Significance
Rab GTPases regulate vesicle traffic within the cell by switching between active (GTP-bound) and inactive (GDP-bound) states. The switch II region of Rab proteins undergoes a significant conformational change to switch between states. Rab1 is hijacked during intracellular Legionella pneumophila infection by bacterial effector-mediated posttranslational modifications of the switch II region, a unique mechanism for regulation of Rab function. We present new evidence that Rab1 is endogenously modified within switch II by TGF-β activated kinase 1 (TAK1), a kinase crucial for responding to infection. We show phosphorylation of Rab1 is necessary for normal Rab1 function. Interestingly, phosphorylation of Rab1 is competed during Legionella infection, adding to evidence that Legionella target substrates of the innate immunity kinase TAK1.
Keywords: chemical genetics, posttranslational modification, kinase substrates, vesicle trafficking, Rab GTPases
Abstract
TGF-β activated kinase 1 (TAK1) is a critical signaling hub responsible for translating antigen binding signals to immune receptors for the activation of the AP-1 and NF-κB master transcriptional programs. Despite its importance, known substrates of TAK1 are limited to kinases of the MAPK and IKK families and include no direct effectors of biochemical processes. Here, we identify over 200 substrates of TAK1 using a chemical genetic kinase strategy. We validate phosphorylation of the dynamic switch II region of GTPase Rab1, a mediator of endoplasmic reticulum to Golgi vesicular transport, at T75 to be regulated by TAK1 in vivo. TAK1 preferentially phosphorylates the inactive (GDP-bound) state of Rab1. Phosphorylation of Rab1 disrupts interaction with GDP dissociation inhibitor 1 (GDI1), but not guanine exchange factor (GEF) or GTPase-activating protein (GAP) enzymes, and is exclusive to membrane-localized Rab1, suggesting phosphorylation may stimulate Rab1 membrane association. Furthermore, we found phosphorylation of Rab1 at T75 to be essential for Rab1 function. Previous studies established that the pathogen Legionella pneumophila is capable of hijacking Rab1 function through posttranslational modifications of the switch II region. Here, we present evidence that Rab1 is regulated by the host in a similar fashion, and that the innate immunity kinase TAK1 and Legionella effectors compete to regulate Rab1 by switch II modifications during infection.
Cellular response to microbial infection is a complex, coordinated process that is initiated by innate immune receptors at the cell surface in response to cytokines or pathogen-associated molecular patterns. Pattern recognition receptors trigger cellular-response cascades culminating in activation of two master transcriptional programs, AP-1 and NF-κB, which drive cytokine production and recruitment of immune cells. In particular, Toll-like receptor (TLR) signaling cascades require the intricate orchestration of activation of downstream pathway components through the formation of diverse complexes and intensive reliance on posttranslational modifications for regulation. Ultimately, these pathways converge on the activation of an essential kinase, TGF-β activated kinase 1 (TAK1), which is responsible for translating receptor activation for the activation of these master transcriptional programs (1).
In the early phase following activation of receptors such as TLR2 or -4, an unusual nondegradative ubiquitin scaffold is assembled, leading to the activation of TAK1, also known as MAP3K7. Once activated, TAK1 serves two roles. First, TAK1 acts as a canonical MAPKKK by phosphorylating the MAPKKs MKK4/7 and MKK3/6. These MKKs phosphorylate and activate p38 and JNK, respectively, initiating AP-1–mediated transcription. Second, TAK1 provides a priming phosphorylation to IKKβ, which colocalizes with TAK1 to M1-poly-Ub chains generated by TRAF6 upon TLR activation (2, 3). Activation of IKKβ leads to the activation IKKα, degradation of IκBα, and finally, activation of NF-κB–driven transcription. The known direct substrates of TAK1 are limited to these protein kinases, TAK1 binding protein 1 (TAB1), and an additional protein kinase, AMPK (4, 5). TAK1 is primarily viewed as an initiator of kinase signaling cascades that lead to transcription factor activation.
Kinases often serve as signaling relays, transferring phosphorylation down a cascade of kinases, but also commonly function as direct effectors of biochemical processes via phosphorylation of enzymes from many classes. Given the importance of TAK1 as the terminal output of pattern-recognition receptor activation, we wondered if TAK1 might possess direct substrates beyond the three characterized classes of downstream kinases. Only one study has characterized a small number of downstream targets of TAK1 using quantitative phosphoproteomics (6). Although no direct TAK1–substrate relationships were established, Gene Ontology term enrichment of TAK1-regulated phosphoproteins suggested involvement in GTPase regulation and membrane organization. Other studies have suggested a role for TAK1 in directly regulating protein degradation to prevent accumulation of reactive oxygen species (7). Thus, we turned to the analog-specific kinase covalent capture methodology (8, 9) to identify direct TAK1 substrates in vitro. Through this method, we identified hundreds of candidate substrates.
We decided to focus, in particular, on a novel phosphorylation site within a dynamic region of small Ras-like GTPase Rab1. Rab proteins are the largest family of small Ras-like GTPases and serve to regulate many steps of membrane trafficking. These proteins act as molecular switches, cycling through active GTP-bound and inactive GDP-bound states. Two regions of the GTPases, termed switch I and switch II, undergo significant conformational shifts between these states, altering the ability of the protein to bind interactors. Rab nucleotide state, and therefore signaling, is tightly regulated by guanine exchange factors (GEFs) and GTPase-activating proteins (GAPs). By binding Rab proteins in the cytoplasm, GDP dissociation inhibitors (GDIs) sequester inactive Rab proteins to further control Rab activation. Although the C-terminal tails of Rab proteins are geranylgeranylated to mediate insertion into membranes, there are few examples of regulation of Rab proteins, or small GTPases in general, by posttranslational modification of the core GTPase domain. Although phosphorylation of Rab proteins, and other Ras-like GTPases, has been previously observed on the C-terminal tail and other outlying regions, there is little consensus on the regulatory effect of these phosphorylations (10–13). The strongest examples of small GTPase regulation by posttranslational modification are when the core GTPase domain is modified, rather than tails. Such modifications are almost exclusively the result of infection by pathogens, such as phosphorylation of switch I of immunity-related GTPases by a secreted Toxoplasma gondii kinase in mice (14) or AMPylation and phosphocholination of the switch region of Rab1 by Legionella pneumophila (15, 16).
Here, we demonstrate that TAK1 phosphorylation of Rab1 within the dynamic switch II region is key to Rab1 signaling. Phosphorylation of Rab1 is necessary for normal Rab1 function in maintaining Golgi structure, and disrupts interaction with GDI1, allowing for activation of Rab1. More interestingly, TAK1-mediated phosphorylation of Rab1 competes with L. pneumophila during intracellular infection. We believe Rab1 is a newly recognized hotspot for regulation via posttranslational modifications, both by a bacterial pathogen and now by TAK1, a host kinase responsible for responding to infection.
Results
Identification and Validation of TAK1 Substrates.
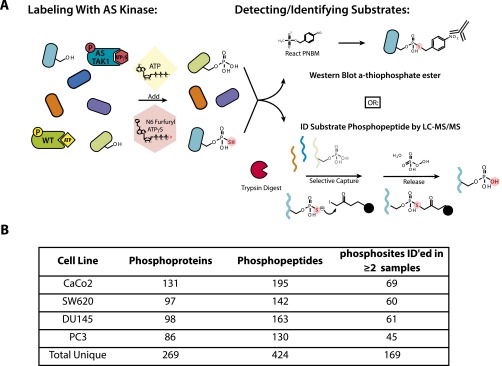
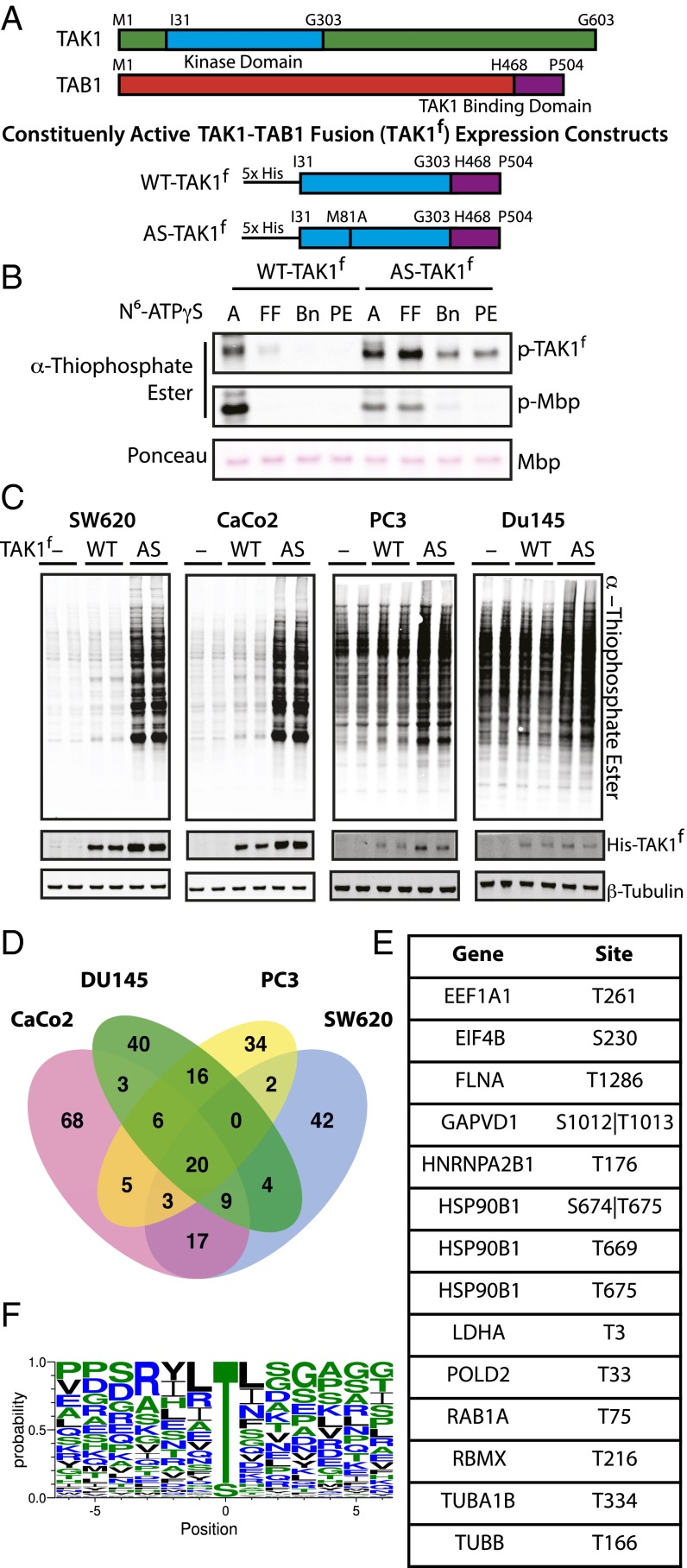
We first sought to identify direct substrates of TAK1 using a chemical genetic, analog-specific (AS) kinase approach (Fig. S1A) (8, 9). Mutation of a single bulky residue within the active site of a kinase, termed the active-site gatekeeper residue, to alanine or glycine expands the native ATP binding pocket. This mutation allows the kinase to accept N6-substituted ATPγS analogs, bulky variants of ATP that fit in the newly expanded active site but not the active sites of WT kinases, creating an AS kinase. The AS-kinase transfers the γ-thiophosphate of the ATP analog to its substrates. This thiophosphorylation acts as a uniquely reactive chemical handle that can be alkylated for detection of substrates by Western blotting or used to affinity-purify and identify substrate proteins by liquid chromatography (LC)-MS/MS. We generated a constitutively active form of TAK1 by expressing and purifying from insect cells a fused TAK1 construct containing the kinase domain of TAK1 fused to the TAK1-activating domain of binding partner TAB1 (Fig. 1A) (17). We will refer to this fusion construct as TAK1f, with WT indicating no mutations to the gatekeeper residue. AS-TAK1f was generated by mutation of gatekeeper methionine 81 to alanine. We tested the specificity and preference of AS-TAK1f for N6-substituted ATPγS analogs through an in vitro kinase assay using myelin basic protein (MBP) as a generic substrate (Fig. 1B). AS-TAK1f used both ATPγS and N6-furfuryl-ATPγS efficiently for autophosphorylation and transphosphorylation of MBP. In contrast, WT-TAK1f was largely incapable of using any bulky ATP analogs. N6-furfuryl-ATPγS was used for lysate-labeling experiments to ensure any detected thiophosphorylation was the result of AS-TAK1f activity.
Fig. S1.
Schematic of AS substrate capture and identification of TAK1 substrates. (A) Schematic of lysate labeling with AS-TAK1 vs. WT-TAK1. Kinase is spiked into cell lysates with a bulky ATP analog, here N6 furfuryl ATPγS, which selectively thiophosphorylates substrates. Thiophosphorylated proteins can be detected two ways: by Western blot after alkylation to form a thiophosphate ester, or by digestion and capture on a thiol-reactive iodoacetyl resin, release and analysis by LC-MS/MS. (B) Numbers of unique phosphoproteins and phosphopeptides identified in each cell line and in total.
Fig. 1.
Characterization of AS-TAK1f and identification of TAK1 substrates. (A) Schematic of full-length TAK1, TAB1, and constituently active fusion constructs of WT-TAK1f and AS-TAK1f used for protein purification. (B) In vitro kinase assay with TAK1f, MBP, and bulky N6-substituted ATPγS analogs (A, ATPγS; Bn, N6 benzyl; FF, N6 furfuryl; PE, N6 phenethyl). Thiophosporylation was evaluated by Western blot. (C) Lysate from SW620, CaCo2, PC3, and Du145 cells were labeled with no kinase (−), His-tagged WT-TAK1f (WT), or His-tagged AS-TAK1f (AS) in biological duplicate. (D) Venn diagrams of phosphoproteins identified in four cell cancer lines, colorectal (SW620, CaCo2) and pancreatic (DU145, PC3). (E) Phosphosites exclusive to and identified in all eight individual AS-TAK1f–labeled samples. (F) TAK1 consensus motif derived from all phosphopeptides identified in ≥2 samples.
We next sought to identify proteins selectively thiophosphorylated by AS-TAK1f in lysates from four cell lines from two cancer types, colorectal (CaCo2, SW620) and pancreatic (PC3, DU145). These cell lines were selected because TAK1 has been shown to be particularly important in colorectal and prostate cancers (18, 19). Lysates were individually labeled by spiking in N6-furfuryl-ATPγS and purified AS-TAK1f, WT-TAK1f, or with no added kinase. A portion of each sample was analyzed by Western blot, where an obvious increase in thiophosphorylation is observed exclusively in the AS-labeled colorectal cell line samples; the contrast between AS and WT or no-kinase conditions is less obvious in the pancreatic cell lines, as these cell lines displayed much higher background thiophosphorylation (Fig. 1C). We attribute this difference, in part, to variability between cell lines in the background proteome activity toward the ATPγS analog. The remainder of the thiophosphorylated lysates were digested, thiophosphorylated peptides covalently captured, converted to phosphorylated peptides upon elution from resin, and analyzed by MS. Despite differences in levels of thiophosphorylation by Western blot, many more phosphopeptides were identified in all AS-TAK1f–labeled samples versus controls. (Datasets S1–S4). Thus, although Western detection of thiophosphorylation is a useful tool, it is limited in comparison with MS results as observed in this study and others (20).
The data were filtered by cell type to exclude background phosphopeptides from the WT-TAK1f and no-kinase conditions (21), leaving phosphopeptides exclusive to AS-TAK1f. In total, 269 phosphoproteins yielding 424 phosphopeptides were identified as candidate TAK1 substrates (Fig. S1B). A list of all candidate substrate phosphopeptides identified is available in Dataset S5. The difference in peptide versus protein number is a result of the identification of multiple phosphopeptides per protein and, in some cases, a single peptide identified multiple times with differing sites of phosphorylation. Whereas all cell lines shared a set of 20 substrate proteins, generally substrates were shared more frequently between cell lines of the same origin, with many substrates uniquely identified in a single cell line (Fig. 1D). Although the stochastic nature of shotgun LC-MS/MS identification may explain some of the lack of overlap, we believe the method of capture used is able to identify cell-type–specific substrates. Conservation of a substrate across cell types may be indicative of a central, conserved function, and therefore a useful means to triage substrates for further study. Fourteen phosphopeptides were identified in all AS-TAK1f samples analyzed (Fig. 1E). To further analyze the substrate preferences of TAK1, we generated a TAK1 consensus sequence from phosphopeptides identified in at least two samples (Fig. 1F) (22). We observed a strong preference for phosphorylation of threonine, with some preference for aliphatic −1 and +1 residues.
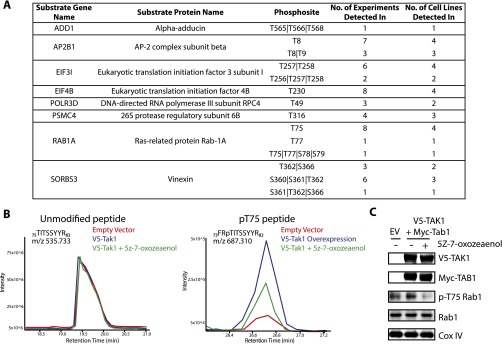
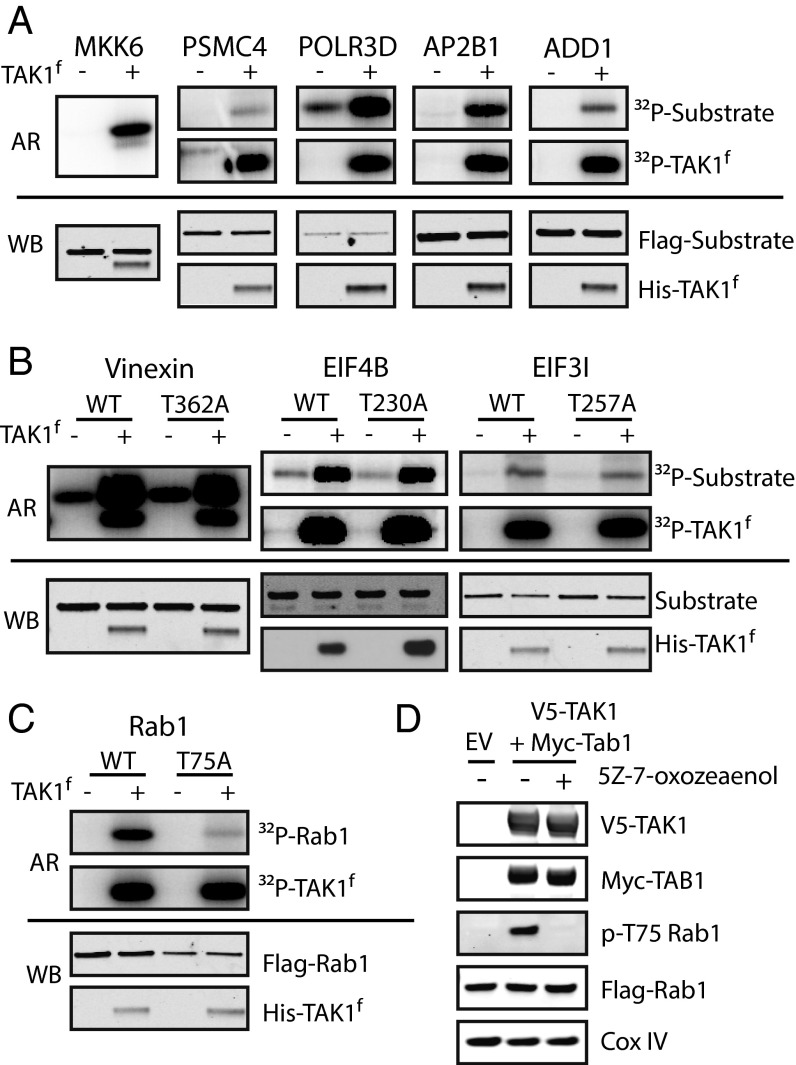
To corroborate our MS results, a subset of substrates identified by MS were selected for further validation (Fig. S2A). We assessed the ability of TAK1 to phosphorylate substrates overexpressed with N-terminal Flag or GST tags and immunoprecipitated from HEK-293Ts by in vitro radioactive kinase assay with WT-TAK1f. As a positive control, we sought to confirm that our WT-TAK1f would strongly phosphorylate a known substrate, MKK6. MKK6 was not identified by MS because of the presence of cysteine in tryptic MKK6 peptides containing TAK1 phosphorylation sites, as Cys-containing peptides are permanently retained on the capture resin (9). TAK1 strongly phosphorylated kinase-dead MKK6 as shown by the incorporation of 32P in the WT-TAK1f–labeled sample (Fig. 2A). Of the eight candidate substrates tested, only PSMC4 was not strongly phosphorylated by WT-TAK1f. The remaining seven substrates were strongly phosphorylated by TAK1 (Fig. 2 A–C). Interestingly, WT-TAK1f was clearly able to phosphorylate more than the single identified site on three substrates (Vinexin, EIF4B, EIF3I), as shown by incorporation of 32P into the nonphosphorylatable T to A mutants (Fig. 2B). It is possible additional phosphosites in these proteins are within regions not amenable for detection by trypsin-based LC-MS/MS and were therefore not detected. Given the high rate of substrate validation, we have high confidence in the validity of the candidate substrates identified by MS.
Fig. S2.
Validation of TAK1 substrates identified by the AS capture method. (A) Table of substrates validated in vitro (assayed in Fig. 2) with number of identifications and phosphosite corresponding to Fig. 2. (B) HEK-293 cells stably expressing Flag-Rab1 were transfected with either empty vector (EV) or full-length V5-TAK1, were treated with DMSO or drugged 2.5 μM TAK1 inhibitor 5z-7-oxozeaenol for 1 h, as indicated. Flag-Rab1 was immunoprecipitated, subject to in-gel digestion, and analyzed by LC-MS/MS. Here, raw abundance, presented as the extracted ion chromatogram, of the pT75 peptide varies across conditions, whereas the unmodified peptide remains constant. Notably, we observed that phosphorylation of T75 forces a missed trypsin cleavage at neighboring R74 and is exclusively detected with this missed cleavage, thus the unmodified and modified peptide have differing sequences. (C) Phosphorylation of endogenous Rab1 increases after overexpression of V5-TAK1 and Myc-Rab1. Inhibition of TAK1 with 5z-7-oxozeaenol for 1 h at 2.5 μM reduces pT75-Rab1 to below basal levels.
Fig. 2.
Validation of TAK1 substrates in vitro and in vivo. (A–C) In vitro radioactive kinase assays with GST- or Flag-tagged TAK1 substrates. Substrates immunoprecipitated from HEK-293Ts were incubated with purified WT-TAK1f (300 nM) and γ[32P]ATP, run on a gel, and imaged by autoradiography (AR). Aliquots removed before γ[32P]ATP addition were used to assess loading by Western blot (WB). MKK6 (kinase dead) is a known substrate and positive control. (D) HEK-293s stably expressing Flag-Rab1 were transfected with either empty vector (EV) or full-length V5-TAK1 and Myc-TAB1 were analyzed by Western blot for pT75 Rab1. One condition was dosed with 2.5-μM TAK1 inhibitor 5z-7-oxozeaenol for 1 h.
TAK1 Selectively Phosphorylated GDP-Bound Rab1.
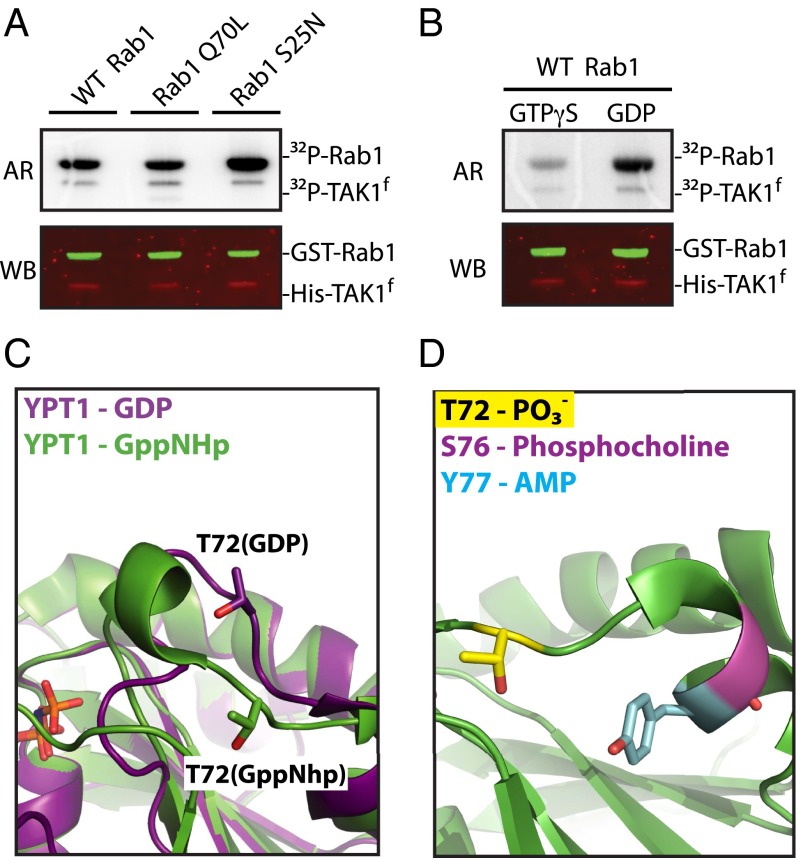
Rab1 was the sole protein tested that was phosphorylated by WT-TAK1f at a single site, T75, within its switch II region, in all four cell lines (Fig. 2C). Switch II is a conserved region within the catalytic domain of GTPases that undergoes a conformational shift between the active (GTP-bound) and inactive (GDP-bound) states (23) and is not a common site of posttranslational modification. This selective phosphorylation motivated us to carry out in vivo validation with a polyclonal antibody raised against pT75 Rab1. In HEK-293 cells stably expressing Flag-Rab1, overexpression of full-length V5-TAK1 and activating partner Myc-TAB1 (24) led to increased phosphorylation of Rab1 T75. Addition of a TAK1-selective inhibitor, 5z-7-oxozeaenol (25), eliminated phosphorylation of Rab1 (Fig. 2D). The increased abundance of pT75 Rab1 following TAK1 overexpression and loss of phosphorylation upon inhibition of TAK1 was also observed by LC-MS/MS analysis of immunoprecipitated Rab1 (Fig. S2B). A similar increase in phosphorylation of endogenous Rab1 is observed upon TAK1 overexpression in normal HEK-293T, as is a decrease in phosphorylation after TAK1 inhibition (Fig. S2C). The modulation of pT75 Rab1 levels upon manipulation of TAK1 catalytic activity by overexpression and inhibition suggests Rab1 is a direct substrate of TAK1 in vivo.
Because switch II occupies two distinct conformations, we hypothesized that the nucleotide state of Rab1 may influence the ability of TAK1 to phosphorylate T75. Radiometric in vitro kinase assays with GST-Rab1 purified from a bacterial expression system and WT-TAK1f showed preferential phosphorylation of the inactive, GDP-locked Rab1S25N (26) and reduced phosphorylation of active-state mimetic Rab1Q70L (27) (Fig. 3A). Correspondingly, TAK1 preferentially phosphorylated Rab1 loaded with GDP by nucleotide exchange versus nonhydrolyzable GTPγS (Fig. 3B). The preference of TAK1 for GDP-Rab1 may be explained by available structural data of the yeast homolog of Rab1, Ypt1. Alignment of the structure of GDP (PDB ID code 2BCG) and GTP mimetic GppNHp- (PDB ID code 1YZN) bound Ypt1 shows the switch II region to be flipped outward from the body of the protein, exposing T75-equivalent residue T72 (Fig. 3C). Our findings suggest that T75 is only accessible for phosphorylation by TAK1 when GDP binding to Rab1 causes the switch II region to become disordered (23). Interestingly, the intracellular pathogen L. pneumophila is well documented to hijack the function of Rab1 in infected cells by posttranslational modification of nearby switch II residues, including adenylylation, also known as AMPylation, of Y80 and phosphocholination of S79 (Fig. 3D) (15, 16, 28).
Fig. 3.
TAK1 preferentially phosphorylates GDP-bound Rab1. (A) In vitro kinase assay of purified GST-Rab1 mutants (10 μM) and WT-TAK1f (100 nM) imaged by autoradiography (AR) and Western blot (WB) as loading control. (B) In vitro kinase assay of WT-TAK1f (100 nM) and purified GST-WT Rab1 (10 μM) loaded with the indicated nucleotide. (C) Alignment of Rab1-homolog Ypt1 structures bound to GDP (PDB ID code 2BCG) or GTP analog GppNHp (PDB ID code 1YZN). Ypt1 T72 aligns to Rab1 T75. (D) Mapping of posttranslationally modified Rab1 residues within switch II where S76 and Y77 (Rab1 S79 and Y80 as aligned to YPT1 structure PDB ID code1YZN) are sites of Legionella-derived modifications.
Phosphorylation of Rab1 Disrupts Interaction with GDI but Not GAP or GEF.
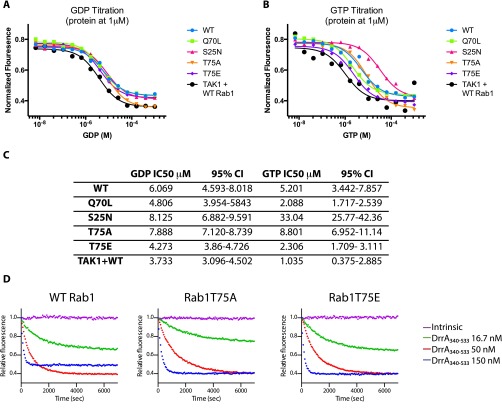
During activation, switch II becomes more ordered and T75 is flipped inward toward the core of Rab1 (Fig. 3C). It is possible that phosphorylation may block Rab1 from binding GTP and localizing to the membrane by sterically hindering the conformational shift of switch II. To determine whether phosphorylation may disrupt GTP binding, we assayed the nucleotide affinities of Rab1 mutants using 2′-deoxy-3′-O-(N-methylanthraniloyl) (mant)-dGDP (mant-GDP) –loaded Rab1, which forms a fluorescent complex. EDTA was used to catalyze nucleotide exchange while titrating unlabeled GDP or GTP (29), with a reduction in fluorescence caused by mant-GDP displacement corresponding to affinity for the titrated nucleotide. All constructs tested, including WT-Rab1 phosphorylated by preincubation with TAK1 and ATP, maintained a similar affinity for GDP (Fig. S3A). A slight increase in affinity for GTP was observed for Rab1Q70L, phosphomimetic Rab1T75E, and WT-Rab1 preincubated with TAK1 and ATP (Fig. S3 B and C). The catalysis of nucleotide exchange by EDTA suggests that phosphorylation does not prevent activation of Rab1, and may in fact enhance GTP affinity.
Fig. S3.
GDP and GTP affinities for GST-Rab1 mutants and GEF catalyzed GDP exchange. (A) mant-GDP is first loaded on GST-Rab1 mutants and excess mant-GDP removed. A single concentration of mant-GDP bound Rab1 (1 μM) is subject to EDTA (5 mM) catalyzed nucleotide exchange in the presence of varying conditions of free GDP (x axis). The reaction is allowed to proceed for 1 h, and the remaining mant-GDP–Rab1 complex is quantified by fluorescence and normalized to an EDTA-free condition (y axis) (n = 1). (B) Similar to A, except with varying concentrations of free GTP (n = 1). (C) Quantification of GDP and GTP titrations in A and B, where IC50 values were obtained from sigmoidal fits (CI, confidence interval). (D) Representative single experiment data for DrrA340-533 catalyzed nucleotide exchange from all mant-GDP–bound Rab1 mutants in the presence of excess GTP (see Fig. 4 A and B).
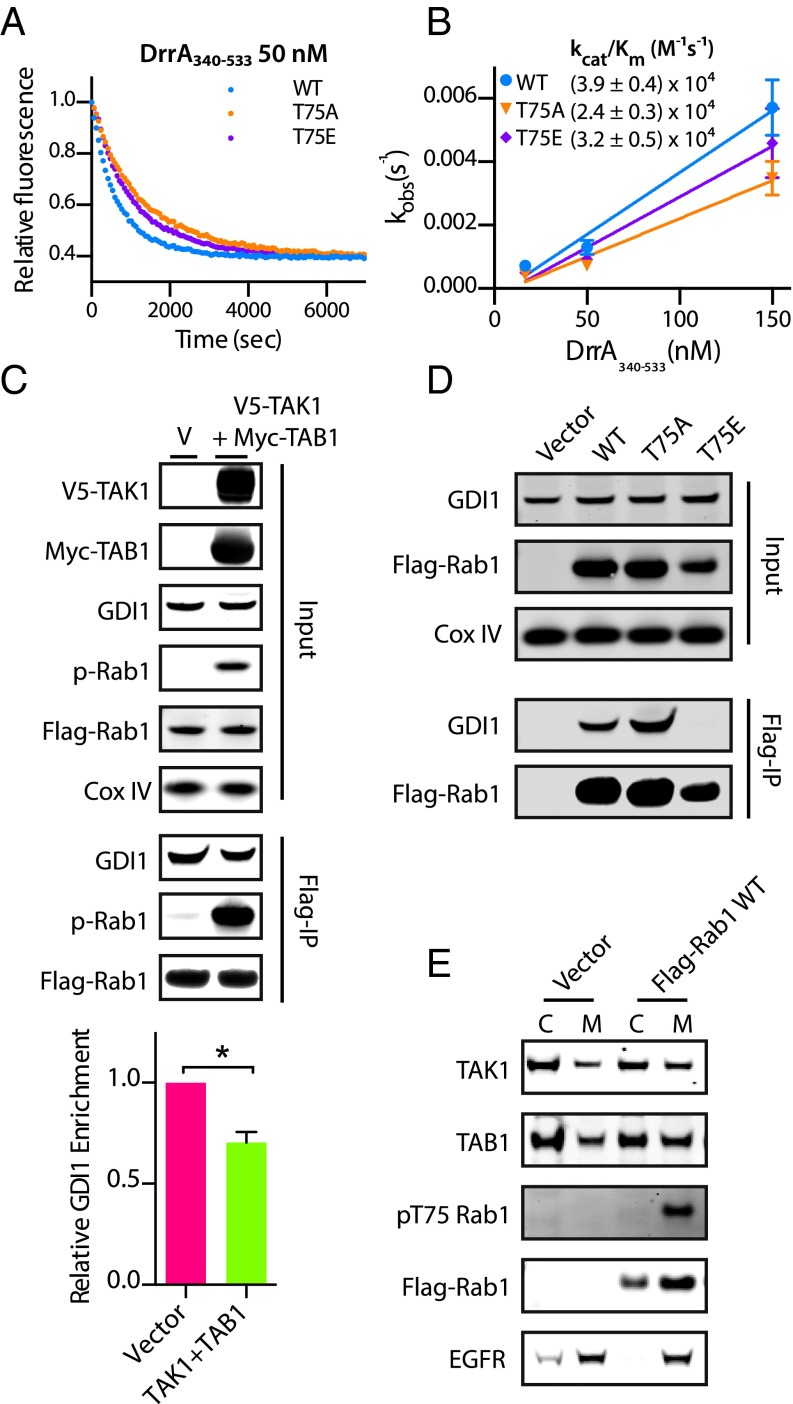
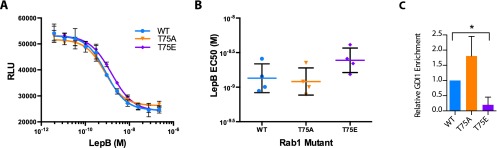
Multiple studies have shown that phosphocholination and AMPylation of Rab1 switch II manipulate Rab1 function by blocking the ability of Rab1 to interact with GEFs and GAPs (16, 28, 30), because switch II serves as the primary interface for these binding events. To investigate whether phosphorylation of Rab1T75 may have a similar effect, we assayed the ability of the Legionella Rab1 GEF DrrA (GEF domain only, residues 340–533) to catalyze the displacement of mant-GDP from WT and mutant Rab1 in vitro (Fig. 4 A and B and Fig. S3D). DrrA340–533 was selected for these assays as a result of numerous publications (30–32) describing similar experiments with this enzyme. We found no significant difference in the kcat/Km of DrrA340–533 toward WT Rab1 and Rab1T75E, and only a slight difference with Rab1T75A; thus, we infer that it is unlikely phosphorylation of T75 prevents Rab1 interaction with GEFs or interferes with activation. Phosphomimetic mutant Rab1T75E yields similarly insignificant effects on the ability of a Legionella GAP, LepB (33), to stimulate Rab1 hydrolysis of GTP, as measured with the Promega GTPase-Glo system (34). Briefly, increasing concentrations of LepB with excess GTP held at a constant concentration are added to wells containing a constant concentration of Rab1 and allowed to react for 1 h. The amount of remaining, unhydrolyzed GTP in each condition is detected by luminescence-coupled assay, plotted against GAP concentration, and a LepB EC50 is determined. We found no difference in LepB activity toward WT, T75E, or T75A (Fig. S4 A and B), which suggests phosphorylation does not interfere with inactivation of Rab1 by GAPs. Considering the GTP/GDP affinity, GEF assay, and GAP assay together, we conclude phosphorylation of Rab1 does not impact the ability of Rab1 to cycle between GDP- and GTP-bound states or interact with GAP and GEFs.
Fig. 4.
Phosphorylation of Rab1 disrupts interaction with GDI but not GEFs. (A) Measurement of mant-GDP dissociation from GST-Rab1 mutants by DrrA340–533 from a single representative experiment where each data point represents the mean of technical replicates (n = 3). (B) Observed rate constants (kobs) for DrrA catalyzed mant-GDP dissociation with error bars for mean ± SD (n = 2) and extrapolated catalytic efficiencies (kcat/Km). (C) HEK-293s stably expressing Flag-Rab1 were transiently transfected with either empty vector (V) or full-length V5-TAK1 and Myc-TAB1. Lysates were subject to immunoprecipitation of Flag-Rab1 using α-Flag antibody-coupled magnetic beads and analyzed by Western blot for coimmunoprecipitation of GDI1. The bar graph represents the ratio of precipitated GDI:Flag-Rab1 (n = 2). *P < 0.05, unpaired t test. (D) HEK-293Ts were transiently transfected with vector, Flag-Rab1 WT, T75A, or T75E and subject to immunoprecipitation of Flag-Rab1. Quantitation is in Fig. S4B. (E) Cell fractionation of HEK-293Ts transiently transfected with empty vector or Flag-Rab1 with cytoplasmic (C) or membrane (M) fractions.
Fig. S4.
GAP-catalyzed hydrolysis of GTP by Rab1 is not disrupted by phosphomemetic mutation T75E, but interaction with GDI1 is. (A) Representative data from a single experiment of a GAP assay using Promega GTPase-Glo. Error bars are mean ± SD for three wells per condition. Purified GST-Rab1 mutants (30 μM) were incubated with excess GTP and varying concentrations of Legionella Rab1 GAP LepB for 1 h. The remaining GTP concentrations were assayed by a luminescence coupled assay and plotted versus LepB concentration. (B) LepB EC50 values calculated from the LepB curves in A for four independent experiments (black lines: mean EC50 ± SD for n = 4). (C) Quantitation related to Fig. 4D, where relative GDI enrichment represents the normalized ratio of immunoprecipitated GDI:Flag-Rab1 (n = 2). *P < 0.05, unpaired t test.
The subcellular location of Rab1 is also generally dictated by its nucleotide association, as recruitment of Rab1 to the membrane of the endoplasmic reticulum (ER) from the cytoplasm is concurrent with displacement of GDP and activation of Rab1 upon binding of GTP (35). Until this displacement occurs, inactive GDP-Rab1 is sequestered in the cytoplasm in complex with a GDI. Similar to their effect on GAPs and GEFs, Legionella-derived posttranslational modifications of Rab1 disrupt association with the GDI (31). We immunoprecipitated Flag-Rab1 from HEK-293 cells stably expressing Flag-Rab1 transfected with either vector or the combination of TAK1 and TAB1. The relative amount of GDI1 coprecipitating with Rab1 (GDI:Flag-Rab1) decreased in cells with highly phosphorylated Rab1 resulting from TAK1 and TAB1 overexpression (Fig. 4C). More strikingly, little to no GDI1 coimmunoprecipitates with Flag-Rab1T75E transiently overexpressed in HEK 293Ts, whereas Flag-Rab1T75A increases association with GDI1 (Fig. 4D and Fig. S4C). Disruption of the GDI:Rab1 complex by phosphorylation suggests pT75-Rab1 is available for activation and recruitment to the ER membrane. Thus, we examined the localization of pT75 Rab1 by cellular fractionation of HEK 293Ts transiently expressing Flag-Rab1 (Fig. 4E). Although total Flag-Rab1 is distributed between cytoplasmic and membrane fractions, pT75 Rab1 is exclusively detected in the membrane fraction, where Rab1 activation occurs. Taken together, these results suggest that phosphorylation of Rab1 by TAK1 may be an important precursor to GTP binding and activation by driving dissociation from the GDI.
Rab1 Phosphorylation Is Required to Maintain Golgi Structure.
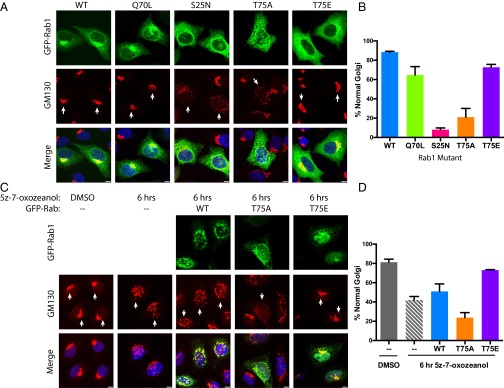
Rab1 is responsible for transporting vesicles from the ER to the Golgi. Disruption of Rab1 activity, by knockdown or overexpression of the dominant-negative Rab1S25N, results in fragmentation of the Golgi apparatus (36, 37). We tested the effect of overexpression of nonphosphorylatable T75A and phosphomimetic T75E Rab1 on Golgi structure to determine if the T75 site was critical for Rab1 function. Immunofluorescence was performed in HeLa cells transiently overexpressing GFP-Rab1 mutants (Fig. 5 A and B) and the Golgi was stained with a cis-Golgi marker, GM130. As previously shown, overexpression of Rab1S25N acts in a dominant-negative fashion, disrupting Golgi structure, whereas Rab1Q70L, the active-state mimic, has no effect on Golgi structure. Similarly, Rab1T75E has no effect on the Golgi. However, Rab1T75A acts in a dominant-negative fashion similar to Rab1S25N to cause extensive Golgi fragmentation.
Fig. 5.
Fragmentation of the Golgi is a result of overexpression of Rab1T75A or inhibition of TAK1. (A) Representative images of HeLa cells transfected with GFP-Rab1 in green and stained for GM130, a cis-Golgi marker in red, and DAPI in blue. (Scale bar, 5 μm.) (B) Quantitation of immunofluorescence experiment (n = 3 replicates, 33 cells per replicate). (C) Representative images of HeLa cells stained for GM130 after dosing with 2.5-μM TAK1 inhibitor 5z-7-oxozeaenol for 6 h and preceding transfection with GFP-Rab1 where indicated. (Scale bar, 5 μm.) (D) Quantitation of immunofluorescence experiment (n = 3 replicates, 33 cells per replicate).
To complement these results, we assayed the effect of inhibiting TAK1 with 5z-7-oxozeaenol (25) on Golgi structure by immunofluorescence. Inhibition of TAK1 for 6 h leads to a marked disruption of normal Golgi structure versus DMSO (Fig. 5 C and D). This effect is largely rescued by overexpression of GFP-Rab1T75E, but not WT or T75A, in the presence of inhibitor. We conclude from these data that the phosphorylation of Rab1, or ability of Rab1 to be phosphorylated, plays an important role in ER to Golgi vesicle transport and in maintaining proper Golgi structure.
Innate Immunity Kinase TAK1 and Pathogen Legionella Compete to Posttranslationally Modify Rab1.
There is a well-established precedent for regulation of Rab1 function by posttranslational modification during microbial infection. The intracellular bacterial pathogen L. pneumophila uses posttranslational modifications of Rab1 in order to establish the Legionella-containing vacuole. Legionella effectors DrrA and AnkX are secreted into the host cell cytoplasm during infection and catalyze the AMPylation at Y80 and phosphocholination at S79 of Rab1, respectively (Fig. 3D) (15, 16, 30, 38). As discussed earlier, these modifications serve as locks on the Rab1 nucleotide state and block interactions with the host enzymes normally responsible for regulating Rab1. Two additional Legionella enzymes, SidD and Lem3, have cognate Rab1-demodifying activities (28, 39). Legionella maintains exquisite and tightly regulated control of Rab1 to mature its replication vacuole by carefully timing the sequential secretion of these effectors and subsequent recruitment of Rab1 to the Legionella-containing vacuole.
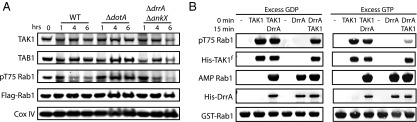
Because TAK1 is a kinase activated by pathogens, such as Legionella, and Legionella extensively modifies the Rab1 switch II region, we examined the interplay between TAK1-mediated phosphorylation of Rab1 and Legionella infection. HEK-293 cells stably expressing FCγIII receptor (to allow for opsonization and endocytosis of Legionella in HEK-293 cells) and Flag-Rab1 were infected with Legionella (WT), an isogenic strain lacking the Dot/Icm type IV secretion system (ΔdotA), an isogenic strain lacking the two known Rab1 posttranslational modifying enzymes, DrrA and AnkX (ΔankX,drrA), or left mock-infected (Fig. 6A). Some basal phosphorylation of Rab1 was detected in the mock-infected (0 h) sample. Phosphorylation of Rab1 increased slightly at 1 h in the WT condition, then tapered to below basal levels at 4 and 6 h. Infection with both ΔdotA and ΔankX,drrA strains lead to increased levels of pT75-Rab1 at 1 and 4 h, with levels remaining high at 6 h in ΔdotA. Deletion of AnkX and DrrA provided a moderate restoration of pT75-Rab1 levels versus WT infection, suggesting these enzymes may be responsible for outcompeting TAK1 for control of Rab1 during WT infection. We next considered the contribution of the AMPylation versus GEF activity of DrrA toward reducing pT75-Rab1 levels during WT infection. GST-Rab1 was incubated for 15 min with ATP and either WT-TAK1f or full-length DrrA in the presence of excess GTP or GDP, then incubated for an additional 15 min after addition of the remaining enzyme. Aliquots were removed and quenched at 0, 15, and 30 min and analyzed by Western blot (Fig. 6B). AMPylation of Rab1 is not affected by preexisting phosphorylation in the presence of either nucleotide. However, phosphorylation of Rab1 by TAK1 is significantly hindered only when excess GTP is present, suggesting that it is the DrrA-catalyzed Rab1-GTP binding, which reduces phosphorylation levels, not the presence of AMPylation, disrupting the TAK1 phosphorylation site. Although the contribution of phosphocholination and other factors to outcompeting Rab1 phosphorylation remain to be determined, these results suggest that TAK1 may be outcompeted in part by Legionella for modification and control of Rab1 during infection by the GEF activity of DrrA.
Fig. 6.
pT75 Rab1 levels decrease during Legionella infection because of the GEF activity of DrrA. (A) HEK-293 cells stably expressing FCγIII receptor and Flag-Rab1 were mock-infected (0 h), infected with WT, secretion deficient (ΔdotA), or AnkX and DrrA-deficient (ΔankX,drrA) Legionella for 1, 4, or 6 h and analyzed by Western blot. (B) Sequential modification of Rab1 by TAK1 and DrrA. Enzymes were added at 0 and 15 min, with aliquots of sample removed at 0 (just before enzyme addition), 15, and 30 min for analysis in the presence of excess GDP or GTP (30 μM).
Discussion
Here, we present an effort to identify a broad set of substrates of TAK1, an S/T kinase with crucial function in the innate immune system. We generated an ATP AS mutant of TAK1, termed AS-TAK1f, to selectively label, isolate, and identify novel substrates. We identified over 200 substrate proteins of TAK1, with a subset validated by in vitro kinase assays. TAK1 demonstrated a striking preference for threonine in the identified phosphosites. A recent study demonstrated that kinases with a β-branched residue in the conserved DFG loop drives specificity for threonine over serine (40). Fittingly, TAK1 contains a threonine at this position, T178. Although only one substrate, Rab1, was studied in depth in this study, a number of these substrates fit with known characteristics of TAK1 signaling. Filamin A functions as a scaffold for MKK4 and JNK association and activation of JNK (41, 42). MKK4 is a direct substrate of TAK1, thus it is possible TAK1 also associates with Filamin A to stimulate MKK4/JNK signaling. Additionally, TAK1 is known to be a client of HSP90 (43). TAK1-mediated phosphorylation of a close relative, endoplasmin (HSP90B1), suggests TAK1 is capable of interacting with additional heat-shock proteins.
We focused in particular on a novel TAK1 substrate, the GTPase Rab1. We demonstrate TAK1 phosphorylates Rab1 at a single site within the dynamic switch II region in vivo. Given the preference of TAK1 for GDP-bound Rab1 in vitro, we believe phosphorylation of Rab1 occurs in the inactive state. However, the GTP affinity of phosphorylated Rab1 is unchanged, GEF DrrA catalyzes nucleotide exchange of Rab1T75E efficiently, Rab1T75E can perform GAP-catalyzed GTP hydrolysis normally, and phosphorylation is present only on membrane-associated Rab1, suggesting phosphorylated Rab1 has a normal catalytic cycle and associates with membranes. We find phosphorylation of Rab1 disrupts interaction with GDI1, an interaction that is stabilized by the residues of switch II (44). Combined, these results suggest phosphorylation of Rab1 may serve to disrupt association with the GDI, and perhaps push Rab1 toward membrane association and activation rather than sequestration. A strong Golgi fragmentation phenotype was observed by immunofluorescence upon overexpression of nonphosphorylatable but not phosphomimetic Rab1, as well as inhibition of TAK1, suggesting that the ability to be phosphorylated is essential for Rab1 function, specifically in maintaining Golgi structure, and perhaps more widely in ER to Golgi vesicle transport. Thus, we propose that TAK1 phosphorylation of Rab1 is a priming step for Rab1 activation and integral component of the Rab1 activity cycle.
We believe these findings are evidence of regulation of Rab1 function by endogenous posttranslational modification within the catalytic domain of the protein. Seen in the context of recent studies and existing data, modification—especially phosphorylation—of switch II may be a widespread endogenous mechanism of Rab family regulation. The Phosphosite.org database contains phosphoproteomic evidence for switch II phosphorylation of at least 15 additional Rab GTPases, and several other small GTPases. Recent work from Mann and colleagues (45) identified phosphorylation by LRRK2 of the switch II regions of Rab3a, Rab8a, Rab10, and Rab12 as a driver of membrane localization. In addition, a few GTPases outside the Rab family, including Cdc42, Rac1, and Ran, are thought to be regulated by modification of switch II (46–48). Switch II has long been recognized for its importance in dictating the Rab activation state; it has now becoming clear that posttranslational modification of this region allows further, external control of Rab function.
Our interest in Rab1 stemmed from the extensive literature describing the ability of L. pneumophila to manipulate Rab1 function by posttranslational modifications to ensure maturation of the Legionella replication vacuole within the host cell (35, 38). Here, we suggest that a similar and endogenous mechanism, TAK1-mediated phosphorylation of Rab1, serves to regulate Rab1 function in normal conditions. We also show that phosphorylation of Rab1 is stimulated by secretion-deficient Legionella (ΔdotA) infection, yet is reduced during WT infection. We believe that TAK1 phosphorylation is outcompeted by secreted Legionella factors, as evidenced by the observed reduction of TAK1 driven phosphorylation of Rab1 exposed to GEF DrrA and the rescue of pT75 Rab1 levels in ΔankX,drrA-infected cells. Interestingly, Legionella also stimulate NF-κB, p38, and JNK signaling through TLR-independent mechanisms during infection (49, 50). TAK1 normally serves to respond to TLR signaling and activate these same pathways during infection. In addition, Yersinia pestis has been shown to inhibit innate immune signaling through acetylation and inactivation of TAK1 (51). Thus, we hypothesize that Legionella has evolved mechanisms to mimic, or perhaps directly manipulate, TAK1 function during infection to control the innate immune response, as evidenced by activation of NF-κB, p38, and JNK, and now by modification of Rab1. The unbiased identification of TAK1 substrates has revealed phosphorylation of Rab1 switch II, a hotspot for posttranslational modification, as a novel regulatory mechanism and potential unique component of innate immunity.
Methods
TAK1 was expressed in SF9 cells and purified as previously described (17). Covalent capture of TAK1 substrates was performed on 2 mg of lysate per sample labeled with 1% (wt/wt) of purified TAK1 and 250 μM N6-furfuryl-ATPγS. Covalent capture of substrates was performed using Sulfolink resin with oxone elution. Samples were analyzed in technical duplicate using HCD or ETD fragmentation on a Thermo Fisher Scientific LTQ-Velos. Purification of Rab1 and DrrA from Escherichia coli was performed as described previously (15, 16). Infection of HEK-293 FCγIII cells stably expressing Rab1 with Legionella was performed as described previously (15), with slight modification as Legionella strains were grown in AYE broth overnight before infection. All experimental procedures are described in detail in SI Methods.
SI Methods
Cell Lines, Plasmids, Antibodies, Other Reagents.
All cell lines were grown using standard conditions in DMEM (Gibco) containing 10% (vol/vol) FBS (Gibco). Cells were harvested at 80–90% confluence. Cell lysis for covalent capture was performed in lysis buffer 1 (20 mM Tris at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.5 mM DTT, 0.25% Nonidet P-40, and 1× Roche Complete protease inhibitor mixture). All other cell lysis was performed in lysis buffer 2 [50 mM Tris at pH 8.0, 200 mM NaCl, 1 mM DTT, 1 mM MgCl2, 0.5% Triton, 1× Roche Complete protease inhibitor mixture, and 1× PhosStop phosphatase inhibitor (Roche)]; 5z-7-oxozeaenol was purchased from Tocris.
WT-TAK1f fusion protein cDNA was synthesized by DNA 2.0 and cloned into pFastBac (Invitrogen) with an N-terminal 6xHis tag. pDONOR plasmids containing cDNA of MKK6, PSMC4, POLR3D, AP2B1, ADD1, Vinexin, EIF4B, EIF3I, TAK1, TAB1, and Rab1 were purchased from DNASU. Rab1, EIF4B, and EIF31 were cloned into gateway vector pDEST with N-terminal GST fusion. TAK1 and TAB1 were cloned into pcDNA3 vector with N-terminal V5 or Myc tags, respectively. The remaining genes were cloned into pcDNA3 with N-terminal Flag tags. Constructs for His-DrrA, His-LepB, pcDNA4/TO-Flag-Rab1, and pGEX-GST Rab-1 were gifts from S.M., as were HEK-293 stably expressing FCγRII receptor and Flag-Rab1. All point mutants, including AS-TAK1f, were generated by site directed mutagenesis of the WT construct.
Antibodies used were as follows, with product number in parenthesis: Cell Signaling Technologies: TAK1 (4505), TAB1 (3226), B-Tubulin (2146), CoxIV (4850); ThermoFisher Scientific: Myc (21316), His (21315), Flag(91878); Sigma: V5 (V8012), GDI1 (AV13086); Abcam: Antithiophosphate ester (ab92570). The pT75 Rab1 rabbit polyclonal antibody was raised by GenScript and is available by request. The anti-AMPylated Rab1 antibody was a gift from the Itzen laboratory, Technical University of Munich, Munich.
Transfection, Western Blotting, and Immunoprecipitation.
Fugene HD (Promega) was used for transfection of HeLa cells for immunofluorescence. For immunoprecipitation before kinase assays in Fig. 2 A–C, HEK-293Ts were transfected with Lipofectamine (Invitrogen). All other HEK-293T transfections were done with Fugene HD. Cells were transfected at 60% confluence for 24 h. For Western blots or immunoprecipitation, cells were lysed in lysis buffer 2 with sonication. Protein concentrations were measured using a BCA assay (Thermo Fisher Scientific). Next, 20–30 μg of protein was separated by SDS/PAGE and transferred to nitrocellulose. Membranes were incubated with antibody per the manufacturer’s guidelines and imaged on a Licor system. All immunoprecipitation of proteins was performed on 500 μg to 1 mg lysate per sample with anti-Flag magnetic beads (Sigma) or glutathione magnetic beads (Thermo Fisher Scientific), as appropriate, for 2 h at 4 °C. For use in in vitro kinase assays, samples were washed three times in 500 μL lysis buffer and eluted with 3xFlag-tide or 10 mM glutathione. For LC-MS/MS analysis, samples were run on SDS/PAGE, Coomassie-stained, subject to a standard in gel trypsin digestion protocol. For coimmunoprecipitations, samples were washed three times in PBS before elution by heating in SDS gel loading buffer.
Purification of TAK1f and Rab1.
TAK1f was purified as previously described (17). Briefly, His-tagged WT-TAK1f and AS-TAK1f were overexpressed using a Baculovirus expression system in SF9 insect cells and purified using Ni/NTA agarose resin (Qiagen). The tagged protein was further purified by size-exclusion chromatography on a Superdex 75 column (Amersham Biotech) and concentrated before the addition of glycerol to 10% and snap freezing.
Rab1 was purified as previously described (52). Purification for DrrA, DrrA340–533, and LepB followed the same conditions. Briefly, Escherichia coli BL21 cells transformed with vectors for GST-Rab1 mutants were grown in LB broth at 37 °C to an OD = 0.4. Next, 0.1 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce protein expression and cells were incubated for 4 h at 30 °C. Cells were harvested by centrifugation and mechanically lysed via microfluidizer in buffer containing 50 mM Tris at pH 8.0, 200 mM NaCl, 1 mM DTT, 1 mM MgCl2, and 1× Roche Complete protease inhibitor mixture. Cleared lysate was incubated with 1 mL/L of culture of glutathione Sepharose resin (GE Healthcare) or Nickel-NTA resin (Qiagen) for 2 h at 4 °C. Bound proteins were washed with lysis buffer and eluted in buffer containing 10 mM glutathione or 50 mM imidazole. All GST-Rab1 proteins were further purified by size-exclusion chromatography on a Superdex 200 column (Amersham Biosciences).
Rab1 Nucleotide Exchange.
Rab1 proteins were loaded with the indicated nucleotides [GTPγS, GDP or mant-GDP (Molecular Probes)] by incubation for 1 h at 37 °C in buffer containing 50 mM Tris at pH 8.0, 200 mM NaCl, 1 mM DTT, 1 mM MgCl2, 5 mM EDTA, and 25 mM excess of the indicated nucleotide. The exchange reaction was quenched by addition of 10 mM MgCl2. For in vitro kinase assays, GST-Rab1 was purified away from excess nucleotide by size-exclusion chromatography. For nucleotide affinity and GEF assays, protein was desalted and excess nucleotide removed on Zebra spin desalting columns (Thermo Fisher Scientific).
Thiophosphorylation and Covalent Capture.
Thiophoshorylation of MBP (dephosphorylated, EMD Millipore) with purified WT-TAK1f was performed in buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 10 mM MgCl2, and 0.5 mM of each ATPγS analog (6-Bn-ATP-γ-S, 6-PhEt-ATP-γ-S or 6-Furfuryl-ATP-γ-S, Axxora). Labeling experiments for covalent capture enrichment were performed on 2 mg of protein lysate per sample. Samples were incubated in lysis buffer 1 supplemented with 250 μM FF-ATPγS, 250 μM ATP, 3 mM GTP, 10 mM MgCl2, and 20 μg of purified TAK1 as indicated. Labeling reactions were left at room temperature for 1 h before quenching with 50 mM EDTA. Thirty-microliter aliquots of each reaction were alkylated with 2 μL of 100 mM p-nitro mesylate (PNBM) for 30 min at room temperature. Thiophosporylation was detected by Western blot with the antithiophosphate ester antibody.
Covalent capture of thiophosporylated proteins was performed as described previously (9). Briefly, lysates were denatured by adding 60% (wt/vol) solid urea, 10-mM final TCEP, and incubating at 55 °C for 30 min. Samples were diluted to 2 M urea with 50 mM ammonium bicarbonate and digested overnight at 37 °C with typsin (Promega) at a 1:20 ratio. Peptides were acidified with triflouroacetic acid, desalted on a SepPak C18 column (Waters), and speed-vacuumed to dryness. Peptides were resuspended in 50 mM Hepes and 50% (vol/vol) acetonitrile and adjusted to pH 7. The peptide solution was incubated overnight rocking with 100 μL of iodoacetyl Sepharose resin in the dark (Thermo Fisher Scientific). Beads were washed by gravity flow with water, 5 M NaCl, 50% (vol/vol) acetonitrile, 5% (vol/vol) formic acid, and 10 mM DTT followed by elution with 1 mg/mL oxone (Sigma). Peptides were desalted with ZipTips (Millipore) and speed-vacuumed to dryness.
LC-MS/MS Analysis and Data Processing.
All desalted peptides were resuspended into 10 μL of 0.1% formic acid. Peptides were loaded on to a nanoACQUITY (Waters) UPLC instrument for reversed-phase chromatography with a C18 column (BEH130, 1.7-μm bead size, 100 μm × 100 mm) in front of an LTQ Orbitrap Velos. The LC was operated at a 600-nL/min flow rate and peptides were separated over an 80-min gradient from 2 to 50% buffer B (buffer A: water and 0.1% formic acid; buffer B: acetonitrile and 0.1% formic acid). Survey scans were recorded over a 350–1,800 m/z range and MS/MS fragmentation was performed using HCD on the top eight peaks. A second injection (i.e., technical replicate) of each sample was performed using ETD fragmentation on the top six peaks. Peak lists were generated with an in-house software called PAVA and searched against the SwissProt Homo sapiens database (downloaded June 27, 2013; 20,264 entries) using Protein Prospector (v5.10.10). Data were searched with a 20-ppm tolerance for parent and fragment ions (HCD or 20 ppm/0.6 Da ETD), allowing for standard variable modifications and S/T/Y phosphorylation. Filtering of background peptides and phosphopeptides was accomplished using an in-house R script described previously (21). Individual phosphopeptide abundance, when noted, was manually extracted using the extracted ion chromatogram functions built into Xcalibur Qual Browser (Thermo Fisher Scientific). Raw LC-MS/MS data and matched search results have been deposited to the ProteomeXchange Consortium via the PRIDE (53) partner repository with the dataset identifier PXD004213. Searched peptide identifications from all runs are also available in Datasets 1–4. Each dataset contains all runs for a single cell type, with individual sheets containing all identifications for a single injection.
In Vitro Kinase Assays with γ[32P]ATP.
Purified kinase to a final concentration of 100–300 nM was added into immunoprecipitated substrate or purified GST-Rab1 in 50 mM Tris pH 7.5, 150 mM NaCl, and 10 mM MgCl2. A small aliquot of this mixture was removed to check loading by Western blot. One to 2 μL of a mixture of cold ATP and γ[32P]ATP, for a final concentration of 100 μM ATP and 1–5 μCi γ[32P]ATP per sample, was added to start each reaction. Reactions were quenched by adding gel loading buffer and heating to 95 °C before SDS/PAGE separation. Gels were washed, dried, and detected by autoradiography.
Cell Fractionation.
Cell fractionation experiments were performed using one confluent well of a six-well plate of HEK-293Ts per sample. Fractionation was performed according to the Subcellular Protein Fractionation Kit protocol (Thermo Fisher Scientific).
Immunofluorescence.
HeLa cells were plated on glass coverslips and grown to 50% confluency. Cells were transfected and grown for 24 h, then fixed. For immunofluorescence experiments with TAK1 inhibition, cells were transfected (or left untransfected), grown for 24 h, then treated with 2.5 μM 5z-7-oxozeanol for 6 h and fixed. Cells were fixed with paraformaldehyde, permiablized with saponin, and stained with rabbit anti-GFP and mouse anti-GM130 (BD Biosciences) for 1 h. Coverslips were washed and stained with anti-rabbit and anti-mouse antibodies conjugated to Alexa-488 and Alexa-568, respectively. Coverslips were then stained with Hoechst reagent, fixed to slides, imaged, and quantified manually.
Legionella Infection Experiments.
HEK-293 expressing FCγRII and Flag-Rab1 were infected as described previously (15). Cells were grown to near confluency in 10-cm dishes and infected with Legionella pneumophila strain LP01 or isogenic mutants at an estimated multiplicity of 100 bacteria to 1 host cell. Cells were harvested and snap-frozen at the indicated time points.
GDP/GTP Preference and GEF Assays.
First, GST-Rab1 mutants were loaded with mant-GDP as described above. The complex of mant-GDP and Rab1 fluoresces significantly more than mant-GDP alone when excited. The GDP/GTP affinity assay was performed as described previously (29). Briefly, mant-GDP loaded Rab1 (1-μM final concentration) was incubated with EDTA (5-mM final concentration) in the presence of varying concentrations of GTP or GDP as indicated. In a 384-well low-volume black plate, 5 μL of EDTA/nucleotide mixture was added to the 10 μL of Rab1 to start the reaction. The presence of EDTA in the reaction mixture helps displace mant-GDP from Rab1, which is then free to bind any nucleotide in solution, and is read out as a decrease in fluorescence. The amount of displacement in the presence of free nucleotide can be used to determine nucleotide preference. Where indicated, WT-TAK1f was also present in the reaction at 100 nM and ATP at and 200 μM, and allowed to react for 15 min before the addition of EDTA and GDP/GTP. This reaction was allowed to reach equilibrium during a 1-h incubation. The amount of remaining mant-GDP bound Rab1 was determined by fluorescence Spectramax M5 plate reader (Molecular Devices; 360-nm excitation, 440-nm emission). For the determination of nucleotide IC50, a sigmoidal curve fit was used for each nucleotide and mutant (Prism).
The GEF assay followed similar conditions to the above nucleotide affinity assay, using the same plate set-up and reader. Briefly, GST-Rab1 mutants were loaded with mant-GDP as described above. Then, 10 μM of Rab1 was added to each well (final concentration, 1 μM). To trigger the reaction, 5 μL of His-DrrA340–533, EDTA, or buffer with excess GTPγS (final concentration 200 μM) was added to the reaction and the fluorescence monitored for 2 h at 60-s intervals. Kobs values were determined using a single exponential decay fit (Prism).
GAP Assay.
The GTPase-Glo system (Promega) (34) was used to measure the GTP remaining after incubation of GST-Rab1 with varying concentrations of His-LepB. Next, 10 μL of a solution containing 2 μM Rab1 in assay buffer (50 mM Tris, 200 mM NaCl, 1 mM DTT, 5 mM MgCl2, 20 mM EDTA) was dispensed into a low-volume white 384-well plate. LepB was serially diluted in assay buffer with 10 μM GTP and 5 μL of this mixture was added to each well to trigger the reaction. The reaction was incubated at room temperature protected from light for 1 h. At 1 h, 10 μL of GTPase-Glo Reagent mix was added to each well and incubated while shaking for 30 min at room temperature. Finally, 20 μL of Detection Reagent was added to each well, incubated for 5 min at room temperature, and luminescence was measured on a Spectramax M5 Plate reader (Molecular Devices; 500-ms read time). For the determination of LepB EC50, concentrations of LepB were plotted against relative fluorescence units and a sigmoidal curve fit was performed (Prism).
In Vitro Labeling of Rab1 by DrrA and TAK1.
For in vitro labeling of Rab1 by DrrA and TAK1, 15 μg of Rab1 was diluted in 30 μL of 50 mM Tris pH 7.5, 150 mM NaCl, and 10 mM MgCl2 with 200 μM ATP and 30 μM GTP or 30 μM GDP. At 0 min, a 10-μL portion was removed and quenched with gel loading buffer. Next, 1 μg of TAK1 or 0.5 μg of DrrA was added to the reaction and allowed to react for 15 min. At 15 min, a 10-μL portion was removed and quenched. Then, 0.25 μg of full-length His-DrrA was added to the TAK1-labeled sample and 0.5 μg of TAK1 to the DrrA-labeled sample and allowed to react for an additional 15 min (30-min total) before quenching with SDS sample loading buffer and analysis by Western blot.
Supplementary Material
Acknowledgments
We thank Dr. Jesse Lipp for guidance regarding data analysis; Dr. Averil Ma for helpful discussions; and two reviewers, who suggested a number of insightful experiments during review, including the addition of GTP to the DrrA/TAK1 experiment in Fig. 5B. Mass spectrometry was conducted at the Bio-Organic Biomedical Mass Spectrometry Resource at the University of California, San Francisco (A.L.B., director), supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences Grant 8P41GM103481. K.M.S. acknowledges support from NIH/National Institute of Allergy and Infectious Diseases (NIAID) Grants U19 AI109622 and R01 CA190408-01. S.M. acknowledges support from NIH/NIAID Grant R01 AI118974-01A1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the PRIDE partner repository (dataset identifier PXD004213).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608355113/-/DCSupplemental.
References
- 1.Shim J-H, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19(22):2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem J. 2014;461(3):531–537. doi: 10.1042/BJ20140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emmerich CH, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA. 2013;110(38):15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai L, Aye Thu C, Liu X-Y, Xi J, Cheung PCF. TAK1, more than just innate immunity. IUBMB Life. 2012;64(10):825–834. doi: 10.1002/iub.1078. [DOI] [PubMed] [Google Scholar]
- 5.Momcilovic M, Hong S-P, Carlson M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem. 2006;281(35):25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 6.Sumiya E, et al. Phosphoproteomic analysis of kinase-deficient mice reveals multiple TAK1 targets in osteoclast differentiation. Biochem Biophys Res Commun. 2015;463(4):1284–1290. doi: 10.1016/j.bbrc.2015.06.105. [DOI] [PubMed] [Google Scholar]
- 7.Kajino-Sakamoto R, et al. TGF-beta-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J Immunol. 2010;185(8):4729–4737. doi: 10.4049/jimmunol.0903587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci USA. 2008;105(5):1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertz NT, et al. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr Protoc Chem Biol. 2010;2(1):15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, Soule G, Overmeyer JH, Maltese WA. Tyrosine phosphorylation of the Rab24 GTPase in cultured mammalian cells. Biochem Biophys Res Commun. 2003;312(3):670–675. doi: 10.1016/j.bbrc.2003.10.171. [DOI] [PubMed] [Google Scholar]
- 11.Bailly E, et al. Phosphorylation of two small GTP-binding proteins of the Rab family by p34cdc2. Nature. 1991;350(6320):715–718. doi: 10.1038/350715a0. [DOI] [PubMed] [Google Scholar]
- 12.Lewandowska A, Macfarlane J, Shaw JM. Mitochondrial association, protein phosphorylation, and degradation regulate the availability of the active Rab GTPase Ypt11 for mitochondrial inheritance. Mol Biol Cell. 2013;24(8):1185–1195. doi: 10.1091/mbc.E12-12-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sluijs P Van Der, et al. (1992) Reversible phosphorylation dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J 11(12):4379–4389. [DOI] [PMC free article] [PubMed]
- 14.Fentress SJ, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 2010;8(6):484–495. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 17.Brown K, et al. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J Mol Biol. 2005;354(5):1013–1020. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 18.Singh A, et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148(4):639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melisi D, et al. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. J Natl Cancer Inst. 2011;103(15):1190–1204. doi: 10.1093/jnci/djr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer BE, et al. Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab. 2015;22(5):907–921. doi: 10.1016/j.cmet.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipp JJ, Marvin MC, Shokat KM, Guthrie C. SR protein kinases promote splicing of nonconsensus introns. Nat Struct Mol Biol. 2015;22(8):611–617. doi: 10.1038/nsmb.3057. [DOI] [PubMed] [Google Scholar]
- 22.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91(1):119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ninomiya-Tsuji J, et al. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398(6724):252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, et al. Mechanism and in vitro pharmacology of TAK1 inhibition by (5Z)-7-oxozeaenol. ACS Chem Biol. 2013;8(3):643–650. doi: 10.1021/cb3005897. [DOI] [PubMed] [Google Scholar]
- 26.Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125(2):225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119(4):749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Y, Arnold RJ, Luo Z-Q. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci USA. 2011;108(52):21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goody PR, et al. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31(7):1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oesterlin LK, Goody RS, Itzen A. Posttranslational modifications of Rab proteins cause effective displacement of GDP dissociation inhibitor. Proc Natl Acad Sci USA. 2012;109(15):5621–5626. doi: 10.1073/pnas.1121161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh H-Y, et al. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J. 2010;29(2):496–504. doi: 10.1038/emboj.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450(7168):365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 34.Mondal S, Hsiao K, Goueli SA. A homogenous bioluminescent system for measuring GTPase, GTPase activating protein, and guanine nucleotide exchange factor activities. Assay Drug Dev Technol. 2015;13(8):444–455. doi: 10.1089/adt.2015.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barr FA. Review series: Rab GTPases and membrane identity: Causal or inconsequential? J Cell Biol. 2013;202(2):191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aizawa M, Fukuda M. Small GTPase Rab2B and its specific binding protein Golgi-associated Rab2B Interactor-like 4 (GARI-L4) regulate Golgi morphology. J Biol Chem. 2015;290(36):22250–22261. doi: 10.1074/jbc.M115.669242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson BS, et al. A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J Cell Biol. 1994;125(3):557–571. doi: 10.1083/jcb.125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardiman CA, Roy CR. AMPylation is critical for Rab1 localization to vacuoles containing Legionella pneumophila. MBio. 2014;5(1):e01035-13. doi: 10.1128/mBio.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Y, Luo Z-Q. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475(7357):506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, et al. Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol Cell. 2014;53(1):140–147. doi: 10.1016/j.molcel.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa K, et al. Filamin associates with stress signalling kinases MKK7 and MKK4 and regulates JNK activation. Biochem J. 2010;427(2):237–245. doi: 10.1042/BJ20091011. [DOI] [PubMed] [Google Scholar]
- 42.Shirakabe K, et al. TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J Biol Chem. 1997;272(13):8141–8144. doi: 10.1074/jbc.272.13.8141. [DOI] [PubMed] [Google Scholar]
- 43.Liu XY, Seh CC, Cheung PCF. HSP90 is required for TAK1 stability but not for its activation in the pro-inflammatory signaling pathway. FEBS Lett. 2008;582(29):4023–4031. doi: 10.1016/j.febslet.2008.10.053. [DOI] [PubMed] [Google Scholar]
- 44.Rak A, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302(5645):646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 45.Steger M, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife. 2016;5(e12813):e12813. doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu S, Wu WJ, Wang J, Cerione RA. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J Biol Chem. 2003;278(49):49293–49300. doi: 10.1074/jbc.M307021200. [DOI] [PubMed] [Google Scholar]
- 47.Kwon T, Kwon DY, Chun J, Kim JH, Kang SS. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J Biol Chem. 2000;275(1):423–428. doi: 10.1074/jbc.275.1.423. [DOI] [PubMed] [Google Scholar]
- 48.de Boor S, et al. Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc Natl Acad Sci USA. 2015;112(28):E3679–E3688. doi: 10.1073/pnas.1505995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge J, et al. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci USA. 2009;106(33):13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin S, et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4(11):e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiefes A, et al. The Yersinia enterocolitica effector YopP inhibits host cell signalling by inactivating the protein kinase TAK1 in the IL-1 signalling pathway. EMBO Rep. 2006;7(8):838–844. doi: 10.1038/sj.embor.7400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8(9):971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 53.Rigden DJ, Fernández-Suárez XM, Galperin MY. The 2016 database issue of Nucleic Acids Research and an updated molecular biology database collection. Nucleic Acids Res. 2016;44(D1):D1–D6. doi: 10.1093/nar/gkv1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.