Significance
A common practice in modern clinics is to identify a match between a mutated oncogenic protein that functions as a “driver” of a particular cancer with a known or new cancer drug from available targeted therapies. To understand mechanisms underlying the differential clinical impact of various targeted therapies on cancers driven by the receptor tyrosine kinase KIT, we analyzed a variety of biochemical and cellular properties of the most common KIT somatic mutations identified in human cancers. Surprisingly, each of the six major KIT oncogenic mutants exhibits distinct properties and responds differently to targeted therapies. These experiments show that detailed biochemical and cellular analyses of oncogenic mutations are required to optimize precision medicine for cancer treatment.
Keywords: receptor tyrosine kinase, KIT, monoclonal antibody, targeted therapy, oncogenic mutant
Abstract
Large genomic sequencing analysis as part of precision medicine efforts revealed numerous activating mutations in receptor tyrosine kinases, including KIT. Unfortunately, a single approach is not effective for inhibiting cancer cells or treating cancers driven by all known oncogenic KIT mutants. Here, we show that each of the six major KIT oncogenic mutants exhibits different enzymatic, cellular, and dynamic properties and responds distinctly to different KIT inhibitors. One class of KIT mutants responded well to anti-KIT antibody treatment alone or in combination with a low dose of tyrosine kinase inhibitors (TKIs). A second class of KIT mutants, including a mutant resistant to imatinib treatment, responded well to a combination of TKI with anti-KIT antibodies or to anti-KIT toxin conjugates, respectively. We conclude that the preferred choice of precision medicine treatments for cancers driven by activated KIT and other RTKs may rely on clear understanding of the dynamic properties of oncogenic mutants.
The rapid growth in large cancer-sequencing efforts using exome and genome sequencing, as well as elucidation of copy number variations of many cancers, has provided important information about the genetic landscapes and somatic mutations that occur in most cancers. The various catalogs of somatic mutations, chromosomal translocations, and aberrant gene expressions have provided valuable insights about distinct patient populations exhibiting gain-of-function mutations that function as causal “driver” genes for subtypes of different cancers (1–3). These studies have revealed numerous oncogenic mutations in receptor tyrosine kinases (RTKs), which result in constitutive ligand-independent tyrosine kinase activation, cell transformation, and oncogenesis. Consequently, more than 20 kinase inhibitors and half a dozen therapeutic antibodies that block the action of RTKs or the action of critical components of their intracellular signaling pathways have been developed during the past decade and successfully applied in the clinic for treatment of many cancers.
A variety of gain-of-function somatic mutations in the receptor tyrosine kinase KIT, including point mutations, in-frame deletions, and in-frame duplications, have been identified in human cancers, including gastro-intestinal-stromal tumors (GISTs), acute myeloid leukemia (AML), mast cell leukemia (MCL), and melanomas (4). Activation of the receptor tyrosine kinase KIT by its ligand stem cell factor (SCF) plays a central role in development of germ cells, hematopoietic cells, interstitial pacemaker cells, and other cells (5). Like other members of the type ΙΙΙ family of RTKs, the extracellular ligand binding domain of KIT contains five Ig-like modules (designated as D1, D2, D3, D4, and D5) connected to a single transmembrane helix, followed by a cytoplasmic region containing a regulatory juxtamembrane (JM) domain, a tyrosine kinase domain (TKD) with a kinase insert region and a C-terminal tail. Tyrosine autophosphorylation sites in the kinase insert region and the cytoplasmic tail are crucial for recruiting activated KIT effectors and mediating cell signaling in response to KIT stimulation (6–8). The major recurrent driver mutations are localized in distinct regions within the extracellular and cytoplasmic domains of KIT (Fig. 1A). The extracellular mutants are mapped to two “hot spots” in KIT-D5 whereas the cytoplasmic domain mutants are mapped to the JM region or the TKD (Fig. 1A). The most common cytoplasmic mutations in the JM domain and the TKD have been proposed to disrupt JM domain-mediated autoinhibition and favor an active kinase domain conformation (9). Oncogenic mutations in D5 of the ectodomain enhance the binding affinity of homotypic contacts, a step necessary for SCF-induced KIT activation (6, 10, 11).
Fig. 1.
Location of the major oncogenic KIT mutations. (A) Schematic presentation of KIT extracellular and cytoplasmic domains showing the positions of oncogenic KIT mutations. The mutations are located in four hot spots. Two are located in D5 of the extracellular domain, one is located in the juxtamembrane (JM) region, and another one is in the TK domain. The extracellular Ig-like domains D1–D5 are marked in blue, green, yellow, brown, and pink, respectively. The transmembrane (TM) is in black, the JM domain and kinase domains are in purple and in red, respectively. The kinase insert (KI) region and the C-terminal tail are in black. (B) Crystal structures depicting the location of the oncogenic mutations in D5, JM, and TK domains within an electron microcopy (EM) image of full-size dimeric KIT dimers fitted with X-ray crystal structures of KIT extracellular and cytoplasmic domains.
Because KIT has been proven to be a dominant oncogene in several cancers, the tyrosine kinase inhibitors (TKIs) imatinib and sunitinib have been successfully used in the clinic to target oncogenic KIT-driven cancers, best exemplified by treatment of GIST patients (4). However, because acquired resistance is often observed in GIST and other cancers treated with TKIs or other targeted therapies, further rationally developed therapies against oncogenic KIT are necessary (12–17). Moreover, it is not clear why anti-KIT mAbs that are ∼1,000-fold more potent than the TKI imatinib and 100-fold more potent than the TKI sunitinib in blocking the tyrosine kinase activity of WT KIT fail to block the activity of the oncogenic V560D KIT mutant. Also, is it possible to block the action of the KIT D816V mutant, a somatic mutation that is resistant to imatinib or sunitinib inhibition? In other words, why is the inhibitory capacity of the tyrosine kinase inhibitors imatinib and sunitinib or the inhibitory capacity of an anti-KIT mAb different toward the various KIT oncogenic mutations?
Here, we describe a detailed analysis of the enzymatic activity, cellular distribution, and dynamic behavior of all major oncogenic mutations found in the extracellular and cytoplasmic domains of KIT, using cells engineered to express WT or KIT oncogenic mutants. Because the TKIs imatinib and sunitinib are not potent inhibitors of WT KIT and of certain oncogenic KIT mutants, our goal is to identify potential therapeutic courses by targeting the regulatory D4 and D5 domains of the KIT ectodomain by using monoclonal antibodies to selectively inhibit cancers driven by WT or certain oncogenic KIT mutants, including the ones that are resistant toward imatinib or sunitinib treatments. We show that certain KIT mutants are constitutively active whereas others are “primed” or “sensitized” for SCF stimulation (i.e., responding robustly at even low ligand concentrations). Next, we demonstrate that activation, proliferation, and cellular transformation of KIT mutants that are confined to D5 are potently inhibited by monoclonal antibodies (mAbs) directed against D4 (anti-D4) or D5 (anti-D5) regions. Cell proliferation and transformation induced by the constitutively activated JM domain and TKD KIT mutants, on the other hand, are not affected by anti-KIT mAb treatment. To target constitutively activated, oncogenic KIT mutants, we exploited their observed rapid endocytosis to serve as a carrier for a toxin-conjugated anti-D4 mAb (αD4-toxin). These experiments established a role of “naked” or toxin-conjugated anti-D4 antibodies in the inhibition of cell proliferation and induction of cell death. Moreover, unveiling the dynamic nature of different oncogenic KIT mutations reveals the advantages of alternative therapeutic approaches for treating tumors driven by different oncogenic KIT mutations.
Results
Analysis of cellular localization using fluorescence microscopy showed that, unlike WT KIT, which is mostly confined to the cell membrane, oncogenic KIT with mutations in the TKD or JM domains is primarily localized inside the cell (18, 19). Because anti-RTK antibodies that have been successfully applied in targeted therapy of cancers (20) exert their inhibitory activity by binding receptor molecules that are located on the cell surface, we compared the cellular localization and dynamic properties of the most common recurrent oncogenic KIT mutations in human cancers.
KIT Cellular Distribution and Dynamic Properties Are Affected by Oncogenic Mutations.
We first compared the cell surface expression of the most frequently described oncogenic KIT mutations, which are located in D5, the JM region, and the tyrosine kinase domain (Fig. 1A). Mutations found in D5 are clustered in two spots that are located in the D5–D5 contact interface (6). One set of mutations, found primarily in AML patients, involves either substitution of D419 or its loss together with one or two flanking amino acids, like the T417IΔ418-419 mutant in which T417 is substituted with Ile and Y418 and D419 are deleted (21, 22). D419 is located in a region connecting strand A with the AB loop of Ig-like D5 (Fig. 1B). The second set of D5 mutations found in GIST patients involves either an N505I substitution, a duplication of amino acids A502 and Y503 (Dup A502Y503), or a duplication of the A502-F506 sequence. These mutations are located on strand G of D5 (Fig. 1B). Approximately 66% of GIST patients bear oncogenic mutations in the JM domain of KIT (23), which usually involves a V560 point mutation. Additional GIST mutations involve a loss of V560, together with one or two flanking amino acids (24). Mutations in the TKD of KIT are frequently found in patients with mastocytoma, such as substitution of Asp-816 by the hydrophobic amino acids Val, His, Phe, or Tyr (25).
We first used fluorescence activated cell sorting (FACS) to analyze surface localization of KIT in NIH 3T3 cells stably expressing WT KIT or an array of representative KIT mutants described above. As shown in Fig. 2 A and B, the presence of WT KIT and the N505I mutant on the cell surface is higher than that of T417IΔ418-419 and Dup A502Y503 mutants. The presence of the cytoplasmic domain mutants V560D, D816V, and V560DY823D on the cell surface was very low, in accordance with previous reports (18, 19). We also used fluorescence microscopy to visualize the cellular distribution of a green fluorescent protein (GFP) fused to WT or each oncogenic KIT mutant expressed in COS7 cells. Comparison of the cellular distribution of GFP-KIT proteins to the cell surface localization of KIT proteins, as revealed by cell surface labeling with phycoerythrin (PE)-conjugated anti-KIT mAb (Fig. S1), provided a consistent picture with the experiments presented in Fig. 2 in which the cellular distribution of WT and the various oncogenic KIT mutants are presented. The conclusions of these experiments are that WT KIT and the N505I oncogenic KIT mutant exhibit pronounced cell membrane distributions whereas reduced surface expression was detected for the T417IΔ418-419 mutant, a significantly lower surface expression was detected for the Dup A502Y503 and the V560D mutants, and negligible surface expression was detected for the D816V mutant (Fig. 2B). In fact, the majority of the D816V mutants exhibited intracellular localization (Fig. S1).
Fig. 2.
Quantification of surface expression and analysis of glycosylation patterns of WT or oncogenic KIT mutants. (A) KIT proteins expressed on the cell surface of NIH 3T3 cells were labeled with phycoerythrin (PE)-conjugated anti-KIT antibodies, and cell surface-bound antibodies were measured using FACS analysis. Black dotted curves represent FACS analyses of control NIH 3T3 cells, and red curves represent NIH 3T3-expressing WT or oncogenic KIT mutants labeled with the PE-conjugated anti-KIT antibodies. (B) The level of WT or oncogenic KIT proteins expressed on the cell surface was quantified from three independent FACS experiments and presented as the percentage of WT ± SEM. (C) Glycosylation patterns of WT or oncogenic KIT mutants. (I) KIT proteins are expressed in a form of fully (145-kDa) and partially (125-kDa) glycosylated proteins. Cell lysates of NIH 3T3 cells expressing WT or oncogenic KIT mutants were subjected to immunoprecipitation with anti-KIT D4 mAb. The washed immunoprecipitates were next treated with PNGase F, an enzyme that cleaves all N-linked sugars, followed by SDS/PAGE analysis and immunoblotting with anti-KIT antibodies. (II) NIH 3T3 cells expressing WT or oncogenic KIT mutants were treated for 16 h with tunicamycin, a drug that blocks protein glycosylation. Cell lysates prepared from tunicamycin-treated or untreated cells were subjected to immunoprecipitation with anti-KIT D4 mAb followed by SDS/PAGE analysis and immunoblotting with anti-KIT antibodies. The core KIT protein seen in the tunicamycin-treated samples migrates as a band with apparent molecular mass of 110 kDa. (D) Cell surface proteins of NIH 3T3 cells expressing WT or oncogenic KIT mutants were labeled with non–cell-permeable biotin probe. Half of the cell lysate was subjected to immunoprecipitation with anti-KIT D4 mAb to determine the total number of KIT proteins (I) and the other half of the lysate was subjected to a pull down experiment with avidin beads to determine KIT proteins located on the cell membrane (II). The samples that were treated with avidin beads were subjected to a second immunoprecipitation with anti-D4 mAb (III). All precipitations were then analyzed by SDS/PAGE and by immunoblotting with anti-KIT antibodies.
Fig. S1.
Cellular localization of WT or oncogenic KIT mutants. GFP fusion proteins of WT or oncogenic KIT mutants were transiently expressed in COS-7 cells grown on glass coverslips. Each plasmid was transfected into the cells at a similar efficiency (∼40–60% by FACS). The cells were fixed with paraformaldehyde and stained with PE-conjugated anti-KIT antibodies (Dako) to visualize the population of KIT molecules expressed on the cell surface. Images were taken using a confocal microscope. (Scale bar: 20 μm.)
Because it was shown that immature, glycosylation-deficient RTKs accumulate inside the cell (26), we next determined the pattern of receptor glycosylation of WT and KIT oncogenic mutants. We and others (18, 19) have shown that glycosylated WT KIT migrates on SDS/PAGE as two distinct bands with apparent molecular masses of 145 kDa and 125 kDa (Fig. 2 C and D). These two populations of KIT represent distinct glycosylation states where the 145-kDa protein is mature and fully glycosylated whereas the 125-kDa is immature and partially glycosylated. KIT oncogenic mutants also migrate on SDS/PAGE as two distinct bands, yet with different abundance of the 145- and 125-kDa KIT forms (Fig. 2C). We confirmed that the two KIT proteins differ only in their glycosylation state by subjecting the immunoprecipitated receptors to treatments with N-glycosidase PNGase F (Fig. 2 C, I), an enzyme hydrolyzing all N-linked carbohydrate residues, or by treating KIT-expressing cells with tunicamycin (Fig. 2 C, II), which prevents the attachment of carbohydrates to the newly synthesized KIT core protein. Both treatments resulted in the production of a similar nascent KIT protein (110 kDa), indicating that the oncogenic forms of KIT migrate differently than WT receptors on SDS/PAGE because they are not equally glycosylated (Fig. 2C).
To examine the possibility that only the mature form of KIT is expressed on the cell surface, we labeled the surface proteins of NIH 3T3 cells expressing the various KIT forms with nonpermeable biotin (Fig. 2D). This experiment showed that avidin beads pulled down mainly the mature 145-kDa KIT form whereas anti-KIT antibodies pulled down both the 145-kDa and the 125-kDa forms from the same cell lysates (Fig. 2D), supporting the notion that only the 145-kDa form of KIT is located on the cell membrane. Moreover, in the lysates that have been depleted of surface proteins by avidin pull down, the remaining KIT protein is the 125-kDa form (Fig. 2 D, III), indicating that this form is intracellular and therefore escaped surface labeling. Quantitative densitometric analysis of the expression level of the 145-kDa KIT protein (Fig. S2 A and B) was consistent with the level of KIT protein expression on the cell surface revealed by FACS analysis (compare Fig. 2B with Fig. S2B), providing further support to the notion that the 145-kDa KIT form is located at the cell surface.
Fig. S2.
Densitometric analysis of KIT protein immunoblots. (A) Five independently developed Western blots with the ChemiDoc XRS+ imaging system (Bio-Rad) were used for quantification. (B) The intensity of upper bands (145-kDa) were quantified using Image Lab software and normalized to WT; the error bar represents the SE of the mean (SEM) from the five blots. (C) The ratio of upper/lower band intensity was calculated from the five blots and expressed as mean ± SEM. A t test was performed between WT and each of the mutants. *P < 0.01, **P < 0.001, ***P < 0.0001.
The low cell surface expression of certain KIT oncogenic mutants may result from their retention inside the cell because of impairment in their membrane localization. Indeed, five out of the six KIT oncogenic mutants we analyzed showed a reduced ratio of surface/intracellular protein level (Fig. S2C), suggesting a potential deficiency in transport to the cell membrane. This notion is supported by the observation that the GFP-tagged V560D and D816V mutants show localization in the endoplasmic reticulum and Golgi apparatus (Fig. S1 and refs. 18 and 19). An alternative but not mutually exclusive mechanism for reduced expression of certain oncogenic activated KIT mutants on the cell surface may be caused by ligand-independent internalization and degradation of the activated KIT mutants.
It is well established that, upon ligand stimulation, KIT, like other RTKs, becomes activated and tyrosine phosphorylated to generate docking sites for effectors that are involved in receptor signaling. Activated KIT also recruits the ubiquitin ligase CBL, resulting in receptor ubiquitination followed by degradation and signal termination (27). We have recently demonstrated that the oncogenic KIT mutants can be categorized into two classes with respect to their activity and responsiveness to ligand stimulation. Class I KIT oncogenic mutants exhibit ligand-sensitized tyrosine kinase activity that is fully activated at low ligand concentration. Class II possess constitutive, ligand-independent tyrosine kinase activity (11). The experiment presented in Fig. 3A shows that the class I mutants DupA502Y503 or N505I can be further activated by SCF stimulation whereas the class II mutants T417IΔ418-419, V560D, and D816V, as well as the V560D/Y823D mutant, are constitutively activated and do not respond to SCF stimulation (Fig. 3A, pTyr). Interestingly, the D5 KIT mutant receptors maintain a greater cell surface localization than the JM domain and TKD mutants (Fig. 2 A and B), which strongly correlates with their stronger response to ligand stimulation, except for the T417IΔ418-419 constitutively activated mutant, which also maintains a notable level of cell surface expression.
Fig. 3.
The stability and ubiquitination of WT KIT or oncogenic KIT mutants in SCF-stimulated or unstimulated cells. (A) Oncogenic KIT mutants are ubiquitinated to become destined for degradation. SCF-stimulated or unstimulated NIH 3T3 cells expressing WT or KIT oncogenic mutants were subjected to immunoprecipitation with anti-KIT mAb followed by SDS/PAGE and immunoblotting with anti-ubiquitin antibodies (Top), anti-phospho tyrosine antibodies (Middle), or anti-KIT antibodies (Bottom). It is noteworthy that only the 145-kDa form is preferentially tyrosine-phosphorylated whereas the 125-kDa form undergoes a weak tyrosine phosphorylation. (B) Basal or ligand-stimulated degradation of WT or oncogenic KIT mutants. NIH 3T3 cells expressing either WT or KIT oncogenic mutants were treated with cycloheximide. Cell lysate from unstimulated cells, cells stimulated with SCF alone, cells stimulated with SCF together anti-D4 mAb, or cells stimulated with SCF together with imatinib were collected at various time points as indicated and subjected to immunoprecipitation with anti-KIT antibodies, followed by SDS/PAGE analysis and immunoblotting using the anti-KIT antibodies.
We next explored the relationship between the receptor’s half-life and the responsiveness to ligand stimulation by treating KIT-expressing cells with cycloheximide, which prevents protein synthesis, and monitoring the degradation of the KIT receptors. NIH 3T3 cells expressing WT or oncogenic KIT mutants were left untreated or treated with SCF for 0 min, 15 min, 30 min, 1 h, 2 h, and 4 h in the presence of cycloheximide (Fig. 3B). SCF stimulation of cells expressing WT KIT resulted in receptor internalization and rapid degradation within 30 min to 1 h whereas receptor levels in unstimulated cells were unchanged after 4 h (Fig. 3B). Interestingly, we observed different receptor dynamics between the different KIT oncogenic mutants. Class I mutants (e.g., D419A, N505I, and Dup A502Y503) that are ligand sensitive have a long basal half-life (2–4 h) and accelerated degradation upon SCF stimulation (half-life 0.5–1 h) whereas class II mutants (e.g., T417IΔ418-419, V560D, and D816V) that are constitutively activated undergo rapid degradation without ligand stimulation, and SCF stimulation does not further accelerate their degradation (half-life of 1–2 h) (Fig. 3B). These experiments also show that the Dup A502Y503 exhibits a basal level of tyrosine phosphorylation (Figs. 3A and 4) and that, upon SCF stimulation, this mutant is degraded more efficiently than the D419A, N505I mutants (Fig. 3B), which exhibit a barely detectable basal tyrosine phosphorylation (Fig. 4). Preventing class I mutant activation with either the TKI imatinib or anti-D4 mAb treatments reverses ligand-stimulated KIT degradation. Also, a kinase-deficient mutant (K623A) has a long receptor half-life (>4 h) and does not respond to SCF stimulation (Fig. S3). Furthermore, introducing a kinase-negative point mutation (K623A or Y823F) on top of class II JM domain mutant V560D prolonged its half-life from <1 h to >2 h (Fig. S3), further confirming that receptor degradation is dependent upon tyrosine kinase activity (28–30).
Fig. 4.
Comparison of the inhibitory effect of anti-D4 mAb, anti-D5 mAb, imatinib, sunitinib, or a combination of monoclonal antibodies with TKIs on the activation of WT or oncogenic KIT mutants. NIH 3T3 cells expressing either WT or KIT oncogenic mutants were stimulated with 25 ng/mL SCF alone, with SCF together anti-D4 mAb, SCF together anti-D5 mAb (A), SCF together with imatinib (B), or SCF together with sunitinib (C). Cell lysates were collected at various time points as indicated and subjected to immunoprecipitation with anti-KIT antibodies, followed by SDS/PAGE and immunoblotting with either anti-pTyr antibodies or anti-KIT antibodies. (A) Anti-D4 or anti-D5 mAbs inhibit the activation of WT and class I oncogenic KIT mutants but have minimal or no effect on the activation of class II oncogenic KIT mutants. The TKI imatinib and sunitinib inhibit the activation of most KIT oncogenic mutants (B and C) but are much less potent than anti-D4 or anti-D5 in inhibiting the activation of WT or class I oncogenic mutants (A). Treatment of cells expressing the Dup A502Y503 D5 mutant with a combination of anti-D4 together with the TKI imatinib (D) or sunitinib (E) shows strong inhibitory effect. Due to the large size of these experiments, some blots that are separated by vertical black lines (D and E) were taken from separate experiments.
Fig. S3.
A kinase-deficient mutant of WT or the V560D KIT mutant exhibit prolonged half-life in SCF-stimulated cells. Two different tyrosine kinase-deficient KIT mutants were generated by mutating a critical amino acid in the nucleotide binding site (K623A) or in the activation loop (Y823F) of the tyrosine kinase domain. KIT degradation in cycloheximide-treated SCF-stimulated or unstimulated cells that were treated with imatinib or anti-KIT D4 mAb was monitored for 4 h. Cell lysates collected at different time points were subjected to immunoprecipitation with anti-KIT mAb, followed by SDS/PAGE and immunoblotting with anti-KIT mAb. The experiment shows that the half-life of the V560D oncogenic mutant containing an additional mutation that compromised tyrosine kinase activity is extended in both unstimulated or SCF-stimulated cells, as well as in cells treated with imatinib or anti-KIT D4 mAb.
We also showed that class II KIT mutants are ubiquitinated in unstimulated cells and therefore are continuously marked for degradation once internalized. The ligand-sensitized class I mutants, on the other hand, become strongly ubiquitinated only after ligand stimulation (Fig. 3A). Thus, we demonstrated that class I KIT mutants, which include the majority of D5 mutants, are ligand-sensitive, which correlates with their increased surface expression as measured through surface labeling and receptor half-life dynamics measurements. In contrast, the majority of class II mutants are constitutively activated and are poorly retained at the cell surface through a mechanism of constitutive internalization and ubiquitination that result in degradation of these mutant receptors.
Monoclonal Antibodies Directed Against KIT D4 or D5 Inhibit the Activity of Ligand-Sensitized Oncogenic Mutants.
We have demonstrated that tyrosine kinase activities of class I oncogenic mutations in D5 rely on the formation of homotypic D4 contacts between Arg381 and Glu386, a salt bridge known to play an important role in SCF-induced KIT activation (6). However, the constitutive kinase activity of class II oncogenic mutants does not depend on formation of D4 homotypic contacts (11).
Because preventing the formation of the D4 homotypic interactions within the KIT dimer by substituting Arg-381 with Ala strongly compromised activation of WT and class I KIT mutants (6), we reasoned that monoclonal antibodies directed against D4 or D5 may exert an inhibitory effect upon cells expressing these KIT mutants. To this point, we have used inhibitory monoclonal antibodies that bind selectively to either D4 (anti-D4) or D5 (anti-D5) of KIT. It is noteworthy that the anti-D5 antibodies recognize the D5 oncogenic KIT mutants because anti-D5 and anti-D4 reveal a similar pattern of KIT proteins immunoprecipitated from cell lysates (Fig. S4). To explore the effect of these antibodies on oncogenic KIT activation, NIH 3T3 cells expressing WT or oncogenic KIT mutants were first incubated with various concentrations of anti-D4 or anti-D5 antibodies for 4 h at 37 °C. The cells were then stimulated with SCF for an additional 5 min, lysed, and subjected to immunoprecipitation with anti-KIT antibodies, followed by immunoblotting with anti-pTyr antibodies or anti-KIT antibodies (Fig. 4A). The tyrosine kinase inhibitors imatinib (Fig. 4B) or sunitinib (Fig. 4C) were also used in parallel experiments to compare their inhibitory effects on KIT activation with those of anti-D4 or anti-D5 mAbs.
Fig. S4.
Oncogenic KIT D5 mutants are recognized by anti-D4 or anti-D5 mAbs. Lysates of cells stably expressing WT or oncogenic KIT mutants were subjected to immunoprecipitation with either anti-D4 or anti-D5 mAbs, followed by SDS/PAGE and immunoblotting with rabbit polyclonal anti-KIT antibodies. A similar pattern of KIT proteins was detected in the anti-D4 or anti-D5 immunoprecipitates.
Indeed, as shown in Fig. 4A, either anti-D4 or anti-D5 antibodies strongly suppressed SCF-induced WT KIT tyrosine phosphorylation at very low antibody concentration (0.5–1 nM) and were at least 1,000- or 100-fold more potent than imatinib (1 µM) (Fig. 4B) and sunitinib (100 nM) (Fig. 4C), respectively. Furthermore, both anti-D4 and anti-D5 antibodies strongly suppressed the kinase activity of oncogenic mutants that are responsive to SCF stimulation, including D419A, N505I, and Dup A502Y503 mutants (Fig. 4A, Class I). Both antibodies inhibited receptor kinase activity at a similar concentration of 0.5–1 nM. However, the antibodies did not suppress tyrosine phosphorylation of class II oncogenic mutants that are constitutively active, including the T417IΔ418-419, V560D, or the D816V mutants (Fig. 4A, Class II) or the double mutant V560D/Y823D, which is found in patients with imatinib or sunitinib acquired resistance. Like anti-D4 and anti-D5 antibodies, neither imatinib nor sunitinib inhibited the activity of the D816V mutant (Fig. 4 B and C).
These experiments demonstrate that targeting ligand-sensitive class I D5 KIT mutants (D419A, N505I, and Dup A502Y503) with anti-D4 or anti-D5 monoclonal antibodies is tractable. Because the tyrosine kinase activity of the Dup A502Y503 D5 mutant was not completely suppressed by antibody treatment (Fig. 4A), we examined the effect of treating the cells with a combination of anti-D4 mAb together with sunitinib or imatinib. The experiment presented in Fig. 4 shows that anti-D4 mAb combined with imatinib (Fig. 4D) or sunitinib (Fig. 4E) treatments significantly inhibited tyrosine phosphorylation of the Dup A502Y503 mutant at lower concentrations than each inhibitor alone (2.5 nM sunitinib or 50 nM imatinib together with 0.5 nM anti-D4), indicating significant inhibitory benefit of combining the two drugs.
Anti-D4 mAb Effectively Inhibits Colony Formation in Soft Agar and Proliferation of Cells Bearing D5 Oncogenic KIT Mutants.
To further define the efficacy of mAbs against oncogenic KIT, we followed the effect of the anti-D4 mAb on colony formation in soft agar of NIH 3T3 cells harboring oncogenic KIT mutants. The results presented in Fig. 5A show that, unlike WT KIT, which forms colonies only in the presence of SCF, KIT D5 mutants, including Dup A502Y503 and T417IΔ418-419, as well as the JM domain mutant V560D, form colonies independent of SCF stimulation, albeit to different extents. Consistent with our biochemical analysis of tyrosine phosphorylation (Figs. 3A and 4A), the number of colonies formed in soft agar by Dup A502Y503 was lower than that of T417IΔ418-419 and V560D mutants. However, in the presence of SCF, this mutant formed a similar number of colonies as the T417IΔ418-419 or the V560D mutants (Fig. 5A). Furthermore, imatinib inhibited colony formation of both D5 and JM domain mutants whereas anti-D4 inhibition was effective only on the D5 mutants.
Fig. 5.
Anti-D4 inhibits colony formation in soft agar of NIH 3T3 cells and proliferation of Ba/F3 cells expressing KIT D5 oncogenic mutants. (A) SCF-stimulated or unstimulated NIH 3T3 cells expressing either WT or KIT oncogenic mutants were grown in the presence or absence of anti-D4 or imatinib in six-well plates containing soft agar medium. After 3 wk, the plates were stained with crystal violet and scanned for colony counting. Only colonies larger than 50 µm were counted, using ImageJ software. Colony number: —, 0–10; *, 10–20; ***, >100. (B) Expression of WT or oncogenic KIT mutants in Ba/F3 cells. Lysates of Ba/F3 cells expressing WT or oncogenic KIT mutants were subjected to immunoprecipitation with anti-D4 mAb, followed by SDS/PAGE analysis and immunoblotting with anti-KIT antibodies. (C) Cell proliferation of Ba/F3 cells expressing WT or oncogenic KIT mutants. Ba/F3 cells expressing WT or oncogenic KIT mutants were grown for 3 d in IL-3–free growth medium supplemented without/with SCF. Cell number was counted from triplicate wells, and cell proliferation was calculated as fold increase in cell number. (D) Inhibition of proliferation of Ba/F3 cells expressing WT or KIT oncogenic mutants by anti-D4, imatinib, or a combination of anti-D4 and imatinib. SCF-stimulated or unstimulated Ba/F3 cells expressing either WT or KIT oncogenic mutants were grown for 3 d in IL-3–free growth media containing different concentrations of anti-D4 mAb, imatinib, or a combination of anti-D4 mAb and imatinib. Cell number was counted from triplicate wells, and cell proliferation was calculated as fold increase in cell number. Anti-D4 exhibits strong inhibition on the proliferation of Ba/F3 expressing WT or the oncogenic D5 mutants (I–III) but does not inhibit the proliferation of Ba/F3 cells expressing either the V560D (IV) or the D816V oncogenic mutants (V and VI). Imatinib alone inhibits the proliferation of the V560D mutant, but anti-D4 mAb has no effect (IV). Dasatinib alone can inhibit D816V, but anti-D4 does not potentiate it (VI). *P < 0.05.
Many KIT-driven cancers, including AML, CML, or mastocytosis, originate in hematopoietic cells. Thus, we examined the effect of the anti-D4 mAb in abrogating cell proliferation in Ba/F3 expressing KIT mutants. Ba/F3 is a fast growing murine pro-B cell line whose growth and survival depend on interleukin (IL)-3. These cells are frequently used in kinase drug discovery programs because their IL-3 dependency can be compensated for by expressing oncogenic, constitutively active RTKs in these cells (31).
We successfully expressed WT KIT, as well as the T417IΔ418-419, DupA502Y503, V560D, and D816V mutants, in Ba/F3 cells. Consistent with the observation in NIH 3T3 cells, the oncogenic KIT mutants expressed significantly less at the cell surface than WT KIT, (Fig. 5B, upper band); surface expression of the D816V mutant was undetectable in Ba/F3 cells (Fig. 5B). Although Ba/F3 cells expressing WT-KIT depend on SCF for proliferation in IL-3–free medium, cells expressing the oncogenic mutants survive and proliferate in the absence of both IL-3 and SCF (Fig. 5C). Again, consistent with our biochemical experiments and the cell transformation analysis in NIH 3T3 cells, SCF can further stimulate the proliferation of cells expressing the KIT Dup A502Y503 mutant, indicating that a population of the Dup A502Y503 mutant is located at the cell membrane and that this sensitized mutant can be further activated by ligand stimulation. Importantly, our observation that anti-D4 mAb is effective against oncogenic D5 mutants activation is corroborated in Ba/F3 proliferation assays shown in Fig. 5D. At a concentration of 5 nM, anti-D4 mAb achieved stronger inhibition than 50 nM imatinib for inhibition of the T417IΔ418-419 and Dup A502Y503 (Fig. 5 D, II and III) mutants. Moreover, treating cells expressing WT or the D5 mutants with anti-D4 mAb in combination with imatinib resulted in stronger inhibition than each treatment alone (Fig. 5 D, I–III). However, anti-D4 mAb treatment inhibited V560D-dependent cell proliferation neither alone nor in cooperation with imatinib to inhibit this mutant. Moreover, both anti-D4 and imatinib failed to inhibit the proliferation of cells expressing the D816V mutant (Fig. 5 D, V). Although a second generation TKI dasatinib seems to have strong inhibitory effect on the D816V mutant, the mAb does not potentiate its effect when used in combination (Fig. 5 D, VI). Interestingly, although the tyrosine phosphorylation of T417IΔ418-419 (a mutant shown to be constitutively activated in AML) (32) was minimally affected by anti-D4 or anti-D5 treatments (Fig. 4A), colony formation in soft agar (Fig. 5A) and the proliferation of Ba/F3 expressing this mutant were clearly suppressed by anti-D4 mAb treatment (Fig. 5 D, II). These results suggest that the KIT inhibitory capacity of anti-D4 mAb treatment is sufficient to reduce the level of downstream effector activation that is essential for cell proliferation and transformation.
Anti-D4 Conjugated to a Toxin (αD4-Toxin) Specifically Kills Cells Expressing Oncogenic KIT Mutants That Undergo Constitutive Internalization.
Although anti-D4 mAb inhibits the activation of WT KIT and oncogenic mutants that are expressed at the cell surface, it failed to block the activation of KIT oncogenic mutants that are primarily localized inside the cell, including V560D and D816V, which constitute the majority of mutations found in GIST and systematic mastocytosis, respectively. Because we observed that anti-D4 and anti-D5 mAbs are able to recognize V560D and D816V in immunoprecipitation experiments, we surmised that these mAbs are not effective inhibitors of these mutants because they do not require formation of D4 or D5 homotypic contacts for their activation. Chase experiments demonstrated that the V560D mutant becomes rapidly degraded after ubiquitination, suggesting that this oncogenic KIT mutant reaches the surface but becomes rapidly internalized because of previous activation. Thus, this mutant receptor may act as a carrier for an anti-D4 antibody conjugated to a cytotoxic molecule (αD4-toxin) that will kill the tumor cells (Fig. S5). We next analyzed the effect of αD4-toxin on the proliferation of Ba/F3 cells that express the D5 mutants or constitutively active JM domain and TKD mutants. An anti-KLH antibody conjugated to the toxin (αKLH-toxin) was used as a control. The same number of cells was incubated with different concentrations of antibody toxin conjugate for 3 d, and cell proliferation was measured by ATP determination using CellTiter Glo (Promega) (Fig. 6A). In cells that express cell membrane-bound KIT, including WT, T417IΔ418-419, Dup A502Y503, and V560D, αD4-toxin exhibited strong cytotoxicity at a very low IC50 concentration (12 pM, 19 pM, 6 pM, and 15 pM, respectively) whereas the control αKLH-toxin required a much higher concentration (3 nM, 3 nM, 1 nM, and 2 nM, respectively) to abrogate cell growth (Fig. 6 A, II–V). The specificity of αD4-toxin over αKLH-toxin was increased by 150- to 250-fold. Not surprisingly, in Ba/F3 cells expressing an empty vector (EV) or mutant D816V that did not show detectable surface localization, αD4-toxin did not show specificity compared with αKLH-toxin (Fig. 6 A, I and VI).
Fig. S5.
Anti-KIT mAb conjugated to toxin. Anti-KIT IgG1ĸ mAb conjugated to the SG3227 pyrrolo[2,1-c][1,4]benzodiazepine antitumor antibiotic. SG3227 is a DNA-interactive pyrrolo[2,1-c][1,4]benzodiazepine dimer that binds to the minor groove of DNA to cross-link DNA strands and produce highly lethal lesions. SG3227 contains a cathepsin-cleavable valine/alanine linker attached at the N10 site of the PBD dimer. Random conjugation occurs via an iodoacetamide functional group in the linker at cysteine residues of the antibody after antibody reduction, with an average drug-to-antibody (DAR) ratio of ∼2.
Fig. 6.
Efficient killing of Ba/F3 cells expressing WT or oncogenic KIT mutants by anti-D4 toxin conjugate (αD4-toxin). (A) Killing Ba/F3 cells expressing WT, each oncogenic KIT mutant or control empty vector. (B) Killing Ba/F3 cells coexpressing WT or each oncogenic KIT mutant together with WT KIT-GFP fusion protein (WG). Ba/F3 cells were plated in 96-well plates to which either αD4-toxin or αKLH-toxin (control toxin conjugate) was added at various concentrations as indicated. Cells were then left to grow for 3 d, and the number of live cells was determined using the CellTiter Glo assay (Promega). The results are presented as raw luminescence units (RLUs), which correlates with the number of live cells, versus logarithm concentration of antibody-toxin conjugates. The IC50 (half maximal inhibitory concentration) was calculated from a sigmoidal dose–response curve fitted using GraphPad Prism software.
Because most tumor cells are heterozygous for KIT mutants, we reasoned that the WT receptor may serve as a target for the αD4-toxin, where the mutant protein is expressed at very low levels at the cell surface. To examine this possibility, we introduced a C terminus GFP-tagged WT KIT into Ba/F3 cells that also expressed various oncogenic mutants to study the cytotoxicity of αD4-toxin in these cells. Cells coexpressing the GFP-tagged full-length KIT allele were established as stable cell lines, and the expression was maintained through functional replacement of IL-3 and different antibiotics, confirmed by immunoblotting experiments and FACS analysis (Fig. S6). These cells were subjected to similar proliferation experiments with various concentrations of αD4-toxin. The results presented in Fig. 6B show that, upon coexpression of full-length KIT, Ba/F3 cells expressing the D816V mutant became sensitive to αD4-toxin treatment with an IC50 of 4 pM, a 100-times lower concentration than the αKLH-toxin control (Fig. 6 B, VI). Coexpression of the WT KIT allele did not change the effectiveness of αD4-toxin in cells expressing the mutant allele that can be targeted by the antibody toxin conjugate when expressed alone (Fig. 6 B, II–V). Taken together, these experiments suggest that anti-KIT antibodies conjugated to a cytotoxic molecule could be applied for targeted therapy of cancer cells expressing even very small populations of oncogenic KIT mutants on their surface, as well as cells expressing the WT allele of oncogenic KIT. Preliminary experiments demonstrate that αD4-toxin inhibits tumor growth in an H526 small cell lung cancer xenograft murine model system (Fig. S7). This experiment showed that treatment of mice with 3 mg/kg αD4-toxin resulted in nearly complete tumor growth inhibition.
Fig. S6.
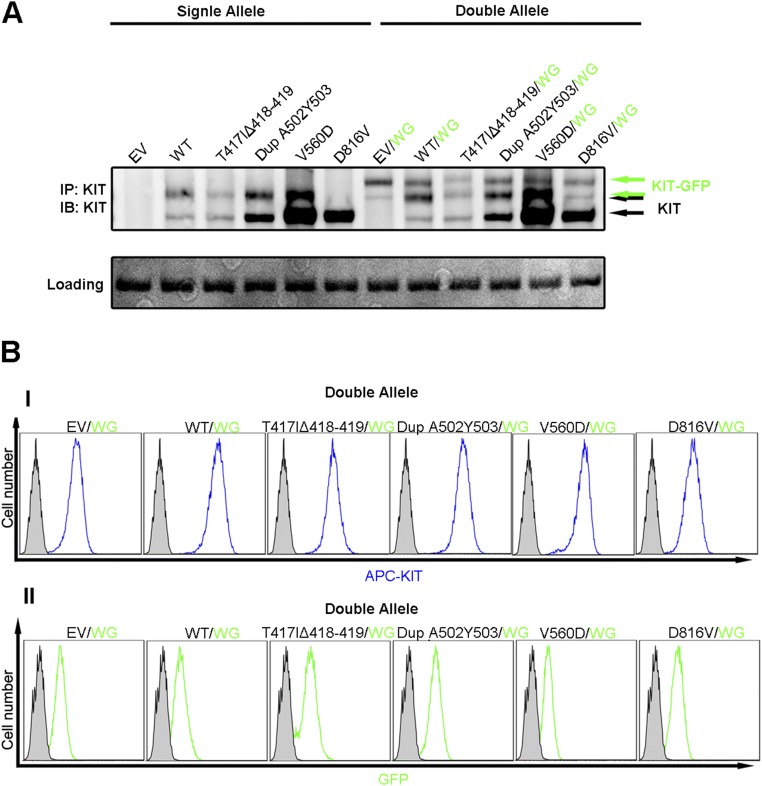
Ba/F3 cells expressing WT or oncogenic KIT mutants alone or together with a GFP fusion full-length KIT fused to GFP. Ba/F3 cells expressing either empty vector (EV), WT, or oncogenic KIT mutants were cotransfected with pMSCVhygro-WT-KIT-GFP (WG). The expression of both alleles was analyzed by immunoblotting with anti-KIT mAb (A) or by FACS analysis (B). WT or oncogenic KIT mutants were analyzed using APC-conjugated anti-KIT mAb (I), and the WG allele was visualized by directly monitoring GFP fluorescence (II).
Fig. S7.
An anti-KIT D4 antibody toxin conjugate (αD4-toxin) inhibits tumor growth in an H526 small cell lung cancer xenograft mouse model. BALB/c mice were s.c. inoculated with 5 × 106 H526 tumor cells expressing WT KIT. Mice were treated with one i.p. injection of control conjugate, anti-KIT conjugate, or PBS after the growth of tumors to 150–200 mm3. Tumor volumes were determined twice weekly. Treatment of mice with 3 mg/kg αD4-toxin resulted in nearly complete tumor growth inhibition whereas treatment with 1 mg/kg resulted in significant tumor inhibition. On the other hand, treatment with αKLH-toxin had no significant impact on tumor growth compared with treatment with PBS alone. For s.c. xenograft studies, H526 small cell lung cancer tumor cells were maintained in RPMI 1640 medium supplemented with 10% (vol/vol) heat-inactivated FBS at 37 °C in 5% CO2. Once growing in an exponential growth phase, cells were harvested for tumor inoculation. Female BALB/c mice were inoculated s.c. in the right flank at 6–8 wk of age with 5 × 106 H526 tumor cells in 0.1 mL of PBS supplemented 1:1 with Matrigel. Upon reaching a tumor size of 150–200 mm3, animals were randomized into treatment groups for the efficacy study. Animals were treated with a single i.p. injection of PBS or conjugated antibody at 10 µL/g. Tumor sizes were measured twice weekly in two dimensions using a caliper, and the volume is expressed in mm3 using the formula: V = 0.5 a × b2, where a and b are the long and short diameters of the tumor, respectively.
Discussion
The determination of the crystal structure of the extracellular domain of KIT before and after ligand stimulation provided insights and opportunities for how to specifically target cancer cells driven by gain-of-function mutations in KIT and other RTKs. Specifically, the crystal structure of the full-length extracellular domain of KIT showed that SCF stimulated dimerization of the three membrane-distal Ig-like domains D1–D3 and induced large changes in the orientation and distance between the two membrane-proximal D4 and D5 domains, which resulted in D4- and D5-mediated homotypic associations between neighboring KIT molecules (6). This lateral movement requires flexible joints at the D3–D4 and D4–D5 hinge regions of each monomer and is crucial for positioning KIT dimers in the correct orientation and distance to enable KIT tyrosine kinase autophosphorylation and activation of cell signaling. Substitution of individual amino acids involved in the formation of salt bridge-mediated homotypic contacts strongly compromises activation and cell signaling via KIT, PDGF receptors, and VEGF receptor 2 (6, 33, 34). Moreover, we have recently determined the crystal structure of D4–D5 of KIT harboring the oncogenic T417IΔ418-419 mutation to demonstrate how strong homotypic contacts taking place between an oncogenic D5 mutant lead to constitutively activated (ligand-independent) KIT stimulation in cancer cells (11).
In this report, we analyzed the biochemical and cellular properties of the most common somatic KIT mutations to determine the feasibility of pharmacologic targeting of oncogenic KIT mutations using monoclonal antibodies that prevent D4 or D5 homotypic contact formation, a step necessary for SCF stimulation of WT KIT. These results indicate that different somatic mutations cause major differences in the cellular distribution and dynamic properties of oncogenic KIT mutants. The differential dynamic properties of the various oncogenic mutants enable the application of alternative therapeutic approaches using site-specific antibodies, small molecule kinase inhibitors, or a combination thereof (Fig. 7).
Fig. 7.
Oncogenic KIT mutations are classified into two main groups, class I and class II. Class I mutants are expressed at the cell surface, albeit at different levels, and exhibit sensitized ligand response. Class I mutations include the D5 point mutations D419A and N505I, deletion of Y418 D419, and duplication of A502Y503. Class II have constitutively activated tyrosine kinase activities and show low or negligible surface expression. Class II mutations include the T417IΔ418-419 D5 mutation and the intracellular V560D and D816V point mutants.
We determined, for the most common KIT mutants found in various cancers, that differential surface localization is correlated with basal and ligand-induced receptor activation. Specifically, oncogenic KIT mutations can be classified into two main groups. Class I mutants exhibit surface localization and sensitized ligand response (in comparison with WT KIT). These mutations include the D5 point mutations D419A and N505I, deletion of Y418 D419, and duplication of A502Y503. Class II mutants exhibit low or negligible surface expression of constitutively activated tyrosine kinase activities. These mutations include the D5 T417IΔ418-419 mutation and the intracellular V560D and D816V point mutants. Interestingly, the effect of anti-D4 mAbs in abrogating oncogenic KIT signaling was seen in mutations localized in D5, which include all class I mutants and the class II T417IΔ418-419 mutation.
Our results show that ligand-induced tyrosine kinase activation, as well as cell proliferation and colony formation in soft agar of D5 KIT mutants, is strongly inhibited by anti-D4 mAb treatment. Interestingly, treating cells expressing the Dup A502Y503 or the T417IΔ418-419 mutants with anti-D4 in combination with a low dose of the small molecule kinase inhibitors imatinib or sunitinib completely abolishes their activation. The low dose of inhibitor used to treat cancer cells in the presence of anti-D4 antibody might delay or reduce the development of drug resistance known to take place in cancers treated with these drugs. Therefore, cancer patients may respond to TKI drugs for longer periods without drug resistance, when administered together with anti-D4 antibody. Interestingly, clinical studies have shown that whereas only 5% of patients bearing mutations in the JM domain of KIT develop resistance to imatinib, 16% of patients bearing mutations in KIT D5 develop imatinib resistance (35). We speculate that patients bearing D5 mutations more frequently develop resistance to imatinib treatment because higher doses of imatinib are required for treating patients harboring D5 mutations.
Although anti-D4 antibody is a potent inhibitor of the kinase activity and proliferation of cells expressing the D5 mutants, they barely influence the activity or proliferation of NIH 3T3 or Ba/F3 cells expressing KIT mutated in the receptor’s cytoplasmic region, including the JM domain and the TKD. It is noteworthy that the kinase and oncogenic activities of the V560D, D816V, or V560DY823D mutants do not rely on formation of D4- and D5-dependent homotypic contacts, and therefore antibodies directed against D4 or D5 will not influence the activation of these oncogenic mutants. Also, we have shown that the constitutively activated V560D KIT mutant resides on the cell membrane only temporarily before it becomes rapidly internalized, and, therefore, insufficient amounts of these mutants will bind the antibody before they become internalized. However, because we observed that these mutant KIT receptors do reach the surface and are internalized constitutively and also that they can be recognized by anti-D4 mAb, we explored the possibility that conjugation of anti-D4 antibodies to a cytotoxic molecule would result in tumor cell killing.
Using as αKIT-toxin conjugate as targeted therapy is a very compelling approach because the toxin is extremely potent in killing tumor cells at pΜ concentrations and its ability to kill cells selectively requires the expression of KIT on the cell surface either continuously or temporarily. The advantage of αKIT-toxin conjugate over naked anti-D4 mAb is its ability to target all KIT mutants, no matter where on the receptor the mutations are localized, and therefore can be used for treatment of all KIT-driven cancers. We initially studied the effect of the toxin-conjugated anti-D4 mAb on Ba/F3 cells expressing D5 mutants, including Dup A502Y503 and T417IΔ418-419, JM domain mutant V560D, and TKD mutant D816V, using WT KIT-expressing cells or empty vector-expressing cells as controls. We generated two sets of cell lines for these studies: one that expressed the mutants alone and a second that coexpressed each mutant receptor together with WT KIT (in a form of GFP tagged to be distinguished from the mutant KIT by size). We used these two sets of cell lines for two reasons. First, most tumors express one normal and one mutant allele of KIT in the same cell. Second, we wished to examine the possibility of whether the presence of full-length WT KIT would augment the killing of KIT mutants that are poorly expressed or even not expressed at all on the cell surface. The experiment presented in the manuscript shows that all oncogenic KIT mutants, except for the KIT D816V mutant, efficiently deliver αKIT-toxin into Ba/F3 cells. Because negligible amounts of the D816V mutant are expressed on the cell surface of Ba/F3 cells, cells expressing this mutant can be killed by αD4-toxin treatment only upon coexpression of WT KIT allele in the same cell. Because new small molecule inhibitors, such as EXEL-0862 and dasatinib, emerge to show an inhibitory effect on D816V mutants (36, 37), multiple choices of treatments for imatinib-resistant KIT mutant will be available.
Through the analysis of the distinct properties of different gain-of-function KIT mutations, it is becoming clear that a single approach cannot be applied for treating cancers driven by all KIT mutants because KIT can be activated by distinct mechanisms. Although identification of somatic mutations provides, to our knowledge, the first insight into the genetic changes that drive cancers, additional functional analysis is required to fully unveil their mode of action and identify the most effective targeted therapy for blocking their activities. Here, we show that, to target all major KIT drivers, we first had to gain insights into the dynamic nature and cellular localization of KIT mutants in the cells, which provided valuable information about how to effectively target the action of the different KIT mutants in cancer cells.
On the basis of the experiments presented in this manuscript, we propose two different regimens for treatment of patients harboring different KIT mutations. Accordingly, treatment regimen I is to apply anti-D4 mAb for treatment of cancers (or other diseases) driven by WT KIT or by oncogenic mutations in D5 alone, as well as in combination with low doses of sunitinib and imatinib. Treatment regimen II is to use toxin anti-D4 mAb conjugates to target oncogenic KIT mutations that rapidly enter the cells and therefore cannot be efficiently inhibited by treatment with naked anti-D4 mAb. Regimen II may also be applied for treating patients who are resistant to, or acquire resistance toward, imatinib or sunitinib treatments or treatment with other TKIs.
Materials and Methods
Cell Culture, Immunoprecipitation, and Immunoblotting.
The retroviral vectors pBABEpuro and pMSCVpuro/hygro were used for generating all KIT-expressing NIH 3T3 and Ba/F3 cell lines, respectively. NIH 3T3 cells were cultured in DMEM supplemented with 10% (vol/vol) bovine serum, and Ba/F3 cells were grown in RPMI 1640 supplemented with 10% (vol/vol) FBS and 1 ng/mL IL-3 (R&D Systems). Ba/F3 cells expressing WT KIT were grown in medium supplemented with SCF instead of IL3. Ba/F3 cells expressing oncogenic KIT mutants survive and proliferate in growth medium deficient of both IL3 and SCF. To generate Ba/F3 cells expressing WT KIT together with a KIT oncogenic allele, cells were transfected with the pMSCVpuro vector expressing the mutants and pMSCVhygro expressing WT KIT in a form of a GFP fusion protein (WG). Before SCF stimulation, cells were starved overnight in serum-free medium as previously described (6, 38).
SCF was expressed in Escherichia coli and was purified as previously described (39). Unstimulated or SCF-stimulated cells were lysed and subjected to immunoprecipitation, followed by immunoblotting with various antibodies (6, 33). Anti-KIT polyclonal antibodies were generated by immunizing rabbits with recombinant KIT ectodomain (6). Anti-phosphotyrosine (pTyr) antibodies were purchased from Upstate Biotechnology and anti-ubiquitin antibodies were purchased from Santa Cruz. Monoclonal anti-D5 KIT antibodies were made as previously described (10).
N-Glycosidase Treatment.
WT and KIT mutants were immunoprecipitated from lysates of NIH 3T3 cells using anti-KIT antibodies. The samples were incubated in glycoprotein denaturing buffer (0.5% SDS, 40 mM DTT) for 10 min at 100 °C to completely denature the KIT proteins and then treated with 500 U of PNGase F (NEB) for 1 h at 37 °C as recommended by the manufacturer. Treated samples were then subjected to SDS/PAGE analysis followed by immunoblotting with anti-KIT antibodies.
Tunicamycin Treatment.
NIH 3T3 cells expressing WT or oncogenic KIT oncogenic mutants were treated with 10 µg/mL tunicamycin for 16 h at 37 °C. Lysates prepared from tunicamycin-treated or untreated cells were subjected to immunoprecipitation, followed by SDS/PAGE and immunoblotting with anti-KIT antibodies.
Cell Proliferation Assay.
Ba/F3 cells (400,000) expressing WT or oncogenic KIT mutants were plated, in triplicate, in six-well plates containing growth medium. Cells were then treated with various concentrations of different stimuli and/or inhibitors for 3 d. Cell growth was monitored by counting living cells using a handheld automated cell counter (Scepter; Millipore).
Treatment of Ba/F3 Cells with Antibody Toxin Conjugates.
Ba/F3 cells (10,000) expressing WT or/and oncogenic KIT mutants were plated in 96-well plates and treated with various concentrations (as indicated in Fig. 6) of anti-D4 toxin conjugate (αD4-toxin) or the control antibody anti-KLH toxin conjugate (αKLH-toxin). Cells were incubated with the toxin-conjugated antibodies for 3 d at 37 °C, and the number of living cells was determined using the CellTiter-Glo Luminescence Assay (Promega) according to the manufacturer’s instructions. In Fig. 6, raw luminescence units (RLUs) are presented on the y axis, and the concentration of toxin conjugate (log[M]) is presented on the x axis. The IC50 was calculated using GraphPad Prism software with sigmoidal dose–response curve fitting.
Colony Formation in Soft Agar.
NIH 3T3 cells (5,000) expressing WT or oncogenic KIT mutants were plated in six-well plates on top of a 0.6% layer of Agarose (Seaplaque). Cells were either left untreated or treated with SCF (100 ng/mL), imatinib (500 nM), and anti-D4 (50 nM) for 21 d. Colony formation was visualized by staining the plates with 0.05% crystal violet and scanning the samples with a high-resolution scanner. Only colonies larger than 100 µm were counted. The results were analyzed using the open source ImageJ software.
Fluorescence-Activated Cell Sorting Analysis.
For FACS analysis, NIH 3T3 and Ba/F3 cells expressing WT and the various KIT mutants were resuspended in PBS containing 1% BSA and 0.01% sodium azide. NIH 3T3 cells were labeled with phycoerythrin (PE)-conjugated anti-KIT monoclonal antibody (R7145; Dako), and Ba/F3 cells were immunostained with allophycocyanin (APC)-conjugated anti-KIT monoclonal antibody (C7247; Dako). Fluorescent-labeled cells were analyzed using a Stratedigm S1000 flow cytometer.
Receptor Degradation.
NIH 3T3 cells expressing either WT or KIT mutants were treated with 10 µg/mL cycloheximide to stop protein synthesis. Cells were then left unstimulated or stimulated with SCF alone, SCF in combination with anti-D4 mAb, or SCF together with TKI. Treated cells were lysed at various time points as indicated and subjected to immunoprecipitation followed by immunoblotting with anti-KIT antibodies.
Surface Biotinylation Assay.
NIH 3T3 cells expressing WT or oncogenic KIT mutants were labeled with nonpermeable Sulfo-NHS-SS-Biotin (Pierce) according to the manufacturer’s recommendation. Biotin-labeled cells were washed with PBS containing 0.1 M glycine, lysed, and subjected to either a pull down with NeutrAvidin agarose beads (Pierce) or immunoprecipitation with anti-KIT antibodies. Also, the supernatants of the avidin agarose pull downs were collected and subjected to a second immunoprecipitation with anti-KIT antibodies (to detect KIT proteins that were not biotin-labeled). All samples were then analyzed using SDS/PAGE, followed by immunoblotting with anti-KIT antibodies.
Footnotes
Conflict of interest statement: E.M.M.-B. and Y.H. are employees of Kolltan Pharmaceuticals.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610179113/-/DCSupplemental.
References
- 1.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 2.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert Opin Investig Drugs. 2013;22(1):103–115. doi: 10.1517/13543784.2013.740010. [DOI] [PubMed] [Google Scholar]
- 5.Ashman LK. The biology of stem cell factor and its receptor C-kit. Int J Biochem Cell Biol. 1999;31(10):1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- 6.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130(2):323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roskoski R., Jr Structure and regulation of Kit protein-tyrosine kinase: The stem cell factor receptor. Biochem Biophys Res Commun. 2005;338(3):1307–1315. doi: 10.1016/j.bbrc.2005.09.150. [DOI] [PubMed] [Google Scholar]
- 9.Foster R, Griffith R, Ferrao P, Ashman L. Molecular basis of the constitutive activity and STI571 resistance of Asp816Val mutant KIT receptor tyrosine kinase. J Mol Graph Model. 2004;23(2):139–152. doi: 10.1016/j.jmgm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Reshetnyak AV, et al. Structural basis for KIT receptor tyrosine kinase inhibition by antibodies targeting the D4 membrane-proximal region. Proc Natl Acad Sci USA. 2013;110(44):17832–17837. doi: 10.1073/pnas.1317118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reshetnyak AV, et al. The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer. Mol Cell. 2015;57(1):191–201. doi: 10.1016/j.molcel.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinrich MC, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 13.Liegl B, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216(1):64–74. doi: 10.1002/path. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, et al. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99(5):1741–1744. doi: 10.1182/blood.v99.5.1741. [DOI] [PubMed] [Google Scholar]
- 15.McLean SR, et al. Imatinib binding and cKIT inhibition is abrogated by the cKIT kinase domain I missense mutation Val654Ala. Mol Cancer Ther. 2005;4(12):2008–2015. doi: 10.1158/1535-7163.MCT-05-0070. [DOI] [PubMed] [Google Scholar]
- 16.Roberts KG, et al. Resistance to c-KIT kinase inhibitors conferred by V654A mutation. Mol Cancer Ther. 2007;6(3):1159–1166. doi: 10.1158/1535-7163.MCT-06-0641. [DOI] [PubMed] [Google Scholar]
- 17.Tamborini E, et al. Functional analyses and molecular modeling of two c-Kit mutations responsible for imatinib secondary resistance in GIST patients. Oncogene. 2006;25(45):6140–6146. doi: 10.1038/sj.onc.1209639. [DOI] [PubMed] [Google Scholar]
- 18.Tabone-Eglinger S, et al. KIT mutations induce intracellular retention and activation of an immature form of the KIT protein in gastrointestinal stromal tumors. Clin Cancer Res. 2008;14(8):2285–2294. doi: 10.1158/1078-0432.CCR-07-4102. [DOI] [PubMed] [Google Scholar]
- 19.Bougherara H, et al. The aberrant localization of oncogenic kit tyrosine kinase receptor mutants is reversed on specific inhibitory treatment. Mol Cancer Res. 2009;7(9):1525–1533. doi: 10.1158/1541-7786.MCR-09-0138. [DOI] [PubMed] [Google Scholar]
- 20.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23(9):1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 21.Cammenga J, et al. Extracellular KIT receptor mutants, commonly found in core binding factor AML, are constitutively active and respond to imatinib mesylate. Blood. 2005;106(12):3958–3961. doi: 10.1182/blood-2005-02-0583. [DOI] [PubMed] [Google Scholar]
- 22.Hussain SR, Naqvi H, Mahdi F, Bansal C, Babu SG. KIT proto-oncogene exon 8 deletions at codon 419 are highly frequent in acute myeloid leukaemia with inv(16) in Indian population. Mol Biotechnol. 2013;54(2):461–468. doi: 10.1007/s12033-012-9584-x. [DOI] [PubMed] [Google Scholar]
- 23.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22(18):3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 24.Hirota S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 25.Verstovsek S. Advanced systemic mastocytosis: The impact of KIT mutations in diagnosis, treatment, and progression. Eur J Haematol. 2013;90(2):89–98. doi: 10.1111/ejh.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contessa JN, Bhojani MS, Freeze HH, Rehemtulla A, Lawrence TS. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008;68(10):3803–3809. doi: 10.1158/0008-5472.CAN-07-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masson K, Heiss E, Band H, Rönnstrand L. Direct binding of Cbl to Tyr568 and Tyr936 of the stem cell factor receptor/c-Kit is required for ligand-induced ubiquitination, internalization and degradation. Biochem J. 2006;399(1):59–67. doi: 10.1042/BJ20060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honegger AM, et al. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell. 1987;51(2):199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- 29.Sorokin A, Mohammadi M, Huang J, Schlessinger J. Internalization of fibroblast growth factor receptor is inhibited by a point mutation at tyrosine 766. J Biol Chem. 1994;269(25):17056–17061. [PubMed] [Google Scholar]
- 30.Moolenaar WH, et al. A point mutation at the ATP-binding site of the EGF-receptor abolishes signal transduction. EMBO J. 1988;7(3):707–710. doi: 10.1002/j.1460-2075.1988.tb02866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warmuth M, Kim S, Gu XJ, Xia G, Adrián F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19(1):55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 32.Gari M, et al. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol. 1999;105(4):894–900. doi: 10.1046/j.1365-2141.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci USA. 2010;107(5):1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105(22):7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corless CL, Heinrich MC. Molecular pathobiology of gastrointestinal stromal sarcomas. Annu Rev Pathol. 2008;3:557–586. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- 36.Pan J, et al. EXEL-0862, a novel tyrosine kinase inhibitor, induces apoptosis in vitro and ex vivo in human mast cells expressing the KIT D816V mutation. Blood. 2007;109(1):315–322. doi: 10.1182/blood-2006-04-013805. [DOI] [PubMed] [Google Scholar]
- 37.Schittenhelm MM, et al. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer Res. 2006;66(1):473–481. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 38.Kouhara H, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89(5):693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Zhang R, Joachimiak A, Schlessinger J, Kong XP. Crystal structure of human stem cell factor: Implication for stem cell factor receptor dimerization and activation. Proc Natl Acad Sci USA. 2000;97(14):7732–7737. doi: 10.1073/pnas.97.14.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]