Significance
Experimental research on mice has yielded tremendous biological insight. However, the ∼140 million y of evolution that separate mice from humans pose a hurdle to direct application of this knowledge to humans. We report here that considerable progress for identifying genetically patterned skeletal phenotypes beyond the mouse model is possible through transdisciplinary approaches that include the anatomical sciences. Indeed, anatomy and paleontology offer unique opportunities through which to develop and test hypotheses about the underlying genetic mechanisms of the skeleton for taxa that are not well suited to experimental manipulation, such as ourselves.
Keywords: paleontology, quantitative genetics, primates, neontology, dental variation
Abstract
Developmental genetics research on mice provides a relatively sound understanding of the genes necessary and sufficient to make mammalian teeth. However, mouse dentitions are highly derived compared with human dentitions, complicating the application of these insights to human biology. We used quantitative genetic analyses of data from living nonhuman primates and extensive osteological and paleontological collections to refine our assessment of dental phenotypes so that they better represent how the underlying genetic mechanisms actually influence anatomical variation. We identify ratios that better characterize the output of two dental genetic patterning mechanisms for primate dentitions. These two newly defined phenotypes are heritable with no measurable pleiotropic effects. When we consider how these two phenotypes vary across neontological and paleontological datasets, we find that the major Middle Miocene taxonomic shift in primate diversity is characterized by a shift in these two genetic outputs. Our results build on the mouse model by combining quantitative genetics and paleontology, and thereby elucidate how genetic mechanisms likely underlie major events in primate evolution.
The relationship between genotype and phenotype is critical to evolutionary biology, because it influences how phenotypes respond to selective pressures and evolve (1, 2). Paleontologists have long sought to incorporate the etiology of the dental phenotype to inform on questions of environmental, dietary, and adaptive change over time (3–6). This research has been advanced significantly by the revolution in developmental genetics over the past few decades (7). Experimental research on mice has yielded tremendous biological insight (8). However, for human phenotypes, ranging from inflammation (9) to placentation (10), the limitations of the mouse model due to the ∼140 million years ago of evolution that have occurred since our last common ancestor ∼70 Ma (11) are starting to be recognized. Here, we demonstrate how to overcome the limitations of the mouse model’s application to the primate dentition by integrating research from quantitative genetics, neontology, and paleontology. The insights gained from this transdisciplinary approach have implications for all of these seemingly disparate subdisciplines of biology.
There was considerable excitement when an inhibitory cascade (IC) mechanism, inferred from experimental manipulation of mouse molar development, showed tremendous explanatory power for the taxonomic variation observed across murines (12). The concept was soon applied to other tripartite skeletal phenotypes (13). However, the extent of the predictive power of the IC for taxa with less similar (14) and more heterodont (15–18) dentitions suggests that either the IC mechanism does not characterize these other organisms’ dental patterning as specifically or the characterization of the mechanistic output (the phenotype) is not accurately assessed. Despite empirical evidence for how primate dental patterning differs from dental patterning of the mouse (19, 20), proponents of the mouse model continue to adhere tightly to it. Most recently, Evans et al. (21) report that the IC model from mice applied uncritically to hominids reveals that the genus Homo is distinct from other hominid genera in having smaller third molars because of a “simple rule.”
Although the developmental mechanisms underlying tooth organogenesis are evolutionarily conserved across mammals (19, 22–24), given the highly derived dentitions of mice (which lack premolars, canines, and replacement teeth) and the vastly different life histories, the finer details of dental patterning are not the same (19). Previous quantitative genetic analyses indicate that the IC model may not work as well for primates as it does for mice because the former’s phenotype can comingle the effect of a genetic patterning mechanism(s) with systemically modulated somatic factors. For example, baboon molar buccolingual width is significantly influenced by the same genes that influence the animal’s crown–rump length (20). Given that crown–rump length is highly sexually dimorphic, tooth size measurements that incorporate buccolingual width are capturing the signal of the systemic effects of body size and sex in addition to the dental patterning mechanism(s) more so than would mesiodistal length alone. Furthermore, we find that the lengths of the fourth premolar and the first through third molars have higher genetic correlations with one another than do the width measurements of these same teeth (19). This pattern of genetic correlation suggests that among primates, variation in mandibular mesiodistal tooth measurements may more directly reflect the genetic patterning mechanisms determining relative tooth sizes than do width measurements. Although Evans et al. (21) interpret their results as indicative of simply the IC, our research (19, 20) shows that the phenotype they have captured for primates includes significant pleiotropic effects with body size. The genus Homo is not only characterized by smaller third molars compared with other hominid genera (21) but also by an ∼30% increase in body size (25). Is the genus Homo different from other hominid genera because of a simple developmental rule or because of all the various factors that lead to a dramatic increase in body size? Evans et al. (21) cannot and do not distinguish between these two very different evolutionary phenomena because their phenotype definition fails to use empirical evidence from primate genetics to modify what we have learned from mouse models.
To extend the mouse model to our own lineage, gene-forward approaches such as are used in mice and other model organisms are not practical. Therefore, the next phase of knowledge will come from phenotype-back methods. To demonstrate this approach, we combined the strength of three datasets—quantitative genetics, neontology, and paleontology—to test hypotheses about how to better define the output of dental patterning mechanisms in primates. If a phenotype definition corresponds directly to a patterning mechanism, it should meet four expectations: (i) the genetically patterned (GP) trait will be significantly heritable, with little to no covariate effects; (ii) it will capture variation that differentiates taxa, representing genetic variance exploited by selection; (iii) different GP traits will be uncorrelated within a species (but may be phenotypically correlated across closely related taxa because of their phylogenetic proximity); and (iv) the fossil record will reveal differing evolutionary histories for GP traits because the genetically independent traits have responded to selection and/or drift relatively independently.
We identify two dental phenotypes that meet these four criteria. These phenotypes characterize the output of two (as-yet-unknown) patterning mechanisms that influence the relative proportions of the various teeth within the two genetic submodules of the primate permanent mandibular postcanine dentition (26). The first GP phenotype is the molar module component (MMC). The MMC follows Kavanagh et al.’s proposed developmental IC model (12), but by using only mesiodistal length, we capture the GP output more directly by removing the same modulating, pleiotropic effects that confounded Evans et al.’s analysis (21):
The second GP phenotype is the premolar/molar module (PMM), defined as
where l is the mesiodistal length of the respective molar crown, and subscripts indicate the position of the tooth crown (M, molar; P, premolar). We use only the length of the second molar in the PMM because it is a stable predictor of the overall length of the molar series [for murines (12) and for Old World monkeys (OWMs) (SI Appendix, Fig. S1)].
Results
Test of Criterion 1.
Quantitative genetic analyses (27) demonstrate that both the MMC and PMM are significantly heritable in an outbred pedigreed breeding colony of baboons. Although all size measurements of the teeth are significantly heritable, MMC and PMM heritability estimates (h2r) indicate little to no covariate effects, in contrast to the large effects of sex (and thus, body size) returned for the h2r estimates of tooth size measurements (20) (Table 1). These results suggest that the MMC and PMM have minimal pleiotropic effects, and may therefore respond readily to selection and/or drift, as previously reported for enamel thickness in this same population (28).
Table 1.
Narrow-sense heritability estimates for dental traits in baboons
| Trait | n | h2 | SE | P value | Percent covariates | Significant covariates |
| Individual dental metrics | ||||||
| P4l | 340 | 0.541 | 0.138 | <0.0001 | 0.418 | Sex |
| P4w | 334 | 0.641 | 0.167 | <0.0001 | 0.159 | Sex |
| P4 area | 299 | 0.707 | 0.133 | <0.0001 | 0.335 | Sex |
| M1l | 310 | 0.832 | 0.146 | <0.0001 | 0.349 | Sex |
| M1w | 303 | 0.697 | 0.178 | <0.0001 | 0.211 | Age, sex, sex * age |
| M1 area | 298 | 0.705 | 0.176 | <0.0001 | 0.312 | Sex, sex * age |
| M2l | 440 | 0.550 | 0.109 | <0.0001 | 0.455 | Sex |
| M2w | 435 | 0.486 | 0.120 | <0.0001 | 0.290 | Sex |
| M3l | 286 | 0.403 | 0.176 | 0.001 | 0.462 | Sex |
| M3w | 438 | 0.560 | 0.132 | <0.0001 | 0.377 | Sex |
| GP dental traits | ||||||
| MMC | 121 | 0.688 | 0.406 | 0.042 | 0.130 | Sex, age2, sex * age2 |
| PMM | 283 | 0.451 | 0.181 | <0.0001 | 0.066 | Age, sex |
Percent covariates indicate the proportion of variance accounted for by significant covariates. Asterisks in significant covariates indicate an interaction of the two covariates. Mesiodistal lengths are indicated with “l”, and buccolingual widths are indicated with “w”. Area is defined as mesiodistal length multiplied by buccolingual width.
Test of Criterion 2.
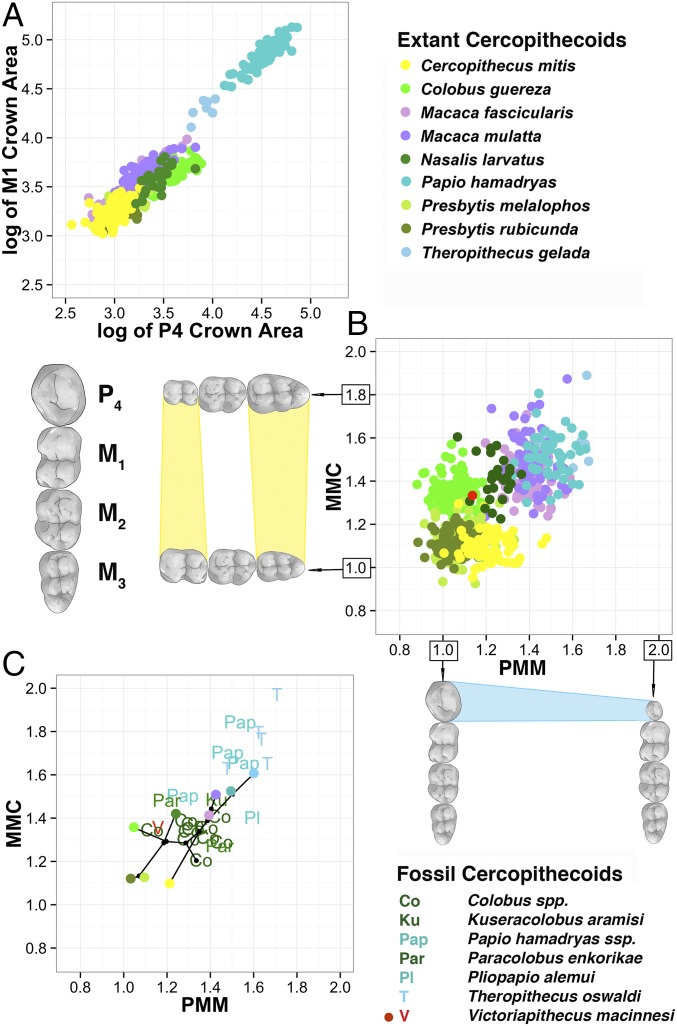
We then assessed the MMC and PMM for 711 individuals representing nine species of OWM, the Cercopithecidae (SI Appendix, Tables S1 and S2). In comparison to the commonly used ratio of P4-to-M1 area (29), MMC and PMM discriminate well between taxa and are not correlated with each other (Fig. 1 A and B and SI Appendix, Fig. S2 and Table S3). Late Miocene and Pliocene fossil OWMs plot either within their subfamily and generic morphospace or are transitional (Fig. 1C and SI Appendix, Tables S4 and S5). The MMC and PMM of Victoriapithecus, the Middle Miocene extinct sister taxon to the Cercopithecidae (30), are situated central to the scatter of both GP traits, providing a sense of the evolutionary trajectories exploited since the cercopithecine/colobine divergence ∼20 Ma.
Fig. 1.
Mandibular MMC and PMM for extant and fossil OWMs. (A) We show the log of mandibular P4 versus M1 areas for extant taxa, a common means through which to describe and assess dental variation in extant and extinct OWMs. We provide this log as a comparison with the next panel. (B) We plot the MMC and PMM for extant cercopithecoids, demonstrating how well these two phenotypes separate the various taxa in comparison to the more traditional method shown in A. The red dot in the center represents where the sister taxon, Victoriapithecidae, plots relative to the extant species. (C) We have plotted the mean for each extant species as a single dot, using the same color coding as in the previous panels. On top of this plotted mean, we plot fossil data from Africa with letter designations, defined to the right of the graph. Colobine fossil taxa are illustrated in green, papionin fossil taxa in blue, and Victoriapithecus in red. Nodes that link taxa are shown in black. More comparisons are provided in SI Appendix, Fig. S2.
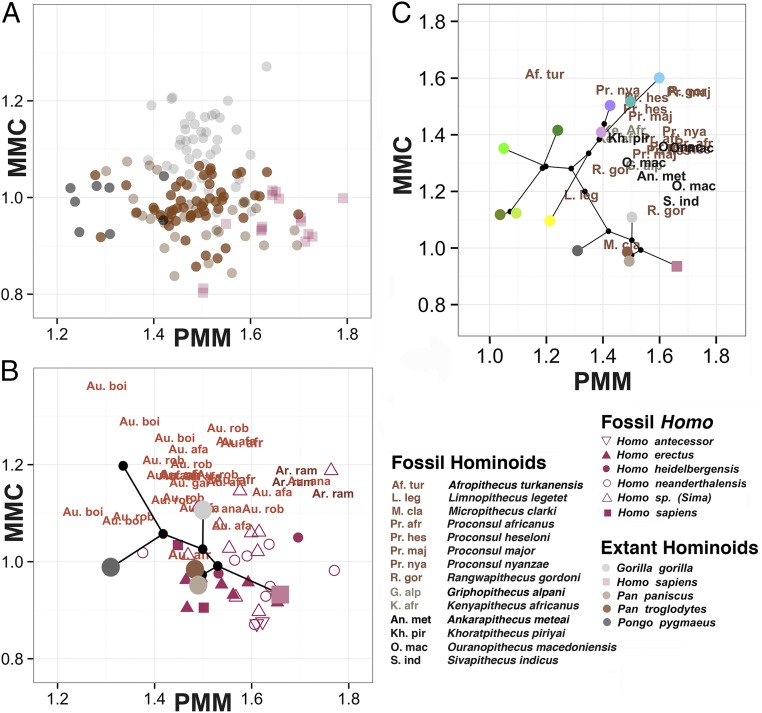
Extending the analysis of the MMC and PMM to our own, more shallowly rooted clade, we assessed 192 individuals from four extant ape species and Homo sapiens (Fig. 2 and SI Appendix, Tables S2 and S6). Gorillas occupy their own MMC range, whereas orangutans have the lowest PMM values and humans have the highest. MMC and PMM values for 13 fossil hominid taxa reflect the evolution of these two GP phenotypes over the past 7 million years ago (our clade postdating the human/chimpanzee common ancestor) (Fig. 2B and SI Appendix, Tables S7 and S8). Remarkably, the PMM and MMC distinguish the three main hominid genera, likely evidence of their differing adaptive regimes. Ardipithecus, the oldest of the three, occupies a MMC and PMM space for which we have no modern analog, further highlighting how distinct it is from living chimpanzees (31, 32). The earliest species of Australopithecus, Australopithecus anamensis, overlaps the ranges of Ardipithecus and later Australopithecus. The overlap between Australopithecus and Gorilla is likely evidence of parallelism in the MMC, given that Ardipithecus shows an ancestral condition for Australopithecus that is distinct from the MMC of Gorilla. Another parallelism in the MMC is between chimpanzees and Homo.
Fig. 2.
Mandibular MMC and PMM for extant hominoids and fossil hominids. (A) We plot individual points for the extant hominoid taxa. Notice that, as we found for the OWMs, these two phenotypes separate the genera and species quite well. (B) We show the mean for each extant hominoid species as a single dot, using the same color coding as in the previous panel. On top of this mean, we plot hominid fossil data with letter designations, save for the genus Homo, which is shown using shapes. Abbreviations are as follows: Au. afa, Australopithecus afarensis; Au. afr, Australopithecus africanus; Au. anam, Australopithecus anamensis; Au. boi, Australopithecus boisei; Au. gar, Australopithecus garhi; Ar. ram, Ardipithecus ramidus; Au. rob, Australopithecus robustus. (C) We combine the extant data for OWMs (as seen in Fig. 1C) and hominoids (as seen in Fig. 2B) with fossil data for Miocene apes (using abbreviations). The fossils are color-coded by age: Early Miocene is shown in brown, Middle Miocene in gray, and Late Miocene in black. The fossil hominoids in the upper MMC range only date to the early part of the Miocene, and by the Plio-Pleistocene, this primate morphospace is occupied exclusively by the papionins (in purple and blue). Extant hominoids occupy only the lowest range of the MMC, far below the morphospace of the Miocene apes, but remain in the same PMM range (compare with B).
Tests of Criteria 3 and 4.
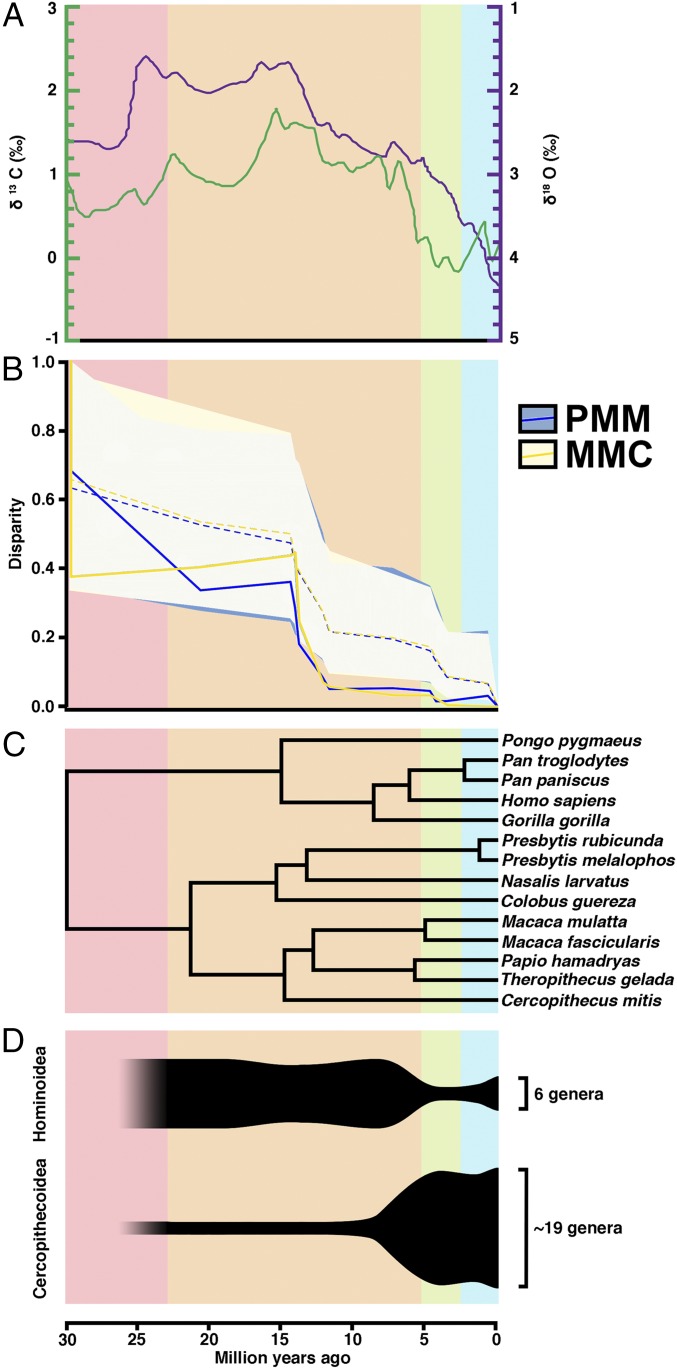
Estimates of Pagel’s λ for the MMC and PMM suggest that phylogenetic relationships explain the observed variation well (SI Appendix, Fig. S3 and Table S9). A disparity-through-time analysis reveals the pattern of diversification for these two phenotypes, showing that disparities in the MMC and PMM fall outside the 95% confidence interval (CI) of a random-walk model, with the significant differences between taxa estimated to have arisen rapidly in the Middle to Late Miocene (Fig. 3 and SI Appendix, Table S10). This pattern indicates that variation since the Middle Miocene is primarily found within the lineages rather than between them, suggestive of an adaptive shift, radiation, or constraints (33). The morphological disparity indices for other dental traits are either below the 95% CI of the random-walk model (indicating that subclade disparity is consistently low across evolutionary time for these traits, given that all of the morphological disparity index values are low; SI Appendix, Table S10) or follow a similar pattern to the MMC and PMM (SI Appendix, Fig. S4). However, in these latter cases, these traits have significant pleiotropic effects with body size; thus, the evolution of disparity through time comingles dental evolution with the evolution of body size.
Fig. 3.
Overview of climate change, primate phylogeny, and primate taxonomic diversity compared with the evolution of the MMC and PMM. (A) Global deep-sea oxygen and carbon isotope records, adapted from a study by Zachos et al. (34). Higher δ18O values indicate cooler temperatures. The δ13C values represent different flora. (B) Our disparity through time results, which are discussed in the main text. Dotted lines indicate the predicted disparity values, and shaded areas indicate their 95% CIs. Solid lines indicate actual trait values. Values near 0 indicate that a particular clade contains little of the overall variation, and that variation in the trait is partitioned between subclades rather than within them. Values near 1.0 suggest that a clade contains a large amount of that variation, and that clades may overlap in trait space. For the PMM and MMC, trait values fall significantly below expected values beginning in the Middle Miocene, indicating trait partitioning between subfamilies within primates. Disparity through time plots of other phenotypes are provided in SI Appendix, Fig. S4. (C) Phylogeny of extant primates based on molecular data. (D) Anthropoid taxonomic diversity through time. Oligocene counts are not included because they precede the cercopithecoid–hominoid split.
Discussion
The dramatic evolution of these two GP dental traits across the Old World higher primates shows a remarkable correlation with global climate and vegetation change as reflected in decreasing δ18O and δ13C values following the Middle Miocene Climatic Optimum (34, 35). Paleobotanical and faunal evidence suggests that the largely mosaic forests and woodlands of Oligocene Africa transitioned in the Middle Miocene to more heterogeneous landscapes (36, 37). This climate change further correlates with the long-recognized shift in species diversity between apes and OWMs (Fig. 3D). Before the Middle Miocene, apes were abundant and diverse across Africa and Eurasia (38), whereas OWMs were still rare. However, by the Pliocene, monkeys had experienced a taxonomic and geographic expansion across the Old World (39) and ape diversity had dramatically reduced to a handful of relict genera that persist today. The origins of the human lineage are embedded within this evolutionary transition (31, 40).
Evolutionary changes in the two GP phenotypes introduced here appear to reflect how their two underlying genetic mechanisms responded to selective pressures acting on the primate postcanine dentition. In turn, these mechanisms may provide a broad and deep evolutionary explanation for primate adaptation during the Neogene. Although Miocene apes occupied the same PMM space that extant apes and all of the hominids demonstrate (Fig. 2C) during the Miocene, the ape MMC phenotype experienced a downward shift as the first and third molars became more equivalent in size. During the Plio-Pleistocene, the papionins, and even some of the larger fossil colobines, evolved MMC and PMM phenotypes that occupy the morphospace of the now-extinct Miocene apes. From the perspective of the patterning of the postcanine dentition, baboon morphology replaced the Miocene ape strategy, which may well underlie and/or reflect the demographic shifts hypothesized for this evolutionary competition (41) (i.e., during the later Miocene, hominoids increasingly invested in parental care of fewer offspring compared to the papionin reproductive strategy). Given that teeth develop and erupt at different times over a long ontogenetic period and that premolars replace deciduous teeth and molars do not (and the mechanisms relating them are unknown), the MMC and PMM may represent two different responses to selective pressures that vary, in part, on the ontogenetic timing of the pressure.
Although the earliest written record of human anatomical studies is 3,000 y old, when cadaver dissection became acceptable in the second century A.D., anatomical sciences became the foundation of medical research (42). However, since the discovery of DNA’s structure and the synthesis of organic compounds from inorganic molecules (43, 44), biology’s focus has progressively shifted from the macroscopic toward the microscopic. The wealth of new insight gained about the genetic and cellular underpinnings of biology might be argued as justification for this relegation of the comparative anatomical sciences, and their associated museum collections, to the disciplinary sidelines. However, because developmental genetics today aims for a genotype/phenotype map that can inform us about our own species’ biology, the many millions of years of evolution that stand between H. sapiens and mice still pose a formidable hurdle (8, 9, 45). We report here that considerable progress for identifying GP skeletal phenotypes beyond the mouse model is possible through transdisciplinary approaches that include the anatomical sciences. Indeed, anatomy and paleontology offer unique opportunities through which to develop and test hypotheses about the underlying genetic mechanisms of the skeleton for taxa that are not well suited to experimental manipulation, such as ourselves.
Materials and Methods
Quantitative Genetics Analyses.
We collected linear measurements of the mandibular postcanine teeth for 632 baboons that are part of a captive, pedigreed breeding colony of Papio hamadryas (46) housed at the Southwest National Primate Research Center (SNPRC) in San Antonio, Texas. Genetic management of the colony allows for data collection from animals that are not inbred. All nonfounder animals in this study resulted from matings that were random with respect to dental, skeletal, and developmental phenotype. The female-to-male sex ratio is ∼2:1. The animals from which linear tooth size measurements were collected [described in detail elsewhere (47)] are distributed across 11 extended pedigrees that are three to five generations deep. The mean number of animals with data per pedigree was 44, and these individuals typically occupied the lower two or three generations of each pedigree. All pedigree data management and preparation was facilitated through use of the computer package PEDSYS (48). The SNPRC Institutional Animal Care and Use Committee, in accordance with the established guidelines (49), approved all procedures related to the treatment of the baboons during the conduct of this study.
Measurements used in these analyses are from the left mandible, except for the measurements of the MMC, which are taken from the right due to sample size limitations on the left. Tooth area is defined as the mesiodistal length of the tooth multiplied by the mesial buccolingual width, where applicable. All trait values were inverse-normalized before analysis to account for residual kurtosis.
Statistical Genetic Analyses.
We conducted our genetic analyses within a maximum likelihood framework, the standard approach in quantitative genetics for finding parameter estimates that maximize the likelihood of the genetic model, conditional on all available data. We used a variance decomposition approach, which, as implemented in the computer package SOLAR (27, 50), makes possible analyses of data from pedigrees of arbitrary size and complexity. Per classical quantitative genetics theory and method, this approach models the phenotypic covariance for each trait of interest within a pedigree as , where Φ is a matrix of kinship coefficients for all relative pairs, is the additive genetic variance, I is an identity matrix, and is the environmental variance. These components of the phenotypic variance are additive, such that . Heritability, or the proportion of the phenotypic variance attributable to additive genetic effects, was estimated as (with 1 − h2 = e2 being variance due to nonadditive genetic factors). We modeled a phenotype of an individual as a linear function of the measurements on the trait, the means of these traits in the population, the covariates and their regression coefficients, plus additive genetic values and random environmental deviations. (The mean effects of sex and age, which served as a proxy for dental wear, were included as covariates when/if they were found to be significant.)
We assessed the significance of the maximum likelihood estimates for heritability and other parameters by means of likelihood ratio tests. Twice the difference of the maximum likelihoods of a general model (in which all parameters are estimated) and a restricted model (in which the value of a parameter to be tested is held constant at some value, usually 0) are compared. This difference is distributed asymptotically approximately as either a 1/2:1/2 mixture of c2 and a point mass at 0 for tests of parameters like h2 for which a value of 0 in a restricted model is at a boundary of the parameter space, or as a c2 variate for tests of covariates for which 0 is not a boundary value (51). In both cases, degrees of freedom are equal to the difference in the number of estimated parameters in the two models (52). However, in tests of parameters like h2, whose values may be fixed at a boundary of their parameter space in the null model, the appropriate significance level is obtained by halving the P value (51).
Extant Phenotypic Data.
We collected standard linear size measurements [following Swindler (53)] from 723 extant OWMs (representing seven genera and nine species) and 199 extant apes (representing four genera and five species) (SI Appendix, Tables S1 and S6). We also collected standard linear size measurements for 56 fossil OWMs (seven genera) and 165 fossil apes (15 genera and 26 species) (SI Appendix, Tables S4 and S7). Data included in our phenotypic analyses are published in Dataset S1. Descriptive statistics are presented in SI Appendix, Tables S2, S5, and S8.
Phylogenetic Analyses.
We used a consensus molecular chronogram from a Bayesian phylogenetic analysis of genetic data downloaded from the web site 10kTrees, version 3 (54). The 10kTrees dataset is based on sequences of 11 mitochondrial and six autosomal genes sampled from the GenBank. Details on tree inference methodology, including fossil calibration and inference of divergence dates, for the consensus chronogram are available through the 10kTrees documentation. One taxon missing from the 10kTrees sample was Presbytis rubicunda, which was added as a sister taxon to Presbytis melalophos in the consensus tree using the R package ape, version 3.0-11 (55). The divergence date for P. rubicunda and P. melalophos has been contentious, in part, because mitochondrial data suggest that populations historically called P. melalophos are paraphyletic with P. rubicunda nested within the historical P. melalophos clade, presumably due to hybridization or incomplete lineage sorting (56). To address this uncertainty, we used two divergence dates for P. rubicunda: 1.31 Ma based on mitochondrial DNA (56) and 2.5 Ma based on nuclear DNA (57, 58). Results were nearly identical between the two analyses, so we chose the date of 1.31 Ma (56) for final analyses. Divergence dates for other taxa as inferred by 10kTrees and referring to the nodes in SI Appendix are provided in SI Appendix, Fig. S3 and Table S11.
Phylogenetic Least Squares Regression.
To understand better how variation in the MMC and PMM is distributed across primates while taking evolutionary relationships into account, we fit each factor within a phylogenetic mixed model (59) using a Bayesian framework in the R package MCMCglmm, version 2.17 (60, 61). The Bayesian framework was in keeping with the tree estimation methodology used to estimate tree structure used by 10kTrees and enable us to incorporate phylogenetic uncertainty into our models. To control for possible sex and body size effects, we included the mesial width of the left M2 (a noted proxy for body size) and sex as random effects along with the consensus molecular chronogram from 10kTrees described above. We used noninformative priors corresponding to an inverse-gamma distribution with shape and scale parameters equal to 0.01. For both the MMC and PMM, we allowed the Markov chains a burn-in period of 4,000 iterations, after which we ran 140,000 iterations while sampling every 20th iteration for the posterior distribution. Pagel’s λ was then estimated from the posterior distribution, along with the 95% credibility intervals. The significance of the deviation of Pagel’s λ from 0 and 1 was tested with mean values for all extant taxa using the gls function in the R package nlme, version 3.1-113 (59). Results are presented in SI Appendix, Tables S9 and S10.
Morphological Disparity Through Time.
Morphological disparity through time was calculated for extant catarrhine taxa measured in this study following the protocol described by Harmon et al. (62) and using the R package geiger, version 2.0.3 (63). For each taxon, the mean value of the MMC and PMM for all extant taxa was used for analysis. Disparity is calculated from average pairwise Euclidian distance between species. Relative disparities for each subclade are represented by the subclade’s (or preceding node’s) disparity divided by the disparity of the entire clade. Values near 0 imply that a particular clade contains little of the overall variation and that variation in the trait is partitioned between subclades rather than within them, whereas values near 1 suggest that a clade contains a large amount of that variation and that clades may overlap in trait space. Results are presented in Fig. 3.
Supplementary Material
Acknowledgments
We thank C. Owen Lovejoy, P. David Polly, Gen Suwa, Tim D. White, and three anonymous reviewers for their feedback on earlier versions of this manuscript. We thank the following people for assistance with data collection and/or project development: Julia Addiss, Stephen Akerson, Sarah Amugongo, Liz Bates, Josh Carlson, Selene Clay, Josh Cohen, Theresa Grieco, Anne Holden, Michaela Huffman, Daniel Lopez, Kurtis Morrish, Jackie Moustakas, Alicia Murua-Gonzalez, Whitney Reiner, Oliver Rizk, Antoine Souron, Risa Takenaka, Kara Timmins, Mallory Watkins, Madsen H. White, Jeffrey Yoshihara, Sunwoo Yu, and Arta Zowghi. We thank the Southwest National Primate Research, supported by NIH National Center for Research Resources Grant P51 RR013986, for access and assistance with the quantitative genetic analysis of baboon dental variation. The following museums provided access to their skeletal collections: American Museum of Natural History, Cleveland Museum of Natural History, Phoebe A. Hearst Museum of Anthropology, University of California Museum of Vertebrate Zoology, and the Smithsonian Institution’s National Museum of Natural History. This work was supported by National Science Foundation Division of Behavioral and Cognitive Sciences Grants 0500179, 0616308, and 0130277 (to L.J.H.). The analyses presented herein relied specifically on the data provided in SI Appendix.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Full Old World monkey phenotypic data are archived online at the Dryad Digital Repository (doi:10.5061/dryad.693j8).
See Commentary on page 9142.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605901113/-/DCSupplemental.
References
- 1.Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat Rev Genet. 2011;12(3):204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 2.Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50(5):1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 3.von Koenigswald W, Rensberger JM, Pretzschner HU. Changes in the tooth enamel of early Paleocene mammals allowing increased diet diversity. Nature. 1987;328(6126):150–152. doi: 10.1038/328150a0. [DOI] [PubMed] [Google Scholar]
- 4.Rensberger JM, Watabe M. Fine structure of bone in dinosaurs, birds and mammals. Nature. 2000;406(6796):619–622. doi: 10.1038/35020550. [DOI] [PubMed] [Google Scholar]
- 5.Figueirido B, Janis CM, Pérez-Claros JA, De Renzi M, Palmqvist P. Cenozoic climate change influences mammalian evolutionary dynamics. Proc Natl Acad Sci USA. 2012;109(3):722–727. doi: 10.1073/pnas.1110246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapaltsyan V, et al. Continuously growing rodent molars result from a predictable quantitative evolutionary change over 50 million years. Cell Reports. 2015;11(5):673–680. doi: 10.1016/j.celrep.2015.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hlusko LJ. Elucidating the evolution of hominid dentition in the age of phenomics, modularity, and quantitative genetics. Ann Anat. 2016;203:3–11. doi: 10.1016/j.aanat.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Fox JG, et al., editors. The Mouse in Biomedical Research. 2nd Ed Academic; New York: 2007. [Google Scholar]
- 9.Seok J, et al. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt A, Morales-Prieto DM, Pastuschek J, Fröhlich K, Markert UR. Only humans have human placentas: Molecular differences between mice and humans. J Reprod Immunol. 2015;108:65–71. doi: 10.1016/j.jri.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Eizirik E, Murphy WJ, O’Brien SJ. Molecular dating and biogeography of the early placental mammal radiation. J Hered. 2001;92(2):212–219. doi: 10.1093/jhered/92.2.212. [DOI] [PubMed] [Google Scholar]
- 12.Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449(7161):427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 13.Young NM, Winslow B, Takkellapati S, Kavanagh K. Shared rules of development predict patterns of evolution in vertebrate segmentation. Nat Commun. 2015;6:6690. doi: 10.1038/ncomms7690. [DOI] [PubMed] [Google Scholar]
- 14.Renvoisé E, et al. Evolution of mammal tooth patterns: New insights from a developmental prediction model. Evolution. 2009;63(5):1327–1340. doi: 10.1111/j.1558-5646.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- 15.Halliday TJD, Goswami A. Testing the inhibitory cascade model in Mesozoic and Cenozoic mammaliaforms. BMC Evol Biol. 2013;13:79. doi: 10.1186/1471-2148-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernal V, Gonzalez PN. Developmental processes, evolvability, and dental diversification of New World monkeys. Evol Biol. 2013;40:532–541. [Google Scholar]
- 17.Asahara M. Unique inhibitory cascade pattern of molars in canids contributing to their potential to evolutionary plasticity of diet. Ecol Evol. 2013;3(2):278–285. doi: 10.1002/ece3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroer K, Wood B. Modeling the dental development of fossil hominins through the inhibitory cascade. J Anat. 2015;226(2):150–162. doi: 10.1111/joa.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hlusko LJ, Sage RD, Mahaney MC. Modularity in the mammalian dentition: Mice and monkeys share a common dental genetic architecture. J Exp Zoolog B Mol Dev Evol. 2011;316(1):21–49. doi: 10.1002/jez.b.21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hlusko LJ, Lease LR, Mahaney MC. Evolution of genetically correlated traits: Tooth size and body size in baboons. Am J Phys Anthropol. 2006;131(3):420–427. doi: 10.1002/ajpa.20435. [DOI] [PubMed] [Google Scholar]
- 21.Evans AR, et al. A simple rule governs the evolution and development of hominin tooth size. Nature. 2016;530(7591):477–480. doi: 10.1038/nature16972. [DOI] [PubMed] [Google Scholar]
- 22.Jheon AH, Seidel K, Biehs B, Klein OD. From molecules to mastication: The development and evolution of teeth. Wiley Interdiscip Rev Dev Biol. 2013;2(2):165–182. doi: 10.1002/wdev.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thesleff I. Current understanding of the process of tooth formation: Transfer from the laboratory to the clinic. Aust Dent J. 2014;59(Suppl 1):48–54. doi: 10.1111/adj.12102. [DOI] [PubMed] [Google Scholar]
- 24.Lesot H, Hovorakova M, Peterka M, Peterkova R. Three-dimensional analysis of molar development in the mouse from the cap to bell stage. Aust Dent J. 2014;59(Suppl 1):81–100. doi: 10.1111/adj.12132. [DOI] [PubMed] [Google Scholar]
- 25.Antón SC. Early Homo: Who, when, and where. Curr Anthropol. 2012;53(Suppl S6):S278–S298. [Google Scholar]
- 26.Grieco TM, Rizk OT, Hlusko LJ. A modular framework characterizes micro- and macroevolution of old world monkey dentitions. Evolution. 2013;67(1):241–259. doi: 10.1111/j.1558-5646.2012.01757.x. [DOI] [PubMed] [Google Scholar]
- 27.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hlusko LJ, Suwa G, Kono RT, Mahaney MC. Genetics and the evolution of primate enamel thickness: A baboon model. Am J Phys Anthropol. 2004;124(3):223–233. doi: 10.1002/ajpa.10353. [DOI] [PubMed] [Google Scholar]
- 29.Fleagle JG, McGraw WS. Skeletal and dental morphology supports diphyletic origin of baboons and mandrills. Proc Natl Acad Sci USA. 1999;96(3):1157–1161. doi: 10.1073/pnas.96.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benefit BR, McCrossin ML. In: The Primate Fossil Record. Hartwig WC, editor. Cambridge Univ Press; Cambridge, UK: 2002. pp. 241–253. [Google Scholar]
- 31.White TD, et al. Ardipithecus ramidus and the paleobiology of early hominids. Science. 2009;326(5949):75–86. [PubMed] [Google Scholar]
- 32.White TD, Lovejoy CO, Asfaw B, Carlson JP, Suwa G. Neither chimpanzee nor human, Ardipithecus reveals the surprising ancestry of both. Proc Natl Acad Sci USA. 2015;112(16):4877–4884. doi: 10.1073/pnas.1403659111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmon LJ, et al. Early bursts of body size and shape evolution are rare in comparative data. Evolution. 2010;64(8):2385–2396. doi: 10.1111/j.1558-5646.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 34.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 35.Behrensmeyer AK, et al. The structure and rate of late Miocene expansion of C4 plants: Evidence from lateral variation in stable isotopes in paleosols of the Siwalik Group, northern Pakistan. Geol Soc Am Bull. 2007;119:1486–1505. [Google Scholar]
- 36.Cerling TE, et al. Global vegetation change through the Miocene/Pliocene boundary. Nature. 1997;389:153–158. [Google Scholar]
- 37.Strömberg CAE. Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci. 2011;39:517–544. [Google Scholar]
- 38.Begun DR, Nargolwalla MC, Kordos L. European Miocene hominids and the origin of the African ape and human clade. Evol Anthropol. 2012;21(1):10–23. doi: 10.1002/evan.20329. [DOI] [PubMed] [Google Scholar]
- 39.Jablonski NG, Frost S. In: Cenozoic Mammals of Africa. Werdelin L, Sanders WJ, editors. Univ of California Press; Berkeley, CA: 2010. pp. 393–428. [Google Scholar]
- 40.Harrison T. Anthropology. Apes among the tangled branches of human origins. Science. 2010;327(5965):532–534. doi: 10.1126/science.1184703. [DOI] [PubMed] [Google Scholar]
- 41.Lovejoy CO. The origin of man. Science. 1981;211(4480):341–350. doi: 10.1126/science.211.4480.341. [DOI] [PubMed] [Google Scholar]
- 42.Persaud TVN. Early History of Human Anatomy: From Antiquity to the Beginning of the Modern Era. Thomas; Springfield, IL: 1984. [Google Scholar]
- 43.Watson JD, Crick FHC. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 44.Miller SL. A production of amino acids under possible primitive earth conditions. Science. 1953;117(3046):528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 45.Jagmag SA, Tripathi N, Shukla SD, Maiti S, Khurana S. Evaluation of models of Parkinson’s disease. Front Neurosci. 2016;9:503. doi: 10.3389/fnins.2015.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jolly CJ. In: Species, Species Concepts, and Primate Evolution. Kimbel WH, Martin LB, editors. Plenum; New York: 1993. pp. 67–107. [Google Scholar]
- 47.Hlusko LJ, Weiss KM, Mahaney MC. Statistical genetic comparison of two techniques for assessing molar crown size in pedigreed baboons. Am J Phys Anthropol. 2002;117(2):182–189. doi: 10.1002/ajpa.10022. [DOI] [PubMed] [Google Scholar]
- 48.Dyke B. PEDSYS: A Pedigree Data Management Software. Southwest Foundation for Biomedical Research; San Antonio, TX: 1996. [Google Scholar]
- 49.National Research Council . Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington, DC: 1996. [Google Scholar]
- 50.Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- 51.Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet. 1982;46(Pt 4):373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 52.Boehnke M, Moll PP, Kottke BA, Weidman WH. Partitioning the variability of fasting plasma glucose levels in pedigrees. Genetic and environmental factors. Am J Epidemiol. 1987;125(4):679–689. doi: 10.1093/oxfordjournals.aje.a114581. [DOI] [PubMed] [Google Scholar]
- 53.Swindler D. Primate Dentition. Cambridge Univ Press; Cambridge, UK: 2002. [Google Scholar]
- 54.Arnold C, Matthews LJ, Nunn CL. The 10kTrees website: A new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- 55.Paradis E, Claude J, Strimmer K. APE: Analysis of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 56.Meyer D, et al. Mitochondrial phylogeny of leaf monkeys (genus Presbytis, Eschscholtz, 1821) with implications for taxonomy and conservation. Mol Phylogenet Evol. 2011;59(2):311–319. doi: 10.1016/j.ympev.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 57.Zain BM. 2001. Molecular systematics of the genus Presbytis. PhD dissertation (Columbia University, New York)
- 58.Meijaard E, Groves CP. The biogeographical evolution and phylogeny of the genus Presbytis. Primate Rep. 2004;68:70–90. [Google Scholar]
- 59.Symonds MRE, Blomberg SP. In: Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology. Garamszegi LZ, editor. Springer; Berlin: 2014. pp. 105–130. [Google Scholar]
- 60.Hadfield JD. MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J Stat Softw. 2010;33(2):1–22. [Google Scholar]
- 61. Pinheiro J, Bates D, DebRoy S, Sarkar D; R Development Core Team (2015) nlme: Linear and nonlinear mixed effects models. R package version 3.1-120. Available at CRAN.R-project.org/package=nlme. Accessed May 2015.
- 62.Harmon LJ, Schulte JAI, 2nd, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301(5635):961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- 63.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24(1):129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.