Significance
We report here that microRNA-309 (miR-309) plays a critical role in ovarian development of female Aedes aegypti mosquitoes. The genetic disruption of miR-309 by CRISPR/Cas9 system displays a failure of ovarian primary follicle formation, and several pathways associated with ovarian development are down-regulated after miR-309 depletion. Comprehensive screening and functional identification have revealed that SIX homeobox 4 protein (SIX4) is a direct target of miR-309. miR-309–targeted degradation of SIX4 mRNA is required for appropriate commencement of the preparatory phase and initiation of the blood feeding-triggered phase of ovarian development. Thus, miR-309 serves as a regulatory switch permitting a stage-specific degradation of the ovarian SIX4 mRNA, allowing a shift from previtellogenic to postvitellogenic phases of ovarian development.
Keywords: Homeobox protein gene, microRNA, CRISPR/Cas9, ovary, fast evolution
Abstract
Obligatory blood-triggered reproductive strategy is an evolutionary adaptation of mosquitoes for rapid egg development. It contributes to the vectorial capacity of these insects. Therefore, understanding the molecular mechanisms underlying reproductive processes is of particular importance. Here, we report that microRNA-309 (miR-309) plays a critical role in mosquito reproduction. A spatiotemporal expression profile of miR-309 displayed its blood feeding-dependent onset and ovary-specific manifestation in female Aedes aegypti mosquitoes. Antagomir silencing of miR-309 impaired ovarian development and resulted in nonsynchronized follicle growth. Furthermore, the genetic disruption of miR-309 by CRISPR/Cas9 system led to the developmental failure of primary follicle formation. Examination of genomic responses to miR-309 depletion revealed that several pathways associated with ovarian development are down-regulated. Comparative analysis of genes obtained from the high-throughput RNA sequencing of ovarian tissue from the miR-309 antagomir-silenced mosquitoes with those from the in silico computation target prediction identified that the gene-encoding SIX homeobox 4 protein (SIX4) is a putative target of miR-309. Reporter assay and RNA immunoprecipitation confirmed that SIX4 is a direct target of miR-309. RNA interference of SIX4 was able to rescue phenotypic manifestations caused by miR-309 depletion. Thus, miR-309 plays a critical role in mosquito reproduction by targeting SIX4 in the ovary and serves as a regulatory switch permitting a stage-specific degradation of the ovarian SIX4 mRNA. In turn, this microRNA (miRNA)-targeted degradation is required for appropriate initiation of a blood feeding-triggered phase of ovarian development, highlighting involvement of this miRNA in mosquito reproduction.
Transmission of mosquito-borne diseases brings enormous human suffering, with more than 1 million deaths worldwide annually. The yellow fever mosquito Aedes aegypti has reemerged as one of the most dangerous vectors of human diseases, transmitting Dengue fever, Yellow fever, Chikungunya, and Zika virus (1–4). Because of the lack of effective vaccines and increasing drug resistance in pathogens, biological control is considered one of the most promising strategies for preventing disease transmission. In particular, female hematophagous mosquitoes rely on acquisition of blood to initiate a series of physiological events promoting egg development (5, 6). Therefore, understanding the molecular mechanisms underlying ovarian activation is of great significance for the development of effective approaches to control mosquito-borne diseases (7, 8).
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression at the posttranscriptional level through translational repression or mRNA decay (9). Hence, miRNAs play significant roles in governing multiple functions in animals and plants via integrating sophisticated miRNA–mRNA regulatory networks (10, 11). In the mosquito, previous studies have shown that several miRNAs manipulate host–pathogen interactions, contributing to maintenance of endosymbiont Wolbachia infection or resistance to DENV (dengue virus) infection (12–14). However, there has been relatively little attention to the roles of miRNAs in reproductive physiology of these essential disease vectors. A study of microRNA-275 (miR-275) revealed its fundamental role in regulation of blood digestion and egg development in female A. aegypti mosquitoes (15). Consequently, accumulated evidence also demonstrated the profound linkage between miRNAs and blood meal-dependent egg activation. The mosquito-specific miR-1174 targets serine hydroxymethyltransferase to control blood intake and digestion in the gut and influences egg development (16). Moreover, the conserved miRNA miR-8 regulates the long-range Wingless signaling that is required for secretion of lipophorin and vitellogenin from the fat body in response to blood feeding, affecting deposition of yolk proteins in oocytes and fecundity of female mosquitoes (17).
Recently, a fast-evolving miRNA cluster mir-309∼6 has been identified, which is undergoing particularly dynamic evolutionary events within the arthropod lineage, including duplication, gain, and/or loss of miRNA members within it (18). In Drosophila, this miRNA cluster is preferentially expressed in eggs and promotes mRNA turnover during the maternal-to-zygotic transition (19). Interestingly, miR-309, a member of this miRNA cluster, persists and duplicates in a lineage-specific manner against the background of loss of many other miRNA cluster members in the mosquito A. aegypti (18). Generally, fast-evolving genes/miRNAs could drive adaptive evolution with respect to lineage-specific physiology and contribute to species divergence. Therefore, from the evolutionary perspective, it can be suggested that miR-309 is associated with an important role in regulation of A. aegypti reproductive physiology. The present study has revealed the essential role of miR-309 in initiation of egg development in female mosquitoes. Genome-wide response to miR-309 depletion has indicated that it participates in regulation of several physiological pathways associated with ovarian development. Importantly, we have identified the target of miR-309, which is the gene encoding the transcription factor homeobox protein SIX4, and elucidated the role of SIX4 in ovarian follicle growth and maturation. miR-309–targeted degradation of SIX4 mRNA is required for appropriate initiation of the blood feeding-triggered phase of ovarian development. Thus, miR-309 serves as a regulatory switch permitting a stage-specific degradation of the ovarian SIX4 mRNA, permitting a shift from previtellogenic to postvitellogenic phases of ovarian development.
Results
Spatiotemporal Expression Profile of miR-309 in the Female Mosquito.
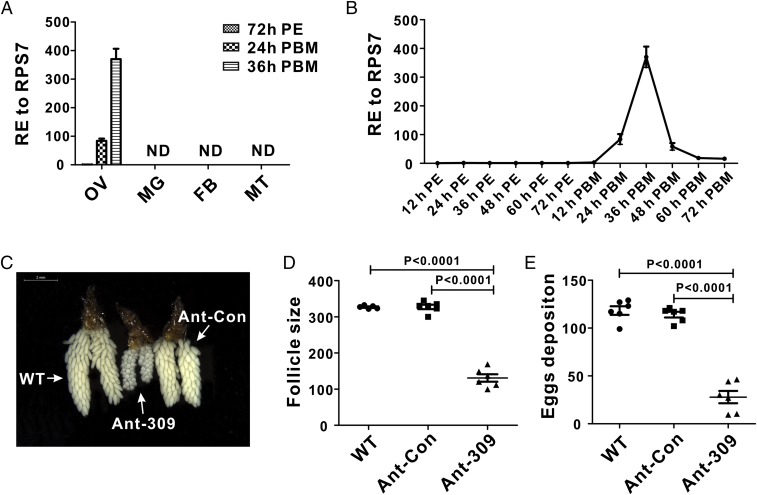
As the first step in exploring possible functions of miR-309 in the mosquito A. aegypti, the tissue distribution of mature miR-309 was determined, and results showed that mature miR-309 was specifically expressed in the ovaries of blood-fed mosquitoes (Fig. 1A). However, it could not be detected in other tissues—fat body, gut, or Malpighian tubule—either before or after blood feeding. We then investigated the time-course expression of this miRNA during ovarian development by means of quantitative real-time (qRT)-PCR analysis. In the ovary, the abundance of mature miR-309 was low during the previtellogenic stage (12–72 h after eclosion, PE), but elevated after blood feeding. It increased 84-fold at 24 h after blood meal (PBM) and peaked at 36 h PBM, with up-regulation of 370-fold in comparison with that of 72 h PE; then it declined sharply at 48 h PBM and later (Fig. 1B). Therefore, specific expression and extremely high induction of miR-309 in the ovary suggest that it plays an important role in mosquito ovarian development.
Fig. 1.
Spatiotemporal expression profile of miR-309 and effect of miR-309 depletion on ovarian development in the female A. aegypti. (A) Relative expression (RE) of miR-309 was analyzed in the ovary (OV), midgut (MG), fat body (FB), and Malpighian tubule (MT) at 72 h PE, and 24 h and 48 h PBM. (B) Relative expression profile of mature miR-309 during ovarian development. The abundance of mature miR-309 was quantified at 12–72 h PE and 12–72 h PBM. (C) Ovaries were dissected from WT (wild-type), Ant-309–, and Ant-Con–injected female mosquitoes at 36 h PBM and were visualized under the Leica M165FC stereo microscope. (Scale bar: 2 mm.) (D) Average follicle size of ovaries isolated from WT, Ant-309–, and Ant-Con–injected female mosquitoes. (E) Egg numbers deposited per female individual from WT, Ant-309–, and Ant-Con–injected female mosquitoes. Data represent six biological replicates with 8–10 individuals in each replication and are shown as mean ± SEM.
Antagomir Silencing of miR-309 Results in the Growth Arrest and Heterogeneous Development of the Ovary.
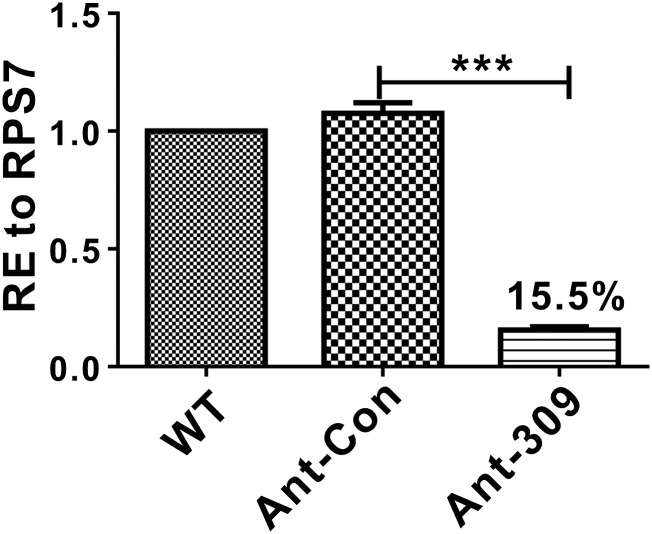
To further investigate the role of miR-309 in the female mosquito, the sequence-specific antagomir for miR-309 (Ant-309), chemically modified antisense oligonucleotides, and a randomly mutagenized “missense” antagomir for control (Ant-con) were designed to knockdown the endogenous miR-309 in vivo. Each female mosquito was injected with 50 pmol Ant-309 or Ant-con at 24 h PE, and the knockdown efficiency was evaluated by using quantitative real-time PCR (qRT-PCR) at 36 h PBM. The results showed that the mature miR-309 level was successfully depleted to 15.5% after Ant-309 injection, in contrast to the Ant-con–injected and wild-type (WT) groups, in which no significant change was observed (Fig. S1).
Fig. S1.
Knockdown of miR-309 expression by injection of antigomir-309. Percent expression of miR-309 was quantified in the WT, antigomir control-, and antigomir-309–injected female mosquito ovarian at 36 PBM. The data presented as mean ± SEM, ***P < 0.001.
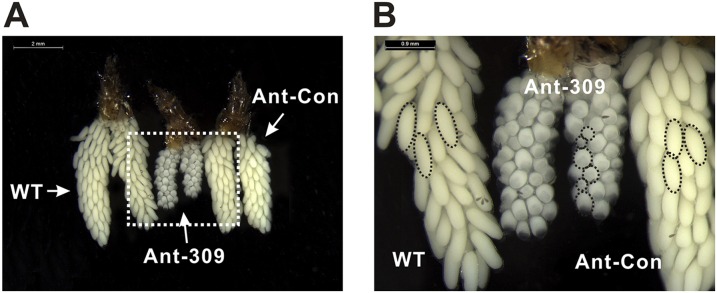
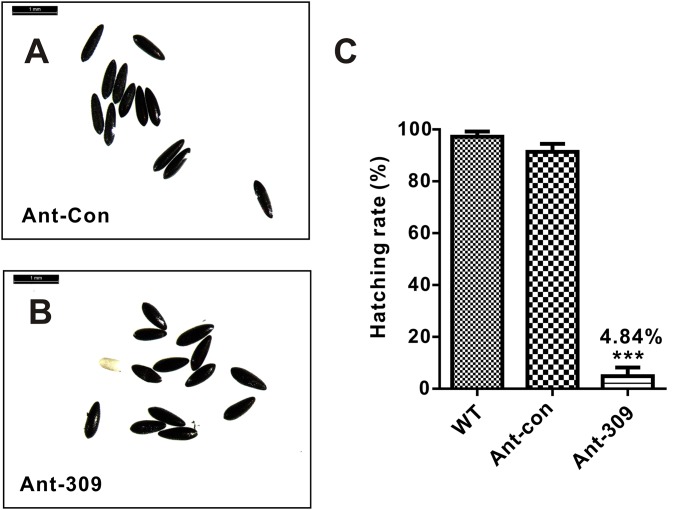
Next, we analyzed the phenotypic manifestations in miR-309–depleted female mosquitoes, which showed multiple defects in ovarian development and embryogenesis. The blood feeding-activated ovarian growth was dramatically arrested after miR-309 depletion (Fig. 1C), and the size of primary follicles in miR-309-depleted mosquitoes was much smaller (130.9 µm on average) than that in the WT or Ant-con–treated mosquitoes (322.8–329.6 µm on average, Fig. 1D) at 36 h PBM. Although not visible in the WT and Ant-con groups, the nurse cells were still developed and occupied approximately one to two quarters of the entire follicle volume in the Ant-309–treated ovary (Fig. S2). In addition, the Ant-309–treated follicles displayed a significant heterogeneity in size, suggesting that miR-309 is associated with regulation of follicle development in ovaries. Furthermore, miR-309 depletion dramatically compromised the egg deposition. Ant-309–treated females laid only approximately 27.8 eggs per mosquito, whereas the WT and Ant-con–treated adults laid 114–118.3 eggs, on average (Fig. 1E). miR-309 depletion affected the egg shape and consequent hatching rate. Eggs from WT and Ant-con–treated mosquitoes exhibited a normal elongated elliptical shape and a hatching rate of 87–95%, whereas the eggs from Ant-309–treated mosquitoes shorted in major axis and only 4.83% of them hatched (Fig. S3).
Fig. S2.
Phenotypic analysis of ovaries dissected from WT, Ant-309–injected, and Ant-Con–injected female mosquitoes at the 36-h PBM with scale bars of 2 mm (A) and 0.9 mm (B), respectively. The dashed circles indicate the follicle shape and size. The nurse cells were visible in the Ant-309–treated ovary.
Fig. S3.
The miR-309 depletion affected the shape and egg hatching rate. (A and B) Morphological comparison of eggs lied from Ant-con– and Ant-309–injected female mosquito. The eggs were visualized by using the Leica M165FC stereo microscope. (Scale bar: 1 mm.) (C) Hatching rate analysis of eggs obtained from WT, Ant-con– and Ant-309–injected female mosquitoes. Data represented three biological replicates with three technician replicates and were shown as mean ± SEM, ***P < 0.001.
Mutation of miR-309 by CRISPR/Cas9 Causes Severe Defects in Ovarian Development.
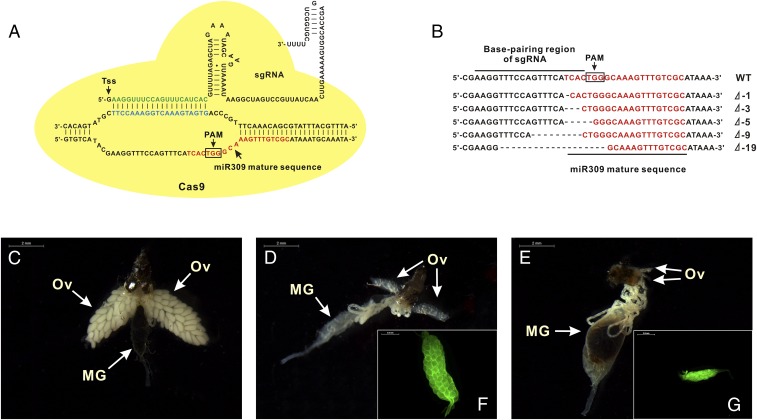
To further decipher the role of miR-309 in ovarian development, miRNA-specific mutations in mosquitoes were generated by means of CRISPR/Cas9 system. As a highly efficient and flexible genome-editing method, CRISPR/Cas9 can achieve accurate targeting specificity through base-pairing of a single guide RNA (sgRNA) to the target genomic region (20, 21). Herein, we developed a miRNA-targeted genomic mutation by designing miR-309–specific sgRNA in A. aegypti. At first, miR-309–specific sgRNA was designed by searching miR-309 mature sequences with protospacer-adjacent motifs (PAMs) of “NGG,” where N refers to any nucleotide. The base-pairing region was required to be 20 bp in length, excluding the PAM, for targeting accuracy. An additional 5′ terminal guanine was included to facilitate transcription by T7 RNA polymerase (Fig. 2A). Next, off-target effects of sgRNA were tested by using several publicly available bioinformatics tools, and no off-target binding sites were found at other gene regions, showing a high specificity of this sgRNA.
Fig. 2.
Genomic disruption of miR-309 by CRISPR/Cas9 blocked ovarian development in mosquitoes. (A) The graphical representation of scaffold of miR309-specific sgRNA. Tss indicated the transcriptional start site at the 5′-terminal end to facilitate in vitro transcription by T7 RNA polymerase. The sequence in green is the base-pairing region of sgRNA that targets miR-309 mature sequence, which is close to protospacer-adjacent motifs (PAMs) of “NGG” nearby. (B) Sequence alignment of sgRNA-targeted genomic region. (C and D) Ovaries were dissected from WT female mosquitoes at 36 h PBM and 72 h PE. (E) Ovaries were dissected from miR-309 mutant female mosquitoes. MG, midgut; Ov, ovary. (F and G) Alexa Fluor 488-labeled phalloidin staining ovaries from WT at 72 h PE and miR-309 mutant at 36 h PBM. The fluorescence imaging was visualized under the Zeiss microscope. (Scale bar: 0.5 mm.)
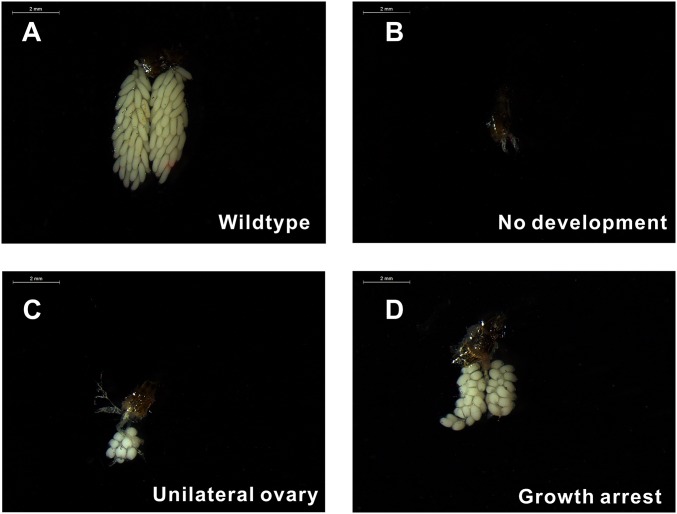
Embryos were injected with a mixture of recombinant Cas9 protein at 333 ng/µL and sgRNA at 40 ng/µL, which has been optimized to compromise between high survival rate and high mutagenesis level in A. aegypti (22). Approximately 51.5% of injected embryos survived to adulthood and were consequently collected to verify whether the miR-309 genomic locus could be effectively disrupted by using the CRISPR/Cas9 technique. We performed the high resolution melting (HRM) to screen for the genomic variation surrounding the sgRNA target site, showing a 64.1% mutagenesis rate (Fig. S4). Our Sanger sequencing further revealed highly polymorphic mutations in sgRNA-injected individuals, where the deletion ranged from 1 bp to 19 bp at the mature sequence and flanking regions of the miR-309 locus (Fig. 2B), demonstrating successful genomic disruption by the CRISPR/Cas9 system. To analyze the possible phenotypic manifestations of ovarian development, ovaries were dissected from miR-309 mutants at 36 h PBM, and results showed that almost half (26 of 53) displayed various defects in their development (Table 1). The phenotypic mutation rate was slightly lower than the genomic mutagenesis rate detected by HRM. Among CRISPR/Cas9 mutants, 41.2% of females displayed a complete block in ovarian development, and their ovaries were observed to be much smaller than the WT PE ovaries (Fig. 2 C–E). Staining with Alexa Fluor 488-labeled phalloidin further revealed that ovaries of miR-309 mutants failed to develop the primary follicles that normally form during the previtellogenic stage in WT (Fig. 2 F and G), strongly suggesting the critical role of miR-309 in initiating formation of primary follicles. Furthermore, we observed other developmental defects in the female miR-309 mutants (Fig. S5). Of all of the mutants, 23.5% showed a delayed developmental phenotype, in which the ovaries contained only a few follicles smaller than those in WT, and 11.8% had a unilateral ovary—one side of the ovary had a few small round follicles and development of the other side was completely blocked. However, gender bias of mosquito (female:male ratio was approximately 53:50) and blood digestion were unaffected after CRISPR/Cas9 disruption of miR-309 (Table 1 and Fig. 2 C–E). We used CRISPR/Cas9 of kynurenine 3-monooxygenase (KMO) as a control. CRISPR/Cas9 mutants of KMO gave rise to the white-eye phenotype but influenced neither the ovarian development nor blood digestion (Fig. S6). Taken together, the miR-309 mutants generated by CRISPR/Cas9 highlight a crucial role of this miRNA in development of ovarian follicles in female mosquitoes.
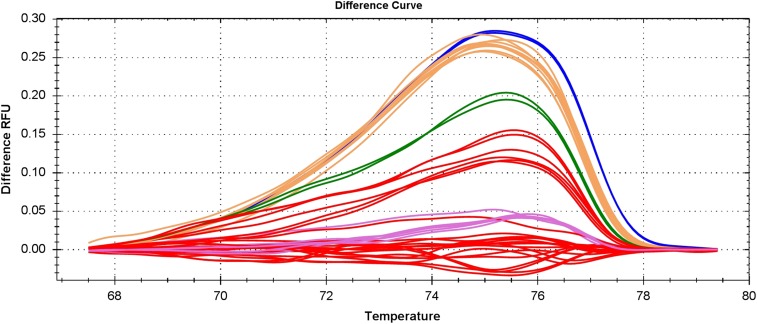
Fig. S4.
HRM analysis of genomic mutation surrounding the sgRNA target site. HRM reactions were performed on Bio-Rad CFX 96 (Bio-Rad), and the difference melting curves were visualized by using Precision Melt Analysis Software.
Table 1.
Individual statistics of the phenotype observed in miR-309 mutants by CRISPR/Cas9
| Phenotype | No. of individuals |
| Normal development | 22 (7) |
| Complete arrest | 14 (14) |
| Delayed development with few round follicles | 8 (7) |
| Unilateral ovary | 4 (4) |
| No blood feeding | 4 (2) |
| Mortality | 1 (0) |
| Total females | 53 (34) |
| Injected embryo | 200 |
| Survived adults | 103 (66) |
The numbers in parentheses indicate the mutants that were detected by HRM.
Fig. S5.
Transgenic disruption of miR-309 by CRISPR/Cas9 system resulted in various severe defects in mosquito ovaries. Ovaries dissected from WT (A) and miR-309 mutants (B–D) were visualized at the 36-h PBM by using the Leica M165FC stereo microscope. (Scale bar: 2 mm.)
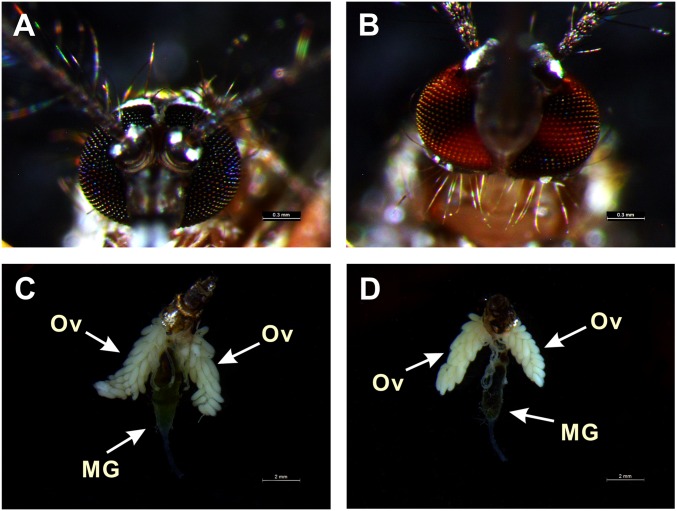
Fig. S6.
CRISPR/Cas9 mutant of KMO had a normal phenotype of ovarian development and blood digestion. White-light photographs of eyes form WT (A) and CRISPR/Cas9 KMO mutants (B) of A. aegypti. Ovaries dissected 36-h PBM from WT (C) and CRISPR/Cas9 KMO mutants (D) are visualized by using the Leica M165FC stereo microscope. (Scale bars: A and B, 0.3 mm; C and D, 2 mm.)
Global Expression Profiling of A. aegypti Ovaries in Response to miR-309 Depletion.
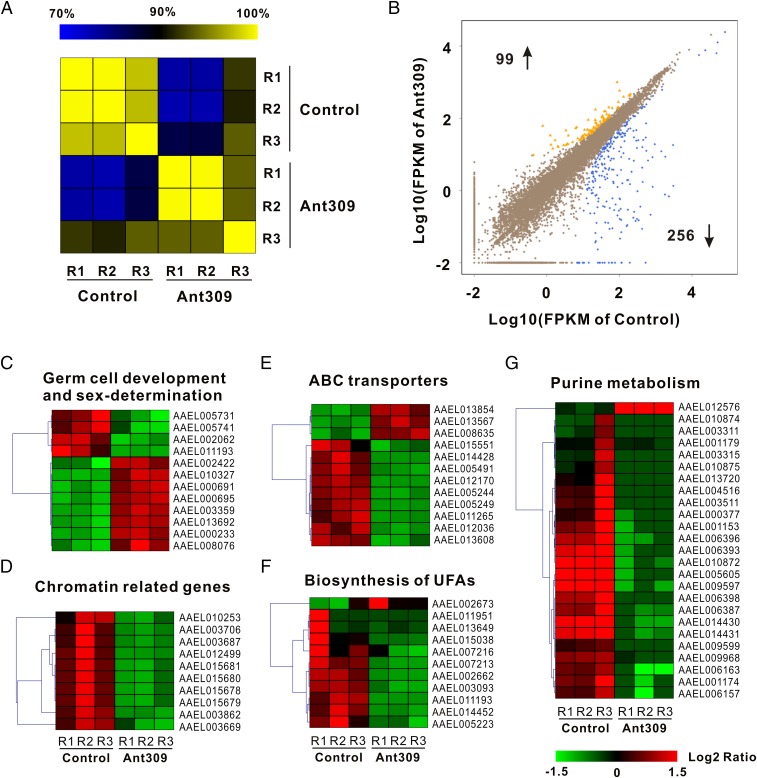
To identify the molecular mechanism underlying control of ovarian development by miR-309, transcriptomic expression profiling of ovaries in response to miR-309 antagomir knockdown was performed by means of RNA-seq quantification. Ovaries were collected at 36 h PBM and Illumina sequencing libraries were constructed by using mRNA from ovaries of Ant-309– and Ant-con–injected mosquitoes, with three biological replicates. We obtained 14,562,516 and 16,911,122 clean reads on average from samples of Ant-309–depleted and control groups, respectively (Table S1). After alignment by Bowtie, 76.14–76.67% and 77.82–78.01% unique reads were mapped into the reference genome of A. aegypti. The mapped reads were used to quantify the expression profiling through methods of FPKM (fragments per kilobase of transcript per million mapped fragments), by which different gene lengths and sequencing discrepancies will be eliminated (Dataset S1). The correlation among three biological replicates in each group was then assessed based on the whole genomic FPKM value by using the Pearson method. The heatmap showed that the square of correlation value ranged from 0.974 to 0.999 and from 0.938 to 0.999 in the Ant-309–depleted and control groups, respectively (Fig. 3A). These values are higher than what the standard Encode plan recommends (≥0.92), indicating reliable replications and reasonable samples. To obtain differentially expressed genes (DEGs) in response to miR-309 depletion, the DEGs were screened according to the Noiseq method, which can be effective in controlling the rate of false discoveries (23). With a cutoff of |log2 (RPKM ratio)|≥ 1 and divergent probability ≥0.8, a total of 355 DEGs were identified in miR-309–depleted ovaries, including 99 up-regulated and 256 down-regulated DEGs (Fig. 3B and Dataset S2).
Table S1.
Summary of alignment and mapping statistics of RNA-seq data
| Sample | Total reads | Total bases | Total mapped reads, % | Unique match, % | Multiposition match, % | Total unmapped reads, % |
| Control_R1 | 15,022,002 | 751,100,100 | 88.01 | 76.67 | 11.35 | 11.99 |
| Control _R2 | 14,828,707 | 741,435,350 | 88.29 | 76.46 | 11.83 | 11.71 |
| Control _R3 | 13,836,840 | 691,842,000 | 88.22 | 76.14 | 12.08 | 11.78 |
| Ant-309_R1 | 19,012,778 | 950,638,900 | 87.63 | 78.01 | 9.62 | 12.37 |
| Ant-309_R2 | 15,155,446 | 757,772,300 | 87.63 | 77.87 | 9.76 | 12.37 |
| Ant-309_R3 | 16,565,142 | 828,257,100 | 87.58 | 77.82 | 9.77 | 12.42 |
Fig. 3.
Global expression profile in response to miR-309 depletion was analyzed by means of RNA-seq quantification. (A) Heatmap showing the square of correlation value from three biological replications in control antagomir and antagomir-309 groups. The square of correlation value was assessed by using the Pearson correlation. R1, R2, and R3 indicate three independent biological replications. (B) Scatter plot presents the DEGs in ovaries after miR-309 depletion. DEGs were calculated based on an algorithm of Noiseq, with a cutoff of |log2 (RPKM ratio) | ≥1 and divergence probability ≥0.8. (C–G) Five functional gene clusters involved in ovarian development were regulated upon miR-309 depletion, including germ cell development and sex-determination, chromatin-related genes, ATP-binding cassette transporter (ABC transporters) system, biosynthesis of unsaturated fatty acids (UFAs) and synthesis, and purine metabolism pathway. The color code indicates the fold change of the gene abundance in the form of a logarithm. The RPKM of genes is normalized in each row, and the dendrograms are constructed based on an algorithm of hierarchical clustering.
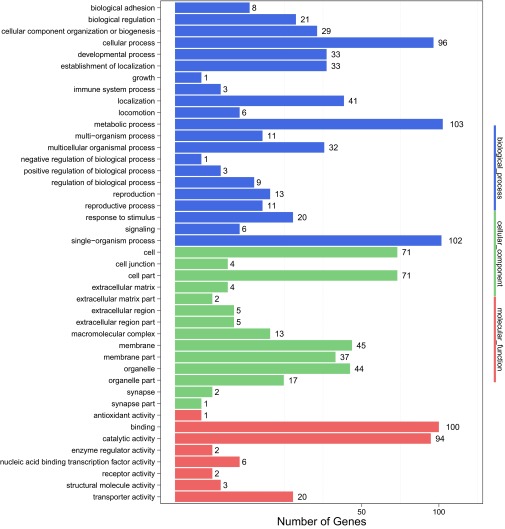
To obtain functional classification, these DEGs were assigned into three main GO (Gene Ontology) categories: molecular functions (156, 36.8%), cellular components (108, 25.5%), and biological processes (160, 37.7%), which were further organized into 43 subcategories (Fig. S7). We also mapped the annotated genes into pathways by using the KEGG Automatic Annotation Server (KAAS). In total, 227 genes (63.9%) could be accessed with a Ko number (Dataset S2) and were significantly enriched in several pathways, including physiological activities associated with ovarian development. Knockdown of miR-309 changed the expression pattern of genes related to germ-cell development and sex determination. For example, forkhead box protein L (FOXL, AAEL005741, and AAEL005731) and nuclear receptor subfamily 5 group A member 3 (NR5A3, AAEL002062), the orthologs of which, in mammals, played a central role in ovarian determination and development (24), were down-regulated in miR-309–depleted ovaries, whereas other genes associated with maintenance of female germ-line stem cells, including homeobox protein SIX4 (AAEL010327), Bruno (AAEL000695 and AAEL000691), oo18 RNA-binding protein (AAEL002422), and Aubergine (AAEL013692 and AAEL008076), were significantly induced after miR-309 depletion, strongly suggesting that miR-309 contributes to the initial differentiation and growth of female germ-line stem cells (Fig. 3C). Meanwhile, 10 chromatin-related genes, including histone H2A (AAEL012499) and histone H2B (AAEL015678), were significantly repressed in ovaries of female mosquitoes after miR-309 depletion (Fig. 3D), suggesting a potential role of miR-309 in controlling the nucleosome structure required for ovarian growth and maturation. Furthermore, RNA-seq results revealed that miR-309 depletion disrupted the ATP-binding cassette transporter (ABC transporters) system, in which many DEGs (9 of 12) were decreased, 6.48-fold on average compared with control group (Fig. 3E). As the largest transporter system, ABC transporters carried out translocation of various substrates across membranes, including metabolic products, lipids, and others (25). Hence, suppression of ABC transporters may reduce nutrient uptake from hemolymph to developing follicle. Moreover, RNA-seq results showed that miR-309 depletion affects genes associated with the biosynthesis of unsaturated fatty acids (UFAs) and purine metabolism. In mature mosquito oocytes, the majority of the fatty acids are present in an unsaturated form, providing the necessary energy storage to fuel the consequent embryogenesis (26). Our results showed that most DEGs involved in regulation of genes involved in biosynthesis of UFAs were down-regulated after miR-309 silence (Fig. 3F), which might lead to insufficient energy storage, also consistent with low hatching rate of these eggs in miR-309–depleted mosquitoes. Similarly, almost all 25 DEGs mapped into the purine metabolism pathway, by which multiple nucleotides could be synthesized, were decreased in the miR-309 knocked-down ovaries, except for the AAEL012576 gene (Fig. 3G), suggesting a strong repressive effect on the nucleotide biosynthesis.
Fig. S7.
GO classification of differentially expressed genes upon miR-309 depletion in ovaries. All DEGs were assigned into three main GO terms, including molecular function, cellular component, and biological process, and further subdivided into 43 subcategories.
The Gene Encoding Homeobox Protein SIX4 Is a Direct Target of miR-309.
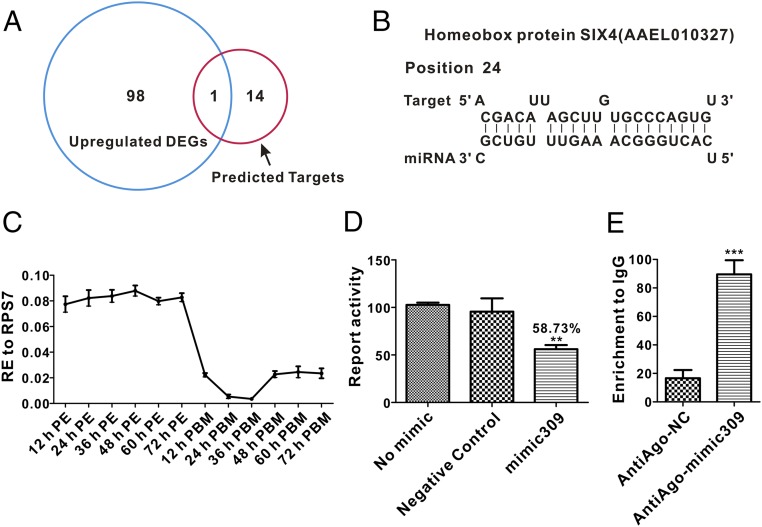
To obtain putative gene targets of miR-309, we compared ovarian up-regulated DEGs after miR-309 knockdown with in silico predicted gene targets. First, multiple bioinformatics tools, including miRanda (27), PITA (28), TargetScan (29), RNAhybird (30), and one program developed “in house” (31), were used to predict putative targets of miR-309, as described (16). Fifteen candidate targets were generated by using these five computational programs (Table S2). Of these genes, only one, Homeobox protein SIX4 (AAEL010327), was found among miR-309–depleted up-regulated DEGs. The sequence alignment showed that SIX4 gene 3′−YTP contained a putative binding site for miR-309 (Fig. 4 A and B). It has been shown that in Drosophila melanogaster, the ortholog of SIX4 is required for the migration of primordial germ cells (PGCs) and somatic gonadal precursors (SGPs) during gonadogenesis (32), also highlighting its regulatory role in gonadal development. Expression profile analysis showed that elevated transcript abundance of SIX4 mRNA in A. aegypti ovaries was maintained during the entire previtellogenic stage of 12–72 h PE, but declined dramatically after blood feeding, with the lowest expression at 36 h PBM (Fig. 4C), displaying strong negative correlation with miR-309 expression.
Table S2.
Multialgorithm prediction of miR-309 target in A. aegypti 3′-UTRs
| Gene ID | Gene name | Species | Algorithm* |
| AAEL004176-RA,RB | Microtubule binding protein | A. aegypti | IN, TS, PITA, R, RH |
| AAEL004513-RA | Neurotransmitter gated ion channel | A. aegypti | IN, TS, PITA, R, RH |
| AAEL004738-RA | Methionine aminopeptidase | A. aegypti | IN, TS, PITA, R, RH |
| AAEL005886-RA | Protein coding | A. aegypti | IN, TS, PITA, R, RH |
| AAEL006880-RA,RB,RC | Rab32 | A. aegypti | IN, TS, PITA, R, RH |
| AAEL008393-RA | Phosphatidylserine synthase | A. aegypti | IN, TS, PITA, R, RH |
| AAEL009189-RA | Encore protein | A. aegypti | IN, TS, PITA, R, RH |
| AAEL010327-RA | SIX/sine homebox transcription factors† | A. aegypti | IN, TS, PITA, R, RH |
| AAEL010814-RC | Isocitrate dehydrogenase | A. aegypti | IN, TS, PITA, R, RH |
| AAEL010823-RA | ATP synthase delta chain | A. aegypti | IN, TS, PITA, R, RH |
| AAEL014379-RA | Adrenodoxin | A. aegypti | IN, TS, PITA, R, RH |
| AAEL017301-RA | Hypothetical protein | A. aegypti | IN, TS, PITA, R, RH |
Algorithms used in this study include IN, in-house; PITA; R, miRanda; RH, RNAhybrid; TS, TargetScan.
Homolog to Drosophila homeobox SIX4.
Fig. 4.
The gene encoding the Homeobox protein SIX4 is a direct target of miR-309. (A) Venn plot shows the overlapping up-regulated DEGs after miR-309 knockdown with in silico-predicted gene targets by using five computational programs. (B) The putative miR-309 binding site in 3′-UTR of AAEL010327 is predicted by using software. (C) The expression profile of the SIX4 mRNA during ovarian development in the mosquito. (D) Luciferase reporter assay showing that miR-309 directly degrades the 3′-UTR of AAEL010327 in vitro. (E) Immunoprecipitation (RIP) assay demonstrating that miR-309 directly targets SIX4 in vivo. The SIX4 mRNA was significantly enriched after immunoprecipitation with antibodies against the Anti-Ago. Data represent the fold change to the IgG-precipitated mRNA with three samples, and each sample in triplicate. The error bars shown in C–E indicate the mean ± SEM, **P < 0.01, ***P < 0.001.
Next, we assessed whether miR-309 could degrade SIX4 mRNA or inhibit its translation in vitro. The SIX4 3′-UTR–fused reporter was constructed by integrating SIX4 3′-UTR into the downstream of Renilla luciferase reporter of psiCheck-2 vector. When cotransfected into Drosophila Schneider (S2) cells with the mimic of miR-309, activity of the SIX4 reporter was observed to yield only 58.73% of the activity of the negative control mimic (Fig. 4D). Moreover, the binding status of miR-309 with SIX4 mRNA in vivo was validated by RNA immunoprecipitation (RIP) assay with antibodies against A. aegypti Argonaute 1 (Anti-Ago), which is the key component of RNA-induced silencing complex mediating miRNA-induced mRNA degradation. The RIP result demonstrated that SIX4 mRNA was significantly enriched in the Anti-Ago immunoprecipitated RNAs from mimic miR-309–injected ovary relative to that in the negative control mimic injected samples (Fig. 4E). Taken together, these data strongly suggest that SIX4 was a direct target of miR-309.
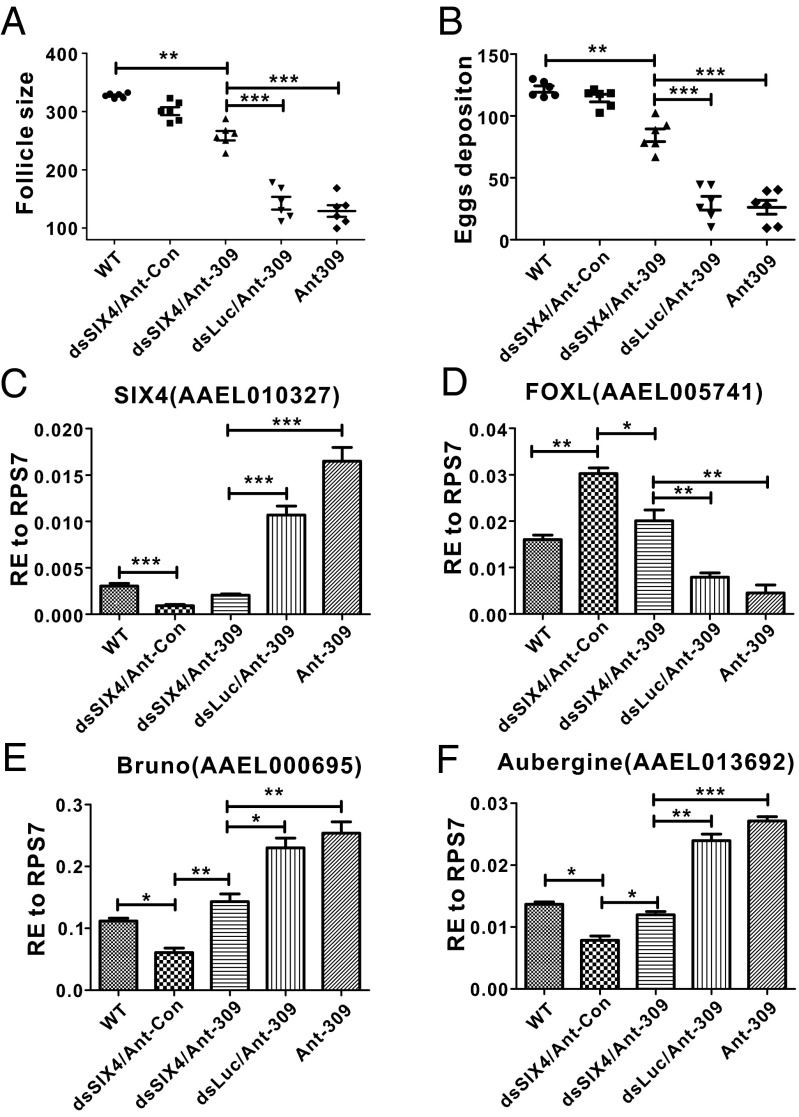
To further verify whether SIX4 is the authentic miR-309 target in the ovary, the phenotypic rescue experiments were conducted by RNA interference (RNAi) silencing of SIX4 in Ant-309–injected female mosquitoes. Adverse phenotypes caused by miR-309 miRNA depletion were expected to be alleviated after silencing its major target gene. Previously, phenotypic defects resulting from miRNA deletions were successfully rescued after RNAi silencing their main target genes in A. aegypti (16, 17). In our rescue experiments, silencing SIX4 was able to significantly recover phenotypic manifestations caused by miR-309 depletion, such as smaller follicle size and significantly reduced egg numbers. The average follicle size from Ant-309 and dsSIX4-coinjected ovaries reached 258.6 µm at 36 h PBM, which was larger than that in Ant-309- or Ant-309/dsLuc–injected mosquitoes (Fig. 5A). Coinjection of Ant-309 and dsSIX4 into female mosquitoes also greatly increased the egg deposition number (84.4 eggs per individual) compared with Ant-309 or Ant-309/dsLuc groups, whereas it was still lower than that in the WT or dsSIX4/Ant-con groups (Fig. 5B). We also tested the expression profiles of SIX4 and several other key genes related to germ cell and ovarian development, including FOXL (AAEL005741), Bruno (AAEL000695), and Aubergine (AAEL013692), to verify whether silencing SIX4 could restore their transcript levels. Our results first showed that, compared with WT or dsLuc/Ant-309 ovaries, SIX4 mRNA dramatically decreased in both dsSIX4/Ant-con and dsSIX4/Ant-309 groups, indicating the effective knockdown of SIX4 through its RNAi (Fig. 5C). Next, it was observed that SIX4 RNAi restored the effect of the miR-309 depletion that had caused FOXL suppression, indicating its negative effect on expression of FOXL (Fig. 5D). As an evolutionary conserved transcription factor in female sex determination in vertebrates (33), FOXL was identified to control mosquito ovarian development and egg laying in A. aegypti (34). Thus, the repression of FOX L by SIX4 supports its constrained role in ovarian development. In Drosophila, Bruno and Aubergine were shown to be essential for maintaining female germ-line cells by forming silencing transcriptional complexes (35, 36). mRNA levels of Bruno and Aubergine were down-regulated after silencing of SIX4, but their proper expressions were observed in the dsSIX4/Ant-309 ovaries (Fig. 5 E and F). Therefore, our results have demonstrated that SIX4 is the major target of miR-309 and plays an important role in maintaining resting status of primary follicles and restricting ovarian growth.
Fig. 5.
RNAi of Homeobox protein Six4 rescues miR-309 depletion phenotype. Average follicle size of ovaries (A) and egg numbers deposited per female individual (B) were analyzed from WT; dsSix4/Ant-Con, coinjection of Six4 dsRNA and control antagomir; dsSix4/Ant-309, coinjection of Six4 dsRNA and antagomir of miR-309; dsLuc/Ant-309, coinjection of Luciferase dsRNA and antagomir of miR-309; and Ant-309 injected female mosquitoes. (C–F) The expression profiles of four genes, including SIX4 (AAEL010327), FOXL (AAEL005741), Bruno (AAEL000695), and Aubergine (AAEL013692), were quantified in different experimental groups, as above. RE used on y axis is relative expression to ribosomal protein S7 (RPS7). Data represent six biological replicates with 8–10 individuals in each replication and are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
It has been shown in Drosophila that the miR-309 cluster could promote mRNA turnover during the maternal-to-zygotic transition and, thus, lead to reduced survival and delayed larval development (18). Here, our results have demonstrated that miR-309 acts to initiate ovarian development appropriately in the mosquito A. aegypti. Using antagomir-based miR-309 knockdown in vivo, severe defects linked to ovarian follicle growth and egg deposition were observed. Characteristically, ovarian primary follicles were dramatically arrested and grew in a nonsynchronized way after miR-309 depletion. Normally, the nurse cells undergo cytoplasmic dumping into the oocyte and cell death at 36 h PBM. However, they were intact and visible in miR-309–silenced ovaries, implicating the critical role of miR-309 in programming correct ovarian development. To further define the regulatory roles of this miRNA, miR-309–specific mutations of mosquitoes were generated by using the CRISPR/Cas9 system, by which accurate genome editing with high efficiency has been achieved in many species, including mosquitoes (22). Herein, we successfully constructed miR-309–specific mutants in A. aegypti, contributing to deciphering its physiological function. miR-309 mutants exhibited more severe defects than observed in Ant-309–injected individuals. Almost half of the G0 mutants were completely sterile and failed to construct the primary follicles that were preformed before blood feeding in WT, revealing that miR-309 promotes the differentiation and formation of primary follicles from PGCs. Meanwhile, several other variations of ovarian development defects, such as developmental delay and unilateral follicle growth, were found in miR-309 mutants, which might be explained by different deletions in the target region or mosaics of G0 progeny. The off-target effects of CRISPR/Cas9 were excluded by genomic comparison, sequencing of putative binding region, indicating the specific effect of miR-309 mutation on female reproduction.
To gain comprehensive insight into how miR-309 affects the development, global gene expression profiling was performed by means of RNA-seq after miR-309 depletion in mosquito ovaries. Our results revealed that many genes associated with ovarian development were affected as a result of miR-309 depletion. FOXL and NR5A, the genes that have been implicated in early female determination and ovary differentiation, were down-regulated in the miR-309–depleted A. aegypti ovaries (24, 37). Conversely, several female germ-line genes, such as oo18 RNA-binding protein (Orb, AAEL002422), Bruno (AAEL000695 and AAEL000691) and aubergine (AAEL013692 and AAEL008076), were activated after miR-309 depletion. Previous studies showed that the germ-line protein Orb was required for oocyte differentiation and restricts oocyte meiosis in Drosophila (38). Hence, these observations support the important role of miR-309 in mosquito oocyte development.
Meanwhile, miR-309 depletion also demonstrated an inhibitory effect on the membrane transport system and purine metabolism. In mosquitoes, lipid reserves are essential for egg maturation, providing ∼90% of the energy required during embryogenesis. However, almost all of the lipids in the egg are imported through a transmembrane transport system, originally synthesized by the fat body (39). A strongly negative effect on transporter systems was observed in miR-309–depleted ovaries—in particular, a dramatic repression of dozens of ABC transporters genes. As the largest and evolutionarily highly conserved cellular transmembrane transport system, ABC transporters have been proven to facilitate the ATP-dependent translocation of lipids or lipid-related substrates across membranes in many species, including insects (25). Indeed, miR-309 depletion was able to suppress lipid uptake in blood feeding-activated oocytes. Meanwhile, the pathway for biosynthesis of UFAs was supposed to be activated because 70% of the fatty acids were unsaturated in the mature oocytes of A. aegypti (40). However, this theory was contradicted by miR-309 depletion, which caused a decrease in many crucial enzymes involved, such as Acyl-CoA dehydrogenases (ACADs, AAEL014452) and Stearoyl-CoA desaturase (SCD, AAEL007213) (41), likely resulting in fewer UFAs in mature oocytes and insufficient energy for embryogenesis. In addition, mature eggs contained large amounts of RNAs in the form of ribosomes, transfer RNA, and maternal mRNA to support programmed protein synthesis during early embryogenesis due to the absence of zygotic transcription (42). Purine metabolism is the one basic de novo synthesis pathway of purine nucleotides, starting from 5-phosphoribosyl-1-pyrophosphate, and finally purine nucleoside triphosphates such as ATP and GTP are produced after a series of steps, providing the basic blocks for RNA synthesis (43). As expected, dozens of genes that take part in the purine metabolism pathway were significantly down-regulated in the miR-309–depleted oocytes, indicating that blocking RNA synthesis might be one way to regulate egg maturation.
Because mRNA levels of hundreds of genes were affected by miR-309, the authentic and principal targets by which it can carry out its crucial role in ovarian development remained unclear. Screening the putative miRNA-309 targets was conducted by comparison of overlapping up-regulated DEGs after miR-309 knockdown with in silico predicted target genes. Surprisingly, only one gene encoding the transcription factor Homeobox protein SIX4 was obtained based on this approach, and its 3′-UTR contained an almost perfectly matched binding sequence with miR-309. Multiple data further confirmed that SIX4 is a direct target of miR-309 in vivo, including their negatively correlated expression during the course of ovarian development, degradation of SIX4 3′-UTR–contained reporter by miR-309 mimic in cell lines, and enrichment of miR-309–SIX4 binding complex by Argonaute immunoprecipitation. Importantly, SIX4 RNAi was able to significantly rescue phenotypic manifestations from miR-309 depletion that caused follicle growth arrest and reduced egg deposition. These results demonstrate that SIX4 is the direct and principal target that mediates the regulatory function of miR-309. Previous studies indicate that SIX4 is crucial for early gonadogenesis. In mice, the knockout of SIX4 ortholog results in a reduced number of gonadal precursor cells and diminished size of gonads (44), whereas in Drosophila, it played an important function in the migration of gonadal precursors (32). Although the exact role of SIX4 in A. aegypti is still unknown, the concomitant reduction in ovaries has indicated its role in follicle growth and maturation. Correspondingly, Bruno and Aubergine, which are essential factors for repression of the germ-line stem cell differentiation in Drosophila by forming a silenced transcriptional complex (35, 36), have been observed to be down-regulated after knockdown of SIX4, being consistent with its role. Meanwhile, SIX4 also depressed expression of FOXL, supporting its constrained role in ovarian development. In mammals, the orthologs of FOXL controlled ovary-specific differentiation and activated female-specific genes (33). Similarly, FOXL RNAi resulted in reduction of egg deposition in A. aegypti (34). Taken together, degradation of endogenous SIX4 might be required to relieve its suppressive effect on oocyte development and activate female-specific development.
In conclusion, we have revealed the essential role of miR-309 in initiation of ovarian development in A. aegypti by means of miRNA knockdowns using Antagomir and CRISPR/Cas9 systems. Depletion of miR-309 disrupted many physiological activities that are necessary for egg development and maturation, including suppression of the membrane transport system and a decrease of biosynthesis of UFAs and purine metabolism. Moreover, genomic screening of miR-309 targets identified that SIX4 is the principal and authentic target in vivo, and functional evidence demonstrated that miR-309-targeted degradation of endogenous SIX4 is required for appropriate ovarian development in mosquitoes. However, miR-309 played a distinct and nonconserved role in Drosophila, because the mutant formed after depletion of miR-309 was still fertile although it failed to turnover maternal mRNAs during embryogenesis. These differences might reflect a divergent evolution in function and an involvement in species-specific reproductive adaptation.
Materials and Methods
Mosquito Rearing.
Adult A. aegypti mosquitoes were raised at 27 °C and 80% humidity, and mosquito larvae were raised at 27 °C in water with a supplemental nutrition mixture, as described (16). Female mosquitoes were blood-fed on the White Leghorn chickens to initiate ovarian development. All of the experiments followed National Institutes of Health-approved conditions at a certified facility at the University of California, Riverside.
Antagomirs and dsRNA Treatment.
Antagomirs were purchased from Dharmacon by using the RNA module for custom single-stranded RNA synthesis. The antagomir to miR-309 was 5′-mU*mG* mGmCmGmAmCmAmAmAmCmUmUmUmGmCmCmCmAmGmU*mG*mA*mA*mU-Chl-3′, in which “*” is a phosphorothioate backbone, and “m” and “Chl” refer to a -OCH3 group on the 2′ end of the base and cholesterol group, respectively. The control antagomir was constructed as described (15). At 24 h PE, mosquitoes were microinjected into the thorax with 50 pmol dsRNA (100 μM in a volume of 0.5 μL) or antagomir (100 μM). Mosquitoes were allowed to recover for 3–4 d before blood feeding and dissected 24 h after blood feeding. dsRNA was synthesized by using the MEGAscript kit (Ambion) with dsDNA template of Homeobox Six4, and then purified by using MEGAclear Transcription Clean-Up Kit (Ambion). For rescue experiments, mosquitoes were coinjected with 0.5 μL of antagomir/dsRNAmixture with a final concentration of 100 μM antagomir and 1 μg/μL dsRNA.
RNA Isolation and Real-Time PCR.
Total RNA was extracted from ovaries or other tissues by using TRIzol (Invitrogen) according to the manufacturer’s instruction. The integrity of the RNA was assessed by means of agarose gel electrophoresis. cDNAs for miRNAs were reverse transcribed from 1 μg of DNase I (Invitrogen)-treated total RNA by using the miScript II RT Kit (Qiagen), in which the HiFlex Buffer was used so that miRNA and mRNA could be quantified in parallel. The qRT-PCR for mature miRNA was measured by using the miScript SYBR Green PCR kit (Qiagen) according to the manufacturer’s protocol. For mRNA quantitation, qRT-PCR was carried out by using the QuantiFast SYBR Green PCR Kit (Qiagen). Each sample was performed in triplicate, and relative expression was calculated by using the 2-ΔΔCt method through normalized with housekeeping gene (RPS7).
sgRNA Design and Embryonic Injection.
To design sgRNAs, miR-309 mature regions were searched with PAMs of “NGG,” where N refers to any nucleotide. The base-paired region was 20 bp in length, excluding the PAM, and one guanine was added at the 5′ terminal end of the sgRNA sequence to facilitate transcription by T7 RNA polymerase. The potential off-target binding was checked by using two online tools: zifit.partners.org/ZiFiT/ and crispr.mit.edu. The dsDNA template for sgRNA synthesis was generated from template-free PCR, where one specific forward primer and one universal reverse primer overlapped (detailed oligonucleotide sequence is listed in Table S3). The negative control sgRNA was obtained from scrambled sgRNA CRISPR/Cas9 All-in-One Lentivirus and KMO targeted sgRNA, respectively. sgRNA was synthesized and purified by using methods as described above. Commercial recombinant Cas9 protein was obtained from PNA Bio. Microinjection into A. aegypti embryos (preblastoderm eggs) was performed with a mixture of recombinant Cas9 protein at 333 ng/μL and sgRNAs at 40 ng/μL according to protocols described (22, 45). The embryos hatched at 4–5 d after injection and were reared to adults based on procedures described (22, 45). Female mosquitoes were blood fed for egg production and phenotypic analysis.
Table S3.
Oligonucleotides were used for synthesis of sgRNA template
| Primer name | Sequence |
| CRISPR forward primer for miR-309 sgRNA | GAAATTAATACGACTCACTATAGGAAGGTTTCCAGTTTCATCACGTTTTAGAGCTAGAAATAGC |
| CRISPR forward primer for KMO sgRNA | GAAATTAATACGACTCACTATAGGCACAGTACAATCCTCGAATCGTTTTAGAGCTAGAAATAGC |
| CRISPR Universal reverse primer* | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
Each specific forward primer can generate one specific sgRNA when overlapped with universal reverse primer.
Microscopy and Staining of Ovaries.
Dissected ovaries were fixed in APS (Aedes physiological saline; 150 mM NaCl, 4 mM KCl, 1.7 mM CaCl2, 25 mM HEPES buffer, pH 7.0) with 4% (vol/vol) formaldehyde (Pierce) for 30 min and then washed three times with APS-T (Aedes physiological saline with 0.3% Triton X-100). The actin cytoskeleton was stained with phalloidin Alexa Fluor 488 (A12379; Invitrogen) for 15 min and washed three times with APS. Then, fluorescence imaging was performed by using a Zeiss microscope, AxioObserverA1.
High-Resolution Melting Analysis.
The sequencing and HRM of amplicons surrounding the putative CRISPR-Cas9 cutting site were performed to analyze CRISPR-Cas9–induced mutations. Genomic DNA was extracted from adult mosquitoes by using the DNeasy Blood & Tissue Kit (Qiagen). HRM primers were designed to generate amplicons of 80–120 bp covering the CRISPR target site. PCR was performed by using Precision Melt Supermix (Bio-Rad) with a genomic DNA template. Reactions were conducted within 96-well plates and run on a BioRad CFX 96 (Bio-Rad). Melting curves were generated and analyzed by using Precision Melt Analysis Software.
RNA-seq Quantification and Bioinformatics Pipeline.
Total RNA was isolated from ovaries in antagomir-309 or antagomir-control injected mosquitoes. The cDNA library for Illumina sequencing was prepared by following the instruction of the TruseqTM RNA sample prep Kit (Illumina). Briefly, mRNA was enriched from total RNA by using poly-T oligo–attached magnetic beads (Illumina) and subsequently disrupted into fragments (200–700 nt). First-strand cDNAs were then synthesized by using SuperScript II reverse transcriptase, and second-strand cDNAs were synthesized by DNA polymerase I. These cDNA fragments were end-repaired and 3′-adenylated and ligated with adaptors, and then these fragments with adaptors were enriched by means of PCR amplification with Phusion DNA polymerase. The cDNA library was quantified by TBS380 (PicoGreen) and then sequenced on the Hiseq 2000 platform by using the single-end sequencing method.
The raw reads produced by Hiseq 2000 were subjected to quality control to remove adapters, unknown biases, and low quality reads. After filtration, the clean reads were aligned with reference genes of A. aegypti (https://www.vectorbase.org/organisms/aedes-aegypti) by using Bowtie (46). Expression levels of genes were quantified by using the FPKM method (fragments per kilobase of transcript per million mapped fragments), by which different gene lengths and sequencing discrepancy were eliminated and used for comparing DEGs. DEGs were calculated based on the Noiseq method with a cutoff of fold change ≥2 and a divergence probability ≥0.8 (23), showing a good performance in finding true positive rate and controlling the rate of false discoveries. The screened DEGs were annotated by GO and KEGG classification with a Bonferroni Correction of P value ≤0.05.
miRNA Target Prediction.
Five different miRNA target prediction programs—PITA (19), miRanda (20), RNAhybrid (21), TargetScan (22), and an in-house miRNA target prediction algorithm—were used to analyze the putative targets. The detailed parameter setting has been described (16).
Cell Culture and Luciferase Report Assay.
Drosophila S2 cells (Invitrogen) were grown in Schneider’s Drosophila medium (Gibco, Life Technologies) containing 10% (vol/vol) heat-inactivated FBS (Gibco) and 1×Antibiotic-Antimycotic (Gibco) at 28 °C in a humidified incubator. The psiCheck-2 reporters were constructed by inserting the miR-309 putative target 3′-UTR into the psiCheck-2 vector (Promega). Thereafter, 100 ng of psiCheck-2 reporters with 100 nM of synthetic aae-miR-309 miScript miRNA Mimic (Qiagen) or AllStars Negative Control siRNA (Qiagen) were cotransfected into Drosophila S2 cells by using FuGENE HD Transfection Reagent (Promega). Cells were collected and lysed at 48 h after transfection, and luciferase activities were measured by using the dual luciferase reporter assay system (Promega). Each sample was performed in triplicate, and transfections were repeated three times.
RIP.
The RIP assay was performed by using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. In brief, dissected ovaries were collected and homogenized in ice-cold RIP lysis buffer. Antibody linked magnetic beads were prepared through preincubation with 5 μg of custom-made A. aegypti Argonaute 1 antibody (GenScript) or normal mouse IgG (Millipore). The immunoprecipitates were pulled down from homogenates with antibody-linked magnetic beads and then digested with protease K to release the RNAs. Finally, the cDNAs were synthesized by using random hexamers, and target genes were quantified by means of qRT-PCR as mentioned above. In each sample, 10% (vol/vol) of supernatant of the RIP lysate was used as “input,” and the enrichment index to IgG controls referred to the binding ability between miRNA and mRNA.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 AI113729 (to A.S.R.) and China Scholarship Council of Chinese Ministry of Education Scholar Fellowship 201408440009.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609792113/-/DCSupplemental.
References
- 1.Barrett ADT, Higgs S. Yellow fever: A disease that has yet to be conquered. Annu Rev Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- 2.Fauci AS, Morens DM. Zika virus in the Americas–Yet another arbovirus threat. N Engl J Med. 2016;374(7):601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 3.Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Mol Biol. 2005;35(7):661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Raikhel AS, Dhadialla TS. Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol. 1992;37(1):217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- 7.Akbari OS, et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science. 2015;349(6251):927–929. doi: 10.1126/science.aac7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17(3):146–159. doi: 10.1038/nrg.2015.34. [DOI] [PubMed] [Google Scholar]
- 9.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Lucas KJ, Myles KM, Raikhel AS. Small RNAs: A new frontier in mosquito biology. Trends Parasitol. 2013;29(6):295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucas KJ, Zhao B, Liu S, Raikhel AS. Regulation of physiological processes by microRNAs in insects. Curr Opin Insect Sci. 2015;11:1–7. doi: 10.1016/j.cois.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci USA. 2011;108(22):9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci USA. 2013;110(25):10276–10281. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain M, Walker T, O’Neill SL, Asgari S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem Mol Biol. 2013;43(2):146–152. doi: 10.1016/j.ibmb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107(52):22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci USA. 2014;111(40):14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas KJ, et al. MicroRNA-8 targets the Wingless signaling pathway in the female mosquito fat body to regulate reproductive processes. Proc Natl Acad Sci USA. 2015;112(5):1440–1445. doi: 10.1073/pnas.1424408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ninova M, Ronshaugen M, Griffiths-Jones S. Fast-evolving microRNAs are highly expressed in the early embryo of Drosophila virilis. RNA. 2014;20(3):360–372. doi: 10.1261/rna.041657.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18(7):501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 20.Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34(1):78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112(49):E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Reports. 2015;11(1):51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139(6):1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013;24(7):342–350. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 30.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark IB, Jarman AP, Finnegan DJ. Live imaging of Drosophila gonad formation reveals roles for Six4 in regulating germline and somatic cell migration. BMC Dev Biol. 2007;7:52. doi: 10.1186/1471-213X-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggers S, Ohnesorg T, Sinclair A. Genetic regulation of mammalian gonad development. Nat Rev Endocrinol. 2014;10(11):673–683. doi: 10.1038/nrendo.2014.163. [DOI] [PubMed] [Google Scholar]
- 34.Hansen IA, et al. Forkhead transcription factors regulate mosquito reproduction. Insect Biochem Mol Biol. 2007;37(9):985–997. doi: 10.1016/j.ibmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci USA. 2011;108(52):21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124(3):521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 37.Allen AK, Spradling AC. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development. 2008;135(2):311–321. doi: 10.1242/dev.015156. [DOI] [PubMed] [Google Scholar]
- 38.Huynh JR, St Johnston D. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development. 2000;127(13):2785–2794. doi: 10.1242/dev.127.13.2785. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler R, Van Antwerpen R. Lipid uptake by insect oocytes. Insect Biochem Mol Biol. 2006;36(4):264–272. doi: 10.1016/j.ibmb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Ziegler R. Lipid synthesis by ovaries and fat body of Aedes aegypti (Diptera: Culicidae) Eur J Entomol. 1997;94(3):385–391. [Google Scholar]
- 41.Kim JJ, Wang M, Paschke R. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc Natl Acad Sci USA. 1993;90(16):7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berry SJ. RNA synthesis and storage during insect oogenesis. Dev Biol (N Y 1985) 1985;1:351–384. doi: 10.1007/978-1-4615-6814-8_8. [DOI] [PubMed] [Google Scholar]
- 43.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto Y, et al. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev Cell. 2013;26(4):416–430. doi: 10.1016/j.devcel.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 45.Kokoza V, et al. Blocking of Plasmodium transmission by cooperative action of Cecropin A and Defensin A in transgenic Aedes aegypti mosquitoes. Proc Natl Acad Sci USA. 2010;107(18):8111–8116. doi: 10.1073/pnas.1003056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.