Significance
Using a social insect model, we tested how supplementing young adult bees with a resident microbiota species affects host physiology and microbiome composition. This supplementation had significant consequences for host development and detoxification responses, parasite susceptibility, and microbiome community structure. Our results show that early perturbation of the microbiota composition can have sustained consequences for hosts. Additionally, this work provides a cautionary tale to the arbitrary use of probiotics in animal health management and highlights the importance of experimental research addressing factors that shape animal microbiome communities.
Keywords: cytochrome P450, dysbiosis, microbial interaction, parasitism, vitellogenin
Abstract
Microbial symbionts living within animal guts are largely composed of resident bacterial species, forming communities that often provide benefits to the host. Gut microbiomes of adult honey bees (Apis mellifera) include core residents such as the betaproteobacterium Snodgrassella alvi, alongside transient parasites such as the protozoan Lotmaria passim. To test how these species affect microbiome composition and host physiology, we administered S. alvi and/or L. passim inocula to newly emerged worker bees from four genetic backgrounds (GH) and reared them in normal (within hives) or stressed (protein-deficient, asocial) conditions. Microbiota acquired by normal bees were abundant but quantitatively differed across treatments, indicating treatment-associated dysbiosis. Pretreatment with S. alvi made normal bees more susceptible to L. passim and altered developmental and detoxification gene expression. Stressed bees were more susceptible to L. passim and were depauperate in core microbiota, yet supplementation with S. alvi did not alter this susceptibility. Microbiomes were generally more variable by GH in stressed bees, which also showed opposing and comparatively reduced modulation of gene expression responses to treatments compared with normal bees. These data provide experimental support for a link between altered gut microbiota and increased parasite and pathogen prevalence, as observed from honey bee colony collapse disorder.
Animal guts contain diverse microbial communities that are often dominated by suites of bacteria but can also include archaea, viruses, protozoans, and fungi (1). Consistently present, or core, bacterial species are often beneficial to their host and are thought to be integral to animal evolution by influencing host growth, development, health, and behavior (2–7). Disrupted proportions among core bacterial communities can alter microbiome homeostasis, causing “dysbiosis” (1, 8, 9) that may alter host interactions. For example, shifts in bacterial population sizes in the mammalian gut have been linked to inflammatory gut disorders, diabetes, and obesity (8). Thus, causes of and consequences from dysbiosis are key to understanding animal health and disease. Whereas the diverse gut communities of mammals contain functional redundancy that may buffer shifts in composition (7, 10), insects usually have much lower microbiota diversity (4, 6, 9, 11) and consequently may be more affected by dysbiosis.
European honey bees, Apis mellifera, are globally significant to agriculture primarily due to their pollination services and honey production. As social insects, they form colonies with tens of thousands of mostly nonreproductive female workers that progressively perform a variety of tasks required for colony maintenance and growth as they age. This division of labor is centrally regulated by the expression of vitellogenin (Vg), a gene under strong positive selection in workers (12, 13) encoding a glycolipoprotein that affects development, lifespan, and immunity (14–18) and serves as a key biomarker for bee health and fitness (13, 19–21).
Gut bacteria acquired by honey bees as larvae are purged during pupation, and new communities are established following adult emergence through contact with nestmates and/or nest surfaces (4, 22–25). Eight bacterial symbionts typify the core adult honey bee gut microbiota, including two abundant Proteobacteria, Snodgrassella alvi (Betaproteobacteria) and Gilliamella apicola (Gammaproteobacteria), as well as two clusters of Firmicutes within the genus Lactobacillus (26–29). Functional data from some Lactobacillus species (30, 31) and from the alphaproteobacterium Parasaccharibacter apium (32, 33) support beneficial roles to larval survival and parasite resistance. However, data from a strain of Frischella perrara (Gammaproteobacteria) is associated with melanization in the bee gut epithelium (34), a response generally associated with host defense against potential pathogens. Roles for S. alvi and G. apicola are not clearly demonstrated, although nutritional and defensive roles have been hypothesized on the basis of genomic sequence data (35). Studies of bumble bee gut microbiota, which similarly include S. alvi and G. apicola, suggest a protective role against trypanosomatid protozoan parasites (36, 37). Related trypanosomatids can be common in honey bee guts (e.g., refs. 28 and 38–43) and have been linked to colony mortalities (39, 40, 44). Other potentially harmful microbes also occur transiently in bee guts and likely interact with other members of the community (45).
Advancing animal health management requires understanding the interplay among stressors, microbiomes, and host genetics. For example, ongoing problems with honey bee health (46–49) are attributed to multiple stress factors including parasites and pathogens, pesticides, suboptimal nutrition, and stress associated with commercial management. Genetic diversity of bee stocks can influence their adaptability to stressors (e.g., refs. 50–53); these effects may involve key metabolic and detoxification genes, such as cytochrome P450s (54–56). Their gut microbiota also likely mediate stress responses, and bacterial strains cultivated from honey bees could be harnessed as probiotic mitigators of stress (30–33, 57).
Here, we used honey bees as a comparatively simple animal model to address the roles of microbial species in gut microbiome homeostasis and in host physiology. We explored how early colonization with a core bacterial symbiont, S. alvi, and a common parasite, Lotmaria passim, affected microbiome composition and host gene expression. We administered S. alvi and/or L. passim to newly emerged worker bees from four genetic backgrounds. We tested the prediction that gut colonization by S. alvi renders the host less susceptible to parasitism by L. passim during “normal” (hive-reared) and “stressed” (laboratory-reared, asocial, protein-deficient) conditions. Further, we tested for impacts of these treatments on host physiology by quantifying expression of vitellogenin and three cytochrome P450 (CYP450) genes, which are known central mediators of development and detoxification capabilities (14–18, 54–56).
Results
Newly emerged adults were inoculated with S. alvi, L. passim, both, or neither, and were then reared in either a normal hive environment (“normal”) or in a nutritionally and socially stressful and largely sterile laboratory environment (“stressed”). At 6 d postemergence, bees were harvested to examine effects on microbiota and expression of key bee genes used as indicators of stress responses and health.
Impacts on the Bacterial Microbiome.
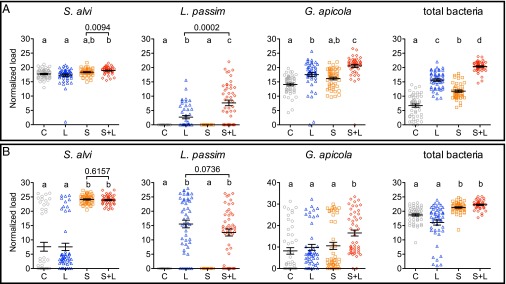
Bees with normal rearing developed high loads of S. alvi (Fig. 1A) whether or not they received experimental S. alvi inocula, as expected via natural social acquisition. Highest loads occurred in bees treated with both S. alvi + L. passim (S+L), larger than untreated controls (C) and L. passim-only (L) bees (one-way ANOVA, P < 0.0001 for both) and larger than direct comparison with bees treated with S. alvi only (S) (Mann–Whitney U test, P = 0.0094; C, 17.71 ± 0.24 SEM; L, 17.31 ± 0.43 SEM; S, 18.34 ± 0.18 SEM; and S+L, 18.92 ± 0.18 SEM). Loads of S. alvi did not vary by host genotype (GH) (Fig. S1A; one-way and two-way ANOVA, P > 0.05).
Fig. 1.
Microbial loads in bees during normal (A) and stressed (B) rearing. Means ± SEM are shown by the black bars and whiskers. Treatments (n = 48 each): sugar water control (C, gray circles); L. passim only (L, blue triangles); S. alvi only (S, orange squares); and S. alvi and L. passim (S+L, red diamonds). Different letters above treatment groups indicate significantly different loads (one-way ANOVA with Dunn’s multiple comparison tests). P values above brackets are from planned comparisons using two-tailed Mann–Whitney U tests. Data presented by each of four colonies in Fig. S1.
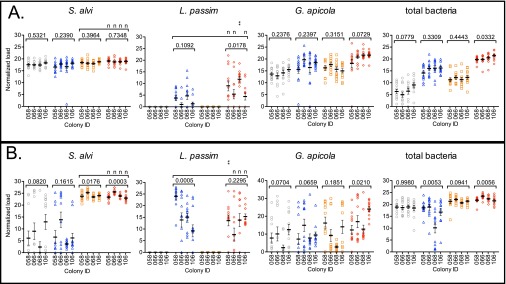
Fig. S1.
Microbial loads (targets noted above each graph) during normal (A) and stressed (B) rearing presented by GH for four parental honey bee colonies. Means ± SEM are shown by the black bars and whiskers. Treatments (n = 12 per colony): sugar water control (gray circles), L. passim (blue triangles), S. alvi (orange squares), and S. alvi + L. passim (red diamonds). P values above brackets are from planned comparisons using nonparametric one-way ANOVA with Kruskal–Wallis tests. Multiple comparisons by colony and treatment are from two-way ANOVA with Bonferroni posttests, shown above bracketed treatment groups: nP > 0.05, **P < 0.05.
Under stressed conditions, bees experimentally inoculated with S. alvi developed distinctly larger mean S. alvi loads (Fig. 1B) than were found in bees not experimentally inoculated (S, 24.11 ± 0.22 SEM and S+L, 23.92 ± 0.2 SEM vs. C, 7.62 ± 1.55 SEM and L, 7.57 ± 1.27 SEM; one-way ANOVA, P < 0.001 for both). The GH-associated variation in these experimentally administered S. alvi loads was significant (Fig. S1B; one-way ANOVA; S, P = 0.0176 and S+L, P = 0.0003) and was independent of L. passim challenge (two-way ANOVA, P > 0.05 for all comparisons). Whereas most (72%) stressed bees not experimentally inoculated with S. alvi had low (below the mean, n = 35/96) or no (n = 34/96) S. alvi, the remainder (28%) had levels that resembled those in experimentally inoculated bees (C, n = 15 and L, n = 12). These incidental S. alvi populations were likely acquired at emergence from brood cell caps contaminated with S. alvi. Such acquisition did not vary by GH (one-way ANOVA, P > 0.05 for both C and L).
A second core gut bacterium, G. apicola, was naturally acquired in the hive by normal bees (99% overall; Fig. 1A). Whereas G. apicola loads in bees from the S. alvi-only treatment did not differ from those in control bees, a significant buildup occurred in bees that received L. passim alone or following S. alvi inoculation (one-way ANOVA, P < 0.001; C, 14.09 ± 0.482 SEM; L, 17.54 ± 0.697 SEM; S, 16.16 ± 0.477 SEM; and S+L, 20.56 ± 0.558). These G. apicola loads did not vary by GH (Fig. S1A, one-way ANOVA, P > 0.05 for all). G. apicola also occurred in stressed bees, but at a lower rate compared with normal bees (Fig. 1B; 80% overall, Fisher’s exact test P < 0.0001). Stressed bees treated with S. alvi + L. passim developed the largest G. apicola loads (S+L, 16.53 ± 1.52 SEM vs. C, 8.20 ± 1.52 SEM; L, 9.85 ± 1.40 SEM; and S, 10.57 ± 1.65 SEM; one-way ANOVA, P < 0.0001), although this was highly influenced by GH (Fig. S1B; one-way ANOVA, P = 0.0210).
Assessment of total bacterial numbers using universal bacterial primers showed that each microbial treatment quantitatively altered the honey bee microbiota in bees reared normally (Fig. 1A; C, 6.72 ± 0.563 SEM; L, 15.61 ± 0.438 SEM; S, 11.76 ± 0.392 SEM; and S+L, 20.37 ± 0.261 SEM; one-way ANOVA, P < 0.01 to < 0.001). Highest total bacterial loads occurred in S. alvi + L. passim-treated bees, which corroborated results observed in this treatment for S. alvi loads and G. apicola loads. In contrast, total bacterial microbiota loads from stressed bees (Fig. 1B) showed variation that corresponded to the presence/absence of S. alvi inoculation (S, 21.33 ± 0.263 SEM and S+L, 22.26 ± 0.226 SEM vs. C, 18.71 ± 0.399 SEM and L, 16.08 ± 0.888 SEM; one-way ANOVA, P < 0.001 for all comparisons). In addition, GH influenced the total bacterial loads in normal bees treated with S. alvi + L. passim (Fig. S1A; one-way ANOVA, P = 0.0332) and in stressed bees treated with L. passim only and S. alvi + L. passim (Fig. S1B; one-way ANOVA, P = 0.0053 and P = 0.0056).
Impacts on L. passim.
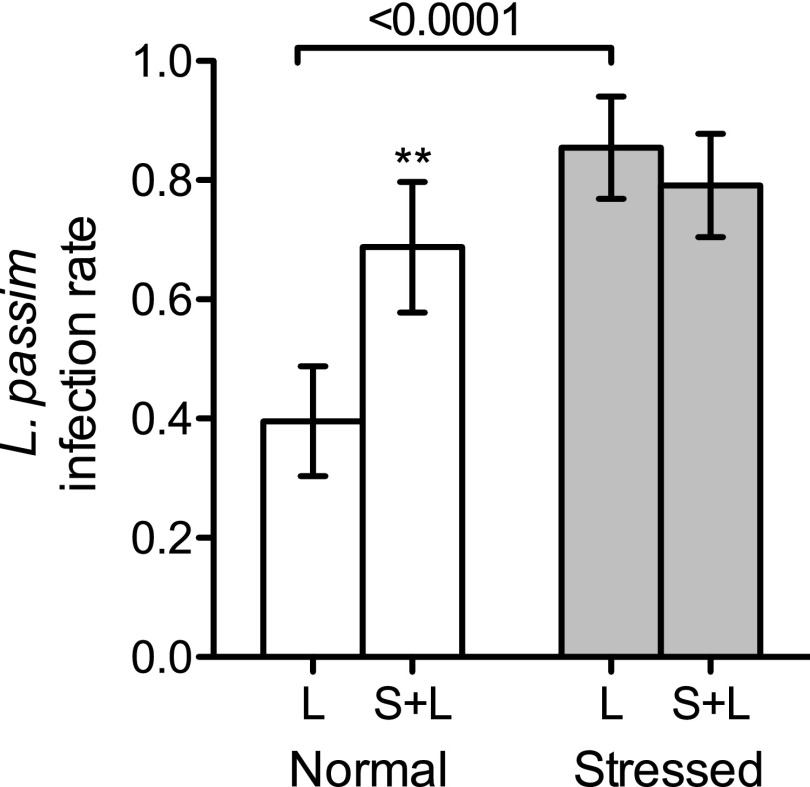
Only bees experimentally inoculated with L. passim had detectable levels of this parasite, whether reared normally (Fig. 1A) or under stress (Fig. 1B). Stressed bees were more susceptible to L. passim infection than bees reared under normal conditions (Fig. 2), with overall infection rates of 82.3% vs. 54.2% (n = 96 each; Fisher’s exact test of independence, P < 0.0001). Infection rates were lower in L. passim-only-treated bees reared under normal vs. stressed conditions (116% lower; Fisher’s exact test, P < 0.0001), whereas the rate difference in S. alvi- pretreated bees was not significant. Unexpectedly, normally reared bees pretreated with S. alvi had higher L. passim loads (Fig. 1A; 185% higher; Mann–Whitney U test, P = 0.0002) and infection rates (Fig. 2; 74% higher; Fisher’s exact test, P = 0.0074) in a GH-dependent manner (Fig. S1A; one-way ANOVA, P = 0.0178).
Fig. 2.
Box plots of L. passim infection rates from bees with normal and stressed rearing. Bar height indicates the mean (n = 48) with SEM (error bars). **Significant difference between L. passim-challenged bees with S. alvi (S+L) vs. without (L); P = 0.0074, Fisher’s exact test of independence.
Pretreatment with S. alvi did not affect susceptibility to L. passim in stressed bees (P > 0.05 for loads, rates, and GH). However, bees from one colony (058) had a significant 44% reduction in L. passim loads when they were pretreated with S. alvi (Fig. S1B; two-way ANOVA, P < 0.01), supporting a GH-dependent dynamic with S. alvi during stress. In addition, L. passim loads varied by GH when bees were not pretreated with S. alvi (one-way ANOVA, P = 0.0005), whereas S. alvi pretreatment abrogated this association (P > 0.05).
Host Gene Expression Responses and Correlations with Microbial Loads.
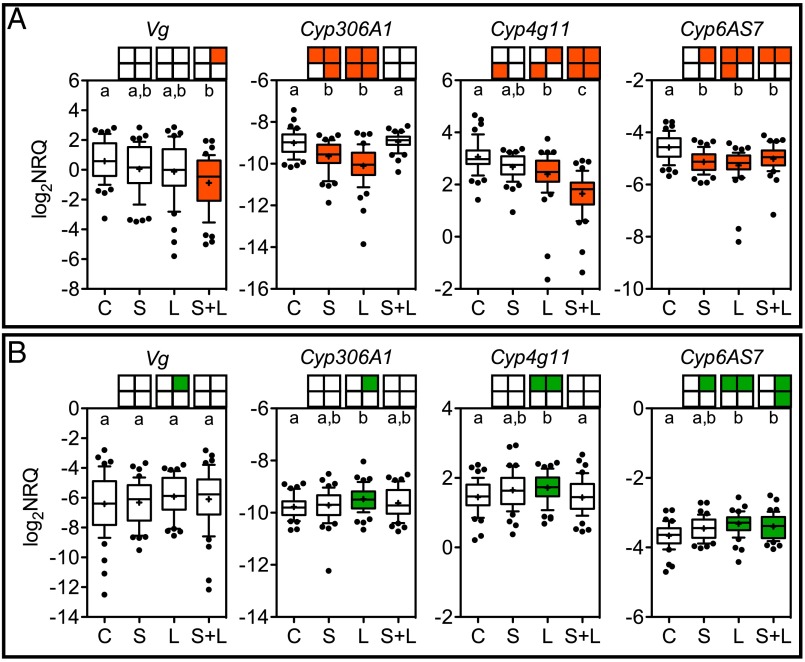
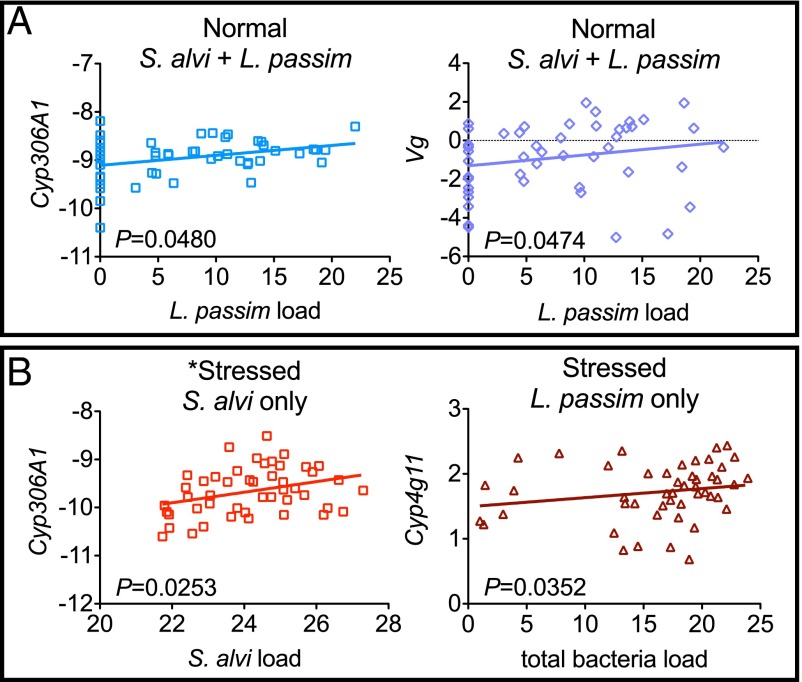
Treatment impacts on host physiology were measured by quantifying the expression of development mediators (Vg and Cyp306A1) and detoxification mediators (Cyp4g11 and Cyp6AS7). Suppressed Vg expression occurred in bees treated with S. alvi + L. passim under normal conditions (Fig. 3A). Although these bees showed suppressed Vg expression overall, individuals that had higher L. passim loads had higher Vg expression (P = 0.0474, r = 0.2877) and had higher Cyp306A1 expression (P = 0.0480, r = 0.2870) (Fig. 4A; Spearman correlation tests). In contrast, stressed bees had no significant treatment-dependent changes in Vg expression overall, but GH-dependent induction was detected in the L. passim-only treatment (Fig. 3B). There were no significant correlations between Vg and microbial loads in stressed bees.
Fig. 3.
Gene expression in bees (n = 48 each) reared under normal (A) or stressed (B) conditions. Boxes above each plot summarize GH data by quadrant: upper left, colony 58; upper right, colony 66; lower left, colony 68; and lower right, colony 106. Significant changes compared with control are shaded: green, induced and red, suppressed. Groups that do not share a letter are significantly different (P < 0.05; Kruskal–Wallis with Dunn’s multiple comparison tests). Data are presented after reference gene normalization and primer efficiency correction. Whiskers span the 10th–90th percentile. Outliers are shown as dots. + indicates expression mean. Treatments: S. alvi (S), L. passim (L), both S. alvi and L. passim (S+L), and sugar water control (C).
Fig. 4.
Significant correlations between microbial loads and host gene expression during normal (A) and stressed (B) rearing. Treatment groups are given above each graph. Best-fit lines are plotted and P values are from two-tailed Spearman nonparametric correlation tests. Data presented are normalized to internal references. One outlier was removed from this dataset (*); removal made the correlation less significant (shown).
Overall, each CYP450 gene was significantly affected by microbial treatments, showing suppression under normal conditions (Fig. 3A) and induction under stressful conditions (Fig. 3B). Bees treated with L. passim only and reared in normal conditions suppressed all CYP450 genes, whereas those reared in stressed conditions induced all CYP450 genes, detectable overall and at GH. Individual expression levels of Cyp4g11 from bees treated with L. passim and reared in stressed conditions were positively correlated with total bacteria load (Fig. 4B; P = 0.0352, r = 0.3047).
Bees treated with S. alvi alone suppressed Cyp6AS7 and Cyp306A1 expression under normal conditions. There was no overall change in CYP450 expression in stressed bees treated with S. alvi, but GH-dependent induction of Cyp6AS7 was detectable (Fig. 3B) and larger S. alvi loads were positively correlated with Cyp306A1 expression (Fig. 4B; P = 0.0253, r = 0.3262). Combined S. alvi + L. passim treatment reduced Cyp6AS7 and Cyp4g11 expression under normal conditions and induced Cyp6AS7 when stressed.
Discussion
Dysbiosis from S. alvi Supplementation and from Stress Increases Susceptibility to Parasitism.
Given that S. alvi is a core microbiota species in both honey bees and bumble bees (29, 35, 58) and that trypanosomatids are common bee parasites (e.g. refs. 28, 38–43, and 59–61), this study provides important insight into the functional roles of prominent microbes in key pollinator species. Experimental supplementation with S. alvi before challenge with L. passim led to measurable perturbation, or dysbiosis, of the core microbiota (i.e., S. alvi, G. apicola, and total bacteria) and, contrary to our hypothesis, increased susceptibility to parasitism by L. passim despite normal rearing conditions within colonies. Presumably, the microbiota acquired naturally in colonies without prior application of S. alvi result in a community that more effectively limits this parasite. These findings offer experimental support linking parasite susceptibility to dysbiosis of the core microbiota, a proposed “biomarker” of colony collapse disorder (CCD) (39). Metagenomic analysis of bee colonies diagnosed with CCD, an enigmatic rapid decline of honey bee colonies, showed “a strong and consistent pattern” of increased Gammaproteobacteria (i.e., G. apicola and/or F. perrara), Betaproteobacteria (i.e., S. alvi), and Firmicutes (i.e., Lactobacillus spp.) along with a decreased relative abundance of Alphaproteobacteria and Actinomycetes (i.e., Bifidobacteria spp.) compared with healthy bee colonies (39). This dysbiosis in CCD colonies was also associated with an overall increase in parasite and pathogen loads, including increased trypanosomatid abundance. Preliminary analysis of CCD bee colonies (28) noted a similar but nonsignificant dysbiosis with a “trend toward increased abundance” of G. apicola (i.e., “Gamma-1”).
All microbial treatments we administered to bees reared normally in hives were associated with a disproportionally large buildup of bacteria. Microbiota buildup in response to S. alvi might indicate that this species facilitates colonization by other microbiota species, as proposed elsewhere (24, 62). Microbiota buildup in response to L. passim may be a protective measure against the parasite, because competitive exclusion of pathogens is a repeatedly identified role of animal microbiota communities (3, 6). Alternatively, because L. passim was more abundant in bees with greatest total bacterial biomass, this may be indicative that L. passim benefits from the gut microbiota in some way, perhaps via provisioned nutrients.
Stress was another factor, in addition to dysbiosis, that affected host susceptibility to parasitism. Stressed bees reared with poor nutrition (no protein), asocial context, and lack of exposure to microbiota from the hive were much more susceptible to L. passim (infection rate and load). Young bees normally consume both protein from pollen and carbohydrates from honey during their early adult development. In addition, bees receive social cues (pheromones) in the hive from the queen, other adults, and larvae that affect worker behavior and physiology (63). Unlike normal conditions, the high susceptibility of stressed bees to L. passim was not altered when their guts were precolonized with S. alvi. The comparatively depauperate microbiota associated with higher L. passim susceptibility in stressed bees suggests that specific and stable proportions of core gut communities are likely important to successfully control this parasite.
These data provide experimental support for significant host genotype-dependent interactions between bacterial microbiota and eukaryotic parasites. Such dynamics are not unprecedented; experimental manipulation of microbiota has been shown to affect eukaryotic gut parasite population sizes in other insect models, such as populations of the apicomplexan parasite Plasmodium in mosquito hosts (64). In bumble bees, correlations between the size and composition of core gut microbiota and trypanosomatid populations have been observed (36, 37), as well as host genotype-dependent variation to trypanosomatid susceptibility (65, 66). Our present results unite the relevance of host microbiota composition, genetic background, and stress as important mediators of parasite susceptibility and insect health.
Gut Microbiome Changes Affect Host Developmental Pathways.
Our analyses suggest an effect on host development from both S. alvi pretreatment and L. passim infection. Worker bees take on various social roles as they age, first within the hive then culminating in a final role as forager for food and other resources outside the hive. Transitioning to these different roles is associated in part with waning levels of vitellogenin, a central hormone of a developmental signaling network in bees (14, 16–18, 20). Cyp306A1 encodes an essential regulator of another hormone in this network, ecdysteroid (55), which can control the production of vitellogenin. Although we found that Vg was suppressed overall in normally reared bees treated with both S. alvi and L. passim, individual analyses showed that bees with larger populations of L. passim had stronger Vg expression. The same positive correlation was observed for Cyp306A1 expression in these bees. Thus, dysbiosis made bees more susceptible to L. passim, and host developmental pathways were altered according to parasite load.
Elevated Vg expression corresponds with a slower onset to the forager stage, preferential foraging for pollen vs. nectar, and longer lifespan (14, 16–18, 20). As such, elevated Vg expression may increase L. passim transmission in the colony by extending potential contact time between infected and naïve individuals. In contrast, the reduced Vg expression we observed in normally reared worker bees inoculated with both S. alvi and L. passim corresponds with precocious development, preferential foraging for nectar, and shortened lifespan. Suppression of vitellogenin in bees parasitized by Varroa destructor mites (ectoparasites) and Nosema ceranae microsporidians (endoparasites) has been reported (19, 67). This phenomenon may be a defensive response to parasitism in social insects causing parasitized individuals to leave the colony sooner and die sooner, predictively mitigating the spread of parasites to other colony members. Our data support this theory and provide the additional observation that L. passim may reverse this response in highly infected individuals, enabling larger parasite population sizes and extended transmission potential to new hosts. We note that in the previously mentioned studies linking parasitism to reduced vitellogenin production bees had access to dietary protein. The absence of protein from the diet of stressed bees in our study likely compromised vitellogenin production and may explain the overall invariance in Vg from this treatment group.
S. alvi supplementation and its subsequent population size were positively correlated with Cyp306A1 expression under stressed conditions. This suggests a potentially important role for S. alvi in developmental pathways when bees experience poor nutrition or have depauperate microbiota.
Microbiome Composition Affects Host Detoxification Responses.
We determined that both normal and stressed bees treated with S. alvi and L. passim showed altered CYP450 gene expression. CYP450s comprise a large superfamily of metabolic enzymes with demonstrated or predicted roles in developmental hormone synthesis (i.e., Cyp306A1 discussed in the prior section) and xenobiotic detoxification (i.e., Cyp4g11 and Cyp6AS7) (54, 56). Because these genes can be highly expressed and locally specific in the adult insect gut (68), the proteins they encode are potentially significant players connecting gut microbial communities with the host. We found that Cyp4g11 expression was altered only in bees treated with L. passim whereas Cyp6AS7 was altered in bees from all microbial treatment groups. Stressed conditions dramatically reversed the regulation patterns of these genes.
As part of the CYP3 clade, Cyp6AS7 is among the most diverse group of CYP450 genes in honey bees (54, 56), but no other experimental data are available for comparison. Cyp4g11 is part of the CYP4 clan, which has the fewest genes in the honey bee CYP450 superfamily (54, 56). It is not known to be responsive to insecticides (56, 69) but is responsive to parasitization by V. destructor (70, 71) and is suppressed in bee guts from CCD colonies (72). It also has potential roles in sensory perception during foraging (73). Our data linking altered Cyp4g11 expression to trypanosomatid infection provide a plausible explanation for modified foraging behavior observed in trypanosomatid-infected bumble bees (74).
Conclusion.
In general, bees are capable of regulating a commonly encountered trypanosomatid gut parasite when provided with adequate nutrition and their normal bacterial microbiota, typically acquired in the hive. Bees dramatically lose their ability to control this parasite, however, when in a state of dysbiosis or when nutritionally and socially stressed. In our experiments, dysbiosis resulted from an early skew in the microbiota composition of young adult bees; application of antibiotics to colonies might also be expected to result in dysbiosis. These findings provide experimental support of past observations linking dysbiosis and high parasite loads in bee colony collapses. Using a core microbiota species from honey bees, S. alvi, we show that probiotic therapy is not always beneficial and has complicated consequences for parasite susceptibility, microbiota homeostasis, and host developmental and detoxification response pathways. This work does not refute the potentially beneficial nature of this core gut symbiont, but it does show that the succession and ratio of individual microbiota species is critical to the future structure and functioning of the microbiota as a whole. Because multiple strains of S. alvi and L. passim occur, it is possible that other strains will have different effects.
Methods
Adult worker bees were removed from four healthy (capped and uncapped brood with normal brood pattern, bee bread and honey stores, queen-right) colonies of A. mellifera ligustica US domestic hybrids upon emergence (n = 960) then separated into four treatment groups (n = 480 each): (i) 5 μL of 1:1 sugar water, (ii) S. alvi only (250,000 cfu, strain wkB2T), (iii) L. passim only (10,000 promastigotes, strain BRL), or (iv) S. alvi + L. passim, and color-coded accordingly. Bees were inoculated with sugar water (treatment i) or S. alvi (treatments ii and iv) within 8 h of emergence and with L. passim (treatments iii and iv) 24 h later. Half of the bees from each colony in each treatment group were returned to their parental colonies 12 h later (“normal rearing”) and the remaining half were isolated according to treatment group within cages held at normal colony temperature (32 °C) within an incubator and maintained on sugar water/no protein diet (“stressed rearing”). Total RNA was extracted individually at 6 d postemergence and 10 μg was converted to first-strand cDNA for use as template to quantify microbes and honey bee genes with primers (Table S1) via quantitative PCR (qPCR) (24, 25, 75, 76). All qPCR data were normalized to empirically determined stable reference genes (Fig. S2) (77–80). Further details are available in SI Methods.
Table S1.
Primer details
| Target (gene name and accession no.) | Sequence (5′ to 3′) | Annealing temperature, °C | Standard curve* | Amplicon size and dissociation | Sources and notes |
| Actin related protein 1 | F: CCAAAGACCCAAGCTCCCTA | 60 | 96, 0.985, | 73 bp | F+R: this study |
| (Arp1, NM_001185146.1) | R: TGGCTTATTGGTTTATGTTTTTCGT | −3.421, 46.645 | 72.0 °C | Reverse spans intron | |
| Ribosomal protein S5a | F: AATTATTTGGTCGCTGGAATTG | 60 | 97, 0.980, | 115 bp | F+R (75) |
| (Rps5a, XM_006570237) | R: TAACGTCCAGCAGAATGTGGTA | −3.387, 46.984 | 75.5 °C | Does not span intron | |
| Vitellogenin | F: TCGACAACTGCGATCAAAGGA | 60 | 99, 0.999, | 164 bp | F+R: this study |
| (Vg, NM_001011578) | R: TGGTCACCGACGATTGGATG | −3.356, 43,033 | 80.5 °C | Amplicon spans intron | |
| Cytochrome P450 4G11 | F: GGCTTGGCGATGGTCTTCTT | 60 | 93, 0.992, | 135 | F+R: this study |
| (Cyp4g11, XM_006559340.1 to XM_006559341.1) | R: AATATGTGGCTGTCGGTGCA | −3.494, 41.967 | 80.5–81.0 °C | Amplicon spans intron; isoforms X1-X2 | |
| Cytochrome P450 306A1 | F: ACAGCATCGTGTTCGGGAAA | 60 | 115, 0.997, | 165 bp | F+R: this study |
| (Cyp306A1, XM_006557824.1 to XM_006557828.1) | R: CGATCGACCGTATCACTCGG | −3.001, 40.707 | 84.0 °C | Amplicon spans intron; isoforms X1-X5 | |
| Cytochrome P450 6AS7 | F: GCCAACCGCGGAATTCCTAT | 60 | 87.1, 0.996, | 120 bp | F+R: this study |
| (Cyp6AS7, XM_006565001 to XM_006565002) | R: GCCAGATGTGAATACTGGCGA | −3.675, 43.821 | 79.0–79.5 °C | Amplicon spans intron; isoforms X1-X2 | |
| Snodgrassella alvi | F: CTTAGAGATAGGAGAGTG | 55 | 99, 0.997, | 128 bp | F+R (24) |
| (SSU, NR_122055) | R: TAATGATGGCAACTAATGACAA | −3.349, 46.367 | 84.5–85.0 °C | ||
| Gilliamella apicola | F: GTATCTAATAGGTGCATCAATT | 55 | 98, 0.998, | 210 bp | F+R (24) |
| (SSU, NR_121727) | R: TCCTCTACAATACTCTAGTT | −3.361, 42.112 | 84.5 °C | ||
| Universal bacteria | F: AGAGTTTGATCCTGGCTCAG | 60 | 108, 0.999, | 328 bp | F+R (25) |
| (SSU, multiple) | R: CTGCTGCCTCCCGTAGGAGT | −3.128, 39.896 | 86.0–87.5 °C | Confirmed CP003325, JQ581978, | |
| HM046576, and NR_121735 | |||||
| Universal trypanosomatid | F: GTGCAGTTCCGGAGTCTTGT | 60 | 118, 0.962, | 103 bp | F+R (76) |
| (LSU, AHIJ01002555) | R: CTGAGCTCGCCTTAGGACAC | −2.933, 41.919 | 83.5–84.0 °C |
In order: efficiency (percent), R2, slope, and y intercept.
Fig. S2.
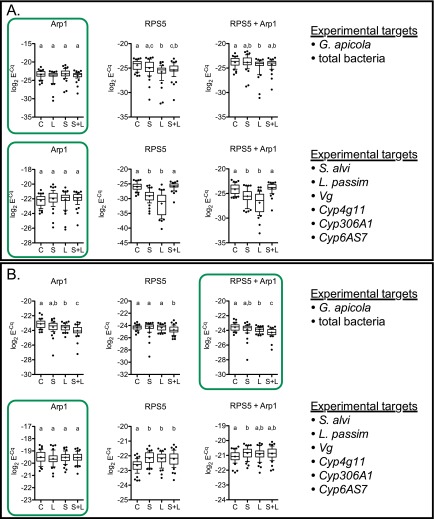
Reference gene data from normal (A) and stressed (B) honey bees. Boxed results in green indicated reference data statistically determined as optimal for normalization of experimental targets (listed at right). Datasets that share a letter above plotted data indicate no significant difference between those treatments. Variability was typically greater in RPS5 expression compared with Arp1. In one case, where both Arp1 and RPS5 expression varied by treatment group, the average of both reference genes was used according to standard protocol for qPCR analysis. Statistics: one-way ANOVA (Kruskal–Wallis, nonparametric) with Dunn’s multiple comparison tests. Whiskers indicate 10th–90th percentile; dots indicate outliers.
SI Methods
Ethics Statement.
Nonendemic parasite strains were not released into the field for this study. The strain of L. passim used (ATCC PRA-422, strain BRL) was originally and recently isolated from the same location where we conducted the hive experiment (42). S. alvi strain wkB2T is a prevalent A. mellifera-specific strain and was originally isolated in 2011 from A. mellifera in West Haven, CT (56), which is 416 km from where it was experimentally introduced into field colonies in Beltsville, MD for this study. Bees were ice-sedated during collection before deep-freezing and tissue analysis. All experimentally infected animal tissues and materials contaminated with parasite cultures were autoclave-sterilized before disposal.
Microbe Cell Cultures.
A lawn of S. alvi strain wkB2T (56) was grown from a single clone picked from a fresh streak on tryptic soy agar with 5% sheep’s blood (Hardy Diagnostics) in a 37 °C + 5% CO2 incubator for 48 h then harvested using 1× PBS. Bacteria cells were centrifuged at 425 × g then washed and pelleted twice in 1 mL of 1× PBS with 425 × g centrifugation. Cells were then suspended in 3 mL of 1× PBS with 1 mL used for OD600 reading and the remainder kept for inoculum. Based on a standard curve generated for these S. alvi cultures, the stock solution contained ∼1 × 109 cfu/mL. A 1:20 dilution of this stock was made using 20% sucrose solution (1:1 vol/vol) in 1× PBS at the time of oral inoculation for a final concentration of ∼50,000 cfu/μL.
Promastigotes of L. passim (strain BRL, American Type Culture Collection PRA-422) were cultured in supplemented DS2 media as described previously (80), pelleted for 10 min at 425 × g, and resuspended in 1× PBS. A 1:100 dilution was used to obtain a cell count on a Neubauer hemocytometer at 400× magnification. Only active promastigotes were counted. A 2,000 cells per μL inoculum was prepared using 20% sucrose solution (1:1 vol/vol) in 1× PBS.
Honey Bees and Treatment Groups.
Frames with many newly emerging workers (NEWs) were collected from four apparently healthy (capped and uncapped brood with normal brood pattern, bee bread and honey stores, queen-right) colonies of A. mellifera ligustica US domestic hybrid within one apiary (“water tower”) maintained at the USDA Beltsville Area Research Center during May of 2013. Three colonies had been started earlier in the year (April) from packages obtained from Georgia (WT058, WT066, and WT068) and one colony was locally overwintered from the previous year (WT106). Frames were cleared of all emerged bees before collection of NEWs. NEWs (n = 960 total, 240 from each colony) were removed as they emerged from brood cells and were temporarily kept in sterile cups with sterile 1:1 sugar water in an incubator at 32 °C + 50% relative humidity until the needed number was reached (<8 h). All NEWs from each colony were then separated according to two experimental groups, “normal rearing” and “stressed rearing” (n = 120 from each colony), and equally distributed to four treatment groups (n = 30 from each colony per experiment): (i) sugar water control, (ii) S. alvi only (strain wkB2T), (iii) L. passim only (strain BRL, ATCC PRA-422), or (iv) S. alvi + L. passim. All bees were color-coded on their thorax according to treatment group and parent colony using paint pens (Mitsubishi Pencil Co.).
On the day of emergence, bees were fed either 5 μL of 1:1 sugar water (group i) or 5 μL of an S. alvi suspension (250,000 cfu; groups ii and iii) and reared in the laboratory incubator for 24 h. Bees from groups iii and iv were then individually fed a 5-μL L. passim suspension (10,000 promastigotes) and all groups were reared in the laboratory for 12 h more. At 36 h postemergence, bees for the normal rearing experiment were released back into their source hives, whereas bees for the stressed rearing experiment were kept in the laboratory incubator and fed with sterile 1:1 sugar water. All bees were collected at 144 h (6 d) postemergence and stored at −80 °C before RNA extraction. Based on preliminary studies in our laboratory, we identified 5 d postinoculation (6 d after bee emergence) as an optimal time point where honey bee trypanosomatid populations, including L. passim, are near or at plateau levels following experimental inoculation. This also encompasses the time at which core gut microbiota climax community sizes occur, under naturally acquired (24) and experimental (34) conditions.
RNA Purification and cDNA Synthesis.
From each experiment, frozen bee abdomens from 48 individuals (n = 12 per colony) of each treatment group (n = 192 per experiment; n = 384 total) were processed individually using 1-mm zirconium beads in 500 μL TRIzol reagent (Life Technologies) with a FastPrep FP120 cell disrupter (Qbiogene, Inc.) followed by the addition of another 500 μL of TRIzol according to the manufacturer’s directions for total RNA isolation. Purified RNA pellets were resuspended in diethylpyrocarbonate-treated water then quantified at 260 nm using a NanoDrop ND8000 UV-visible spectrophotometer (Thermo Fisher Scientific). Each RNA sample was individually reverse-transcribed to cDNA from 1.5 μg of total RNA using one master mix for each experimental group to avoid batch-to-batch variation (n = 12 per colony; n = 192 per experiment) in a two step process: (i) DNA degradation (1 U RNase-free DNase I; Life Technologies) at 37 °C for 30 min with 5 mM EDTA followed by 75 °C for 10 min to inactivate the enzyme and denature secondary RNA structure and (ii) first-strand cDNA synthesis using 100 U SuperScript II (Invitrogen) primed with 100 ng random heptamers and 50 ng poly(dT)12–18 in the presence of 20 U RNase OUT (Invitrogen) at 25 °C for 10 min, 42 °C for 50 min, and 70 °C for 15 min. Each sample (∼1,500 ng cDNA total) was diluted to a final concentration of 10 ng/μL with nuclease-free water and stored at −20 °C until use. Intron-spanning primers have confirmed the effectiveness of this procedure in removing residual DNA contamination (80), and a newly developed intron-spanning reference primer pair (Arp1) detected no gDNA contamination. All sample preparation was conducted by the same individual to minimize handler variation.
qPCR Analysis.
Primers developed for this study (Table S1) followed MIQE guidelines (minimum information for publication of quantitative real-time PCR experiments) (77). These included primer pairs targeting honey bee actin-related protein 1 (Arp1), vitellogenin (Vg), and three cytochrome P450 genes (Cyp4g11, Cyp306A1, and Cyp6AS7). Previously published qPCR primers given in Table S1 (24, 25, 75, 76) were used to determine microbial loads based on 16S rRNA (G. apicola, S. alvi, and universal bacteria) and 28S rRNA (L. passim). Arp1 largely proved to be a stable qPCR reference (Fig. S2) except in the G. apicola and universal bacteria experiments from incubator-reared bees, where some variation occurred across treatment groups. For these data, the average of Arp1 and a second previously developed reference primer pair targeting honey bee ribosomal protein subunit 5 (RPS5) (80) were used for normalization.
qPCR analyses were performed in white 384-well Microseal PCR plates using a CFX384 real-time system (Bio-Rad; G. apicola and total bacteria) or in clear 96-well plates using a CFX96 real time system (Bio-Rad; S. alvi and L. passim), with samples distributed in a randomized block pattern to avoid plate-effect error. Plate-effect error was further minimized using interplate calibration of baseline threshold across plates. Optically clear Microseal ‘B’ plate sealers (Bio-Rad) were used for both platforms. The qPCR protocol and analysis for this study was based on recommended optimal guidelines (77–79). A single batch of master mix was prepared for each primer pair within an experiment from which all samples were run consecutively in a single day on the same machine to avoid batch variation error. Each cDNA sample (n = 192 per experiment) was run in triplicate. No template control (NTC) reactions (n = 6) to monitor for contamination and positive controls using serial dilutions (1.00E+01–1.00E+08) of sequence-verified recombinant clones were run from each master mix to monitor amplification efficiency and to generate standard curves and primer efficiencies for each target, given in Table S1. All reactions contained equal amounts (10 ng) of template cDNA (or nuclease-free water for NTC), 150 nM each of a forward and a reverse primer, and 1× SsoFast EvaGreen supermix (Bio-Rad). Thermal cycling conditions were as follows: initial denaturing step of 97 °C for 2 min, 50 amplification cycles of 95 °C for 2 s and 55 °C to 60 °C annealing for 5 s, and melt curve analysis from 65 °C to 95 °C at 0.5 °C/5 s increments to confirm expected dissociation curves. All qPCR experiments were performed by the same individual to minimize handler variation.
Normalized relative quantity (NRQ) of cDNA (target) was determined using the mean Cq normalized to the reference template average (Ref is Arp1 or Arp1 + RPS5/2) adjusted for primer efficiency:
where and .
Data were log2-transformed back to a Cq scale (where 1 Cq = twofold change in target template abundance) and gene expression was plotted as log2NRQ. For graphical clarity, microbial loads were linearly transformed to a positive scale, plotted as “normalized load.” Data across treatment groups were typically non-Gaussian-distributed; thus, variation in load was analyzed using one-way ANOVA with nonparametric Kruskal–Wallis test and Dunn’s multiple comparison tests at α = 0.05. Planned paired comparisons were made on microbial loads in single vs. mixed species treatment groups with two-tailed Mann–Whitney U tests (α = 0.05). Data means were analyzed according to treatment and colony of origin variables with two-way ANOVA and Bonferroni posttests. Spearman correlations of two measured variables (microbial load and gene expression) were performed using nonparametric two-tailed tests with 95% confidence intervals. Statistics were performed with Prism 5 version 5.0b for Mac OSX (GraphPad Software, Inc.).
Acknowledgments
We thank Kim Hammond, Waldan Kwong, Eli Powell, and Allen Smith for providing S. alvi cultures, culturing advice, and equipment and the anonymous reviewers who improved the manuscript. This work was supported by National Science Foundation Grants 1046153 and 1415604 (to N.A.M. and J.D.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606631113/-/DCSupplemental.
References
- 1.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 2.Eleftherianos I, Atri J, Accetta J, Castillo JC. Endosymbiotic bacteria in insects: Guardians of the immune system? Front Physiol. 2013;4:46. doi: 10.3389/fphys.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37(5):699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6(2):a016113–a016113. doi: 10.1101/cshperspect.a016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas AE. Multiorganismal insects: Diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huttenhower C, et al. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sartor RB. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc Natl Acad Sci USA. 2008;105(43):16413–16414. doi: 10.1073/pnas.0809363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamdi C, et al. Gut microbiome dysbiosis and honeybee health. J Appl Entomol. 2011;135:524–533. [Google Scholar]
- 10.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109(27):11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent CF, Issa A, Bunting AC, Zayed A. Adaptive evolution of a key gene affecting queen and worker traits in the honey bee, Apis mellifera. Mol Ecol. 2011;20(24):5226–5235. doi: 10.1111/j.1365-294X.2011.05299.x. [DOI] [PubMed] [Google Scholar]
- 13.Harpur BA, et al. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci USA. 2014;111(7):2614–2619. doi: 10.1073/pnas.1315506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto LZ, Bitondi MM, Simões ZL. Inhibition of vitellogenin synthesis in Apis mellifera workers by a juvenile hormone analogue, pyriproxyfen. J Insect Physiol. 2000;46(2):153–160. doi: 10.1016/s0022-1910(99)00111-0. [DOI] [PubMed] [Google Scholar]
- 15.Amdam GV, Norberg K, Hagen A, Omholt SW. Social exploitation of vitellogenin. Proc Natl Acad Sci USA. 2003;100(4):1799–1802. doi: 10.1073/pnas.0333979100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amdam GV, et al. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp Gerontol. 2004;39(5):767–773. doi: 10.1016/j.exger.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5(3):e62. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmela H, Amdam GV, Freitak D. Transfer of immunity from mother to offspring is mediated via egg-yolk protein vitellogenin. PLoS Pathog. 2015;11(7):e1005015. doi: 10.1371/journal.ppat.1005015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amdam GV, Hartfelder K, Norberg K, Hagen A, Omholt SW. Altered physiology in worker honey bees (Hymenoptera: Apidae) infested with the mite Varroa destructor (Acari: Varroidae): A factor in colony loss during overwintering? J Econ Entomol. 2004;97(3):741–747. doi: 10.1093/jee/97.3.741. [DOI] [PubMed] [Google Scholar]
- 20.Münch D, Amdam GV, Wolschin F. Ageing in a eusocial insect: Molecular and physiological characteristics of life span plasticity in the honey bee. Funct Ecol. 2008;22(3):407–421. doi: 10.1111/j.1365-2435.2008.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dainat B, Evans JD, Chen YP, Gauthier L, Neumann P. Predictive markers of honey bee colony collapse. PLoS One. 2012;7(2):e32151. doi: 10.1371/journal.pone.0032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilliam M. Microbial sterility of intestinal content of immature honey bee, Apis mellifera. Ann Entomol Soc Am. 1971;64:315–316. [Google Scholar]
- 23.Gilliam M. Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol Lett. 1997;155:1–10. [Google Scholar]
- 24.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78(8):2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol. 2014;80(23):7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeyaprakash A, Hoy MA, Allsopp MH. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J Invertebr Pathol. 2003;84(2):96–103. doi: 10.1016/j.jip.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Mohr KI, Tebbe CC. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol. 2006;8(2):258–272. doi: 10.1111/j.1462-2920.2005.00893.x. [DOI] [PubMed] [Google Scholar]
- 28.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 29.Martinson VG, et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20(3):619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 30.Vásquez A, et al. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS One. 2012;7(3):e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killer J, Dubná S, Sedláček I, Švec P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int J Syst Evol Microbiol. 2014;64(Pt 1):152–157. doi: 10.1099/ijs.0.053033-0. [DOI] [PubMed] [Google Scholar]
- 32.Corby-Harris V, et al. Parasaccharibacter apium, gen. nov., sp. nov., improves honey bee (Hymenoptera: Apidae) resistance to Nosema. J Econ Entomol. 2016;109:537–543. doi: 10.1093/jee/tow012. [DOI] [PubMed] [Google Scholar]
- 33.Corby-Harris V, et al. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl Environ Microbiol. 2014;80(24):7460–7472. doi: 10.1128/AEM.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engel P, Bartlett KD, Moran NA. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. MBio. 2015;6(3):e00193–e15. doi: 10.1128/mBio.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci USA. 2014;111(31):11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108(48):19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cariveau DP, Elijah Powell J, Koch H, Winfree R, Moran NA. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus) ISME J. 2014;8(12):2369–2379. doi: 10.1038/ismej.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Runckel C, et al. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One. 2011;6(6):e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornman RS, et al. Pathogen webs in collapsing honey bee colonies. PLoS One. 2012;7(8):e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravoet J, et al. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS One. 2013;8(8):e72443. doi: 10.1371/journal.pone.0072443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morimoto T, et al. Molecular detection of protozoan parasites infecting Apis mellifera colonies in Japan. Environ Microbiol Rep. 2013;5(1):74–77. doi: 10.1111/j.1758-2229.2012.00385.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang B, Peng G, Li T, Kadowaki T. Molecular and phylogenetic characterization of honey bee viruses, Nosema microsporidia, protozoan parasites, and parasitic mites in China. Ecol Evol. 2013;3(2):298–311. doi: 10.1002/ece3.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz RS, et al. Characterization of two species of Trypanosomatidae from the honey bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J Eukaryot Microbiol. 2015;62(5):567–583. doi: 10.1111/jeu.12209. [DOI] [PubMed] [Google Scholar]
- 44.Schwarz RS, Huang Q, Evans JD. Hologenome theory and the honey bee pathosphere. Curr Opin Insect Sci. 2015;10:1–7. doi: 10.1016/j.cois.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Evans JD, Schwarz RS. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011;19(12):614–620. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Carreck N, Neumann P. Honey bee colony losses. J Apic Res. 2010;49:1–6. [Google Scholar]
- 47.Lee KV, et al. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie (Celle) 2015;46:292–305. [Google Scholar]
- 48.Guzmán-Novoa E, et al. Varroa destructor is the main culprit for the death and reduced populations of overwintered honey bee (Apis mellifera) colonies in Ontario, Canada. Apidologie (Celle) 2010;41:443–450. [Google Scholar]
- 49.Pirk CWW, Human H, Crewe RM, van Engelsdorp D. A survey of managed honey bee colony losses in the Republic of South Africa–2009 to 2011. J Apic Res. 2014;53:35–42. [Google Scholar]
- 50.Tarpy DR. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc Biol Sci. 2003;270(1510):99–103. doi: 10.1098/rspb.2002.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seeley TD, Tarpy DR. Queen promiscuity lowers disease within honeybee colonies. Proc Biol Sci. 2007;274(1606):67–72. doi: 10.1098/rspb.2006.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desai SD, Currie RW. Genetic diversity within honey bee colonies affects pathogen load and relative virus levels in honey bees, Apis mellifera L. Behav Ecol Sociobiol. 2015;69:1527–1541. [Google Scholar]
- 53.Johnson RM. Honey bee toxicology. Annu Rev Entomol. 2015;60:415–434. doi: 10.1146/annurev-ento-011613-162005. [DOI] [PubMed] [Google Scholar]
- 54.Claudianos C, et al. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15(5):615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa R, et al. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279(34):35942–35949. doi: 10.1074/jbc.M404514200. [DOI] [PubMed] [Google Scholar]
- 56.Berenbaum MR, Johnson RM. Xenobiotic detoxification pathways in honey bees. Curr Opin Insect Sci. 2015;10:51–58. doi: 10.1016/j.cois.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Audisio MC, Sabaté DC, Benítez-Ahrendts MR. Effect of Lactobacillus johnsonii CRL1647 on different parameters of honeybee colonies and bacterial populations of the bee gut. Benef Microbes. 2015;6(5):687–695. doi: 10.3920/BM2014.0155. [DOI] [PubMed] [Google Scholar]
- 58.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63(Pt 6):2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 59.Meeus I, Brown MJF, De Graaf DC, Smagghe G. Effects of invasive parasites on bumble bee declines. Conserv Biol. 2011;25(4):662–671. doi: 10.1111/j.1523-1739.2011.01707.x. [DOI] [PubMed] [Google Scholar]
- 60.Popp M, Erler S, Lattorff HMG. Seasonal variability of prevalence and occurrence of multiple infections shape the population structure of Crithidia bombi, an intestinal parasite of bumblebees (Bombus spp.) MicrobiologyOpen. 2012;1(4):362–372. doi: 10.1002/mbo3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitehorn PR, Tinsley MC, Brown MJF, Darvill B, Goulson D. Genetic diversity, parasite prevalence and immunity in wild bumblebees. Proc Biol Sci. 2011;278(1709):1195–1202. doi: 10.1098/rspb.2010.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moran NA. Genomics of the honey bee microbiome. Curr Opin Insect Sci. 2015;10:22–28. doi: 10.1016/j.cois.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Traynor KS, Le Conte Y, Page RE., Jr Queen and young larval pheromones impact nursing and reproductive physiology of honey bee (Apis mellifera) workers. Behav Ecol Sociobiol. 2014;68(12):2059–2073. doi: 10.1007/s00265-014-1811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gendrin M, et al. Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat Commun. 2015;6:5921. doi: 10.1038/ncomms6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barribeau SM, Sadd BM, du Plessis L, Schmid-Hempel P. Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system. Proc Natl Acad Sci USA. 2014;111(9):3496–3501. doi: 10.1073/pnas.1318628111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baer B, Schmid-Hempel P. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature. 1999;397:151–154. [Google Scholar]
- 67.Alaux C, Crauser D, Pioz M, Saulnier C, Le Conte Y. Parasitic and immune modulation of flight activity in honey bees tracked with optical counters. J Exp Biol. 2014;217(Pt 19):3416–3424. doi: 10.1242/jeb.105783. [DOI] [PubMed] [Google Scholar]
- 68.Chung H, et al. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc Natl Acad Sci USA. 2009;106(14):5731–5736. doi: 10.1073/pnas.0812141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boncristiani H, et al. Direct effect of acaricides on pathogen loads and gene expression levels in honey bees Apis mellifera. J Insect Physiol. 2012;58(5):613–620. doi: 10.1016/j.jinsphys.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 70.Le Conte Y, et al. Social immunity in honeybees (Apis mellifera): Transcriptome analysis of varroa-hygienic behaviour. Insect Mol Biol. 2011;20(3):399–408. doi: 10.1111/j.1365-2583.2011.01074.x. [DOI] [PubMed] [Google Scholar]
- 71.Gregorc A, Evans JD, Scharf M, Ellis JD. Gene expression in honey bee (Apis mellifera) larvae exposed to pesticides and Varroa mites (Varroa destructor) J Insect Physiol. 2012;58(8):1042–1049. doi: 10.1016/j.jinsphys.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 72.Johnson RM, Evans JD, Robinson GE, Berenbaum MR. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera) Proc Natl Acad Sci USA. 2009;106(35):14790–14795. doi: 10.1073/pnas.0906970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao W, Schuler MA, Berenbaum MR. Task-related differential expression of four cytochrome P450 genes in honeybee appendages. Insect Mol Biol. 2015;24(5):582–588. doi: 10.1111/imb.12183. [DOI] [PubMed] [Google Scholar]
- 74.Gegear RJ, Otterstatter MC, Thomson JD. Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proc Biol Sci. 2006;273(1590):1073–1078. doi: 10.1098/rspb.2005.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans JD. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J Invertebr Pathol. 2006;93(2):135–139. doi: 10.1016/j.jip.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 76.Vanengelsdorp D, et al. Colony collapse disorder: A descriptive study. PLoS One. 2009;4(8):e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bustin SA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 78.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 79.Rieu I, Powers SJ. Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell. 2009;21(4):1031–1033. doi: 10.1105/tpc.109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwarz RS, Evans JD. Single and mixed-species trypanosome and microsporidia infections elicit distinct, ephemeral cellular and humoral immune responses in honey bees. Dev Comp Immunol. 2013;40(3–4):300–310. doi: 10.1016/j.dci.2013.03.010. [DOI] [PubMed] [Google Scholar]