Significance
The origin recognition complex (ORC) binds sites from which DNA replication is initiated. By mapping binding sites in human cells, we show that ORC binds selectively to open (DNase I-hypersensitive) regions containing active chromatin marks. There are far more ORC sites in early replicating regions of the genome, and computational simulation based on ORC binding indicates that replication timing is due primarily to ORC density and stochastic initiation of DNA replication from origins. Large genomic regions with a paucity of ORC sites are strongly associated with common fragile sites and recurrent deletions in cancers. Thus, replication origins, replication timing, and replication-dependent chromosome breaks are determined ultimately by the genomic distribution of activator proteins at enhancers and promoters.
Keywords: DNA replication, replication origins, chromatin, replication timing, ORC
Abstract
The origin recognition complex (ORC) binds sites from which DNA replication is initiated. We address ORC binding selectivity in vivo by mapping ∼52,000 ORC2 binding sites throughout the human genome. The ORC binding profile is broader than those of sequence-specific transcription factors, suggesting that ORC is not bound or recruited to specific DNA sequences. Instead, ORC binds nonspecifically to open (DNase I-hypersensitive) regions containing active chromatin marks such as H3 acetylation and H3K4 methylation. ORC sites in early and late replicating regions have similar properties, but there are far more ORC sites in early replicating regions. This suggests that replication timing is due primarily to ORC density and stochastic firing of origins. Computational simulation of stochastic firing from identified ORC sites is in accord with replication timing data. Large genomic regions with a paucity of ORC sites are strongly associated with common fragile sites and recurrent deletions in cancers. We suggest that replication origins, replication timing, and replication-dependent chromosome breaks are determined primarily by the genomic distribution of activator proteins at enhancers and promoters. These activators recruit nucleosome-modifying complexes to create the appropriate chromatin structure that allows ORC binding and subsequent origin firing.
Replication origins are established by the assembly of the prereplication complex at discrete sites of the genome. The first step of this process involves binding of the highly conserved six-subunit origin recognition complex (ORC), which serves as a loading platform for the subsequent assembly of helicases, DNA polymerases, and cofactors required for DNA synthesis (1, 2). In the yeast Saccharomyces cerevisiae, ORC binds DNA in an ATP-dependent manner and recognizes a specific DNA sequence (3). In Drosophila, ORC localizes to regions of open chromatin with contributions from activating histone modifications, DNA sequence, DNA binding proteins, and nucleosome remodelers (4–6). In mammals, the mechanism(s) through which ORC is localized and establishes a functional origin remains unclear.
A great deal of effort and a variety of experimental approaches have been devoted to describing the nature and position of replication origins in mammalian genomes. DNA combing technology, replication timing analysis, short nascent strand (SNS) enrichment, and bubble trapping approaches suggest that DNA replication initiation sites are enriched in CpG-rich regions, open chromatin domains, and transcriptional regulatory elements (7–17). However, these methods lack the necessary resolution to investigate important relationships of ORC binding with other features of the genome. In addition, the divergence in protocols and bioinformatic pipelines between laboratories has led to some controversial and nonreproducible observations. Finally, these studies assume that the identified replication initiation sites are comparable to ORC binding sites.
In addition to the issue of how replication origins are selected, genomic regions are replicated at distinct times within S phase. Some regions are replicated early, whereas others such as heterochromatic regions are replicated late (18–20). One possible explanation for the replication timing pattern is that origins are programmed to generate the same pattern in all cells. Alternatively, origins could fire stochastically so that the pattern varies from cell to cell. Early versions of the stochastic firing model invoked functional differences between early versus late origins. However, the finding of more ORC binding sites in early replicating regions of the Drosophila genome suggested the possibility that the timing pattern might arise from stochastic firing from all origins (4, 5). In addition, the replication timing pattern in Drosophila could be simulated computationally by stochastic firing from DNase I-hypersensitive sites (21). The basis of replication timing in human cells is not understood.
Using chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq), ∼13,000 ORC1 binding sites were identified in HeLa cells, essentially all of which were associated with transcription start sites (TSSs) of coding and noncoding RNAs (22). Transcription levels correlated with replication timing, and it was suggested that there are two classes of origins. Early firing origins were associated with moderate/high transcription of coding RNAs, whereas later firing origins were associated with low transcription of noncoding RNAs (22). It should be noted that these ORC1 ChIP-seq experiments were performed on partially purified, “low-density” chromatin that may selectively enrich for certain types of genomic regions. In addition, unlike other ORC subunits that remain associated with origins throughout the cell cycle, ORC1 only transiently associates with origins in G1 and is released from chromatin as cells enter S phase (23, 24).
To acquire a genome-wide high-resolution map of ORC binding sites in the human genome, we used unfractionated chromatin for ChIP-seq analysis of ORC2, a subunit of ORC that binds origins throughout the cell cycle (25–28). We show that the ORC2 binding profile is similar to that of ORC1 and that selectivity of ORC binding in human cells is similar to that in Drosophila. We suggest that selectivity of ORC binding in vivo involves nonspecific interaction with accessible DNA and recognition of modified histones. A computational simulation of DNA replication based on stochastic firing from our mapped ORC sites is in excellent accord with the replication timing pattern in vivo. Lastly, large genomic regions with a paucity of ORC2 binding sites are strongly associated with common fragile sites (CFSs) and recurrent deletions in cancers. We suggest that origin specificity, replication timing, and delay-induced errors of DNA replication arise from the genomic distribution activator proteins that recruit histone-modifying complexes to create the appropriate chromatin structure that permits ORC binding.
Results
Mapping ORC Binding Sites in the Human Genome.
Using ChIP-seq, we map binding sites of ORC2, a subunit of the ORC complex, in asynchronous K562 human erythroid cells (Fig. 1A and SI Appendix, Fig. S1 A–C). This cell line has been extensively analyzed by ChIP-seq and other functional genomic experiments as part of the Encyclopedia of DNA Elements (ENCODE) Project Consortium (29), thus facilitating comprehensive studies of mechanisms underlying ORC recruitment. We identify ∼52,000 ORC2 binding sites (SI Appendix, Table S1), including most ORC sites in human cells identified in single-locus studies (SI Appendix, Fig. S1D). ChIP quantitative PCR (ChIP-qPCR) analysis validates 39 out of 40 of these sites (SI Appendix, Fig. S1E and Table S2), indicating that the vast majority of identified ORC2 sites are true positives. As further validation, an independent ChIP-seq experiment using an ORC2 antibody raised against another region of the protein identifies similar ORC2 binding sites (r = 0.74) whose peak summits are indistinguishable (SI Appendix, Fig. S2).
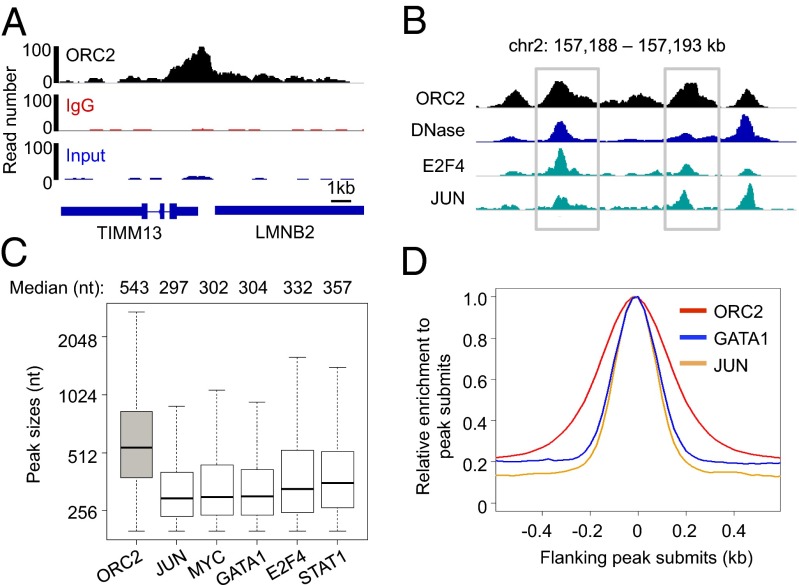
Fig. 1.
ORC2 binding peaks are broader than those of typical transcription factors. (A) DNAs enriched in ORC2 ChIPs were analyzed by massive sequencing using the Illumina’s Solexa technology and DNA-tag enrichment at the LMNB2 replication origins visualized using IGV software. Input (blue), IgG (red), and ORC2 (black) are presented on similar y-axis scales. (B) Example genomic region showing binding peaks of ORC2 and transcription factors including E2F4 and JUN. Example ORC2 binding peaks are highlighted in gray boxes. (C) Distribution of peak sizes of ORC2 and example transcription factors. (D) Read distribution around summits of ORC2, GATA1, and JUN peaks.
ORC Binding Is Not Determined by Sequence Motifs and It Occurs Over a Broad Region Linked to DNase I Hypersensitivity.
When analyzed by ChIP, transcription factors that bind to short recognition sequences (i.e., point sources) give a characteristic peak profile that is related to the size of the chromatin (30, 31). Interestingly, the profile of ORC2 binding sites has a median size of 550 nt, which is ∼200 nt broader than typical peak transcription factors (e.g., JUN, MYC, GATA1, E2F4, and STAT1) derived from the same chromatin sample (Fig. 1 B–D). This observation suggests ORC behaves differently than typical transcription factors whose binding is limited to short sequence motifs.
ORC2 binds to DNA in open chromatin regions as defined by DNase I hypersensitivity, and the ORC2 peak summits are highly colocalized with those of DNase I-hypersensitive sites (Fig. 2A). ORC2 site profiles in early replicating domains (G1 or S1) are slightly broader and overall binding levels slightly greater than ORC2 site profiles in later replication domains (S4 and G2) (SI Appendix, Fig. S3A). Similarly, DNase I-hypersensitive regions associated with ORC2 sites in early replicating regions are slightly larger and more open than those located in later replicating regions (SI Appendix, Fig. S3B). Thus, the ORC2 binding profile is very strongly correlated with the DNase I-hypersensitivity profile.
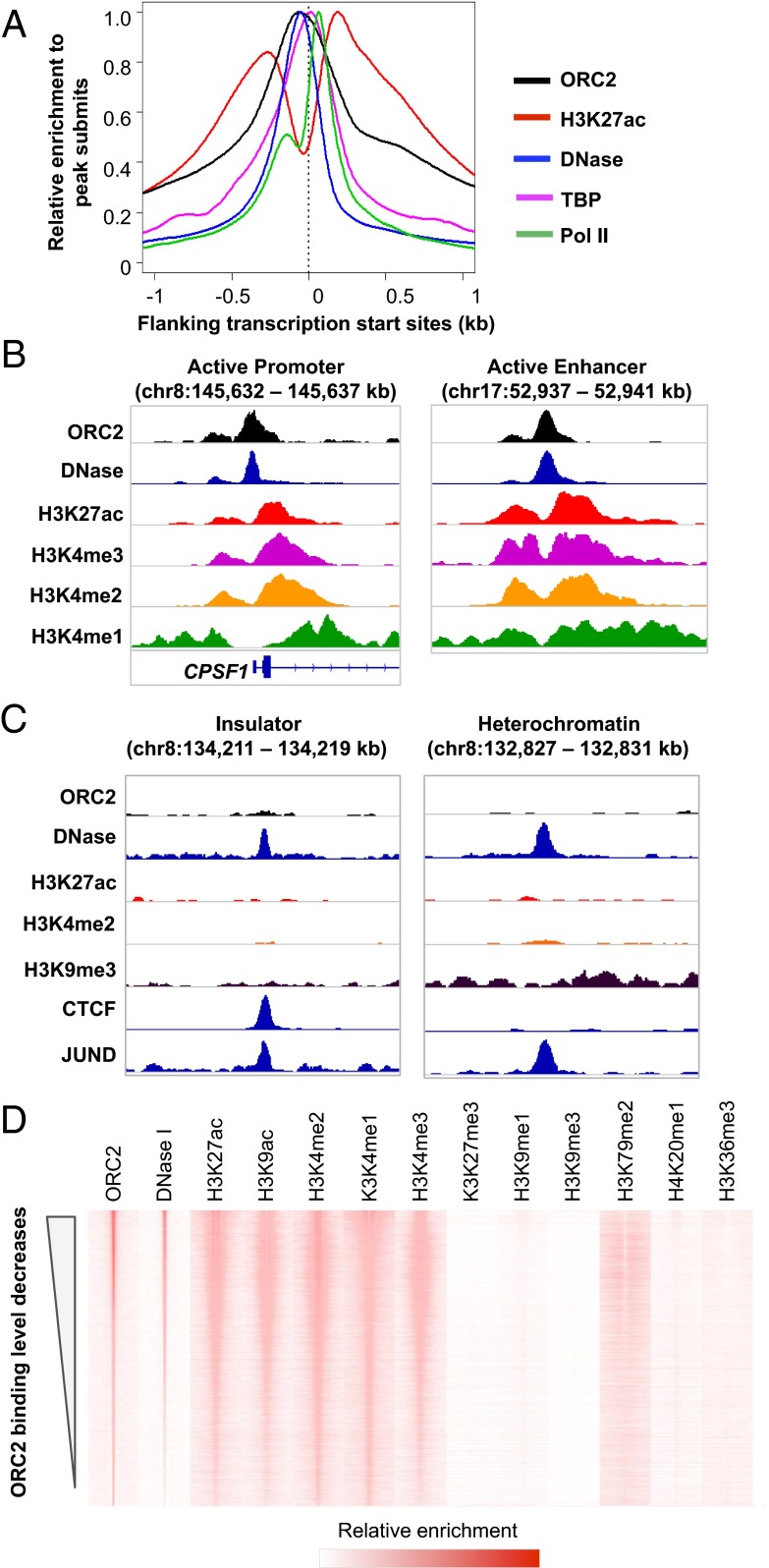
Fig. 2.
Epigenetic features of ORC2 binding sites. (A) Distribution of ORC2, H3K27ac, DNase-seq, TBP, and Pol II reads around gene promoter regions. (B) Examples of ORC2 binding sites in the promoter of the CPSF1 gene and in an active enhancer region. (C) Examples of insulator and heterochromatin regions do not have ORC2 binding. (D) The DNase-seq and ChIP-seq read distribution around ORC2 binding sites, ranked by ORC2 binding levels.
In accord with previous indications, open chromatin regions bound by ORC2 are more likely to be located within CpG islands and/or contain G-quadruplex motifs (32) (SI Appendix, Fig. S4 A–C) than open chromatin regions not bound by ORC2. However, only 31% of ORC binding sites contain G-quadruplex motifs and 26% are located in CpG islands, indicating that neither of these features is necessary for ORC binding. Similarly, many transcription factor binding sites (e.g., E2F, MYC, NF-κB, GATA, and AP-1) are enriched in ORC2 binding regions compared with open chromatin regions that are not bound by ORC2 (SI Appendix, Fig. S4D). However, ORC2 binding sites do not completely overlap sites with any individual transcription factor or two-way or three-way combinations of transcription factors, and ORC2 peak summits do not coincide with those of RNA polymerase II or the general transcription factor TBP (Fig. 2A). Lastly, using global run-on sequencing (GRO-seq) data in K562 cells to measure active transcription levels (33), we observe only a modest relationship (Pearson correlation 0.33) between ORC2 binding and transcription (SI Appendix, Fig. S5). Taken together, these observations suggest that ORC is not recruited to their target sites by transcription factors or the basic transcription machinery, and it is only modestly linked with transcription per se. However, we cannot exclude the possibility that, in some cases, ORC could be directly recruited by a transcription factor.
ORC2 Recognizes Active Open Chromatin Regions.
Based on chromatin states classified according to the histone modification pattern (34), ORC2 tends to bind to active promoters, weak promoters, and active enhancers but not insulators and heterochromatin regions (Fig. 2 B and C and SI Appendix, Fig. S6). To address the contribution of chromatin states to ORC recruitment in a systematic and unbiased manner, we examined the correlation between ORC2 binding levels and individual histone modifications based on all measurements from the ENCODE Project Consortium (Fig. 2D). ORC2 binding level is most correlated with the degree of chromatin accessibility measured by DNase-seq (R = 0.79; Fig. 2D and SI Appendix, Fig. S7A). In addition, ORC2 binding regions are enriched with histone modifications representing active chromatin, such as H3K27ac, H3K9ac, H3K4me2, H3K4me1, and H3K4me3 (Pearson correlation values 0.66, 0.63, 0.68, 0.64, and 0.64, respectively; SI Appendix, Fig. S7 B–E), but are depleted with histone markers representing heterochromatin and/or suppressed transcription, such as H3K27me3 and H3K9me3 (Fig. 2D). Under stringent cutoffs of peak calling [model-based analysis of ChIP-Seq (MACS) P value < 10−12 for H3K27ac and ORC2 peaks], about 83% of acetylated, open chromatin regions show ORC2 binding. For the remaining 17% of regions, ORC2 binding levels are significantly higher than random genomic regions (SI Appendix, Fig. S8), although they did not pass our arbitrary threshold for calling ORC2 peaks.
To address the chromatin states that determine ORC binding, we randomly picked open chromatin regions with similar accessibility but with or without ORC2 binding. As expected, ORC2 binding regions have significantly higher H3K27ac and H3K4me2 levels than nonbinding regions (P < 10−200) (Fig. 3 A–D). Conversely, we randomly selected open chromatin regions with a similar H3K27ac level but with or without ORC2 binding. ORC2 binding sites have significantly higher chromatin accessibility and H3K4me2 levels (SI Appendix, Fig. S9). These results indicate that both chromatin accessibility and active histone modifications are important for ORC2 recruitment.
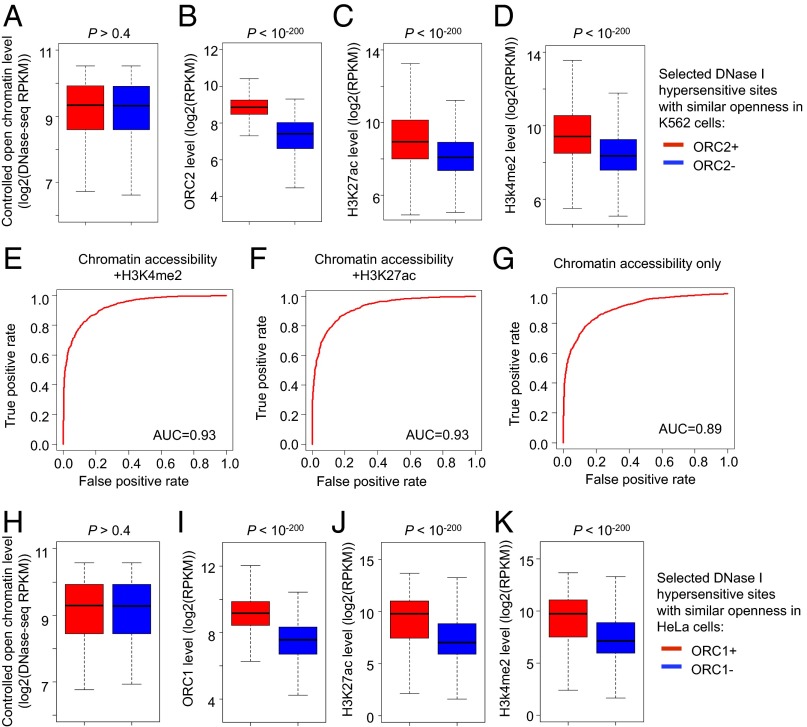
Fig. 3.
ORC2 binds to active open chromatin regions. (A–D) Randomly selected 6,000 open chromatin regions with similar accessibility (A) but with or without ORC2 binding sites (B) were examined for H3K27ac (C) and H3K4me2 (D) levels in K562 cells. The Wilcoxon rank sum test P values comparing two groups of open chromatin regions are shown. (E–G) The AUC values measuring performances of logistic regression classifiers predicting ORC2 binding status, based on indicated training parameters. (H–K) Randomly selected 6,000 open chromatin regions with similar accessibility (H) but with or without ORC1 binding sites (I) were examined for H3K27ac (J) and H3K4me2 (K) levels in HeLa cells. The Wilcoxon rank sum test P values comparing two groups of open chromatin regions are shown.
We then built a logistic regression model to predict ORC2 binding states based on DNA accessibility and histone modifications (see Materials and Methods for details). The combination of chromatin accessibility and H3K27ac/H3K4me2 levels predicts ORC2 binding status with high accuracy [area under receiver operating characteristic (ROC) curve (AUC) = 0.93; Fig. 3 E and F]. As expected, when predicted binding probabilities are higher, ORC2 binding levels estimated from ChIP-seq using two different antibodies increase (SI Appendix, Fig. S10 A and B). This level of predictive accuracy is remarkable given the cutoff issues involved in peak calling and the fact that the experimental data were generated from different laboratories. Predictive accuracy is lower when considering chromatin accessibility or histone modifications alone (AUC ≤ 0.89; Fig. 3G and SI Appendix, Fig. S10 C and D), but this effect is modest because open chromatin and histone acetylation is strongly correlated (Pearson correlation = 0.71; Fig. 2D). Thus, ORC binds to the vast majority of active, open chromatin regions. The lack of detectable ORC2 binding in heterochromatin and insulator regions that are accessible (DNase I hypersensitivity) and capable of binding CTCF, JUND, and other factors suggests that one or several histone modifications representing active/permissive transcription are important for ORC2 recruitment.
Although we detect very few ORC2 binding sites in heterochromatin, immunofluorescence experiments have revealed some ORC binding (35). In addition, ORC and CBX5 (also known as HP1α) can interact physically, and they appear to be mutually important for association with heterochromatin (35). Taken together with our results, we suggest that ORC association with heterochromatin is weak and/or diffuse over the entire heterochromatic region, thereby explaining the near absence of localized ORC2 binding sites. We cannot exclude the formal possibility that ORC might not directly bind DNA in heterochromatin, which would likely reduce cross-linking efficiency.
ORC2 and ORC1 Have Similar Binding Profiles.
It is difficult to directly compare our ORC2 binding profile to that of the published ORC1 binding data (22), because these studies were performed in different cell lines. However, the ORC1 binding profile can be determined by integrating the ORC1 ChIP-seq data (22) with extensive histone modification data (29) in the same cell line (HeLa). This makes it possible to indirectly compare ORC1 and ORC2 binding profiles via their chromatin state preferences in the relevant cell lines. As is the case for ORC2, ORC1 binding site profiles are well colocalized with DNase I-hypersensitive sites, and the ORC1 peak sizes are larger than those of typical transcription factors such as JUN and MYC (SI Appendix, Fig. S11 A and B). In addition, as is the case for ORC2 sites, the level of ORC1 binding is strongly correlated with the level of active histone modifications (H3K27ac and H3K4me2; SI Appendix, Fig. S11C). Furthermore, ORC1 binding levels defined by the published ChIP-seq data (22) are in excellent accord with the predicted binding probability (SI Appendix, Fig. S11D), based on the model derived for ORC2 binding determined here. These observations indicate that ORC1 and ORC2 recognize similar chromatin states and hence are likely to have similar binding profiles. This conclusion is consistent with the observation that ORC1 interacts in a cell cycle-specific manner with the core ORC complex, and it suggests that there are few, if any, genomic regions bound selectively by ORC1 or ORC2. Furthermore, the similarity of the ORC1 and ORC2 binding profiles provide mutual validation for the location of ORC binding sites in human cells.
ORC2 Binding Is Enriched in Early Replicating DNA Domains.
The Repli-seq technique permits the mapping of newly replicated (BrdU-labeled) DNA in synchronized cells during consecutive phases (G1, S1, S2, S3, S4, and G2) of the cell cycle (10). Using Repli-seq data in K562 cells (29), we investigate the relationship between ORC2 binding and replication timing. Early replicated DNA regions are highly enriched for ORC2 binding compared with late replicated regions (Fig. 4A), and the density of ORC binding sites decreases progressively in accord to when the regions are replicated (Fig. 4B). There are 2.6 ORC binding sites per 100 kb of genomic DNA that replicates in G1 and S1 phases, whereas there are only about 0.2 ORC binding sites per 100 kb of genomic DNA that replicates in S4 and G2 phases (Fig. 4B). Thus, early DNA replication initiates preferentially in regions with high numbers of ORC binding sites.
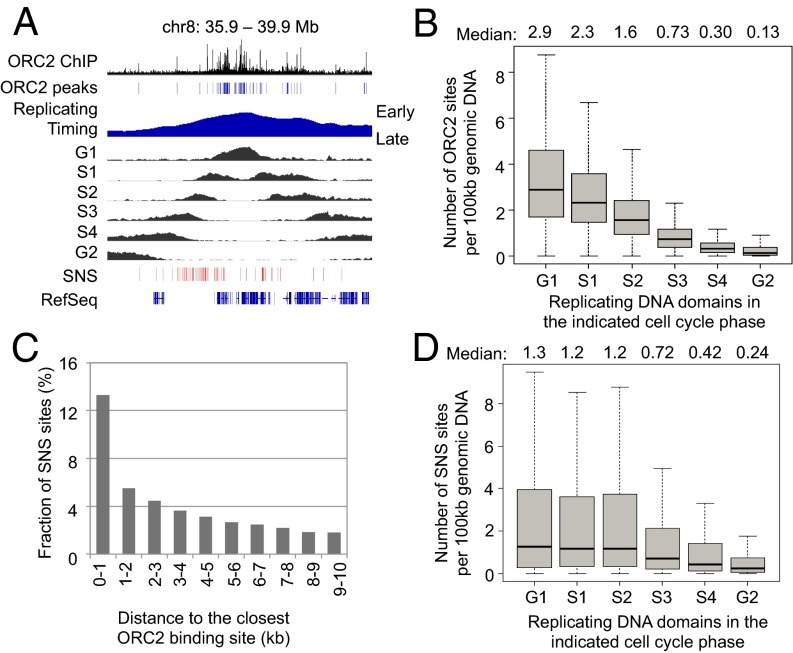
Fig. 4.
Concordance among ORC2 binding sites, newly synthesized DNA domains measured by Repli-seq, and SNS sites. (A) An example genomic location showing ORC2 binding, replicating DNA domains in G1, S1, S2, S3, S4, and G2 phases and SNS sites. (B) Density of ORC2 binding sites in per 100 kb replicating DNA regions in G1, S1, S2, S3, S4, and G2 phases. (C) Distribution of distances between SNS sites and closest ORC2 binding sites. (D) Density of SNS sites in per 100 kb replicating DNA regions in G1, S1, S2, S3, S4, and G2 phases.
Mapping SNSs of DNA in nonsynchronized cells has been used to identify replication initiation sites (14). There is a modest correlation between ORC2 and SNS sites, with 41% of SNS sites in K562 cells being located within 10 kb and 13% located within 1 kb of ORC2 binding sites (Fig. 4C). The discordance between ORC2 binding and SNS sites at many genomic locations is not an artifact of cutoffs used to define these functional entities. Although there is a very mild enrichment of SNS sites in early replicating DNA domains (Fig. 4D), the enrichment is much less dramatic than that of ORC2 binding sites (Fig. 4B). In particular, ORC2 density decreases considerably as cells pass through the G1, S1, and S2 phases (Fig. 4B), whereas the SNS site densities during these times are comparable (Fig. 4D). Analysis of an independently generated SNS dataset (17) yields similar results (SI Appendix, Fig. S12).
Lineage-Specific Early DNA Replication Is Correlated with Predicted ORC Binding.
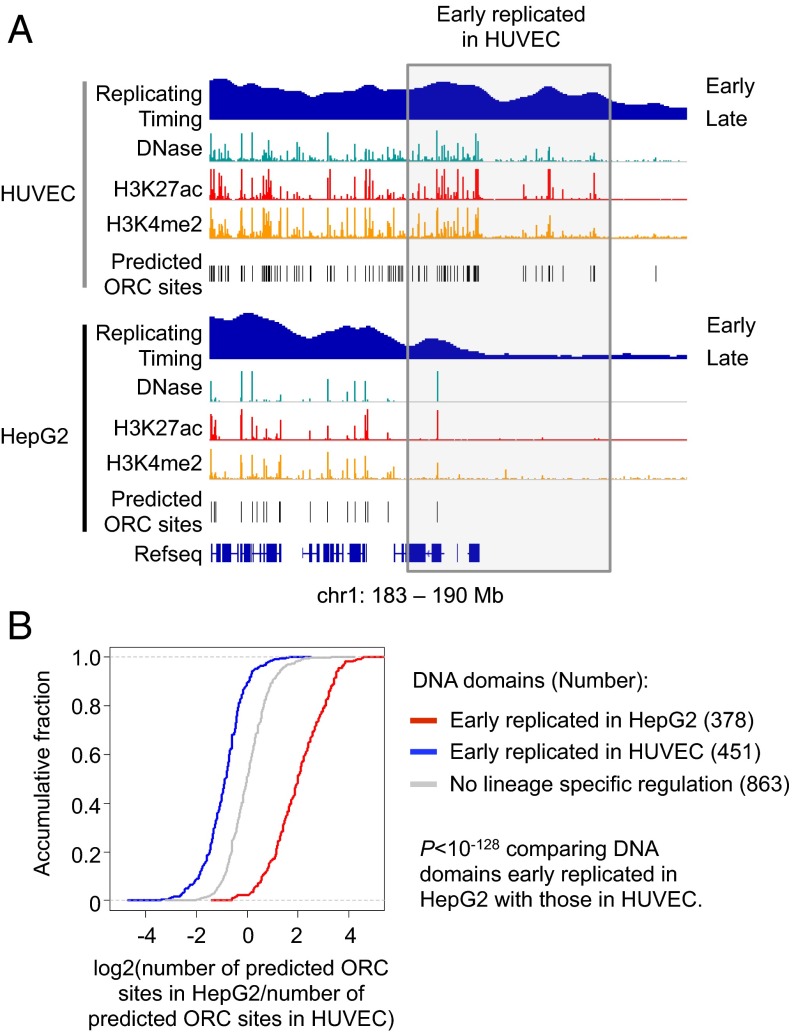
We applied our logistic regression classifier learned in K562 cells to predict ORC binding sites in HUVEC and HepG2 cells, using DNase-seq and H3K27ac and H3K4me2 ChIP-seq data from the ENCODE Project Consortium. Importantly, genomic domains showing lineage-specific early replication have more predicted ORC binding sites in the relevant cell type (Fig. 5). This observation provides independent confirmatory evidence that ORC binding is determined by chromatin accessibility and active histone modifications and that replication timing is linked to the density of ORC binding sites.
Fig. 5.
Lineage-specific early DNA replication timing is correlated with predicted ORC binding. (A) An example genomic region showing predicted lineage-specific ORC binding sites are correlated with differential early DNA replication. (B) Correlation between lineage-specific DNA replication and number of predicted ORC binding sites.
A Model of Stochastic Firing from ORC Binding Sites Consistent with the Replication Timing Profiles.
ORC sites are far more prevalent in early replicating genomic regions, and ORC binding regions in early and late replicating portions of the genome share similar properties (SI Appendix, Fig. S3). From these observations, we considered the possibility that the replication timing profile is determined simply by a mechanism involving stochastic firing of replication origins. In this model, origin firing is inefficient (18, 20, 21, 36), and the choice of which origin to fire at any given time is random, provided it has not been previously replicated in that S phase. Thus, on a population basis, origin firing in early replicating regions at the beginning of S phase is favored simply because there are far more ORC sites, whereas firing from relatively few ORC sites in late replication regions is due to increased time and the unavailability of ORC sites previously replicated. As such, the precise pattern of replication firing varies from cell to cell, including a minority of cells where origins in late replicating are fired early. Replication origins fire inefficiently and stochastically in fission yeast (18, 37), but this is not linked to replication timing.
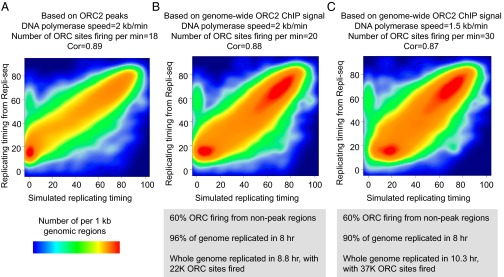
To test this model, we first performed a computational simulation of DNA replication process based directly on our 52,000 identified ORC2 binding sites and on stochastic firing from ORC sites not already replicated during the simulated S phase. Specifically, we set the S phase length at 8 h, the speed of DNA polymerase at 2 kb/min, and the relative probability that an ORC site can fire by its observed binding levels and then varied the “number of ORCs firing per minutes.” We then calculated Pearson correlation coefficient values between simulated and Repli-seq measured values. We observe an optimal correlation value of 0.89 with the parameter that 18 ORC sites fire per minute (Fig. 6A). At the optimized correlation value, ∼83% of the genome is replicated during S phase, with the unreplicated sequences primarily located in large regions essentially devoid of detectable ORC2 peaks (see ORC2-Poor Regions Are Enriched for CFSs and Genomic Regions Frequently Deleted in Cancers).
Fig. 6.
A simulated model for stochastic replication initiation. (A) Simulation based on the assumption that DNA replication initiates only at ∼52,000 ORC2 ChIP peaks. Shown is the correlation between replicating timing from simulation and that measured by Repli-seq choosing the value of 18 ORC sites firing per minute. (B and C) Simulations based on the assumption that DNA replication initiates at both ORC2 ChIP peaks and low-affinity, nontargeted ORC binding at nonpeak regions. Shown is the correlation between replicating timing from simulation and that measured by Repli-seq and with the indicated parameters for replication speed and ORC sites fired per minute.
As the above simulation is based strictly on the 52,000 identified sites, “ORC-poor” regions can only be replicated passively from ORC sites located outside these regions. However, two lines of evidence strongly suggest some ORC-dependent firing from within these ORC-poor regions. First, weak, nonlocalized ORC binding in large heterochromatin regions is observed by immunofluorescence (35). Second, as shown from Repli-seq experiments (e.g., Fig. 7A), the replication timing pattern of these ORC-poor regions is distinct from the classic pattern in which DNA synthesis initiates from an ORC site(s) within a localized region and then spreads bidirectionally. Instead, replication of these ORC-poor regions is relatively uniform (Fig. 7A), suggesting initiation from “nonspecific” positions (i.e., nonlocalized ORC sites that are not detected by ChIP). Thus, the unreplicated 17% of the genome in the above simulation is likely to arise from the constraint that firing could only occur from the 52,000 ORC binding sites identified by ChIP.
Fig. 7.
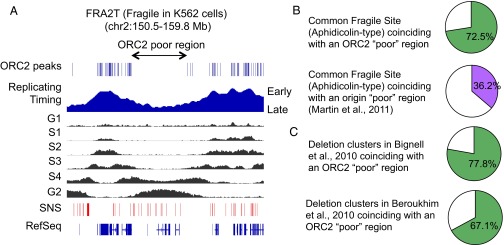
Genomic regions lacking ORC2 binding sites are strongly linked to CFSs and recurrent deletions in cancer. (A) Snapshot of the genomic region of chromosome 2 encompassing fragile site FRA2T. The position of genes, the replication timing profile, as well as the position of ORC2 binding sites are reported. The location of the ORC2-poor regions, overlaying with a late replicating domain, is indicated. (B) Graph indicating the proportion of CFSs, aphidicolin-sensitive type, overlapping with ORC2-poor and Origin-poor regions of the genome. A detailed list is provided in SI Appendix, Table S3. (C) Graph indicating the proportion of recurrent deletions in cancer (43, 44) overlapping with ORC2-poor regions of the genome. A detailed list is provided in SI Appendix, Table S4.
To address this issue, we performed new simulations that include an estimate for “nontargeted, low-affinity” ORC binding throughout the genome including ORC-poor regions. We assumed that firing from such “low-affinity” sites would represent 60% of the ORC firing events, a number consistent with a 104 specificity ratio (targeted vs. low-affinity sites) typical for transcription factors and Pol II (38). Based on background-subtracted ChIP-seq reads, we calculated the frequency of ORC binding for all genomic regions. We performed two simulations that differ in firing rate and replication speed and obtained comparable correlation values, but with a larger proportion of the genome (90% or 96%) being replicated during S phase (Fig. 6 B and C). Although changing the parameters will affect the correlation value and the fraction of the genome that is replicated, the very high correlation between our computational simulations and the Repli-seq data provides strong evidence for the stochastic firing model. We note that our simulations are concerned with the distinction between early versus late replicating regions and not replication boundaries. As such, these simulations ignore potential contributions of topologically associating domains (39).
ORC2-Poor Regions Are Enriched for CFSs and Genomic Regions Frequently Deleted in Cancers.
Many large genomic regions are devoid of detectable ORC2 peaks (Fig. 7A), an observation not anticipated by SNS data analysis (14, 17). Because ORC2-poor regions often correspond with late replicating domains of the genome and large heterochromatic regions, we speculated, like others (40), that these domains might delineate CFSs. Strikingly, 73% of CFSs, often defined only by a chromosomal band (41, 42), overlapped with ORC2-poor regions (Fig. 7B and SI Appendix, Table S3). In contrast, only 36% of CFSs span a region deficient in origins defined by SNSs (14) (Fig. 7B). Thus, ORC2 binding is a better predictor of CFS location than SNSs, and our data should be useful in refining the boundaries of known or unknown CFSs.
In a related vein, we examined whether recurrent deletions in cancer (43, 44) were linked to ORC2-poor regions. Again, recurrent deletions in cancer show a striking overlap (78% or 67% depending on the dataset) with ORC2-poor regions in K562 cells (Fig. 7C and SI Appendix, Table S4). Thus, irrespective of cell type, the lack of ORC2 sites over an extended region is strongly associated with genomic recurrent breaks, gaps, and rearrangements in response to replication stress and in cancer.
Discussion
How Is ORC Targeted to Specific Locations in the Genome?
By definition, the selective binding of ORC to genomic regions in vivo is due to common biochemical features of these regions. In the simplest case, ORC binding sites contain all these features, all genomic regions with these features are bound by ORC, and ORC does not bind to regions that lack all these features. Although ORC binding correlates with many individual genomic features, no individual feature is sufficient to account for ORC binding. In addition, the pattern of ORC binding is distinct from all of the many other factors analyzed in K562 cells by the ENCODE Project Consortium. We therefore looked for combinations of features that predict ORC binding with high accuracy.
The combination of open chromatin regions (DNase I hypersensitivity), acetylated H3, and methylated H3K4 is highly predictive of ORC binding. No other combinations of factors are as predictive. For example, although most sequence-specific transcription factors bind to target sites in open chromatin regions, it is difficult to find combinations of transcription factors that predict ORC binding. Similarly, although chromatin-modifying complexes are recruited to and help create/maintain open chromatin regions, individual complexes are recruited only to a subset of open regions (45). Together with the observation that the ORC binding profile is broader than that observed for transcription factors, these considerations strongly argue that ORC is not directly recruited to genomic regions by direct interactions with transcription factors, coactivators, or chromatin-modifying complexes.
Many studies have linked transcriptional activity to replication origins and ORC1 binding, and indeed we also observe this correlation with ORC2 binding. However, on a quantitative basis, the correlation between transcriptional activity (nascent RNA levels) within 0.5 kb of the ORC binding site with ORC binding levels is only modest. In addition, the location and peak profile of ORC2 sites is clearly different from those of any factor involved directly in Pol II transcription including Pol II and TBP, but it is remarkably similar to that of DNase I sensitivity. More generally, it is difficult to come up with a plausible molecular model for how transcription is mechanistically linked to ORC binding that accounts for the experimental results presented here. Thus, we argue that the correlation between ORC binding and transcriptional activity reflects not a direct link but rather the independent strong correlation of Pol II transcription with open chromatin regions flanked by acetylated nucleosomes.
Our results suggest that selectivity of ORC binding in vivo is determined by a mechanism involving nonspecific interaction with accessible DNA and recognition of modified histones with active marks. Both the open chromatin regions and the modified histones are due to the action of chromatin-modifying activities recruited by sequence-specific transcription factors and the Pol II machinery. However, ORC recognizes the physical features generated by these chromatin-modifying activities, not the activities themselves. In this regard, ORC resembles the Pol III machinery (46, 47) and the V(D)J recombinase (48), whose binding in vivo is strongly influenced by specific histone modifications in the vicinity of the recognition motif. ORC differs from these complexes in that it does not appear to have significant DNA sequence specificity.
We do not know which histone modification(s) or ORC subunit (or ORC-interacting protein) mediates site selectivity in vivo. Given their near-universal presence at ORC2 sites, acetylated H3 and/or dimethylated H3K4 are excellent candidates to be recognized by ORC. H3K4 dimethylation promotes replication origin function in yeast (49), and H34K4me3 demethylation promotes replication origin firing (50), although the mechanism is unknown. However, the ORC subunits do not contain previously identified domains for interacting with these histone modifications. Thus, if these modifications are involved in ORC binding selectivity, the interaction would be mediated by a novel domain and/or a conventional domain of an ORC-interacting protein.
ORC could also recognize a histone modification not examined by the ENCODE Project Consortium. In vitro, the BAH domain of ORC1 binds dimethylated H4K20 with high specificity (51), and the WD domain of the ORC-interacting protein LRWD1 (also known as ORCA) interacts preferentially with trimethylated H4K20 (52). Both of these interactions are linked to replication licensing. In addition, artificial recruitment of KMT5A (also known as SET8), the enzyme that catalyzes monomethylation of H4K20, can recruit ORC1 and ORCA to a target site in a manner dependent on KMT5C (previously known as Suv420H2), the enzyme that mediates di- and trimethylation of H4K20 (52, 53). Although ORC binding specificity via methylated H4K20 is attractive, it is unknown whether the genomic pattern of di- and/or trimethylated H4K20 is in accord with ORC binding. Furthermore, the temporal specificity of ORC1 binding and increased H4K20 methylation does not simply explain the binding of ORC2 and other ORC subunits throughout the cell cycle. We note that ORC specificity via H4K20 methylation is not mutually exclusive with H3 acetylation or dimethylated K4, and SET8 and/or Suv420H2 might preferentially bind chromatin in which H3 is acetylated and dimethylated at K4.
Lastly, it is possible that ORC does not interact directly with acetylated histones but rather is indirectly affected by the effect of acetylated histones on nucleosome stability and/or remodeling. Acetylated nucleosomes are less stable and more accessible to proteins than nonacetylated proteins (54–56). In addition, many nucleosome remodeling complexes contain subunits with bromodomains that directly interact with acetylated histones. Open chromatin regions flanked by acetylated nucleosomes are more dynamic and hence may be more accessible to ORC.
Our results are in excellent accord with similar studies in Drosophila that mapped ORC2 binding sites with respect to numerous chromatin properties and transcription factor binding sites (4–6). Although there are some differences in the analyses performed and in the molecular interpretation, the overall similarities of results and conclusions are striking. As such, the prior work in Drosophila provides independent validation of the results in human cells presented here. More importantly, the similarities of ORC binding in Drosophila and human cells strongly suggest that the mechanism of ORC binding is conserved among metazoans.
Relationship of ORC Binding to Replication Origins.
Although we identify 52,000 ORC2 binding sites with high confidence, our results do not bear on the issue of whether ORC binding per se is sufficient to initiate DNA replication. This is a difficult issue to address, because there is no definitive assay for a replication origin in vivo. Many studies have used SNS mapping to identify replication initiation sites (14). As observed here and in Drosophila (4, 5), there is a relationship between ORC2 binding and SNS sites, but the majority of ORC sites are not SNS sites and vice versa. One possible explanation for this discrepancy is that origin firing occurs from only a subset of ORC sites. Alternatively, the SNS sites that do not appear to bind ORC might arise by DNA repair and/or other mechanisms including nuclease activity during sample preparation. In addition, the stability, and hence detection, of SNSs might vary and depend on mechanisms unrelated to DNA replication per se.
We suggest that several observations are consistent with the idea that ORC sites generally correspond to origins. First, ORC binding is an easily interpreted assay, and ORC sites have similarly broad binding profiles and common chromatin patterns; hence, differences in origin firing would have to occur by an unknown molecular property. Second, as discussed below, the computational simulation based on stochastic firing from all identified ORC sites is in excellent accord with replication timing data. Third, compared with SNS sites, ORC sites are distributed more nonrandomly in the genome, and they are more highly correlated to replication timing, fragile sites, and recurrent cancer deletions. The increased nonrandomness of ORC sites compared with SNS sites for these properties and the likelihood that some SNSs may arise by mechanisms other than origin firing suggest that ORC binding is a better indicator of replication origins than SNS sites. Consistent with this suggestion, a very recent paper that maps origins via the pattern of Okazaki fragments is broadly consistent with our ORC2 binding data and less consistent with SNS sites (57). However, these observations are only suggestive, and it is possible that the level of ORC binding does not strictly determine the level of origin firing.
Relationship of ORC2 Binding and Replication Timing: Evidence for Stochastic Initiation.
Genomic regions differ with respect to when they are replicated in S phase, but the mechanism for differential replication timing in mammalian cells is poorly understood. From work in yeast and humans, it has been suggested that replication origins fire stochastically but at varying efficiency (18, 20, 21, 36). Here, we show that early replicating regions in human cells have far more ORC binding sites than late replicating regions. However, ORC binding sites in both early and late replicating regions have very similar chromatin environments and similar levels of ORC2 binding. The only difference we observed between these ORC binding sites is that those in late replicating regions have slightly shorter open chromatin regions. These observations suggest that ORC binding sites throughout the genome are recognized and function in a mechanistically similar manner.
We therefore suggest that replicating timing is due to stochastic initiation from ORC sites. This model is strongly supported by a computational simulation based on identified ORC binding sites that is in excellent accord with Repli-seq data performed in the same cell line. In this model, initiation is limiting such that only a small fraction of ORC binding sites is capable of firing at any given time. As a consequence, at the beginning of S phase, replicating initiation is strongly biased to genomic regions that have many ORC sites. This bias is maintained as S phase progresses, thereby generating early replicating regions of the genome. Late replicating regions arise for two reasons. First, they may be passively replicated via origins located far away in early replicating regions. Second, late replicating regions, by definition, are available for a longer time to permit stochastic firing from ORC sites within these regions. The stochastic initiation model can also explain why there are more ORC sites than SNS sites in early replicating regions. In particular, may ORC sites in early replicating regions might not be fired and hence not appear as SNS sites due to passive replication from nearby ORC sites. In late replicating regions, where ORC sites are further apart, passive replication of ORC sites would occur at lower frequency.
This model of stochastic firing due to limiting initiation factors, as well as the experimental definition of early and late replicating regions, is based on the average behavior of the entire cell population. In a small fraction of individual cells, the stochastic firing model predicts that some “late” ORC sites actually fire early. In addition, this model suggests that only a subset of “early” ORC sites initiate replication in individual cells; that is, firing from a given ORC site will lead to replication of regions that contain neighboring ORC sites that do not initiate replication in that cell. Our model does not exclude the possibility that some ORC sites are inherently better at firing than others.
Large ORC2-Poor Segments Coincide with Late Replicating Domains, CFSs, and Recurrent Deletions in Cancer.
It has been suggested that the spacing between strong replication initiation sites could predict fragile sites (58) and that genomic deletions encountered in cancer are enriched in late replicating domains (59). Here, we show that large genomic regions, often several megabases in length, are nearly devoid of ORC2 binding sites. These ORC-deficient regions are strongly associated with both CFSs and recurrent deletions in cancer. The origin firing frequency that best fits the proposed stochastic initiation model suggests that large ORC-deficient regions might be difficult to be fully replicated in S phase. Indeed, many of these regions are replicated primarily very late in S phase (defined as G2 phase in the Repli-seq experiments). Thus, we suggest that the paucity of ORC sites leads to regions of late replicating DNA that are especially sensitive to chromosomal breaks that occur upon replicative stress.
Replication Origins, Timing, and Consequences Are Ultimately Determined by the Genomic Distribution of DNA-Binding Activator Proteins.
The only known mechanism for generating open chromatin regions with active histone involves DNA-binding activator proteins that are bound at enhancers and promoters and recruit nucleosome remodelers and histone acetylases. Once generated, such open and active regions are necessary and sufficient for ORC binding. The nonrandom genomic distribution of ORC binding sites, together with stochastic firing of origins, can account for the replication timing pattern. Lastly, the existence of large genomic regions with a paucity of such open active regions, and hence ORC binding sites, results in late and perhaps incomplete replication that is likely to generate fragile sites and common deletions in cancer. As such, the genomic distribution of transcriptional activator proteins, via its effects on chromatin structure (but not necessarily transcription per se), ultimately determines many aspects of the DNA replication process in vivo.
Materials and Methods
Cell Culture and Chromatin Fragmentation.
Human K562 erythroid cells were obtained from American Type Culture Collection (lot no. 4607240) and cultured according to ENCODE Project Consortium procedures. Cells were cross-linked in 1% formaldehyde, nuclei isolated, and the chromatin shredded by sonication in buffer to obtain chromatin fragments of ∼200–300 bp. Note that our samples were prepared using the same set of chromatin and procedures as those used for transcription factors mapping and presented on the genome browser as YaleTracks (29).
Antibody Validation.
All antibodies were tested by Western blot and were validated by immunoprecipitation followed by Western blot (IP/Western) (SI Appendix, Figs. S1 and S2). A standard ChIP experiment using a rabbit antibody that specifically recognizes an epitope in the C-terminal domain of ORC2 shows enrichment for known origins (SI Appendix, Fig. S1), thereby validating the antibody and the sample.
ChIP.
After preclearing with protein A Sepharose beads (4 °C for 1 h), the chromatin from an equivalent of 5 × 107 cells was used for IP with an ORC2 antibody (Santa Cruz, sc-28742 or BD Biosciences, bd-6875) or IgG as a control. After 4 °C overnight incubation, beads were washed, eluted in buffer E (25 mM Tris·HCl, pH 7.5; 5 mM EDTA; 0.5% SDS) and cross-links reversed at 65 °C with proteinase K for 6 h. Resulting naked DNAs were then purified using the QIAquick PCR purification kit (Qiagen) and DNA eluted in 100 μL distilled water. Quantitative real-time PCR was performed using SYBR Green I. Enrichment for a specific DNA sequence was calculated using the comparative Ct method as previously described (28). Primer pairs are listed in SI Appendix, Table S5.
Preparation of Sequencing Libraries and Illumina Sequencing.
ChIP libraries were created using 15 cycles of amplification according to the Illumina manufacturer’s protocol. Libraries were run on a 2% (wt/vol) agarose gel, and the 150–450- bp fraction was extracted and purified using DNA gel extraction kit (QIAGEN). To estimate the yield of library and its relative amplification value, library DNA was quantitated using a Nanodrop. Quality of the library was assessed by qPCR monitoring at known origins (see primers above).
The library sequence reads obtained were aligned to the University of California, Santa Cruz (UCSC) human genome assembly hg19 using the Eland application (Illumina), allowing no more than two mismatches per sequence. For the ORC2 C-terminal datasets, 14,658,932 and 16,434,328 total reads were generated with 53.05% and 48.24% unique match, respectively. For the ORC2 N-terminal dataset, 23,291,113 total reads were generated with 19.04% unique match.
Integrative Analysis of ChIP-Seq, DNase-Seq, and Repli-Seq Data.
ORC2 binding sites were mapped by combining the sequencing reads from two independent biological replicates that were highly reproducible. Corresponding ChIP and input samples were analyzed with MACS software (60) to call binding peaks for ORC2 and histone modifications using the cutoff P value < 10−12. At this cutoff, only 70 peaks were observed in the input sample on their own. ChIP-seq for histone modifications and transcription factors, DNase-seq, and Repli-seq data were obtained from the ENCODE Project Consortium (29). DNase I-hypersensitive sites were defined as by the ENCODE Project Consortium. Replicating DNA domains during phases of cell cycles were defined by Repli-seq peaks using the cutoff MACS P value < 10−300.
We examined G quadruplex motifs in ORC2 binding peaks by searching for the occurrence of the G(3,10)N(1,7)G(3,10)N(1,7)G(3,10)N(1,7)G(3,10) motif. The symbol N indicates a position where any nucleotide is accepted, and repetition of an element is indicated by a numerical value; for example, G(1,3) corresponds to G, GG, and GGG. In addition, we also included G quadruplex motifs identified by an experimental approach (61). Locations of CpG islands in hg19 were downloaded from the UCSC genome browser (62).
To study lineage-specific regulation of DNA replication, we used the replicating timing defined by Wave Signal files from the ENCODE Project Consortium. Lineage-specific early replicating DNA domains were defined as regions showing differential signal values >10 in each 1-kb region.
Logistic Regression Classifiers to Predict ORC2 Binding.
For each DNase I-hypersensitive site, the chromatin accessibility level was measured as read per million (RPM) values of DNase-seq reads. Histone modification levels were measured as ChIP-seq reads in the open chromatin region and 500 nt extended. For the model training, we randomly picked 2,000 DNase I-hypersensitive sites with ORC2 binding as positive examples and 4,000 sites without ORC2 binding as negative examples. The model incorporates various features, including chromatin accessibility and/or histone modification levels, and we used the “glm” function (“binomial”) in R (R core team, 2015) (63) to build the logistic regression classifier. For testing, we randomly picked another 2,000 positive examples and 4,000 negative examples. We used DNase-seq+H3K27ac and DNase-seq+H3K4me2 to predict ORC binding probability in K562, HeLa, HepG2, and HUVEC cells. We take the averaged values from two classifiers as the predicted probabilities for ORC binding. ORC binding sites in HepG2 and HUVEC cells in Fig. 5 were defined if the predicted binding P value > 0.5 using parameters DNase-seq+H3K27ac and DNase-seq+H3K4me2.
Analyses of CFSs.
CFS coordinates were recovered from DNA repository databanks or from Genecards when only the chromosome band was known (64). The coordinates of the frequently deleted regions of the genome in tumors were directly obtained from the authors (43, 44). The final data were manually curated to take into account copy number variation and chromosomal rearrangement in K562 cells.
Estimation of the Density of ORC2 Binding Sites in the Genome.
The density of ORC2 binding sites in the genome was computed by calculating the distance between two adjacent ORC2 binding sites. For this analysis, we considered that the distribution of ORC2 binding sites represents a homogenous distribution in the population and therefore that the distances calculated actually represent the distances onto the DNA between two ORC2 binding sites. The final datasets of ORC2–ORC2 segments were manually edited to eliminate ORC2–ORC2 segments encompassing (i) centromeres, (ii) homozygous deletions over 100 kb present in K562 cells (UCSC Genome Browser/Common Cell CNV), and (iii) gaps of poor sequence uniqueness over 25 kb (UCSC Genome Browser/Mapability). The remaining ORC2–ORC2 segments were used to evaluate the relative distance between ORC2 sites on chromosome arms and thus the density of ORC2 sites in a given genomic segment. Regions over 700 kb with no detectable ORC2 binding sites were considered ORC2-poor regions. “ORC2-rich” segments are defined as the complement segments on the genome. A similar method was used to identify “origin-poor” and “origin-rich” regions of the genome from the datasets of replication initiation sites in K562 cells (14).
Motif Analysis.
Transcription factor binding site matrixes were defined by JASPAR Vertebrates and UniPROBE mouse databases (65, 66). We used the tool Find Individual Motif Occurrences (FIMO) to search binding sites in open chromatin regions using a cutoff P < 1E-4 (67).
A Simulated Model for Stochastic Replicating Initiation.
We took two approaches to simulate the DNA replication process in S phase, both of which involve setting the length of S phase at 8 h, setting the speed of DNA polymerase speed at 2 kb/min, and then obtaining optimal values for the variable number of ORC sites firing per minute. Approach 1 is based strictly on the assumption that DNA replication initiates only from the 52,000 ORC2 sites identified by ChIP-seq. Individual ORC sites have different relative firing probabilities (f) based on read number (n), using the following definition: f = 1, if n < 40; f = 2, if n ≥ 40 and n < 80; f = 3, if n ≥ 80 and n < 120; and f = 4, if n ≥ 120.
Approach 2 includes the possibility that ORC firing can occur at low efficiency from nontargeted regions throughout the entire genome. ORC firing from such nontargeted regions is conceptually similar to nonspecific binding activity characteristic of sequence-specific DNA-binding proteins. Based on measurements of 75,000 ORC complexes per cell (68) and a typical specificity of 104 for targeted versus nontargeted ORC binding (38), we estimated that firing from nonpeak locations represented 30% of the total firing and hence is equivalent to 25,000 ORC binding sites. We calculated the frequency of ORC firing from nontargeted regions in the following manner. Using the entire ORC2 ChIP-seq dataset, we extended each mapped sequencing read 200 nt downstream of the 5′ end, which represents the average fragment length in the ChIP-seq library. We counted overlapping tags to represent the read coverage at each nucleotide position in the genome. For every 100-nt region, we counted read tags located in the region. We then subtracted 200 tags in every genomic location, which represents the read background distribution (nonspecific ChIP signal). The number of 200 tags was chosen to optimize the replication firing probability at 40% from ORC2 peaks and 60% to nonpeak regions. For locations with positive ORC signals after the normalization, we assigned different relative firing probabilities (f) based on tag number (n), using the following definition: f = 1, if n > 50 and n ≤ 100; f = floor(n/100), if n > 100 and n ≤ 4,000; and f = 41, if n > 4,000.
DNA replication in a locus can be passively replicated via origins located far away in early replicating regions or be due to a new firing origin located nearby. And ORCs in already replicated regions cannot fire anymore. The final simulated replicating timing was normalized to the value representing earliest replicating. The early replicating region has the value close to 100, and the late replicating region has the value close to 0. We tested different values for the variable number of ORC sites firing per minute, simulated the DNA replication process for 100 times, and calculated the Pearson correlation coefficient values between mean replicating timing from simulations and that measured by Repli-seq. The optimal value of number of ORC sites firing per minute has the highest coefficient value.
Supplementary Material
Acknowledgments
We thank Michael Snyder, Mark Gerstein, and members of their laboratories for DNA sequencing and generating the raw sequencing files; Johannes Walter and Steve Bell for useful discussions; and Nick Rhind for suggesting why ORC sites might be more frequent than SNS sites in early, but not late, replicating regions. The ORC2 ChIP-sequencing data were generated as part of the ENCODE Project Consortium. This work was supported by National Institutes of Health Grants GM30186 and HG4458 (to K.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE70165).
See Commentary on page 9136.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609060113/-/DCSupplemental.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6(6):476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20(2):201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton ML, et al. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21(2):164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubelsky Y, et al. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24(7):1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadoret JC, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA. 2008;105(41):15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequeira-Mendes J, et al. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5(4):e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell. 2010;21(3):393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA. 2010;107(1):139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cayrou C, et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21(9):1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesner LD, et al. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 2011;21(3):377–389. doi: 10.1101/gr.111328.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valenzuela MS, et al. Preferential localization of human origins of DNA replication at the 5′-ends of expressed genes and at evolutionarily conserved DNA sequences. PLoS One. 2011;6(5):e17308. doi: 10.1371/journal.pone.0017308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin MM, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21(11):1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombraña R, et al. High-resolution analysis of DNA synthesis start sites and nucleosome architecture at efficient mammalian replication origins. EMBO J. 2013;32(19):2631–2644. doi: 10.1038/emboj.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besnard E, et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19(8):837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 17.Picard F, et al. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10(5):e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhind N. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol. 2006;8(12):1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhind N, Yang SC, Bechhoefer J. Reconciling stochastic origin firing with defined replication timing. Chromosome Res. 2010;18(1):35–43. doi: 10.1007/s10577-009-9093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012;28(8):374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellino GI, et al. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23(1):1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C-J, DePamphilis ML. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol Cell Biol. 2002;22(1):105–116. doi: 10.1128/MCB.22.1.105-116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25(22):5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J Biol Chem. 2002;277(35):31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 26.Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol Cell Biol. 2002;22(4):1036–1048. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdurashidova G, et al. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22(16):4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22(19):2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18(9):5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong K-H, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 2011;25(23):2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshina S, et al. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem. 2013;288(42):30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46(12):1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA. 2010;107(34):15093–15098. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das SP, et al. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015;25(12):1886–1892. doi: 10.1101/gr.195305.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17(1):308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 39.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515(7527):402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Tallec B, et al. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18(12):1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- 41.Glover TW. Common fragile sites. Cancer Lett. 2006;232(1):4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 42.Le Tallec B, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Reports. 2013;4(3):420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44(3):410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17(5):635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17(5):620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450(7172):1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rizzardi LF, Dorn ES, Strahl BD, Cook JG. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics. 2012;192(2):371–384. doi: 10.1534/genetics.112.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rondinelli B, et al. H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing. Nucleic Acids Res. 2015;43(5):2560–2574. doi: 10.1093/nar/gkv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo AJ, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484(7392):115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck DB, et al. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012;26(23):2580–2589. doi: 10.1101/gad.195636.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tardat M, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12(11):1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 54.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268(1):305–314. [PubMed] [Google Scholar]
- 55.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 56.Bauer WR, Hayes JJ, White JH, Wolffe AP. Nucleosome structural changes due to acetylation. J Mol Biol. 1994;236(3):685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 57.Petryk N, et al. Replication landscape of the human genome. Nat Commun. 2016;7:10208. doi: 10.1038/ncomms10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: Mechanisms of instability revisited. Trends Genet. 2012;28(1):22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 59.De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol. 2011;29(12):1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam EY, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 63.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna) 2015 Available at https://www.R-project.org/. Accessed June 1, 2015. [Google Scholar]
- 64.Safran M, et al. GeneCards 2002: Towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18(11):1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 65.Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42(Database issue):D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43(Database issue):D117–D122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong PG, et al. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS One. 2011;6(3):e17533. doi: 10.1371/journal.pone.0017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.