Significance
Malignant cells of breast carcinoma and nonmalignant epithelia of branching mammary glands share the ability to migrate through their surroundings. To form the mammary tree-like architecture, nonmalignant epithelia must migrate in a controlled fashion, integrating cues from their microenvironment, notably, the glycan appendages on extracellular proteins and lipids. Here, we show that Galectin-1, a glycan-binding protein, is able to sense glycan signatures on mammary gland epithelia, transmit this information to epithelial nuclei by direct translocation, and drive branching migration. Nuclear galectin-1 is regulated by the relative levels of α2,6–sialic acids and N-acetyllactosamine on extracellular glycans. Similar lectin–glycan signatures were observed in malignant breast cells and suggest cancer cells use this pathway during their invasion.
Keywords: galectin-1, sialic acid, mammary gland, breast cancer, glycobiology
Abstract
Branching morphogenesis in the mammary gland is achieved by the migration of epithelial cells through a microenvironment consisting of stromal cells and extracellular matrix (ECM). Here we show that galectin-1 (Gal-1), an endogenous lectin that recognizes glycans bearing N-acetyllactosamine (LacNAc) epitopes, induces branching migration of mammary epithelia in vivo, ex vivo, and in 3D organotypic cultures. Surprisingly, Gal-1’s effects on mammary patterning were independent of its glycan-binding ability and instead required localization within the nuclei of mammary epithelia. Nuclear translocation of Gal-1, in turn, was regulated by discrete cell-surface glycans restricted to the front of the mammary end buds. Specifically, α2,6–sialylation of terminal LacNAc residues in the end buds masked Gal-1 ligands, thereby liberating the protein for nuclear translocation. Within mammary epithelia, Gal-1 localized within nuclear Gemini bodies and drove epithelial invasiveness. Conversely, unsialylated LacNAc glycans, enriched in the epithelial ducts, sequestered Gal-1 in the extracellular environment, ultimately attenuating invasive potential. We also found that malignant breast cells possess higher levels of nuclear Gal-1 and α2,6–SA and lower levels of LacNAc than nonmalignant cells in culture and in vivo and that nuclear localization of Gal-1 promotes a transformed phenotype. Our findings suggest that differential glycosylation at the level of tissue microanatomy regulates the nuclear function of Gal-1 in the context of mammary gland morphogenesis and in cancer progression.
Transmission of information between neighboring cells and their tissue microenvironment is essential for organ morphogenesis and homeostasis. The process of transmission can be spatially separated into an extracellular component, which includes cell–ECM adhesion and soluble ligand binding, and an intracellular component, encompassing phosphorylation networks and transcription programs. This strict division is spanned by transmembrane proteins that relay molecular and mechanical cues through both outside-in and inside-out mechanisms. Recently, a growing number of proteins with distinct functions inside and outside cells have been recognized to subvert this conventional mode of cellular communication via alternative secretion (1). Noncanonically secreted proteins can potentially integrate intracellular and extracellular information, in effect influencing tissue specificity and organogenesis (2).
Galectin-1 (Gal-1), a soluble lectin, lacks a signal peptide but is secreted to the extracellular environment through unconventional transport. Outside the cell, Gal-1 interacts with glycoconjugates, modulating their surface organization and mediating cell–cell and cell–ECM contact (3–5). Within the cell, Gal-1 is found in the cytosol and nucleus, where it has been proposed to play roles in signaling (6) and transcription (7, 8), respectively, that are unrelated to glycan-binding activity (9). To date, no connection has been made between the distinct functions of Gal-1 in different subcellular contexts.
We and others have shown that tissue architecture is a dominant regulator of cancer cell phenotype (10–12). Although glycomic changes such as hypersialylation have long been shown to correlate with cancer cell metastasis (13), how glycans and lectins mechanistically drive the invasive processes during cancer progression remains obscure. Gal-1 is up-regulated in invasive breast cancer (14), which involves epithelial proliferation accompanied by a radical alteration in glandular architecture (10). Normal mammary epithelia also proliferate and migrate within their surrounding stroma during the branching stage of glandular development, albeit in a more controlled manner relative to their transformed counterparts. Accordingly, we sought to determine whether Gal-1 levels are also modulated in this developmental process.
In this paper, we demonstrate that endogenous Gal-1 induces branching of mammary epithelia. By engineering the localization of Gal-1 and the glycan microenvironment in 3D, we show that Gal-1’s function in the mammary gland requires nuclear localization, which in turn is regulated by the glycomic signatures of the epithelial microenvironment. Our findings indicate that Gal-1 can directly transmit glycan-encoded information of its surroundings to the nucleus, where it assists in executing a branching program. We also confirm our results in transformed breast cells and argue that the spatiotemporal signatures of cell-surface glycans can play crucial and analogous roles in glandular ontogeny and oncogeny.
Results and Discussion
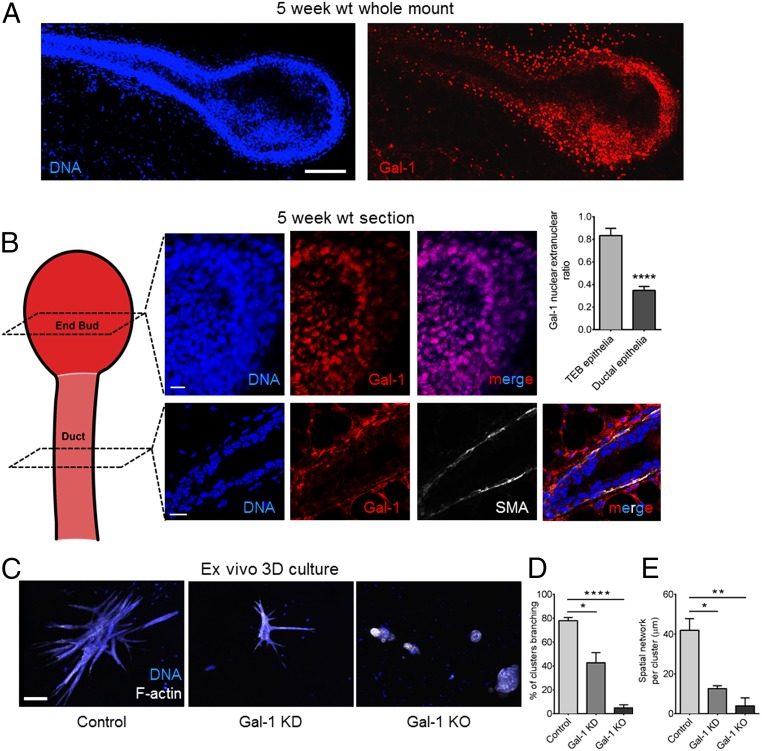
Using immunofluorescence, we assayed for Gal-1 protein in murine mammary glands at distinct stages of development. Gal-1 levels were found to be highest during the early stages of branching morphogenesis (5 wk; 35 d postpartum) (Fig. 1A and SI Appendix, Fig. S1 show low to moderate levels of Gal-1 at other developmental stages of the mammary gland). Within the glands of 5-wk old mice, Gal-1 expression was highest in epithelial cells at the terminal end bud (TEB), which represents the invading front of the mammary arbor during the branching stage of development (Fig. 1A). Gal-1 levels were lower in the quiescent, noninvasive epithelia of mammary ducts even at this stage. Unexpectedly, we found a major difference in Gal-1’s subcellular localization in the two microenvironments: high levels of Gal-1 were observed within the nuclei of end-bud epithelia, whereas, in the ductal cells, Gal-1 was largely depleted from their nuclei (Fig. 1B).
Fig. 1.
Gal-1 is essential for mammary branching morphogenesis. (A) Immunofluorescence micrographs of a 5-wk murine mammary gland stained with α-Gal-1 antibody (Right) and DAPI (Left). (See SI Appendix, Fig. S1 for control.) (Scale bar, 100 µm.) (B) In the end-bud epithelia, Gal-1 is enriched in the nucleus (Top), whereas Gal-1 staining in the mammary duct epithelia is mainly extranuclear (Bottom). Ductal epithelia were costained for α smooth muscle actin (SMA), a marker of mammary myoepithelial cells. (See SI Appendix, SI Materials and Methods for quantification). (Scale bar, 15 µm.) (C) Micrographs of organoids cultured in 3D CL-1 and stained for F-actin (depicted in white) and DNA (depicted in blue): control organoids from wild-type (WT) mice (Left), organoids from WT mice after Gal-1 knockdown (KD) (Middle), and organoids from Gal-1−/− mice (Right). (Scale bar, 100 µm.) (D and E) Quantification of branching clusters and spatial network per organoid shows impaired branching upon Gal-1 depletion (20 organoids analyzed per culture). Spatial network is defined as the sum of the branch lengths for each organoid or branching structure. For all bar graphs, error bars represent SEM. Statistical significance is given by *P < 0.05; **P < 0.01; ****P < 0.0001.
Culturing primary mammary cells ex vivo within 3D Type-1 collagen (CL-1) scaffolds is an organotypic assay well suited to delineate the roles of proteins expressed predominantly within glandular epithelia during branching and polarization (15). We performed shRNA-based lentiviral knockdown of Gal-1 within organoids from wild-type C57BL/6 and Balb/C mice. In addition, we compared branching in organoids from Gal-1−/− mice with their wild-type counterparts. In both cases, knockdown and knockout, we found a significant decrease in branching (Fig. 1 C–E). We also observed a significant decrease in branch-point number in carmine-stained mammary glands from 35 d postpartum Gal-1−/− mice compared with wild-type controls (SI Appendix, Fig. S2). The branching defect was less pronounced in vivo than in ex vivo cultures, which could be explained by the fact that the loss of expression or function of a single protein, no matter how important, is often compensated by other proteins with similar biochemical function (mammals have 15 galectins) and, more importantly, by the maintenance of the architectural integrity.
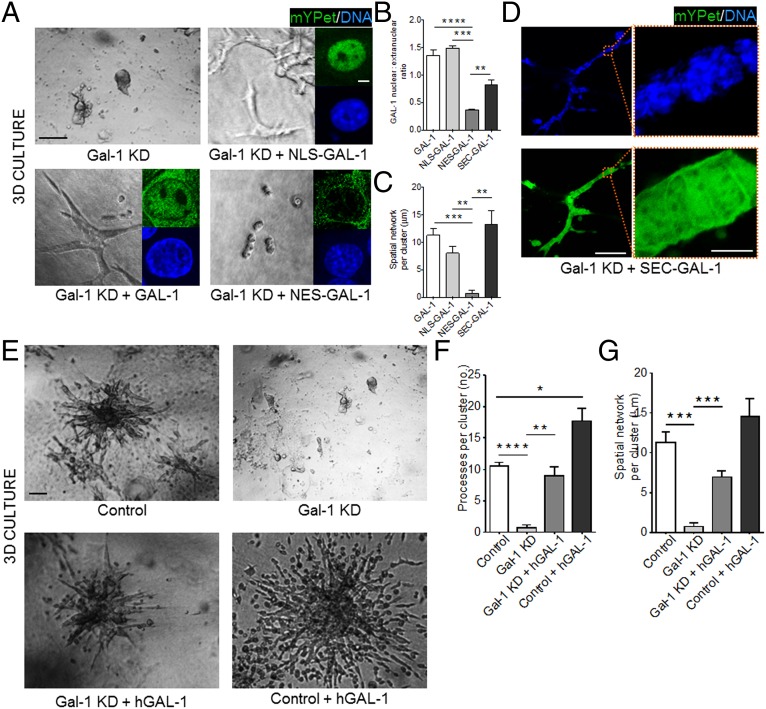
We next took advantage of a different organotypic 3D culture model that relies on the mammary epithelial cell line, EpH4 (16–19), which is more amenable to genetic manipulations. This culture system was used to probe the influence of the nuclear pool of Gal-1 on epithelial migration and branching. Upon addition of epidermal growth factor, EpH4 cells embedded in CL-1 invade into the gel and form branched structures with high levels of Gal-1 in the extensions, similar to primary mammary epithelia grown ex vivo (SI Appendix, Fig. S3). Depletion of Gal-1 by shRNA (SI Appendix, Fig. S4 A and B) abrogated branching in 3D cultures (Fig. 2A). We rescued the invasive phenotype by adding recombinant human Gal-1 (GAL-1) to EpH4 cells (Fig. 2 E–G). To probe the effect of Gal-1 subcellular localization on phenotypic rescue, we overexpressed GAL-1 constructs tagged either with a nuclear localization or a nuclear export signal (NLS and NES, respectively; SI Appendix, Fig. S5 A and B). Whereas nuclear resident NLS–GAL-1 rescued branching (Fig. 2 A–C and SI Appendix, Fig. S5C for control), cytoplasmic NES–GAL-1 did not (Fig. 2 A–C and SI Appendix, Fig. S5C for control).
Fig. 2.
Nuclear Gal-1 drives mammary epithelial branching and migration in 3D. (A) Gal-1 KD EpH4 cells (Top, Left) ectopically expressing either NLS–GAL-1 (Top, Right), GAL-1 (Bottom, Left), or NES–GAL-1 (Bottom, Right) were cultured in 3D CL-1 gels. Branching was observed only upon expression of GAL-1 or NLS–GAL-1. (Scale bar, 50 µm.) (Inset) Fluorescence micrographs indicate the subcellular localization of each mYPet fusion construct [DNA (blue) and mYPet (green)] (see SI Appendix, Fig. S5 for construct map and construct controls). (Scale bar, 5 µm.) (B) Quantification of GAL-1 nuclear:extranuclear ratio for each of the GAL-1 constructs. (C) Quantification of the spatial network per cluster for each of the GAL-1 constructs in 3D (20 clusters analyzed per culture). (D) Gal-1 KD EpH4 cells ectopically expressing SEC–GAL-1 branch when cultured in a 3D CL-1 gel (Left). (Scale bar, 100 µm.) mYPet fluorescence of SEC–GAL-1 fusion construct is distributed between the extracellular space and the nucleus (Right) (see SI Appendix, Fig. S5 for construct map and construct controls). (Scale bar, 10 µm.) (E) Brightfield micrographs of EpH4 cells (Top, Left) with Gal-1 KD (Top, Right), with Gal-1 KD and treatment with recombinant human GAL-1 (Bottom, Left), and with recombinant human GAL-1 (Bottom, Right) cultured in 3D CL-1 gels. (Scale bar, 50 µm.) (F and G) Quantification of the number of processes per branching cluster and the spatial network of each cluster upon endogenous Gal-1 KD and/or treatment with recombinant human GAL-1. For all bar graphs, error bars represent S.E.M. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Endogenous Gal-1 is known to translocate to the extracellular space through unconventional secretion (2, 20). We wondered whether, once extracellular, Gal-1 could traffic back to the nucleus and exert its influence on mammary migration. To address this, we designed a GAL-1 construct bearing a secretion signal peptide (SEC–GAL-1) that should transit to the extracellular space through the classical secretory pathway and, once there, be poised to reenter the cell through Gal-1–dependent uptake. Indeed, overexpressed SEC–GAL-1 was secreted and was able to relocalize to the nucleus and rescue branching (Fig. 2D; see SI Appendix, Fig. S5 for control and SI Appendix, Fig. S6 for nuclear accumulation of SEC–GAL-1 in NLS–mCherry-expressing EpH4 cells). In addition, recombinant Gal-1 added exogenously to mammary cells in 3D culture was detected in nuclei as well as other compartments (SI Appendix, Fig. S5D). Although these experiments do not suggest a mechanism of Gal-1 reentry into the cell, there are many possible routes by which Gal-1 could traverse the cell membrane. For instance, internalization of Gal-1 by endocytosis, similar to Gal-3 (21), or by flippase activity of glycolipid–Gal-1 complexes (22) are possible candidates for lectin translocation. Collectively, these data suggest that nuclear Gal-1 is necessary for migration and branching and that Gal-1 is able to translocate from the extracellular space to the nucleus.
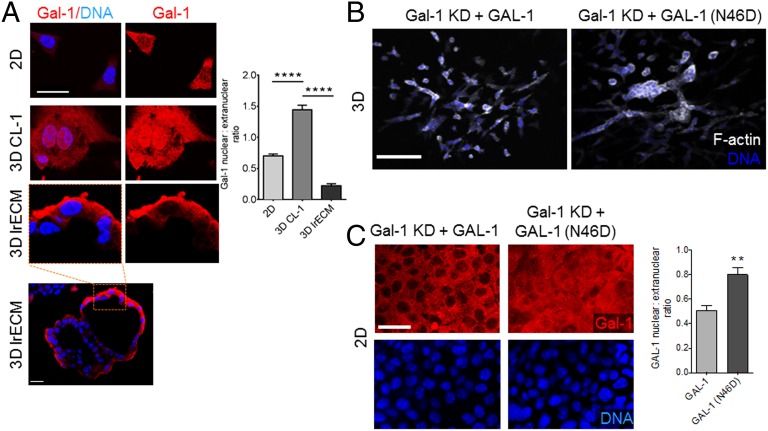
To pinpoint the microenvironmental context in which Gal-1 localizes to the epithelial nucleus, we cultured EpH4 cells (i) in 2D, (ii) on CL-1 gels, and (iii) on laminin-rich ECM (lrECM) (Fig. 3A). In this experiment, CL-1 and lrECM gels approximate an in vivo branching and ductal microenvironment, respectively. In both 2D and on lrECM, where cells form a lumen-containing acinar-like structure, the epithelial nuclei showed sparse Gal-1, whereas, on top of CL-1, mammary epithelia displayed high nuclear Gal-1 levels. Therefore, Gal-1 nuclear localization strongly correlates with a microenvironmental context that is associated with invasive epithelia, i.e., TEB epithelia, and not their quiescent counterparts, i.e., ductal epithelia.
Fig. 3.
Glycan recognition by Gal-1 is dispensable for epithelial branching. (A) Immunofluorescence micrographs of EpH4 cells cultured in 2D (Top), on top of 3D CL-I gel (Middle), and on top of 3D lrECM gel (Bottom Left: acinar-like architecture with lumen. Scale bar, 20 µm) and stained for Gal-1 (depicted in red) and DNA (depicted in blue). Quantification of Gal-1 nuclear:extranuclear ratio for EpH4 cells cultured in 2D and 3D conditions. (Scale bar, 25 µm.) (B and C) Fluorescence micrographs of Gal-1 KD EpH4 cells ectopically expressing GAL-1 (Left) or GAL-1 (N46D) (Right) fusion proteins in 3D (Top) or 2D (Bottom) and stained for F-actin (depicted in white) and DNA (depicted in blue). Cells expressing GAL-1 (N46D), a mutant with attenuated glycan binding, invade and branch when cultured in 3D. Quantification of GAL-1 nuclear:extranuclear ratio shows GAL-1 (N46D) is concentrated in the nucleus. (Scale bar, 100 µm.) For all bar graphs, error bars represent S.E.M. Statistical significance is given by **P < 0.01; ****P < 0.0001.
Galectin–glycan binding has previously been reported to influence cell invasion and migration (23). We investigated whether mammary epithelial morphogenesis also requires this activity using a GAL-1 mutant, N46D, which attenuates glycan binding (24). Overexpression of GAL-1 (N46D) in Gal-1–silenced EpH4 cells rescued the branching phenotype in 3D culture similar to wild-type GAL-1 (Fig. 3B). Thus, nuclear Gal-1’s ability to drive morphogenesis is independent of its sugar-binding activity. Interestingly, GAL-1 (N46D) showed a greater degree of nuclear localization than wild-type Gal-1 when expressed in cells cultured in 2D (Fig. 3C).
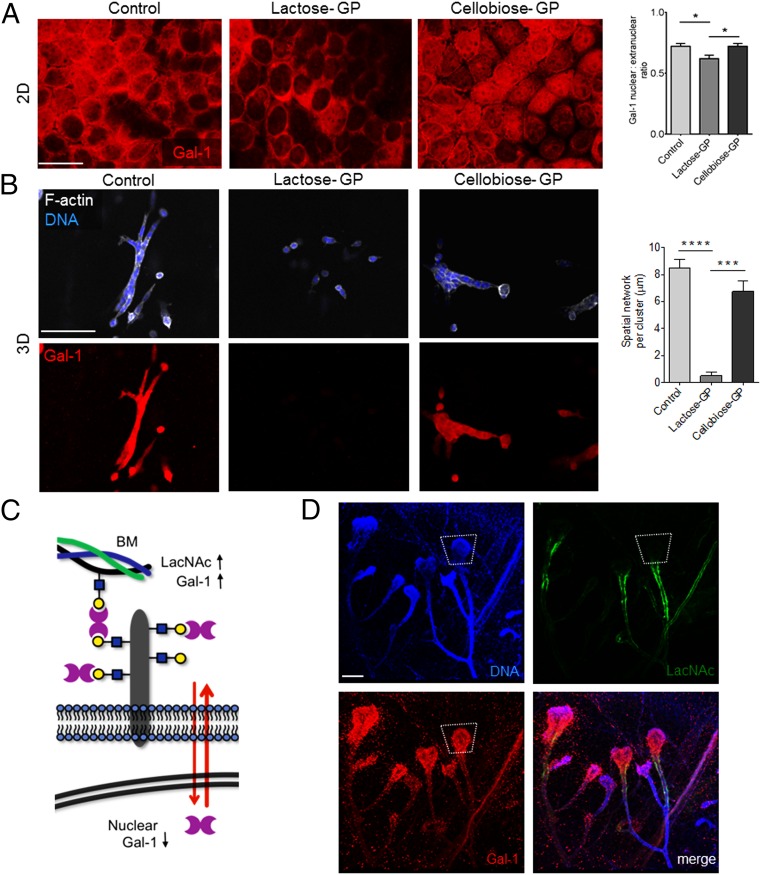
Our observation above prompted us to consider whether Gal-1’s glycan-binding activity might regulate the protein’s function by altering its distribution between the nucleus and extracellular microenvironment. To test this idea, we reengineered the microenvironment of the mammary epithelia by adding glycopolymers that mimic ECM glycoproteins to the exterior of the cells (25). We synthesized glycopolymers (GPs) functionalized with multiple Gal-1 ligands (lactose) or control glycan structures (cellobiose) that do not associate with Gal-1. When lactose–GP was added to mammary epithelial cells cultured on top of CL-1 gels, we found a marked decrease in nuclear Gal-1 after 1 d (Fig. 4A). In contrast, untreated cells and cells treated with cellobiose–GP showed higher nuclear levels of Gal-1. When added to 3D CL-1 cultures, lactose–GP abrogated branching (Fig. 4B). After washing lactose–GP-treated cells, nuclear Gal-1 was undetectable, suggesting that the glycopolymer redistributed Gal-1 to the extracellular space. In contrast, Gal-1 staining was unaltered in branching cultures of cells treated with cellobiose–GP. Similar results were obtained with glycopolymer treatment of EpH4 Gal-1 knockdown cells overexpressing fluorescent GAL-1 fusion protein (SI Appendix, Fig. S7).
Fig. 4.
Extracellular glycans control the nuclear localization of Gal-1. (A) Immunofluorescence micrographs of EpH4 cells grown on top of CL-1 gels treated with either soluble lactose–GP, which binds Gal-1, or soluble cellobiose–GP, which does not interact with Gal-1, and stained with an α-Gal-1 antibody (see SI Appendix, Fig. S7 for glycopolymer treatment on Gal-1 KD EpH4 cells overexpressing full-length GAL-1). Quantification of the Gal-1 nuclear:extranuclear ratio for EpH4 cells treated with either the lactose–GP or the cellobiose–GP. (Scale bar, 50 µm.) (B) Immunofluoresence micrographs of EpH4 cells cultured in 3D CL-1 gel in the presence or absence of either lactose–GP or cellobiose–GP and then washed and stained for F-actin and with DAPI and an α-Gal-1 antibody. Quantification of the spatial network per cluster of EpH4 cells in the presence or absence of GPs. (Scale bar, 100 µm.) (C) Model of extracellular glycan patterns regulating nuclear Gal-1 in mammary epithelial cells. In a LacNAc-rich environment containing intact ECM proteins, Gal-1 is mainly concentrated in the extracellular space. (D) Immunofluorescence micrographs of murine mammary gland stained with Erythrina Crystagalli lectin (ECL) (Top, Right), which is specific for terminal LacNAc disaccharides, an α-Gal-1 antibody (Bottom, Left), and DAPI (Top, Left). LacNAc appears to line the ductal epithelia, whereas Gal-1 is heavily concentrated in the invasive end bud. (SI Appendix, Fig. S8 shows colocalization of LacNAc residues and laminin on the surface of ductal epithelia). (Scale bar, 150 µm.) For all bar graphs, error bars represent S.E.M. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Our data therefore point to a dynamic reciprocity between the glycan microenvironment and nuclear Gal-1 levels (Fig. 4C). When N-acetyllactosamine (LacNAc) epitopes are abundant in the extracellular environment, Gal-1’s distribution reequilibrates in that direction. In this model, extracellular glycans act as a molecular sink, trapping Gal-1. On the other hand, when Gal-1 is unable to bind extracellular glycan ligands, e.g., GAL-1 (N46D), the partition shifts to a higher abundance of nuclear Gal-1, promoting epithelial invasiveness.
To determine whether this mechanism operates during mammary branching morphogenesis in vivo, fixed mammary gland whole mounts from 35 d postpartum C57BL/6 mice were stained for terminal LacNAc residues using FITC–Erythrina Crystagalli lectin (ECL) (26). Fluorescence micrographs revealed strong levels of extracellular LacNAc in ductal regions and relatively sparse levels in TEB epithelia (Fig. 4D). Sections of 35-d postpartum mammary gland ducts stained for LacNAc showed strong colocalization with the basement membrane and low levels of extracellular Gal-1 (SI Appendix, Fig. S8).
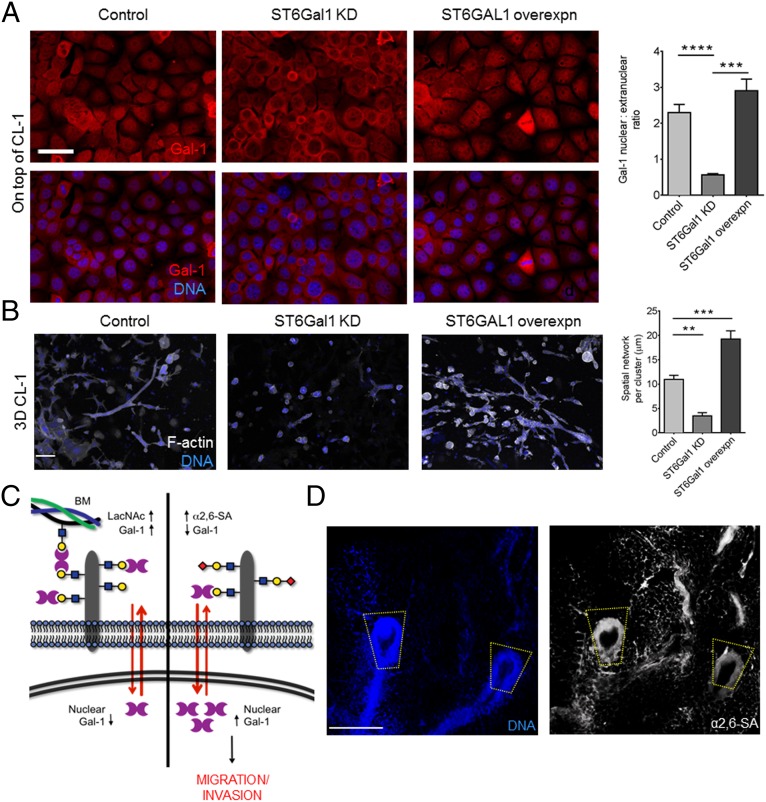
The addition of α2,6–sialic acid (α2,6–SA) residues is known to block Gal-1’s binding to LacNAc epitopes (27, 28). To test the effects of sialylation on Gal-1 nuclear localization and mammary branching, we elevated sialoside levels by exogenous addition of peracetylated N-acetylmannosamine (Ac4ManNAc) (29), a metabolic precursor of sialic acid, or by overexpression of UDP–N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE), the rate-limiting enzyme in sialic acid synthesis (SI Appendix, Fig. S9 A–C) (30). Both approaches led to an increase in α2,6–SA epitopes, as measured by staining with Sambucus nigra agglutinin (SNA) (30), as well as an increase in nuclear Gal-1 levels (SI Appendix, Fig. S9 A–C). shRNA-mediated depletion of GNE in EpH4 cells caused a decrease in branching (SI Appendix, Fig. S9D). To specifically test the role of α2,6–SA regiochemistry, we either knocked down or overexpressed β-galactoside α2,6–sialyltranferase 1 (ST6Gal1) in EpH4 cells (SI Appendix, Fig. S9E). We observed lower levels of nuclear Gal-1 in ST6Gal1–knockdown cells and higher levels of nuclear Gal-1 in cells overexpressing ST6GAL1, relative to control cells (Fig. 5A). Finally, EpH4 cells depleted in ST6Gal1 and those overexpressing ST6GAL1 showed attenuated and exacerbated branching morphogenesis relative to their control counterparts, respectively (Fig. 5B).
Fig. 5.
α2,6–SA regulates Gal-1’s nuclear abundance and induction of mammary epithelial morphogenesis. (A) Fluorescence micrographs of EpH4 cells cultured on top of CL-1 and stained for Gal-1 (red) and DNA (blue): wild-type cells (Left), cells with ST6Gal1 knockdown (Middle), and cells overexpressing ST6GAL1 (Right) (see SI Appendix, Fig. S9E Extended Data Fig 9e for α2,6–SA levels of EpH4 cells with ST6Gal1 depletion and overexpression). Quantification of Gal-1 nuclear:extranuclear ratio of EpH4 cells with varying levels of ST6Gal1. (Scale bar, 25 µm.) (B) Fluorescence micrographs of EpH4 cells cultured in 3D within CL-1 gels and stained for F-actin (depicted in white) and DNA (depicted in blue): wild-type cells (Left), cells with ST6Gal-1 knockdown (Middle), and cells overexpressing ST6Gal-1 (Right). Quantification of the spatial network per cluster of EpH4 cells with varying levels of ST6Gal1 (50 clusters counted per culture). (Scale bar, 25 µm.) (C) Complete model of glycan signatures regulating nuclear Gal-1 in mammary epithelial cells. α2,6–sialylation of LacNAc structures causes Gal-1 to accumulate in the nucleus, resulting in an invasive phenotype (Right). (D) Fluorescence micrographs of murine mammary gland stained with SNA (Right; white) and DAPI (Left; blue) show high levels of α2,6–SA residues in the invasive end bud of the mammary gland. (Scale bar, 200 µm.) For all bar graphs, error bars represent S.E.M. Statistical significance is given by **P < 0.01; ***P < 0.001; ****P < 0.0001.
Within the whole mounts, α2,6–SA epitopes were enriched at the end buds and absent in the ducts (Fig. 5D). These results suggest that LacNAc residues on invading mammary epithelia are capped by α2,6–SA, which negatively regulates binding to Gal-1 (31). Notably, the transcript levels for ST6Gal1, an enzyme that adds α2,6–SA to terminal LacNAc residue, have previously been found to be higher in terminal end buds compared with ductal epithelia (32).
We conclude from the above data that mammary epithelial branching morphogenesis is driven by the dynamics of Gal-1 subcellular localization, which in turn is a sensor of the glycan signature in the epithelial microenvironment. LacNAc, the cognate glycan ligand for Gal-1, acts as a sink to retain Gal-1 in the extracellular milieu. We found high levels of unmodified LacNAc and low nuclear Gal-1 levels in the quiescent ductal epithelia of mammary glands and, reciprocally, extracellular α2,6–SA and nuclear Gal-1 were abundant in the proliferating epithelia at the invading edge of mammary end buds (Fig. 5C). Thus, α2,6–sialylation acts as a switch to potentiate Gal-1–mediated mammary morphogenesis.
The molecular mechanism(s) by which nuclear Gal-1 promotes branching and invasion is an intriguing question. In vitro studies have shown that nuclear Gal-1 is involved in pre-mRNA splicing and coexists in a complex with Gemin-4 (7). In accordance with these findings, we observed that Gal-1 localizes to the Gemini bodies of mammary epithelia (SI Appendix, Fig. S10A), indicating that it may be part of the Gemin-4–containing transcription-regulating complexes (33). shRNA knockdown of Gemin-4 in mammary epithelia (SI Appendix, Fig. S10B) also abrogated branching. The cells remained alive and formed noninvasive spherical clusters, phenocopying Gal-1 depletion (SI Appendix, Fig. S10C). These data suggest that the interaction of Gemin-4 and Gal-1 within the nucleus plays a functional role in mammary epithelial morphogenesis. As well, we found that overexpression of nuclear Gal-1 leads to up-regulation in gene expression of Erk1/2 (SI Appendix, Fig. S11), a key signaling node in mammary gland branching (34) and possible target of transcriptional regulation.
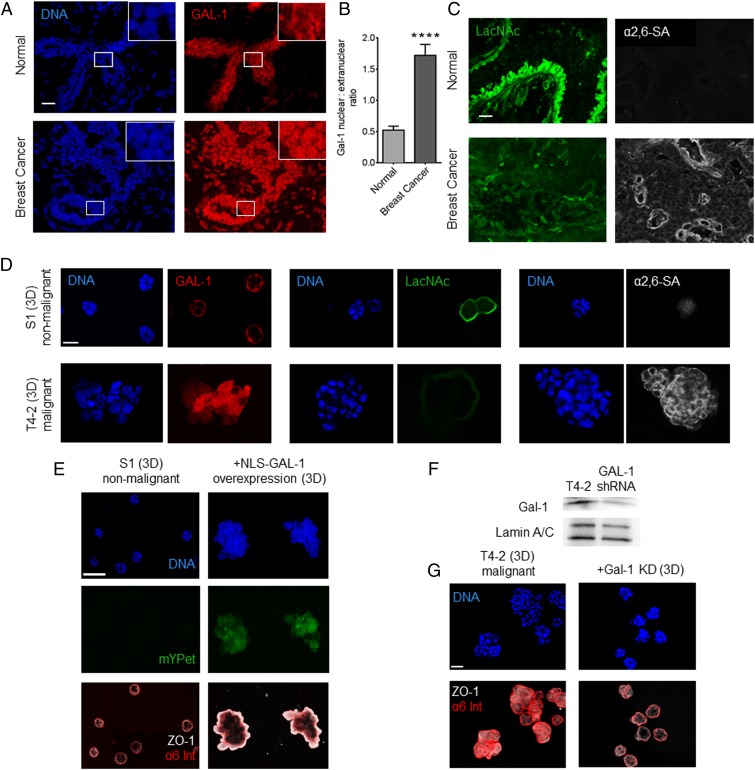
Finally, we speculated that our findings were relevant to the acquisition of epithelial invasiveness in breast cancer. We found higher levels of nuclear Gal-1 in malignant epithelia from human invasive ductal carcinoma sections relative to nonmalignant tissues (Fig. 6 A and B). To examine if our glycan-dependent model may explain this overlooked feature of Gal-1 in breast cancer, we proceeded to stain the tissue for cognate glycan epitopes of Gal-1. We observed low levels of LacNAc and high levels of α2,6–SA in the malignant epithelia, relative to nonmalignant tissue sections (Fig. 6C). Other sialylated structures, such as truncated O-glycans like sialyl TN or α2,3–SA on core 1, have previously been linked to breast cancer (35–38). Some of these glycans may also contribute to blocking Gal-1 binding within the malignant glycocalyx by preventing extension of O-glycans to terminal LacNAc repeats. The invasive lectin–glycan signatures were also observed by fluorescence in 3D cultures of malignant breast cells (T4-2) in comparison with their isogenic nonmalignant (S1) counterparts (Fig. 6D). Moreover, overexpression of NLS–Gal-1 in S1 cells impaired their growth-arrested basoapical polarity (Fig. 6E), whereas GAL-1 depletion in malignant T4-2 cells (Fig. 6F) arrested their growth and partially restored their polarity (Fig. 6G). As previously documented, invasive breast cancer cells show high levels of both Gal-1 (14, 39) and α2,6–SA (40), the modification that masks the ligand of Gal-1 and is associated with mammary epithelial invasiveness (41). Our results seem to reconcile all these observations by linking the nuclear localization of Gal-1 and its ability to induce migration to extracellular α2,6–SA. As well, our finding that Gal-1 translocates to the nucleus of malignant breast cancer cells due to hyper-α2,6–sialylation may be relevant for development of Gal-1 specific inhibitors in breast cancer treatment (42).
Fig. 6.
Malignant breast epithelia possess high levels of nuclear Gal-1 and α2,6–SA and low levels of LacNAc. (A) Fluorescence micrographs of sections of normal breast tissue (Top) and invasive ductal carcinoma (Bottom). The set shows staining for Gal-1 (depicted in red) and DNA (depicted in blue) with insets highlighting single cells for subcellular staining distribution. (Scale bar, 20 µm.) (B) Graph showing quantification of nuclear:extranuclear ratio of Gal-1 in breast epithelia from normal and invasive ductal carcinoma tissues. (C) Micrographs of normal breast tissue (Top) and invasive ductal carcinoma (Bottom) showing staining for terminal LacNAc residues (depicted in green, Left) and α2,6–SA (depicted in white, Right). (Scale bar, 20 µm.) (D) Fluorescence micrographs of 3D cultures formed by nonmalignant S1 cells (Top) and isogenic malignant T4-2 cells (Bottom). Staining for Gal-1 (depicted in red) and DNA (depicted in blue) (Left). Staining for terminal LacNAc residues (depicted in green) and DNA (depicted in blue) (Middle). Staining for α2,6–SA (depicted in white) and DNA (depicted in blue) (Right). (Scale bar, 20 µm.) (E) Fluorescence micrographs of 3D cultures of S1 cells, wild-type (Left) and overexpressing NLS–GAL-1 (Right). S1 cells overexpressing NLS–GAL-1 show loss of growth-arrested phenotype seen in wild-type S1 cells and aberrant basoapical polarity of basal (α6 integrin; depicted in red) and apical (zonula occludens, ZO-1; depicted in white) markers. (Scale bar, 40 µm.) (F) Immunoblot showing levels of GAL-1 (Top) and Lamin A/C (internal control, Bottom) in T4-2 cells with and without shRNA-based knockdown. (G) Fluorescence micrographs of 3D cultures of T4-2 cells (Left) and T4-2 cells with GAL-1 knockdown (Right) showing a recovery of acinar phenotype and basoapical polarity upon GAL-1 knockdown. For bar graphs, error bars represent S.E.M. Statistical significance is given by ****P < 0.0001. (Scale bar, 20 µm.)
Supplementary Material
Acknowledgments
We thank Jason Hudak, Chin Thi, and Angélica Maciel Gomes for helpful discussions and for critical reading of the manuscript. The work from the M.J.B. laboratory has been supported by grants from the US Department of Energy, Office of Biological and Environmental Research, and Low Dose Scientific Focus Area; by multiple grants from the National Cancer Institute; by a grant from the Breast Cancer Research Foundation; and by two “Innovator awards” from the US Department of Defense. The work from the C.R.B. laboratory was funded by a grant from the NIH (GM059907). The work from the Q.-T.L. laboratory was supported by a grant from the NIH (R01CA161585-05). R.B. was supported by a postdoctoral fellowship from Susan G. Komen for the Cure (KG111229), and B.B. was supported by a National Science Foundation predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609135113/-/DCSupplemental.
References
- 1.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10(2):148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 2.Radisky DC, Stallings-Mann M, Hirai Y, Bissell MJ. Single proteins might have dual but related functions in intracellular and extracellular microenvironments. Nat Rev Mol Cell Biol. 2009;10(3):228–234. doi: 10.1038/nrm2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barondes SH, Cooper DN, Gitt MA, Leffler H. Galectins. Structure and function of a large family of animal lectins. J Biol Chem. 1994;269(33):20807–20810. [PubMed] [Google Scholar]
- 4.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2(5):238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 5.Bhat R, et al. A regulatory network of two galectins mediates the earliest steps of avian limb skeletal morphogenesis. BMC Dev Biol. 2011;11:6. doi: 10.1186/1471-213X-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elad-Sfadia G, Haklai R, Ballan E, Gabius HJ, Kloog Y. Galectin-1 augments Ras activation and diverts Ras signals to Raf-1 at the expense of phosphoinositide 3-kinase. J Biol Chem. 2002;277(40):37169–37175. doi: 10.1074/jbc.M205698200. [DOI] [PubMed] [Google Scholar]
- 7.Vyakarnam A, Dagher SF, Wang JL, Patterson RJ. Evidence for a role for galectin-1 in pre-mRNA splicing. Mol Cell Biol. 1997;17(8):4730–4737. doi: 10.1128/mcb.17.8.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vyakarnam A, Lenneman AJ, Lakkides KM, Patterson RJ, Wang JL. A comparative nuclear localization study of galectin-1 with other splicing components. Exp Cell Res. 1998;242(2):419–428. doi: 10.1006/excr.1998.4111. [DOI] [PubMed] [Google Scholar]
- 9.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: A small protein with major functions. Glycobiology. 2006;16(11):137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 10.Beliveau A, et al. Raf-induced MMP9 disrupts tissue architecture of human breast cells in three-dimensional culture and is necessary for tumor growth in vivo. Genes Dev. 2010;24(24):2800–2811. doi: 10.1101/gad.1990410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: The role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17(3):320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan S. Sialic acid as a tumor marker. Ann Clin Lab Sci. 1994;24(4):376–384. [PubMed] [Google Scholar]
- 14.Dalotto-Moreno T, et al. Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease. Cancer Res. 2013;73(3):1107–1117. doi: 10.1158/0008-5472.CAN-12-2418. [DOI] [PubMed] [Google Scholar]
- 15.Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17(2):103–110. doi: 10.1007/s10911-012-9251-7. [DOI] [PubMed] [Google Scholar]
- 16.Reichmann E, Ball R, Groner B, Friis RR. New mammary epithelial and fibroblastic cell clones in coculture form structures competent to differentiate functionally. J Cell Biol. 1989;108(3):1127–1138. doi: 10.1083/jcb.108.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montesano R, Schaller G, Orci L. Induction of epithelial tubular morphogenesis in vitro by fibroblast-derived soluble factors. Cell. 1991;66(4):697–711. doi: 10.1016/0092-8674(91)90115-f. [DOI] [PubMed] [Google Scholar]
- 18.Hirai Y, et al. Epimorphin functions as a key morphoregulator for mammary epithelial cells. J Cell Biol. 1998;140(1):159–169. doi: 10.1083/jcb.140.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori H, et al. Transmembrane/cytoplasmic, rather than catalytic, domains of Mmp14 signal to MAPK activation and mammary branching morphogenesis via binding to integrin β1. Development. 2013;140(2):343–352. doi: 10.1242/dev.084236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper DN, Barondes SH. Evidence for export of a muscle lectin from cytosol to extracellular matrix and for a novel secretory mechanism. J Cell Biol. 1990;110(5):1681–1691. doi: 10.1083/jcb.110.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepur A, et al. Galectin-3 endocytosis by carbohydrate independent and dependent pathways in different macrophage like cell types. Biochim Biophys Acta. 2012;1820(7):804–818. doi: 10.1016/j.bbagen.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Seelenmeyer C, et al. Cell surface counter receptors are essential components of the unconventional export machinery of galectin-1. J Cell Biol. 2005;171(2):373–381. doi: 10.1083/jcb.200506026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizqiawan A, et al. Autocrine galectin-1 promotes collective cell migration of squamous cell carcinoma cells through up-regulation of distinct integrins. Biochem Biophys Res Commun. 2013;441(4):904–910. doi: 10.1016/j.bbrc.2013.10.152. [DOI] [PubMed] [Google Scholar]
- 24.Voss PG, et al. Dissociation of the carbohydrate-binding and splicing activities of galectin-1. Arch Biochem Biophys. 2008;478(1):18–25. doi: 10.1016/j.abb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belardi B, O’Donoghue GP, Smith AW, Groves JT, Bertozzi CR. Investigating cell surface galectin-mediated cross-linking on glycoengineered cells. J Am Chem Soc. 2012;134(23):9549–9552. doi: 10.1021/ja301694s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu AM, et al. Differential affinities of Erythrina cristagalli lectin (ECL) toward monosaccharides and polyvalent mammalian structural units. Glycoconj J. 2007;24(9):591–604. doi: 10.1007/s10719-007-9063-y. [DOI] [PubMed] [Google Scholar]
- 27.Hirabayashi J, et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim Biophys Acta. 2002;1572(2-3):232–254. doi: 10.1016/s0304-4165(02)00311-2. [DOI] [PubMed] [Google Scholar]
- 28.Stowell SR, et al. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 2008;283(15):10109–10123. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs CL, et al. Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry. 2001;40(43):12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 30.Shibuya N, et al. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2-6)Gal/GalNAc sequence. J Biol Chem. 1987;262(4):1596–1601. [PubMed] [Google Scholar]
- 31.Amano M, Galvan M, He J, Baum LG. The ST6Gal I sialyltransferase selectively modifies N-glycans on CD45 to negatively regulate galectin-1-induced CD45 clustering, phosphatase modulation, and T cell death. J Biol Chem. 2003;278(9):7469–7475. doi: 10.1074/jbc.M209595200. [DOI] [PubMed] [Google Scholar]
- 32.Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dyn. 2006;235(12):3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152(1):75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fata JE, et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306(1):193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julien S, et al. ST6GalNAc I expression in MDA-MB-231 breast cancer cells greatly modifies their O-glycosylation pattern and enhances their tumourigenicity. Glycobiology. 2006;16(1):54–64. doi: 10.1093/glycob/cwj033. [DOI] [PubMed] [Google Scholar]
- 36.Recchi MA, Harduin-Lepers A, Boilly-Marer Y, Verbert A, Delannoy P. Multiplex RT-PCR method for the analysis of the expression of human sialyltransferases: application to breast cancer cells. Glycoconj J. 1998;15(1):19–27. doi: 10.1023/a:1006983214918. [DOI] [PubMed] [Google Scholar]
- 37.Picco G, et al. Over-expression of ST3Gal-I promotes mammary tumorigenesis. Glycobiology. 2010;20(10):1241–1250. doi: 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burchell JM, Mungul A, Taylor-Papadimitriou J. O-linked glycosylation in the mammary gland: Changes that occur during malignancy. J Mammary Gland Biol Neoplasia. 2001;6(3):355–364. doi: 10.1023/a:1011331809881. [DOI] [PubMed] [Google Scholar]
- 39.Kreunin P, Yoo C, Urquidi V, Lubman DM, Goodison S. Proteomic profiling identifies breast tumor metastasis-associated factors in an isogenic model. Proteomics. 2007;7(2):299–312. doi: 10.1002/pmic.200600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recchi MA, et al. Multiplex reverse transcription polymerase chain reaction assessment of sialyltransferase expression in human breast cancer. Cancer Res. 1998;58(18):4066–4070. [PubMed] [Google Scholar]
- 41.Lin S, Kemmner W, Grigull S, Schlag PM. Cell surface alpha 2,6 sialylation affects adhesion of breast carcinoma cells. Exp Cell Res. 2002;276(1):101–110. doi: 10.1006/excr.2002.5521. [DOI] [PubMed] [Google Scholar]
- 42.Ingrassia L, et al. Anti-galectin compounds as potential anti-cancer drugs. Curr Med Chem. 2006;13(29):3513–3527. doi: 10.2174/092986706779026219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.