock implements a custom normalization method, and (ii) it was constructed using training data that were normalized in different ways.
Epigenetic AgeAccel.
We used the DNAm age-based biomarker of aging from ref. 15 because (i) its accurate measurement of age across tissues is unprecedented (and it applies to both blood and buccal epithelium); (ii) it is prognostic for all-cause mortality (27, 28); (iii) it correlates with measures of cognitive and physical fitness in the elderly (27, 40); and (iv) it has been found useful for studying aging effects in Down syndrome (41), PD (42), neuropathological variables (40), obesity (37), and HIV infection (43). DNAm age was defined using the 353 CpGs and coefficient values reported in ref. 15. These CpGs and coefficient values were chosen in independent data using the Elastic Net penalized regression model to regress age on CpGs, resulting in DNAm age measures defined as predicted age, in years. Our measure of AgeAccel, which applies to all sources of DNA, was defined as residual resulting from a linear model that regressed DNAm age on chronological age. Thus, a positive value for AgeAccel indicates that the observed DNAm age is higher than expected. AgeAccel has only a weak correlation with blood cell counts (43).
Genome-Wide SNP Data from the WHI Study.
Genotyping was performed for all participants on Affymetrix 6.0, Illumina HumanOmni1-Quad v1.0, or Illumina HumanOmniExpressExome-8v1.0. Imputation was performed using MaCH with haplotypes phased in Beagle or Minimac (44, 45). The reference panel for imputation was based on the 1000 Genome haplotypes (released in June 2011). The quality of imputed markers was assessed by MaCH R2 > 0.3. Additional quality-control filters that were used included Hardy–Weinberg Equilibrium P values <10−3 and minor allele frequency (MAF) >0.01. Finally, to account for population structure, principal component analysis (PCA) was performed to generate sample eigenvectors, the first two of which were included as covariates in all genome-wide complex trait (GCTA) analysis.
We related the two highly significant SNPs from the recent large-scale metaanalysis of age at menopause (31) to AgeAccel using the WHI data. Association analysis was conducted in the two subsets of individuals stratified by platform. All women were of European ancestry, identified by multidimensional scaling analysis in PLINK. We combined the results into a single estimate by fixed-effects models weighted by inverse variance, as implemented in R metaphor (Meta-Analysis Package for R). For association analysis, we regressed each of the three AgeAccel trait values on expected genotype dosage, adjusted for the first two principal components when necessary.
Ethics.
All participants of the WHI, InCHIANTI, PEG, and NSHD gave written informed consent for their samples to be used in genetic studies of health. This study was reviewed by the Institutional Review Board of the University of California, Los Angeles (IRB nos. 13-000671 and 14-000061) and the Central Manchester Research Ethics Committee approved the use of the NSHD samples for epigenetic studies of health in 2012.
Software Code and Data Availability.
We used the online version of the epigenetic clock software, which is freely available at https://dnamage.genetics.ucla.edu/. The R source code is publicly available from additional file 20 in ref. 15. The WHI and inCHIANTI data are available through the National Heart, Lung, and Blood Institute (NHLBI) (https://biolincc.nhlbi.nih.gov/studies/bhs/). The PEG data are available from Gene Expression Omnibus (accession no. GSE72775).
SI Methods
Dataset 1: WHI Sample Description (Blood).
Participants were part of a subsample from the WHI, who were enrolled in an integrative genomics study with a primary aim of identifying novel genomic determinants of coronary heart disease. This sample included women age 50–79 y, with an overrepresentation of racial/ethnic minorities. The integrative genomics subsample used a case-control sampling design. All cases and controls were required to have undergone genome-wide genotyping at baseline and profiling of seven cardiovascular biomarkers, as dictated by the aims of other ancillary WHI studies. As shown in Table S1, the mean age at baseline for the 1,864 women in the WHI sample was 65.31 y (SD = 7.1 y). AgeAccel ranged from −22.6 to 42.9, with a mean of 0.08 and a SD of 5.3, and age at menopause ranged from 25–60 y, with a mean of 47.43 y (SD = 6.9 y). Overall, approximately half of our sample were non-Hispanic white (47.1%), one-third (32.2%) were African American, and about 20% were Hispanic. The majority of our sample reported never smoking (53.4%), 36.4% reported past smoking history, and about 10% reported they were current smokers at the time of blood draw.
Dataset 2: InCHIANTI Sample Description (Blood).
The InCHIANTI study is a population-based prospective cohort study of residents age 30 y or older from two areas in the Chianti region of Tuscany, Italy. Sampling and data collection procedures have been described elsewhere (46). Briefly, participants were enrolled between 1998 and 2000 and were examined at 3-y intervals. Overall 1,326 participants donated a blood sample at baseline (1998–2000), 784 of whom also donated a blood sample at the 9-y follow-up (2007–2009). DNAm was assayed using the Illumina Infinium HumanMethylation450 platform for participants with sufficient DNA at both baseline and year 9 visits (n = 499). Our study focused only on women (n = 200). Ages for the 200 women in InCHIANTI ranged from 50–91 y, with a mean age of 70.64 y (Table S1); baseline AgeAccel ranged from −12.9 to 12.05, with a mean of 0. Age at menopause ranged from 26–60 y, with a mean of 49.1 y. Overall, ∼75.5% of the women in the InCHIANTI study had never smoked, 12.0% were former smokers, and about 12.5% were current smokers.
Dataset 3: Women from the PEG Cohort (Blood and Saliva).
We used two types of tissues from the PEG study cohort: blood and saliva. The PEG study is a large, population-based, case-control study of PD in rural and township residents of California's central valley (47). Our blood data came from subjects from wave 1 (PEG1). PD status did not confound the relationship in blood, because it was not associated with age at menopause; however, we adjusted for it in multivariate analyses. The 256 women in the PEG study ranged in age from 35–91 y, with a mean age of 67.9 y (Table S1). Only three participants self-identified as non-Hispanic black, and 23 self-identified as Hispanic. Average age at menopause was 46.4 y. Overall, ∼4% of the women from PEG study were current smokers at the time of blood draw, and 38% were former smokers.
The saliva methylation data were collected at a later time point than the blood data. We had both blood and saliva methylation data for about half the women, but epigenetic AgeAccel of blood tissue was not correlated with AgeAccel in saliva.
Dataset 4. NSHD (Buccal Epithelium).
The buccal samples came from a subsample of 790 women participants in a British birth cohort, the UK Medical Research Council NSHD as described in ref. 39. The women were all 53 y old at the time of sample collection in 1999. At that time, 419 women were postmenopausal, and 371 women were premenopausal. MHT status was coded as “yes” only if MHT started before the age of sample collection (i.e., 53 y). Our results regarding the relationship between age at menopause and epigenetic AgeAccel were largely unchanged after excluding women who experienced surgical menopause.
All women gave written informed consent for their samples to be used in genetic studies of health, and the Central Manchester Research Ethics Committee approved the use of these samples for epigenetic studies of health in 2012. Women were selected from those who provided a buccal and blood sample at age 53 y in 1999, who had not previously developed any cancer, and for whom there was complete information on epidemiological variables of interest. Smoking status did not confound the reported relationships, because smoking was not significantly associated with our measure of epigenetic age acceleration.
Acknowledgments
This study was supported by NIH/NHLBI Grant 60442456 BAA23 (to T.L.A., D.A., and S.H.), NIH/National Institute on Aging Grant 1U34AG051425-01 (to S.H.), and NIH/National Institute of Neurological Disorders and Stroke Grant T32NS048004 (to M.E.L.). The WHI program is funded by the NHLBI of the NIH, US Department of Health and Human Services through Contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. Part of this work was funded by the Eve Appeal (https://www.eveappeal.org.uk/) and was done at University College London Hospital/University College London, which received a proportion of its funding from the Department of Health National Institute for Health Research Biomedical Research Centres funding scheme (M.W.). The NSHD is funded by UK Medical Research Council Grant MC_UU_12019/1. The PEG study was funded by NIH/National Institute of Environmental Health Sciences Grants R21-ES024356 (to S.H. and B.R.R.) and R01-ES10544 (to B.R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604558113/-/DCSupplemental.
References
- 1.Finch CE. The menopause and aging, a comparative perspective. J Steroid Biochem Mol Biol. 2014;142:132–141. doi: 10.1016/j.jsbmb.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ossewaarde ME, et al. Age at menopause, cause-specific mortality and total life expectancy. Epidemiology. 2005;16(4):556–562. doi: 10.1097/01.ede.0000165392.35273.d4. [DOI] [PubMed] [Google Scholar]
- 3.de Bruin JP, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16(9):2014–2018. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 4.van Asselt KM, et al. Heritability of menopausal age in mothers and daughters. Fertil Steril. 2004;82(5):1348–1351. doi: 10.1016/j.fertnstert.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 5.Murabito JM, Yang Q, Fox C, Wilson PW, Cupples LA. Heritability of age at natural menopause in the Framingham Heart Study. J Clin Endocrinol Metab. 2005;90(6):3427–3430. doi: 10.1210/jc.2005-0181. [DOI] [PubMed] [Google Scholar]
- 6.Hardy R, Kuh D. Social and environmental conditions across the life course and age at menopause in a British birth cohort study. BJOG. 2005;112(3):346–354. doi: 10.1111/j.1471-0528.2004.00348.x. [DOI] [PubMed] [Google Scholar]
- 7.Hardy R, Kuh D. Does early growth influence timing of the menopause? Evidence from a British birth cohort. Hum Reprod. 2002;17(9):2474–2479. doi: 10.1093/humrep/17.9.2474. [DOI] [PubMed] [Google Scholar]
- 8.Kuh D, et al. Childhood cognitive ability and age at menopause: Evidence from two cohort studies. Menopause. 2005;12(4):475–482. doi: 10.1097/01.GME.0000153889.40119.4C. [DOI] [PubMed] [Google Scholar]
- 9.Mishra G, Hardy R, Kuh D. Are the effects of risk factors for timing of menopause modified by age? Results from a British birth cohort study. Menopause. 2007;14(4):717–724. doi: 10.1097/GME.0b013e31802f3156. [DOI] [PubMed] [Google Scholar]
- 10.Richards M, Kuh D, Hardy R, Wadsworth M. Lifetime cognitive function and timing of the natural menopause. Neurology. 1999;53(2):308–314. doi: 10.1212/wnl.53.2.308. [DOI] [PubMed] [Google Scholar]
- 11.Day FR, et al. PRACTICAL Consortium kConFab Investigators AOCS Investigators Generation Scotland EPIC-InterAct Consortium LifeLines Cohort Study Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294–1303. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perls TT, Fretts RC. The evolution of menopause and human life span. Ann Hum Biol. 2001;28(3):237–245. doi: 10.1080/030144601300119052. [DOI] [PubMed] [Google Scholar]
- 13.Bocklandt S, et al. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10) R115:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weidner CI, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15(2):R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollati V, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130(4):234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakyan VK, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vivithanaporn P, et al. Neurologic disease burden in treated HIV/AIDS predicts survival: A population-based study. Neurology. 2010;75(13):1150–1158. doi: 10.1212/WNL.0b013e3181f4d5bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath S, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90(2):260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alisch RS, et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22(4):623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS One. 2013;8(6):e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day K, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013;14(9):R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marioni RE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16(1):25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiansen L, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15(1):149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 2015;7(12):1159–1170. doi: 10.18632/aging.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marioni RE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolk L, et al. LifeLines Cohort Study Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44(3):260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GD, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 33.Shuster LT, Gostout BS, Grossardt BR, Rocca WA. Prophylactic oophorectomy in premenopausal women and long-term health. Menopause Int. 2008;14(3):111–116. doi: 10.1258/mi.2008.008016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kok HS, et al. Heart disease risk determines menopausal age rather than the reverse. J Am Coll Cardiol. 2006;47(10):1976–1983. doi: 10.1016/j.jacc.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 35.Rose MR. Evolutionary Biology of Aging. Oxford Univ Press; New York: 1991. [Google Scholar]
- 36.Mason JB, Cargill SL, Anderson GB, Carey JR. Transplantation of young ovaries to old mice increased life span in transplant recipients. J Gerontol A Biol Sci Med Sci. 2009;64(12):1207–1211. doi: 10.1093/gerona/glp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teschendorff AE, et al. Correlation of smoking-associated DNA methylation changes in buccal cells with DNA methylation changes in epithelial cancer. JAMA Oncol. 2015;1(4):476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 40.Levine M, Lu A, Bennett D, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging (Albany NY) 2015;7(12):1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvath S, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14(3):491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany, NY) 2015;7(12):1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrucci L, et al. Subsystems contributing to the decline in ability to walk: Bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48(12):1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 47.Costello S, Cockburn M, Bronstein J, Zhang X, Ritz B. Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am J Epidemiol. 2009;169(8):919–926. doi: 10.1093/aje/kwp006. [DOI] [PMC free article] [PubMed] [Google Scholar]