Significance
Wnt signaling is an important orchestrator of embryonic development. We provide evidence that Wnt scaffolding protein Dishevelled (DVL) contributes to the dissolution of centrosomal linker, preceding separation of centrosomes. We show that DVL accumulates toward mitosis, and its centrosomal function is controlled by NEK2. Our data demonstrate that DVL is required for the removal of linker proteins from centrosome, an event necessary for the correct formation of a bipolar spindle and cell cycle transition. This surprising function of DVL creates a mechanistic basis for the novel crosstalk between Wnt signaling pathways and the centrosome cycle.
Keywords: Wnt signaling, centrosome, Dishevelled, NEK2, linker proteins
Abstract
Dishevelled (DVL) is a key scaffolding protein and a branching point in Wnt signaling pathways. Here, we present conclusive evidence that DVL regulates the centrosomal cycle. We demonstrate that DVL dishevelled and axin (DIX) domain, but not DIX domain-mediated multimerization, is essential for DVL’s centrosomal localization. DVL accumulates during the cell cycle and associates with NIMA-related kinase 2 (NEK2), which is able to phosphorylate DVL at a multitude of residues, as detected by a set of novel phospho-specific antibodies. This creates interfaces for efficient binding to CDK5 regulatory subunit-associated protein 2 (CDK5RAP2) and centrosomal Nek2-associated protein 1 (C-NAP1), two proteins of the centrosomal linker. Displacement of DVL from the centrosome and its release into the cytoplasm on NEK2 phosphorylation is coupled to the removal of linker proteins, an event necessary for centrosomal separation and proper formation of the mitotic spindle. Lack of DVL prevents NEK2-controlled dissolution of loose centrosomal linker and subsequent centrosomal separation. Increased DVL levels, in contrast, sequester centrosomal NEK2 and mimic monopolar spindle defects induced by a dominant negative version of this kinase. Our study thus uncovers molecular crosstalk between centrosome and Wnt signaling.
Signaling pathways activated by Wnt ligands are evolutionary conserved drivers of proliferation, differentiation, and morphogenesis (1). Their deregulation leads to numerous developmental abnormalities and contributes to the pathogenesis of several cancers (2).
Dishevelled (DVL), in mammals with three isoforms DVL1, DVL2, and DVL3, is a crucial scaffolding component present at the crossroads of both Wnt/β-catenin and noncanonical Wnt/planar cell polarity pathways. DVL acts as a signal integrator, which determines subsequent downstream signaling. Its domain structure provides sites for binding of regulatory proteins and its function is further modulated by phosphorylation (3). DVL was recently found to also localize to the centrosome-derived structures, where it regulates functioning of basal body (4), ciliary disassembly (5), and mitotic spindle orientation in tandem with Plk1 (6).
Centrosomes are evolutionary conserved structures, which serve as centers for microtubule nucleation, mitotic spindle organization, and formation of basal bodies of cilia and flagella (7). The two connected orthogonal centrioles that each cell inherits after division go through several steps of structural changes (8, 9), which are tightly controlled and coupled to cell cycle progression (10). From G1 phase onward, a loose proteinaceous linker, containing centrosomal Nek2-associated protein 1 (C-NAP1) (11), CDK5 regulatory subunit-associated protein 2 (CDK5RAP2) (12, 13), and fibrous Rootletin (14), is established and continues to anchor centrioles until it is degraded during the G2/M phase (15). As the cell approaches mitosis, linker proteins are phosphorylated by the Plk1-Mst2-NIMA-related kinase 2 (NEK2) cascade (16, 17), which in turn causes cleavage of Rootletin (14) and displacement of C-NAP1 from centrosomes (18). Interference with linker proteins or their phosphorylation has a drastic effect on cell division (19) and on genome integrity caused by chromosome segregation errors (20). Structural or numerical centrosomal abnormalities are linked to chromosomal instability, cancer progression, and various developmental diseases (7, 21).
In this work, we present evidence that DVL dynamically associates with the proteins of centrosomal linker and is an essential part of the centrosomal cycle. These DVL functions are mediated by NEK2, which phosphorylates DVL at several residues, and induces interaction of DVL with the linker proteins CDK5RAP2 and C-NAP1. We reveal a mechanism of DVL-dependent linker protein release from centrosome and show that DVL is a crucial regulator of centrosomal cycle.
Results
Dishevelled Localizes to the Centrosome via Its DIX Domain and Is a NEK2 Target.
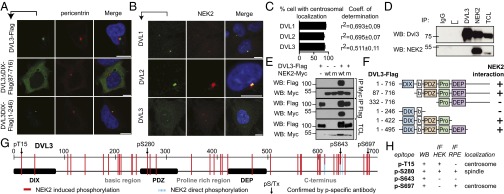
Previous research revealed that DVL localizes to centrosome (6, 22), which we confirmed by endogenous immunocytochemistry and biochemical fractionation (SI Appendix, Fig. S1 A–C). Building on these data, we aimed to define which DVL domain is required for its centrosomal localization. By expressing low levels of DVL in HEK293 cells, we were able to analyze its colocalization with the centrosomal marker pericentrin. Exogenously expressed DVL3 localized to centrosome vicinity (Fig. 1A) in the same fashion as endogenous protein (SI Appendix, Fig. S1D), and the DVL3 dishevelled and axin (DIX) domain was both sufficient and required for its centrosomal localization (Fig. 1A; for expression levels see SI Appendix, Fig. S1E). The DIX domain confers to DVL the ability to polymerize in a head-to-tail manner (23). Interestingly, the DVL2 M1 (F43S) mutant, which is multimerization-defective but can still form dimers with endogenous DVL (23), also localized to the centrosome (SI Appendix, Fig. S1F). We conclude that the DIX domain, and not the ability of DVL to polymerize, is required for its centrosomal localization. The requirement for the DIX domain is in line with the previously described requirement of the Axin DIX domain for centrosomal attachment (24, 25).
Fig. 1.
DIX domain is required for DVL centrosomal localization, and NEK2 is a DVL kinase. (A) HEK293 cells were transfected with a low amount of DVL3, DVL3ΔDIX, and DVL3 DIX domain (green), and colocalization with pericentrin (red) was assessed. (B) NEK2 (red) colocalizes with DVL1, DVL2, and DVL3 (green) on an endogenous level in RPE cells. (C) Quantification (% of cells where proteins colocalized and coefficient of determination R2). (D) Endogenous DVL3 was co-IPed, using NEK2 as bait; anti-DVL3/IgG isotype was used as a positive and negative control. (E) HEK293 cells were cotransfected with DVL3 and NEK2, as indicated, and subjected to coimmunoprecipitation. DVL3 interacts with and shows an electrophoretic mobility shift, as a result of posttranslational modifications, only after coexpression with the wild-type form of the NEK2 kinase. (F) Summary of interaction of individual DVL3 deletion mutants with NEK2 (raw data, SI Appendix, Fig. S2 A and B). Domain structure is schematized. (b, basic region; Pro, proline-rich region). (G) HEK293 cells were transfected with DVL3, cotransfected with empty vector or wild-type NEK2, coimmunoprecipitated, and subjected to MS analysis. Schematic view of identified phosphorylated residues of DVL3 by MS. Red solid lines represent sites whose phosphorylation was induced by NEK2. Blue dashed lines represent sites directly phosphorylated by NEK2; purified DVL3 domains were incubated with recombinant kinase in presence of ATP and subjected to MS analysis. Arrows, induction of phosphorylation by NEK2 coexpression confirmed by phospho-specific antibody. For a complete list of residues, see SI Appendix, Table S1. (H) Panel indicating the successful use of phospho-specific antibodies using different methods and cell lines. (Scale bars, 10 μm.)
It has been previously reported by functional screens in Drosophila that the centrosomal kinase NEK2 phosphorylates fly Dishevelled (Dsh) (26). We confirmed the colocalization of endogenous DVL1, DVL2, and DVL3 with NEK2 in retinal pigment epithelium (RPE) cells (Fig. 1 B and C) and showed that NEK2 coimmunoprecipitated DVL3 in HEK293 cell lysates (although DVL3 was unable to pull down NEK2) (Fig. 1D). Interestingly, DVL3 coimmunoprecipitated only with wild-type (wt), but not with kinase-dead mutant K37R (mut), of NEK2, suggesting that kinase activity of NEK2 is required for efficient binding (Fig. 1E). Domain mapping experiments performed either by coimmunoprecipitation or as relocalization of cytoplasmic NEK2 to nucleus with DVL3 mutants lacking nuclear export sequence identified a critical role for the PDZ domain in the interaction of DVL and NEK2 (Fig. 1F; raw data: SI Appendix, Fig. S2 A and B).
MS analysis of DVL3 revealed a variety of specific NEK2-induced phosphorylation events (Fig. 1G and SI Appendix, Table S1), and in vitro kinase assay confirmed that several serine residues are phosphorylated directly by NEK2 (Fig. 1G and SI Appendix, Fig. S2C). We verified induction of phosphorylation by NEK2 of some of the DVL3 phosphorylation sites (T15, S280, S643, and S697) independently using novel phospho-specific antibodies and point S-A mutants (Fig. 1G and SI Appendix, Fig. S3). Phosphorylation at S643 was associated with homogenous cytoplasmic localization of DVL3 (27), and indeed NEK2, similar to casein kinase (CK)1ε coexpression (28, 29), triggered subcellular relocalization of DVL from punctate to even cytoplasmic distribution (SI Appendix, Fig. S2D). Moreover, pT15- and pS697-DVL3 phospho-specific antibodies showed a centrosomal signal, which for pS697 accumulated throughout the cell cycle, whereas pS280-DVL3 was found only in mitotic spindle and was absent in the interphase centrosome (Fig. 1H and SI Appendix, Fig. S3 E–G). These data suggest that NEK2 controls accumulation and subsequent release of DVL3 from the centrosome.
DVL Is Required for Efficient Separation of Centrosomes and Interacts with Linker Proteins After NEK2-Mediated Phosphorylation.
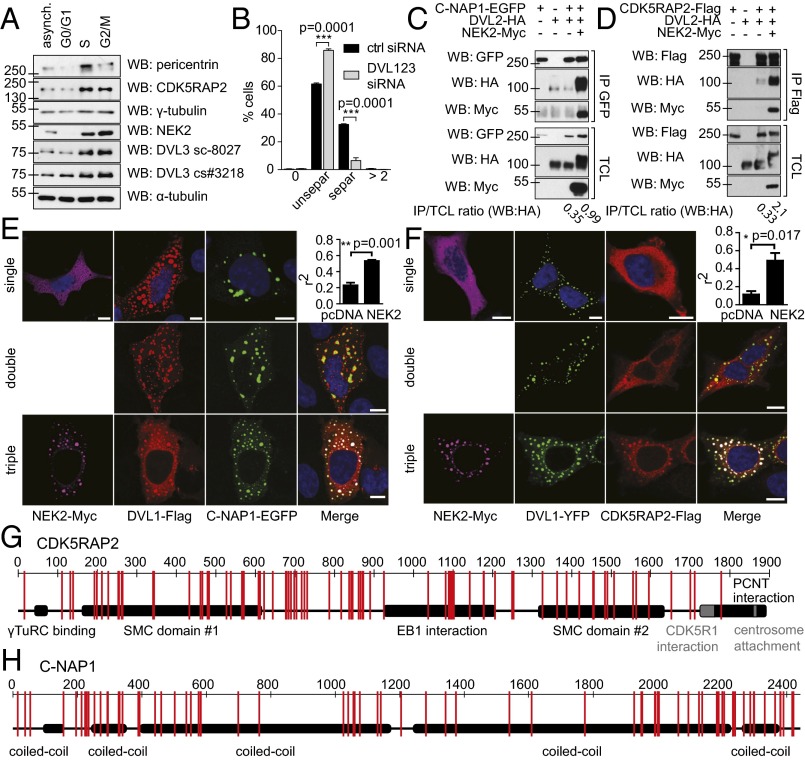
NEK2 expression levels peak at G2/M phase of the cell cycle. Protein analysis in individual cell cycle phases using sorted HeLa S. Fucci cells (30) showed that DVL3 expression gradually increases throughout the cell cycle. This was reminiscent of numerous centrosomal components, which increase when centrosomes duplicate (γ-tubulin, CDK5RAP2, pericentrin), or regulators of G2/M cell cycle phases such as NEK2 (Fig. 2A and quantification, SI Appendix, Fig. S4A, and sorting control, SI Appendix, Fig. S4B).
Fig. 2.
DVL influences centrosome separation and associates with linker proteins CDK5RAP2 and C-NAP1. (A) HeLa S. Fucci cells were sorted according to the cell cycle phase, subjected to Western blotting, and probed with antibodies against DVL3, centrosomal proteins (pericentrin, CDK5RAP2, γ-tubulin, NEK2), and α-tubulin as loading control. (B) RPE cells were stained for pericentrin and glutamylated tubulin, and distribution of centrosomal signals (SI Appendix, Fig. S4G) was quantified. Cells transfected with DVL1-2-3 siRNA show increased proportion of centrosomes before separation. (C and D) HEK293 cells were transfected, as indicated, and subjected to coimmunoprecipitation and Western blotting. Components of proteinaceous centrosomal linker (C) C-NAP1 and (D) CDK5RAP2 show enhanced interaction with DVL2 after coexpression of NEK2 kinase. (E and F) HEK293 cells were transfected as indicated, and colocalization was inspected by confocal microscopy and quantified. Colocalization of both C-NAP1 (E) and CDK5RAP2 (F) with DVL1 is visibly enhanced after NEK2 coexpression. (G and H) HEK293 cells were transfected with CDK5RAP2 or C-NAP1, cotransfected with empty vector or wild-type NEK2, coimmunoprecipitated, and subjected to analysis by mass-spectrometry. Differentially phosphorylated residues are indicated (schematic view), for a complete list of residues, see SI Appendix, Table S2. Graphs represent mean ± SEM of three independent replicates. *P < 0.05; **P < 0.01; ***P < 0.001; Student t test, E and F. DAPI (blue). (Scale bars, 100 μm.)
We were thus intrigued by the possibility that DVL could be involved in regulation of centrosome number and configuration. After DVL1/DVL2/DVL3 (DVL1-2-3) knockdown (for knockdown efficiencies and controls, see SI Appendix, Fig. S4 C–H), we observed marked reduction of cells with the clearly separated centrosomes and an increase of cells with closely connected centrosomes (Fig. 2B and SI Appendix, Fig. S4I). This suggests that the process of centrosomal separation is defective in the absence of DVL. Knockdown of individual DVL isoforms caused significant centrosomal separation, as well as proliferation phenotypes, but never reached the levels of triple DVL1-2-3 knock-down, suggesting the functional redundancy of individual DVL isoforms (SI Appendix, Fig. S4 J and K). DVL knockdown in RPE cells did not affect the localization of structural centrosomal components (pericentrin, γ-tubulin, CDK5RAP2, C-NAP1, Rootletin, and CEP164) (SI Appendix, Fig. S5A) and did not cause any obvious ultrastructural aberrations in centriolar morphology analyzed by electron microscopy (SI Appendix, Fig. S5B). Moreover, it did not block the ability to duplicate centrioles seen after overexpression of PLK4, which can trigger procentriole formation (SI Appendix, Fig. S5C). We conclude that DVL contributes to centrosomal separation but is not involved in other centrosomal processes, either as structural or scaffolding component.
These results prompted us to test the connection of DVL to linker proteins. We analyzed the amount of DVL3 in CDK5RAP2, C-NAP1, and Rootletin pulldowns in the presence or absence of exogenous NEK2. Interestingly, levels of DVL associated with CDK5RAP2 and C-NAP1, but not Rootletin, were significantly increased when NEK2 was coexpressed (Fig. 2 C and D and SI Appendix, Fig. S5D). When visualized by immunofluorescence, colocalization of DVL with either C-NAP1 or CDK5RAP2 also improved when NEK2 was coexpressed (Fig. 2 E and F).
C-NAP1 is a known NEK2 substrate, in which multisite phosphorylation introduces bulk negative charge and facilitates its release from centrosome and triggers centrosome disjunction (18). We tested whether the interaction of DVL with C-NAP1/CDK5RAP2 is a prerequisite for or consequence of NEK2-mediated phosphorylation by performing MS analysis of NEK2-triggered phosphorylation of the linker proteins in wt and triple-knockout DVL1/DVL2/DVL3 cells. We have identified more than 80 amino acid residues of C-NAP1 and CDK5RAP2 specifically phosphorylated by NEK2, with the pattern comparable in both cell lines (Fig. 2 G and H and SI Appendix, Table S2). This suggests that the role of CDK5RAP2 in centrosome cohesion can be regulated in a way similar to C-NAP1, and that DVL is not required for NEK2-mediated phosphorylation of linker proteins. We cannot, however, rule out that DVL is important for low stoichiometry phosphorylation events below the detection limit of our methods.
Dishevelled Mediates NEK2-Triggered Displacement of Linker Proteins from Centrosome.
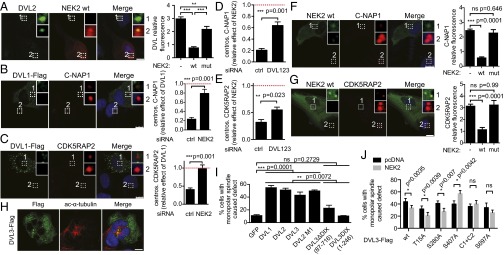
To get deeper insight into the role of DVL in the NEK2-controlled linker dissolution, we performed a series of experiments in RPE cells, where we quantified levels of endogenous DVL, C-NAP1, or CDK5RAP2 at the centrosome. NEK2 was able, in a kinase activity-dependent manner, to displace endogenous DVL from the centrosome (Fig. 3A). This NEK2-dependent displacement from centrosome resembles behavior of linker proteins such as C-NAP1 (31). We therefore hypothesized that DVL may be involved with NEK2 in removal of linker proteins. Indeed, overexpressed DVL was able to displace both C-NAP1 and CDK5RAP2, but not centrin, from the centrosome, and this phenotype was rescued by NEK2 knockdown (Fig. 3 B and C; raw data: SI Appendix, Fig. S5 E–H; controls: SI Appendix, Fig. S4F and SI Appendix, Fig. S6 A–C). Similarly, DVL depletion by DVL1-2-3 triple knockdown was able to rescue centrosomal displacement of C-NAP1 and CDK5RAP2 observed upon overexpression of NEK2 (Fig. 3 D and E). The role of NEK2 in the centrosomal displacement of CDK5RAP2 has not been described yet, but our data clearly demonstrate that only wt NEK2, not the inactive kinase, is able to remove CDK5RAP2 from centrosome (Fig. 3G and SI Appendix, Fig. S6B), similar to the previously described effects on C-NAP1 (Fig. 3F and SI Appendix, Fig. S6A) (18). These data are in agreement with the hypothesis that DVL serves as a temporally regulated scaffold required for the efficient NEK2-controlled release of both C-NAP1 and CDK5RAP2 in the process of centrosomal separation.
Fig. 3.
DVL mediates NEK2-triggered displacement of linker proteins from the centrosome. (A) RPE cells with overexpressed NEK2 show significantly reduced fluorescence intensity of endogenous DVL2 at centrosome in a kinase activity-dependent manner. (B and C) Overexpressed DVL1 displaces linker proteins from centrosome manifested by reduced fluorescence of CDK5RAP2 and C-NAP1. This phenotype is rescued by NEK2 siRNA. (D and E) Overexpressed NEK2 wt displaces linker proteins from centrosome manifested by reduced fluorescence of CDK5RAP2 and C-NAP1. This phenotype is rescued by DVL1-2-3 siRNA. Red dashed line shows baseline fluorescence of linker proteins. (F and G) C-NAP1 and CDK5RAP2 are removed from the centrosome after transfection of wt but not mut NEK2. (H) DVL overexpression (green) in HEK293 cells leads to monopolar spindle phenotype; acetylated-α-tubulin (red). (I) Extent of monopolar spindle formation in full-length DVLs, DVL3 with deleted DIX domain (DVLΔDIX87-716), DVL3DIX1-246, and nonpolymerizable mutant of DVL2 (DVL2 M1). (J) Quantification of the monopolar spindle-related phenotypes in S–A mutants at the DVL3 sites phosphorylated by NEK2 in the presence and absence of exogenous NEK2 was quantified. Graphs represent mean ± SEM of three independent replicates. *P < 0.05; **P < 0.01; ***P < 0.001 (ANOVA, Bonferroni's posttest: A, F, G, I; Student t test: B–E; multiple comparison adjusted Student t test: J). DAPI (blue). (Scale bars, 10 μm.)
RPE cells possess undisturbed cell cycle checkpoints and stop dividing when centrosome is affected (e.g., by DVL depletion). To study consequences of centrosomal defects caused by DVL, we therefore performed experiments in transformed HEK293 cells, which continue cycling even with severe centrosomal defects. Loss of DVL in HEK293 caused increased proportion of cells with aberrant nuclei and longer cell cycle (SI Appendix, Fig. S6 D and E). Overexpression of each DVL isoform in HEK293 cells caused a multinuclear phenotype (Fig. 3H and SI Appendix, Fig. S6F), which is a consequence of the monopolar spindle defect connected with the failure of centrosome function (32). The ability of DVL isoforms and its mutants to trigger formation of a monopolar spindle (Fig. 3I) correlated well with the ability to localize to the centrosome (compare with Fig. 1A). This is exemplified by DVL3ΔDIX deletion mutant, which fails to induce monopolar spindle-induced defects, in contrast to the multimerization defective mutant (DVL2 M1) (Fig. 3I). One of the well-described inducers of monopolar spindle is dominant-negative NEK2 (19). A possible explanation for DVL-induced monopolar spindle is thus sequestration of active NEK2 from centrosome by overexpressed DVL. We failed to observe monopolar spindle-caused defects on overexpression of a DVL deletion mutant lacking the PDZ domain (DVL3DIX 1-246) (Fig. 3I), which can associate with centrosome (Fig. 1F), but cannot bind and sequester NEK2 (SI Appendix, Fig. S2B). In line with this assumption, NEK2 overexpression can partially rescue DVL3-induced monopolar spindle formation (Fig. 3J). We took advantage of this phenomenon to identify which of the sites phosphorylated by NEK2 (Fig. 1G and SI Appendix, Table S1) are required for the centrosomal DVL3 function. Analysis of monopolar spindle phenotype in DVL3 mutants showed the special importance of cluster of phosphorylations sites C1 and C2 (SI Appendix, Fig. S3H) and S697 residue of DVL3 (Fig. 3J). This suggests that the phosphorylation of the C-terminal part of DVL is functionally critical for the centrosomal roles of DVL. We, however, do not exclude the importance of other sites.
NEK2-Mediated Displacement of DVL from the Centrosome Increases Cell Response to the Activation of the Wnt/β-Catenin Pathway.
A novel role of DVL controlled by NEK2 opens the question of how the centrosomal function of DVL coordinated with other well-defined roles of DVL; namely, with its function in the basahe l body docking, required for primary cilia formation, and in the Wnt/β-catenin pathway (33). To address the first issue, we tested the ability of NEK2 to affect the interaction of DVL3 with Chibby, CEP164, and Inversin (Inv), three proteins with the important role in ciliogenesis and links to DVL or other Wnt pathway components (34–36). SI Appendix, Fig. S6 shows that NEK2 can interact with all three proteins and potentiates the DVL3–Inversin interaction (SI Appendix, Fig. S6G). It, however, cannot trigger the DVL3–Chibby interaction (SI Appendix, Fig. S6H) and has no positive effect on DVL3–CEP164 binding (SI Appendix, Fig. S6I; for controls, see SI Appendix, Fig. S6J). This suggests that players other than NEK2 are involved in the DVL role in ciliogenesis, and that the issue requires further investigation.
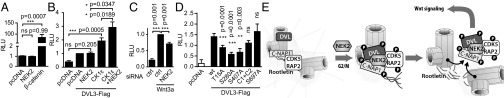
To address the role of NEK2-mediated phosphorylation of DVL in the canonical Wnt pathway, we performed gain-of-function experiments using TopFlash reporter assays. NEK2 was unable to trigger Wnt/β-catenin pathway activation, neither on its own (Fig. 4A) nor when coexpressed with DVL3 (Fig. 4B, columns 1–3). However, we have observed a synergy of NEK2 with CK1ε when DVL was present (Fig. 4B, columns 2–5), and knockdown of NEK2 reduced reporter activation induced by exogenously supplied Wnt-3a (Fig. 4C). These results are in agreement with the possibility that NEK2 controls (via displacement from centrosome) size of the DVL3 pool available for the Wnt/β-catenin pathway. This assumption is confirmed by the analysis of DVL3 S-A mutants, which in numerous cases show lower ability to trigger Wnt/β-catenin signaling (Fig. 4D). In combination with other data, we conclude that NEK2 is able to positively influence the output of the Wnt/β-catenin pathway by increasing the proportion of the DVL pool available for Wnt/β-catenin pathway.
Fig. 4.
DVL phosphorylation by NEK2 affects Wnt signaling. Proposed mechanism of DVL action during centrosome separation. HEK293 cells were transfected as indicated and analyzed for Topflash reporter activity. (A) NEK2 overexpression shows no significant activation of Wnt/β-catenin pathway, unlike β-catenin. (B) NEK2 alone is not able to potentiate Wnt/β-catenin activation induced by DVL3, but synergy is observed in the presence of CK1ε. (C) Knockdown of NEK2 by siRNA decreases Wnt/β-catenin pathway activation by recombinant Wnt-3a. (D) Topflash reporter activity after transfection of Ser to Ala mutants at the DVL3 sites phosphorylated by NEK2. (E) DVL accumulates during cell cycle at the centrosome. In G2/M, when NEK2 activity is peaking, DVL is phosphorylated on several residues, which increases its affinity toward linker proteins. DVL works as a scaffold that facilitates and is required for NEK2-induced release of C-NAP1 and CDK5RAP2 from centrosomes. DVL released from the centrosome by the activity of NEK2 is than available for and increases the Wnt/β-catenin pathway activation. Graphs represent mean ± SEM of at least three independent replicates. *P < 0.05; **P < 0.01 (ANOVA, Bonferroni's posttest, A–D).
Discussion
Here we provide evidence for a function of DVL in the interphase centrosome, where it serves as an indispensable regulator of the loose centrosomal linker. Our work provides further insight into the centrosome-associated function of DVL, in addition to the recently described role of DVL in the microtubule-kinetochore attachment and spindle orientation (6), as well as in the basal body docking and related functions in ciliogenesis/ciliary disassembly (4, 5, 35).
Our data suggest that DVL is an important regulator of linker protein displacement during the process of centrosomal separation. The proposed mechanism of action is summarized in Fig. 4E. During interphase, DVL slowly accumulates at the centrosome, together with other proteins of centrosomal linker such as C-NAP1 and CDK5RAP2. We propose that at G2/M, when the centrosomal kinase NEK2 reaches its maximal activity (37), it phosphorylates DVL at multiple positions, which increases DVL affinity toward linker proteins. Phosphorylated DVL binds CDK5RAP2 and C-NAP1 and helps release them from centrosome.
The exact mechanism that leads to DVL-mediated release of linker protein complexes from centrosome is not entirely clear. Electrostatic repulsion or sterical exclusion were proposed for NEK2-driven removal of C-NAP1 from the centrosome (18, 19). Given that most linkers proteins are heavily phosphorylated [we identified NEK2-induced phosphorylation on 82 (C-NAP1), 81 (CDK5RAP2), or 41 (DVL3) unique Ser/Thr sites], we believe that electrostatic repulsion can represent a key mechanism explaining centrosomal release of CDK5RAP2 and DVL3. Our work adds CDK5RAP2 onto the list of NEK2 substrates, which are present in the centrosomal linker and are required for centrosomal cohesion (13, 38).
One intriguing possibility raised by our results is the role for the centrosome as an organelle coordinating the cell cycle and Wnt signaling. We propose that NEK2-mediated release of DVL from centrosome increases the availability of cytoplasmic DVL for Wnt/β-catenin pathway, where it has a crucial function as a component of signalosomes (39). Similarly, retention of DVL at centrosome as a result of depletion of NEK2 by siRNA manifested itself as attenuation of Wnt/β-catenin signaling, despite the fact that NEK2 overexpression did not activate Wnt/β-catenin pathway per se. This hypothesis reconciles the data observed by us and by Schertel and colleagues (26). Interestingly, NEK2 can phosphorylate S33/S37/T41 of β-catenin (40), which is inhibitory. This suggests that NEK2 phosphorylation of β-catenin and DVL have distinct effects on Wnt/β-catenin signaling.
Novel function of DVL in centrosomal separation and extensive phosphorylation by NEK2 reopens the question of how DVL can perform its multiple roles. DVL is needed for Wnt/β-catenin and Wnt/planar cell polarity pathways, as well as basal body docking and proper cilia function. These functions are controlled mainly by the activity of several kinases (for review, see ref. 41). The efforts to explain DVL’s individual “fates” by single-point mutations were not, so far, successful. For example, NEK2 resembles in many aspects the best-described DVL kinase CK1ε: both kinases are capable of inducing a dramatic DVL phosphorylation shift, overlap in many target residues (e.g., S643 and S280 in hDVL3) (27), and are capable of promoting even cytoplasmic localization of DVL, but only CK1ε can induce downstream Wnt/β-catenin signaling. It is likely that only identification and functional characterization of complex phosphorylation barcodes of individual DVL subcellular pools will fully reconcile the issue. We believe that unique reagents generated in this study, mainly a panel of phospho-specific antibodies, will pave the way for the ultimate understanding of multiple functions of DVL in the near future.
Materials and Methods
Cell Culture and Transfection.
HEK293, RPE, and HeLa S. Fucci cells were grown at 37 °C and 5% (vol/vol) CO2 in DMEM, 10% (vol/vol) FBS, and 1% antibiotics (penicillin/streptomycin).
Cells were transfected 24 h after seeding, using polyethyleneimine in a stoichiometry of 2.5:1 (polyethyleneimine:DNA). For immunofluorescence, HEK293 or RPE cells were seeded on 24-well plates with gelatin-coated coverslips and transfected, as indicated, with 0.3 μg of each corresponding plasmid. For coimmunoprecipitation, HEK293 cells were seeded on 10-cm dishes and transfected with 3 μg of each corresponding plasmid. For MS/MS-based identification of phosphorylations, HEK293 cells were seeded on 15-cm dishes and transfected with 15 μg of each corresponding plasmid. Used plasmids are summarized in SI Appendix, Table S4.
RNA Interference.
RPE or HEK293T cells were transfected according to the manufacturer's instructions (Ambion). In brief, control and DVL siRNAs (Ambion, Eurogentec; SI Appendix, Table S5) were mixed with Lipofectamine RNAiMAX (Invitrogen) in serum-free DMEM in a ratio of 1:1 and incubated for 30 min at room temperature. The transfection mixture was added to the 12- or 24-well plate and mixed with a suspension of freshly trypsinized cells, resulting in a final concentration of 130 nm siRNA. When a combination of different siRNAs was used, each siRNA was used at 130 nM, and the amount of control siRNA was scaled accordingly.
Immunofluorescence, Cell Sorting, and Centrosomal Preparation.
HEK293 and RPE were seeded on 0.1% gelatin-coated coverslips and harvested as follows. Samples without fluorescently tagged proteins were fixed and stained as previously described (42). Samples containing fluorescently tagged proteins were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton-X100, blocked with PBS/BSA/Triton/Azide buffer (PBTA) [3% (wt/vol) BSA, 0.25% Triton, 0.01% NaN3] and incubated overnight with antibodies in PBTA. Next day, samples were washed in PBS, incubated in fluorescent secondary antibodies AlexaFluor-405, AlexaFluor-488, AlexaFluor-568 (Invitrogen), or DyLight-647 (Jackson ImmunoResearch); nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/mL). Cells were visualized using a Leica SP5 confocal microscope using 63× (oil) objective with Leica Application Suite software. Counting of mitotic cells, centrosomal patterns, monopolar spindles, and quantification of centrosomal proteins was performed on Olympus IX51 fluorescent microscope using 40× (air) or 100× (oil) objectives with QuickPHOTO Camera software. For quantification workflow details, see SI Appendix.
HeLa S. Fucci cells were sorted according to the cell cycle phase on an Aria II cell sorter (BD Biosciences). Both mKO2 and mAG were excited by a 488-nm laser, and fluorescence signals were collected at 530 nm (530/28 BP) for mAG, at 575 nm (575/26 BP) for mKO2. The same amount of cells was directly lysed in Laemmli sample buffer and subjected to Western blotting analysis. Crude centrosomal fraction was prepared as previously described (43); for procedure, see SI Appendix.
Dual Luciferase Assay, Coimmunoprecipitation, MS, and Western Blotting.
Dual luciferase was performed according to manufacturer’s instruction and as described previously (29). Immunoprecipitation protocol used was performed as described previously (27). Expanded protocol can be found in SI Appendix. Immunoblotting and sample preparation was performed as previously described (44) and developed using either light-sensitive films (Agfa, GE Healthcare) or chemiluminescence documentation system FusionSL (Vilber-Lourmat). MS for posttranslational modifications was performed as previously described (27), for full experimental details, see SI Appendix. Antibodies used for immunoprecipitation are summarized in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Randy Moon, Madelon Maurice, Jeff Wrana, Jonathan M. Graff, Erich Nigg, Andrew M. Fry, Ken Takemaru, and S. Yanagawa for providing plasmids. This study was supported by the Czech Science Foundation (13-31488P, 13-32990S, 15-21789S), Masaryk University (MUNI/M/1050/2013, MUNI/A/1398/2014), the European Regional Development Fund (KI-MU; CZ.1.07/2.3.00/20.0180), Marie Curie International Training Network WntsApp (Grant 608180), South Moravian Center for International Mobility (Brno PhD Talent, IC), Swedish Cancer Society (CAN2011/690, 2014/659), and the Canadian Institutes for Health Research (Grant MOP-84273 to S.A.). The MS/MS part was carried out with the support of Proteomics Core Facility of Central European Institute of Technology (CEITEC) and supported by project CEITEC 2020 (LQ1601). D.P. (MS/MS analysis) especially thanks the Czech Science Foundation (CSF) project (P206/12/G151). Transmission electron microscopy analysis was supported by European Social Fund in the Czech Republic, project HistoPARK, reg. no. CZ.1.07/2.3.00/20.0185.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608783113/-/DCSupplemental.
References
- 1.Hikasa H, Sokol SY. Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol. 2013;5(1):a007955. doi: 10.1101/cshperspect.a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010;22(5):717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40(7):871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee KH, et al. Identification of a novel Wnt5a-CK1ε-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 2012;31(14):3104–3117. doi: 10.1038/emboj.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuchi K, Niikura Y, Kitagawa K, Kikuchi A. Dishevelled, a Wnt signalling component, is involved in mitotic progression in cooperation with Plk1. EMBO J. 2010;29(20):3470–3483. doi: 10.1038/emboj.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Tsou MFB, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17(3):344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442(7105):947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 11.Fry AM, et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141(7):1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120(Pt 24):4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 13.Barrera JA, et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18(6):913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171(1):27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’regan L, Blot J, Fry AM. Mitotic regulation by NIMA-related kinases. Cell Div. 2007;2:25. doi: 10.1186/1747-1028-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12(12):1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardin BR, Agircan FG, Lange C, Schiebel E. Plk1 controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr Biol. 2011;21(13):1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 18.Hardy T, et al. Multisite phosphorylation of C-Nap1 releases it from Cep135 to trigger centrosome disjunction. J Cell Sci. 2014;127(Pt 11):2493–2506. doi: 10.1242/jcs.142331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14(7):2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23(3):401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27(8):307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24(6):664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz-Romond T, et al. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14(6):484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 24.Alexandrova EM, Sokol SY. Xenopus axin-related protein: A link between its centrosomal localization and function in the Wnt/beta-catenin pathway. Dev Dyn. 2010;239(1):261–270. doi: 10.1002/dvdy.22125. [DOI] [PubMed] [Google Scholar]
- 25.Fumoto K, Kadono M, Izumi N, Kikuchi A. Axin localizes to the centrosome and is involved in microtubule nucleation. EMBO Rep. 2009;10(6):606–613. doi: 10.1038/embor.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schertel C, et al. Systematic screening of a Drosophila ORF library in vivo uncovers Wnt/Wg pathway components. Dev Cell. 2013;25(2):207–219. doi: 10.1016/j.devcel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Bernatík O, et al. Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1ε and frizzled5. J Biol Chem. 2014;289(34):23520–23533. doi: 10.1074/jbc.M114.590638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalley MJ, et al. Dishevelled (Dvl-2) activates canonical Wnt signalling in the absence of cytoplasmic puncta. J Cell Sci. 2005;118(Pt 22):5279–5289. doi: 10.1242/jcs.02647. [DOI] [PubMed] [Google Scholar]
- 29.Bernatik O, et al. Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. J Biol Chem. 2011;286(12):10396–10410. doi: 10.1074/jbc.M110.169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 31.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci. 2002;115(Pt 16):3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 32.Tillement V, et al. Spindle assembly defects leading to the formation of a monopolar mitotic apparatus. Biol Cell. 2009;101(1):1–11. doi: 10.1042/BC20070162. [DOI] [PubMed] [Google Scholar]
- 33.Wallingford JB, Mitchell B. Strange as it may seem: The many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 2011;25(3):201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takemaru K, et al. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422(6934):905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 35.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150(3):533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37(5):537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz SJ, Fry AM, Sutterlin C, Ried T, Nigg EA. Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 1994;5(6):625–635. [PubMed] [Google Scholar]
- 38.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151(4):837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilic J, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316(5831):1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 40.Mbom BC, Siemers KA, Ostrowski MA, Nelson WJ, Barth AI. Nek2 phosphorylates and stabilizes β-catenin at mitotic centrosomes downstream of Plk1. Mol Biol Cell. 2014;25(7):977–991. doi: 10.1091/mbc.E13-06-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryja V, Bernatik O. Dishevelled at the crossroad of pathways. In: Hoppler S, Moon RT, editors. Wnt signaling in Development and Disease: Molecular Mechanisms and Biological Functions. John Wiley & Sons, Inc.; Hoboken, NJ: 2014. [Google Scholar]
- 42.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114(Pt 20):3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 43.Meigs TE, Kaplan DD. 2008. Isolation of centrosomes from cultured Mammalian cells. CSH Protocols 2008:pdb prot5039.
- 44.Bryja V, et al. Increased apoptosis in differentiating p27-deficient mouse embryonic stem cells. Cell Mol Life Sci. 2004;61(11):1384–1400. doi: 10.1007/s00018-004-4081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.