Significance
Pyrin, encoded by the MEFV gene, is causative for familial Mediterranean fever (FMF), an autoinflammatory disease. Pyrin responds to bacterial modifications/inactivation of Rho GTPases by assembling an inflammasome complex for activating caspase-1. Pyrin is a unique immune sensor because it senses bacterial virulence rather than recognizing microbial products. We found that Pyrin is phosphorylated on two serine sites which keep Pyrin inactive through binding by 14-3-3 proteins. Toxin stimulation and bacterial infection trigger Pyrin dephosphorylation and 14-3-3 dissociation, allowing Pyrin inflammasome activation. Colchicine, a microtubule-disrupting drug used to treat FMF, inhibits Pyrin activation downstream of dephosphorylation and 14-3-3 dissociation. These findings not only help us understand FMF pathogenesis/treatment but also shed mechanistic insights into cytosolic immunity.
Keywords: inflammasome, Pyrin, phosphorylation, FMF, Rho toxins
Abstract
Pyrin, encoded by the MEFV gene, is best known for its gain-of-function mutations causing familial Mediterranean fever (FMF), an autoinflammatory disease. Pyrin forms a caspase-1–activating inflammasome in response to inactivating modifications of Rho GTPases by various bacterial toxins or effectors. Pyrin-mediated innate immunity is unique in that it senses bacterial virulence rather than microbial molecules, but its mechanism of activation is unknown. Here we show that Pyrin was phosphorylated in bone marrow-derived macrophages and dendritic cells. We identified Ser-205 and Ser-241 in mouse Pyrin whose phosphorylation resulted in inhibitory binding by cellular 14-3-3 proteins. The two serines underwent dephosphorylation upon toxin stimulation or bacterial infection, triggering 14-3-3 dissociation, which correlated with Pyrin inflammasome activation. We developed antibodies specific for phosphorylated Ser-205 and Ser-241, which confirmed the stimuli-induced dephosphorylation of endogenous Pyrin. Mutational analyses indicated that both phosphorylation and signal-induced dephosphorylation of Ser-205/241 are important for Pyrin activation. Moreover, microtubule drugs, including colchicine, commonly used to treat FMF, effectively blocked activation of the Pyrin inflammasome. These drugs did not affect Pyrin dephosphorylation and 14-3-3 dissociation but inhibited Pyrin-mediated apoptosis-associated Speck-like protein containing CARD (ASC) aggregation. Our study reveals that site-specific (de)phosphorylation and microtubule dynamics critically control Pyrin inflammasome activation, illustrating a fine and complex mechanism in cytosolic immunity.
Cytosolic innate immunity is immerging as a critical host defense against infections. Inflammatory caspases, including the early identified caspase-1 and the recently characterized murine caspase-11 (caspase-4/5 in human), represent a major arm of this immune axis (1, 2). Excessive activation of inflammatory caspases leads to autoinflammatory conditions and septic shock and also is associated with autoimmune and metabolic diseases (3, 4). Both caspase-1 and caspase-4/5/11 induce pyroptosis, a lytic inflammatory cell death, through cleavage of the GSDMD protein (5, 6). Caspase-1 also processes IL-1β/18 to stimulate the inflammation. Different from caspase-4/5/11 that function as the receptor for bacterial LPS (7, 8), caspase-1 is activated by a cytosolic complex termed the “inflammasome.” In the original proposal (9), a nucleotide-binding domain leucine-rich repeat (LRR) protein (NLR) uses its LRR to recognize a microbial product or danger signal; the NLR then binds to the apoptosis-associated Speck-like protein containing CARD (ASC) adaptor through heterotypical PYRIN domain (PYD) interactions to form the inflammasome complex. However, well-characterized inflammasomes feature a much more diversified assembly mechanism than originally proposed (2). The neuronal apoptosis inhibitory protein (NAIP)–NOD-like receptor, CARD-domain containing 4 (NLRC4) inflammasomes for bacterial flagellin and type III secretion apparatus use NAIP as the receptor and NLRC4 as an adaptor for signal amplification (10–15). Inflammasome recognition of cytosolic DNA is mediated by a non-NLR protein, AIM2 (16). Moreover, detection of cytosolic LPS requires no NLR, and caspase-4/5/11 bear both the sensor and execution functions (7, 8).
Mutations in the Pyrin-encoding gene MEFV cause a human autoinflammatory disease, familial Mediterranean fever (FMF) (17, 18), characterized by acute peritoneal inflammation, skin rash, and pleural and arthritic attack (19). The N-terminal PYD in Pyrin can bind ASC and activate caspase-1 (20). Mice engineered to harbor FMF-associated mutations suffer from FMF-like inflammation, which requires IL-1β and ASC (21). We recently established the Pyrin inflammasome and uncovered its physiological function in sensing bacterial modifications of Rho GTPases (22). Various Rho-modifying toxins, including the glucosyltransferase cytotoxin TcdB from Clostridium difficile, FIC-domain adenylyltransferases (VopS from Vibrio parahaemolyticus and IbpA from Histophilus somni), and Clostridium botulinum ADP ribosylating C3 toxin, which modify different switch-I residues and inactivate Rho, can activate the Pyrin inflammasome. A Burkholderia cenocepacia type VI effector, TecA, triggers Pyrin inflammasome activation by deamidating Asn-41 in RhoA, which plays an important role in B. cenocepacia-induced lung inflammation (22, 23). Thus, Pyrin, a non-NLR protein, senses bacterial virulence rather than directly recognizing a microbial molecule; this unique mode of immune detection further highlights the mechanistic diversity in cytosolic innate immunity.
Pyrin was shown to interact with actin in an early study (24). A recent report suggests that an inactivating mutation in WDR1, an actin-depolymerizing cofactor, induces Pyrin-dependent IL-18 secretion and drives an autoinflammatory disease in mice (25). This observation supports the postulate that Pyrin functions by sensing a signaling event downstream of Rho modification/inactivation in the actin cytoskeleton pathway (22). Here we show that Pyrin is phosphorylated on Ser-241/205 and is inhibited by 14-3-3 binding. Rho modification triggers Pyrin dephosphorylation and therefore the activation of the Pyrin inflammasome. Colchicine, a microtubule-disrupting drug used clinically for FMF treatment, inhibited Pyrin-mediated ASC aggregation but not Pyrin dephosphorylation and 14-3-3 dissociation.
Results
C. difficile Cytotoxin TcdA Can also Trigger Pyrin Inflammasome Activation.
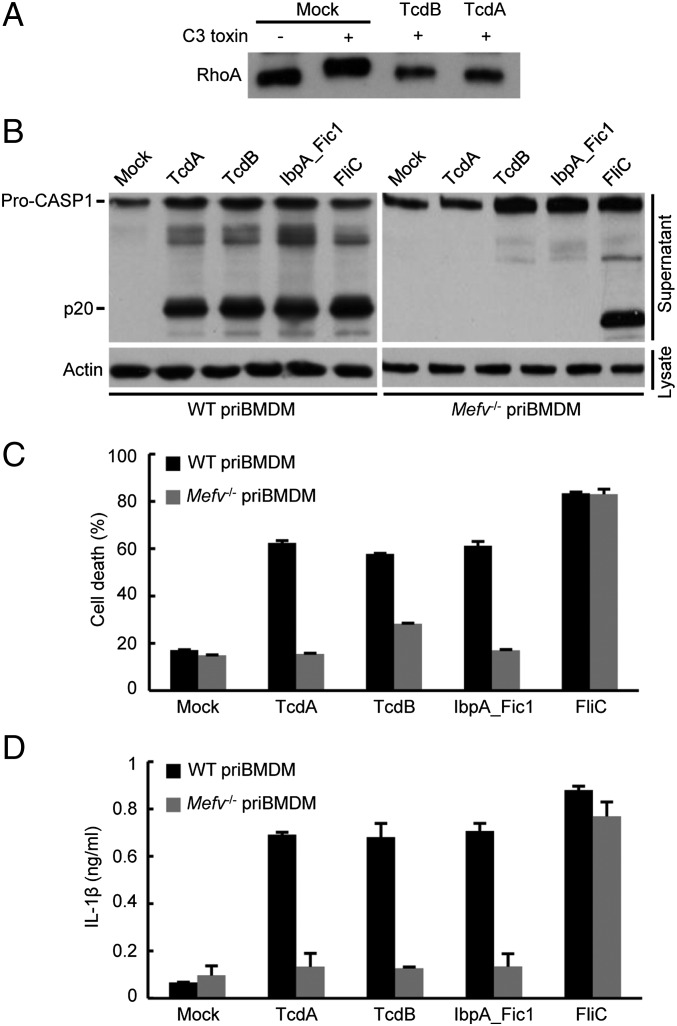
C. difficile TcdB is the prototypical stimulus that can potently activate Pyrin by glycosylating Thr-37 in RhoA (22). C. difficile is the leading cause of nosocomial diarrhea (26). This Gram-positive organism also releases TcdA that resembles TcdB in structure and function. The two toxins share similar substrate spectra in monoglucosylating small GTPases in vitro (27) despite their different cell-entry mechanisms. TcdA and TcdB, belonging to the large clostridial glycosylating cytotoxin family (28), are the major virulence determinant for C. difficile (29, 30). C. difficile harboring either TcdA or TcdB is sufficient to cause disease in the hamster model (31). However, there is evidence indicating different cellular and biological functions of TcdA and TcdB. TcdA and TcbB could modify RhoA similarly in mammalian cells (Fig. 1A). In murine bone marrow-derived macrophages (BMDMs), purified TcdA could induce caspase-1 autoprocessing to a similar extent as TcdB, IbpA_Fic1 (the first Fic domain of IbpA), and cytosolic flagellin (Fig. 1B). Consistently, all the stimulated cells underwent extensive pyroptotic cell death, accompanied by marked IL-1β secretion (Fig. 1 C and D). When the same assay was performed in Mefv−/− BMDMs, TcdA, TcdB, and IbpA_Fic1 failed to trigger detectable caspase-1 autoprocessing; pyroptosis and IL-1β secretion were also diminished (Fig. 1 B–D). In contrast, cytosolic flagellin, an agonist of the NAIP5-NLRC4 inflammasome (10, 11), induced the same extent of caspase-1 inflammasome activation in Mefv-deficient and -proficient BMDMs (Fig. 1 B–D). Thus, the Rho-modifying activities of TcdA and TcdB are detected similarly by the Pyrin inflammasome in macrophages.
Fig. 1.
TcdA activates the Pyrin inflammasome in mouse BMDM cells. (A) TcdA could modify RhoA as TcdB does. Lysates of TcdA- or TcdB-treated DC2.4 cells were subjected to in vitro ADP ribosylation reaction by purified C3 toxin. Anti-RhoA immunoblotting shows that RhoA resisted further modification by C3 toxin after TcdA treatment, as it did after TcdB treatment. (B–D), Assays of inflammasome activation by TcdA in comparison with TcdB, IbpA_Fic1, and FliC (from Pseudomonas aeruginosa) in primary BMDM (priBMDM) cells from wild-type C57BL/6 or Mefv−/− mice. TcdA and TcdB were added directly to the cell medium. IbpA_Fic1 and FliC were delivered by the LFn-PA system. The supernatants of BMDMs following stimulation were collected and used for the assays. Anti–caspase-1 immunoblotting of the supernatants is shown with anti-actin immunoblots serving as the loading control (B). Lactate dehydrogenase (LDH) release-based cell death (C) and ELISA of IL-1β release (D) are expressed as mean values ± SD from three technical replicates. Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. Data shown are representative of three independent experiments.
Pyrin Is Bound to Cellular 14-3-3s and the Binding Is Disrupted by Toxin Stimulation.
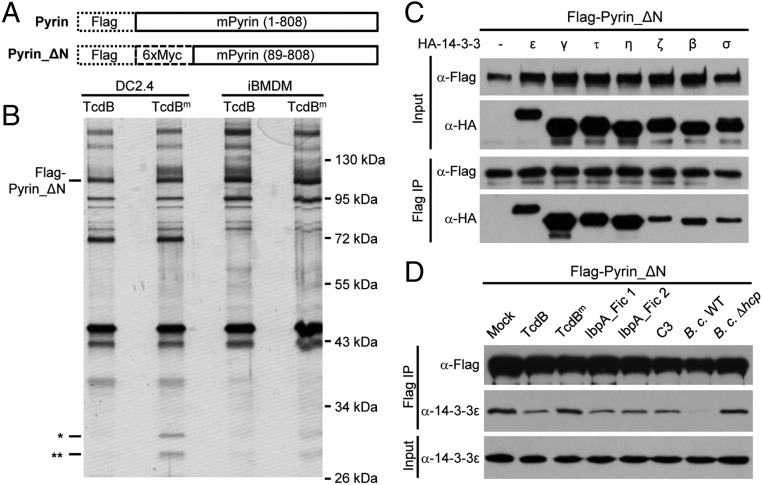
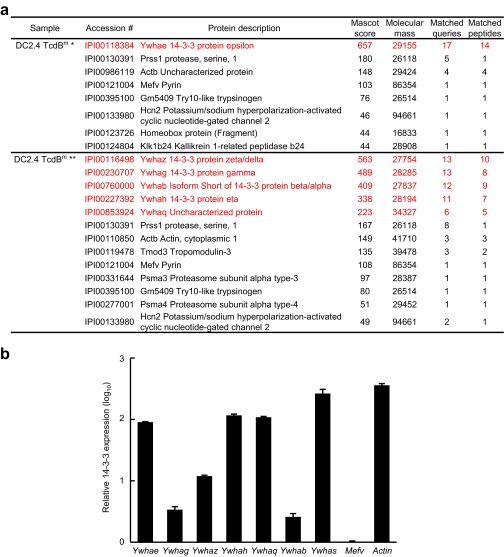
Pyrin senses Rho modifications with diverse chemical moieties but does not directly recognize Rho GTPases (22). Pyrin likely perceives a signaling downstream of Rho inactivation through interaction with a cellular protein. Early studies reported a cadre of proteins, identified from yeast two-hybrid screens, to be Pyrin-binding proteins, but few turned out to be relevant to its inflammasome function (19). Full-length Pyrin, when overexpressed in mammalian cells, is prone to aggregate because of PYD-mediated oligomerization. To overcome this aggregation, we constructed a Pyrin mutant with the PYD deleted, Pyrin_ΔN, with one Flag tag and six copies of Myc tags at the N terminus (Flag-6xMyc-Pyrin_ΔN) (Fig. 2A) and generated DC2.4 and immortalized BMDM (iBMDM) cells stably expressing the tagged Pyrin_ΔN mutant. Affinity purification of Pyrin complex in cells stimulated with TcdB or TcdBm (the W102A/D288N mutant devoid of the glucosyltransferase activity) identified two proteins of around 30 kDa, which, along with several other cellular proteins, were copurified with Pyrin_ΔN in unstimulated and TcdBm-stimulated cells (Fig. 2B). These two proteins, unlike other copurified proteins, disappeared upon TcdB stimulation (Fig. 2B). Mass spectrometry identified the two bands as 14-3-3 proteins, with the upper band being 14-3-3ε and the lower one being other 14-3-3 isoforms (Fig. S1A). Interestingly, 14-3-3 was identified more than 10 y ago in a Pyrin yeast two-hybrid screen, but its functional significance has not been known (32).
Fig. 2.
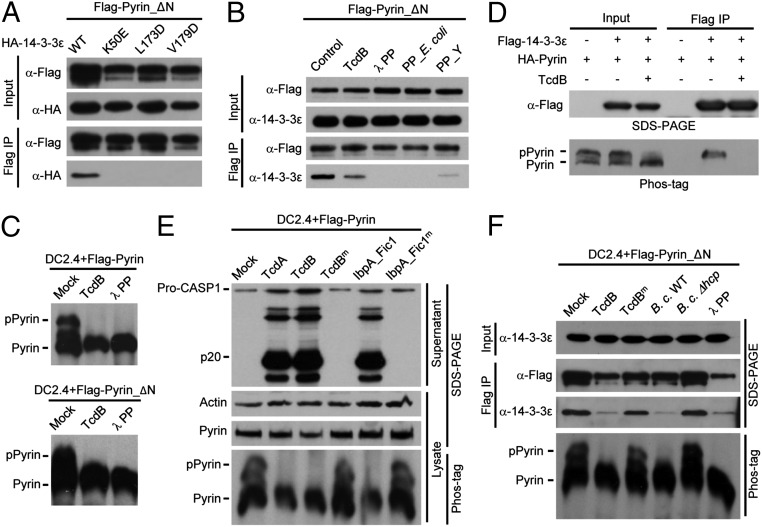
Association of Pyrin with 14-3-3 and dissociation of the complex during activation of the Pyrin inflammasome. (A) Schematic representation of Flag-Pyrin_ΔN used for immuno-purification of the Pyrin complex and subsequent experiments in this study. (B) DC2.4 and iBMDM cells stably expressing Flag-Pyrin_ΔN were stimulated with wild-type TcdB or catalytically inactive mutant TcdB (TcdBm). Cell lysates were subjected to immuno-purification using the anti-Flag M2 resin. The samples were analyzed by SDS/PAGE followed by silver staining. The single and double asterisks indicate two protein bands coeluted with Pyrin_ΔN in the presence of TcdBm. (C and D) Coimmunoprecipitation assays of Pyrin and 14-3-3 association. Flag-Pyrin_ΔN and the indicated HA-tagged 14-3-3 isoforms were cotransfected into 293T cells (C). To examine the stimulus-dependent interaction of Pyrin with endogenous 14-3-3ε, DC2.4 cells stably expressing Flag-Pyrin_ΔN were stimulated with TcdB or other indicated toxins or were infected with B. cenocepacia (B. c.) (D). Total cell lysates (Input) and the anti-Flag M2 immunoprecipitates (Flag IP) were blotted with antibodies against the indicated epitope or 14-3-3ε. –, empty vector; Mock, nonstimulation control. All data shown are representative of three independent experiments.
Fig. S1.
Mass spectrometry identification of 14-3-3 as the Pyrin-binding protein and the expression of different 14-3-3 isoforms in primary BMDM cells. (A) Mass spectrometry identification data for the two protein bands that were coeluted with Flag-6xMyc-Pyrin_ΔN purified from stably expressing DC2.4 cells shown in Fig. 2A. The two proteins disappeared upon stimulation of the cells with TcdB. (B) Quantitative real-time PCR (qRT-PCR) analyses of 14-3-3 expression in primary BMDMs. The mRNA level of 14-3-3 was normalized to that of Mefv. Sequences of the qRT-PCR primers used for the seven different 14-3-3 isoforms are available upon request.
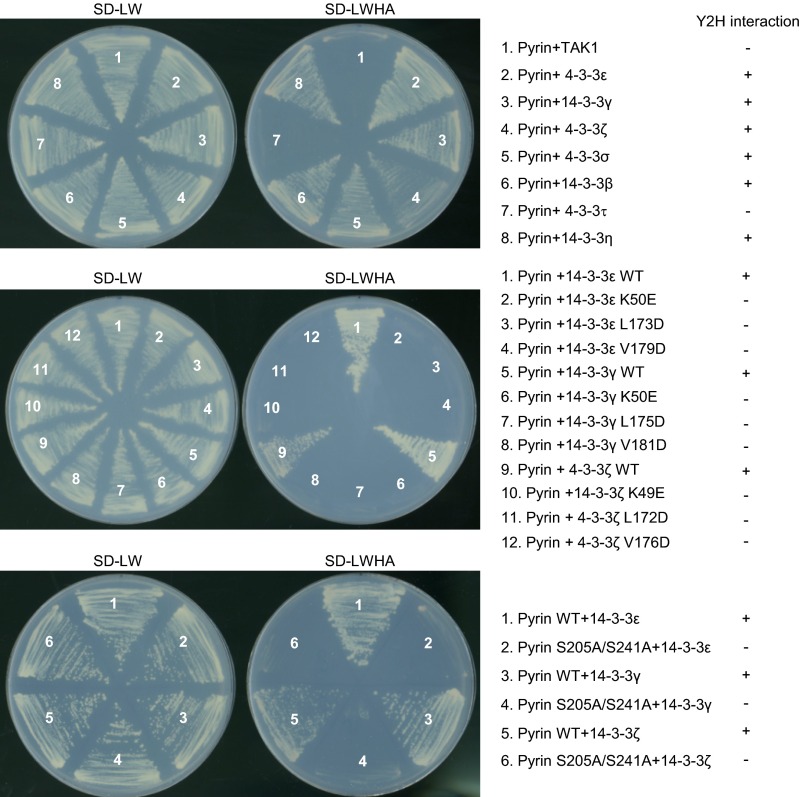
The 14-3-3 proteins are highly conserved from yeast to mammals and are abundantly expressed in eukaryotic cells (33). In human and mouse, there are seven distinct 14-3-3 genes, denoted β, γ, ε, η, σ, τ, and ζ. Transcripts of all seven 14-3-3 isoforms could be detected in mouse primary BMDMs (Fig. S1B), suggesting a functional redundancy in regulating Pyrin. In 293T cells, Pyrin_ΔN could coimmunoprecipitate all seven 14-3-3 isoforms, with a slight preference for the ε, γ, τ, and η isoforms (Fig. 2C). The interaction between Pyrin and the different 14-3-3 isoforms could also be recapitulated in the yeast two-hybrid system (Fig. S2). This finding is consistent with the highly similar structures and biochemical functions shared by the 14-3-3 proteins (34) and with most known 14-3-3–binding proteins showing little selectivity among the seven isoforms.
Fig. S2.
Yeast two-hybrid assay of the interaction between Pyrin and 14-3-3. Pyrin (wild type or the S205A/S241A double mutant) was cloned into the pGAD-GH bait vector; 14-3-3 (different isoforms or the indicated point mutations) was cloned into the pGBK-T7 prey vector. TAK1 in the prey vector was included as the negative control. The chart on the right summarizes the interaction results.
In addition to TcdB, IbpA_Fic1, and IbpA-Fic2, C. botulinum C3 toxin and B. cenocepacia infection could trigger the dissociation of endogenous 14-3-3ε from Pyrin in Pyrin_ΔN-expressing DC2.4 cells (Fig. 2D). TcdBm and B. cenocepacia Δhcp, which are deficient in Rho modification and Pyrin inflammasome activation (22), showed little effect on the in vivo interaction between Pyrin and 14-3-3ε (Fig. 2D). These data strongly indicate that the interaction with and dissociation from 14-3-3 are likely of functional significance for Pyrin activation.
14-3-3 Binding Requires Pyrin Phosphorylation.
The 14-3-3 family is the first identified phosphoserine/phosphothreonine (pS/pT)-binding protein (33). Structural studies have revealed that the conserved Lys-49, Leu-173, and Val-179 of 14-3-3ε, located in the inner surface of the ligand-binding groove, play a critical role in chelating the phosphate (35, 36). The K49E, L173D, and V179D mutants of 14-3-3ε were completely deficient in binding to Pyrin_ΔN in 293T cells (Fig. 3A). The three mutants as well as equivalent mutants of two other 14-3-3 isoforms were also defective in binding to Pyrin in the yeast two-hybrid system (Fig. S2). Furthermore, treatment of Pyrin_ΔN with λ phosphatase or other bacterial phosphatases (PP_E. coli and PP_Y, λ phosphatase-like phosphatases from E. coli and Y. enterocolitica, respectively) also disrupted the Pyrin–14-3-3ε association (Fig. 3B). Pyrin and Pyrin_ΔN stably expressed in DC2.4 cells migrated as two major bands on the phos-tag gel (37); the slower-migrating species disappeared upon λ phosphatase treatment (Fig. 3C). Similar results were obtained in 293T cells (Fig. 3D). Thus, the upper and lower bands represent phosphorylated Pyrin (pPyrin) and nonphosphorylated Pyrin, respectively. These analyses support the notion that 14-3-3 specifically recognizes pPyrin in resting cells.
Fig. 3.
Phosphorylation of Pyrin mediates its interaction with 14-3-3. (A and B) Coimmunoprecipitation assays of Pyrin and 14-3-3ε interaction in 293T cells. (A) Flag-Pyrin_ΔN was expressed into 293T cells. HA-tagged 14-3-3ε or its phospho-binding–deficient mutants were cotransfected with Flag-Pyrin_ΔN. (B) Cell lysates were subjected to anti-Flag immunoprecipitation. Cells were stimulated with TcdB before immunoprecipitation, or the immunoprecipitates were treated with the indicated recombinant phosphatases. PP_E. coli and PP_Y are two λ phosphatase-like phosphatases from E. coli and Y. enterocolitica, respectively. (C and D) Phos-tag gel analyses of Pyrin phosphorylation and its response to TcdB or λ phosphatase treatment. (C) DC2.4 cells expressing full-length Pyrin (Upper) or Pyrin_ΔN (Lower) were assayed. (D) Flag-tagged 14-3-3ε was cotransfected with HA-tagged full-length Pyrin into 293T cells. In C and D, live cells were stimulated with TcdB; λ phosphatase was added into the cell lysates before analyses in C. Flag-14-3-3ε also was immunoprecipitated and analyzed by immunoblotting as indicated. (E and F) Phos-tag gel analyses of Pyrin phosphorylation and its effect on caspase-1 activation and14-3-3ε interaction. DC2.4 cells expressing full-length Pyrin (E) or Pyrin_ΔN (F) were stimulated with the indicated Rho-modifying toxins or were infected with B. cenocepacia (wild type or the Δhcp mutant strain). Cell supernatants were analyzed by anti–caspase-1 immunoblotting (E), and cell lysates were also subjected to anti-Flag immunoprecipitation (F). The phos-tag gel analyses were performed with total cell lysates. The total lysates (Input) and the immunoprecipitates (Flag IP) were analyzed by immunoblotting using indicated antibodies. Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. All data shown are representative of three independent experiments.
Pyrin Dephosphorylation Correlates with 14-3-3 Disassociation and Inflammasome Activation.
TcdB and other Pyrin inflammasome agonists all triggered disassociation of 14-3-3 from Pyrin. Consistently, TcdB stimulation caused the disappearance of pPyrin (Fig. 3 C and D). Reciprocally, only pPyrin, but not the unphosphorylated form, was present in Flag–14-3-3 immunoprecipitates, which were sensitive to TcdB stimulation (Fig. 3D). TcdA and IbpA_Fic1 showed the same effect (Fig. 3E). Toxin-induced dephosphorylation of Pyrin correlated well with caspase-1 activation in DC 2.4 cells. In contrast, inactive mutants of these toxins that are incapable of inducing caspase-1 activation did not affect Pyrin phosphorylation (Fig. 3E). The disappearance of pPyrin was not caused by protein degradation, because the total level of cellular Pyrin did not change (Fig. 3E). B. cenocepacia-induced Pyrin dephosphorylation also correlated with the dissociation of 14-3-3ε (Fig. 3F). Pyrin_ΔN, which cannot signal caspase-1 activation, showed an intact response to Rho-inactivating toxins (Fig. 3 C and F), suggesting that dephosphorylation is independent of caspase-1 activation. Thus, 14-3-3ε interacts with phosphorylated Pyrin, and toxin triggers Pyrin dephosphorylation and 14-3-3ε dissociation during inflammasome activation.
Phosphorylation of Ser-205 and Ser-241 Is Important for 14-3-3 Binding.
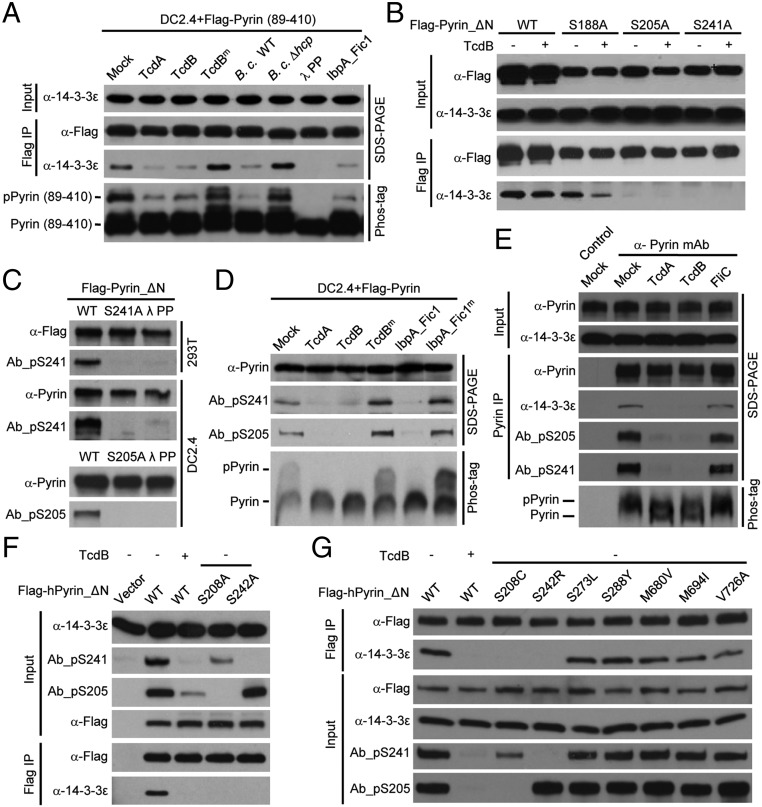
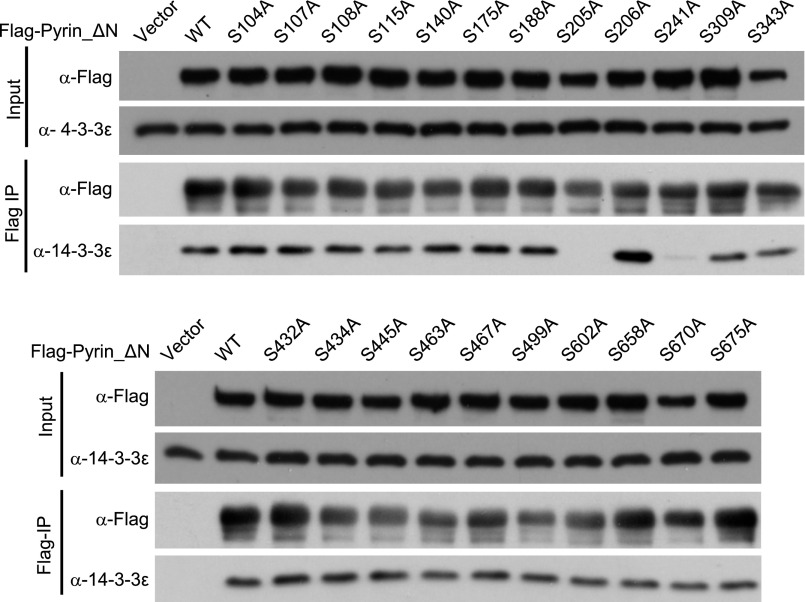
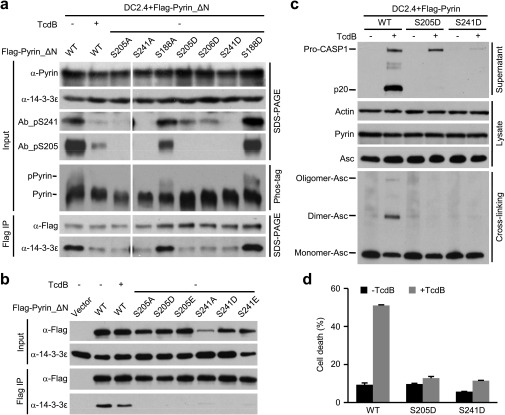
To identify the exact phosphorylation sites in Pyrin, we performed truncation analyses and identified a mouse Pyrin fragment (residues 89–410) that was fully responsive to TcdA/TcdB, IbpA_Fic1, and B. cenocepacia infection (Fig. 4A). Phos-tag gel analyses confirmed the phosphorylation of Flag-Pyrin (89–410) and its dephosphorylation in response to the Rho-modifying agents (Fig. 4A). This finding suggests that the functional phosphorylation site(s) are located within residues 89–410. We then performed tandem mass spectrometry (MS/MS) analysis of Flag-Pyrin (89–410) purified from unstimulated DC2.4 cells. The analyses reached ∼95% sequence coverage and consistently identified three phosphorylated residues, Ser-188, Ser-205, and Ser-241 (Fig. S3). When the same experiment was performed with Pyrin_ΔN, an additional phosphorylated residue, Ser-467, was identified. Among the four serine residues, only S205A and S241A mutations could abolish the coimmunoprecipitation between Pyrin_ΔN and 14-3-3ε (Fig. 4B and Fig. S4). Ser-188 was further excluded because Pyrin S188A showed an intact response in TcdB-induced 14-3-3ε dissociation (Fig. 4B). A yeast two-hybrid assay confirmed that Ser-205 and Ser-241 are critical for Pyrin interaction with different 14-3-3 isoforms (Fig. S2). Individual mutation of the other 19 serine residues that are conserved in human and mouse Pyrin did not affect the interaction between Pyrin and 14-3-3ε (Fig. S4).
Fig. 4.
Dephosphorylation of Pyrin on Ser-205 and Ser-241 correlates with activation of the Pyrin inflammasome.(A and B) Assays of Pyrin (89–410) truncation and serine mutants of Pyrin_ΔN in signal-induced dephosphorylation and 14-3-3ε disassociation. DC2.4 cells stably expressing Flag-Pyrin (89–410) (A) and 293T cells expressing Flag-Pyrin_ΔN (or the indicated serine mutants) (B) were stimulated with TcdB or with the other indicated Pyrin inflammasome stimuli. (C) Flag-Pyrin_ΔN or its phosphorylation-site mutants were transiently expressed in 293T cells or stably expressed in DC2.4 cells. Total cell lysates were either directly subjected to immunoblotting analyses or were treated first with λ phosphatase. (D and E) Assays of toxin-induced Ser-241/205 dephosphorylation and 14-3-3ε disassociation from Pyrin. DC2.4 cells stably expressing Flag-Pyrin (D) or intact primary BMDMs (E) were stimulated with the indicated Rho-modifying toxins. (F and G) Assays of human Pyrin phosphorylation on Ser-242/Ser-208 and effects of point mutations of serine residues. 293T cells transiently transfected with indicated human Pyrin_ΔN (hPyrin_ΔN) point mutations were treated with or without TcdB. The point mutants analyzed in F are mutants found in FMF patients. Exogenous Flag-tagged Pyrin was immunoprecipitated using anti-Flag antibody (A, B, F, and G), and endogenous Pyrin was immunoprecipitated using a monoclonal anti-Pyrin antibody (E). The immunoprecipitates and total lysates were analyzed by anti-Flag, anti-Pyrin, anti–14-3-3ε, or the phospho-specific antibodies (Ab_pS241 and Ab_pS205) immunoblotting as indicated. Ab_pS241 and Ab_pS205 denote rabbit phospho-S241– and phospho-S205–specific monoclonal antibodies, respectively. The total lysates also were subjected to the phos-tag gel immunoblotting (A, D, and E). All data shown are representative of three independent experiments.
Fig. S3.
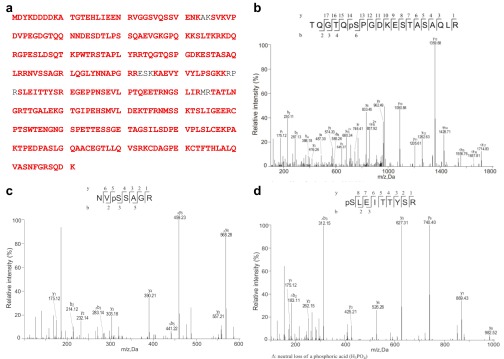
Identification of Pyrin phosphorylation sites by mass spectrometry. Flag-Pyrin (89–410) was affinity-purified from the stable DC2.4 cell line and was subjected to mass spectrometry analysis. (A) Red-highlighted sequences are tryptic peptides identified from the spectra obtained. The sequence coverage is about 95%, among which three serine residues, Ser-188, Ser-205, and Ser-241, were found to be phosphorylated. (B–D) The MS/MS spectra of tryptic peptides containing Ser-188, Ser-205, and Ser-241 are shown in B, C, and D respectively. The β- and γ-type product ions are marked in the spectra and are illustrated along with the corresponding peptide sequence above spectra. Data shown are representative of two independent experiments.
Fig. S4.
Mutation screen of candidate serine residues in Pyrin that are important for 14-3-3 binding. 293T cells transiently transfected with constructs of the single serine mutation in Flag-Pyrin_ΔN were subjected to anti-Flag immunoprecipitation. Anti-Flag and anti–4-3-3ε immunoblots of the total cell lysates (Input) and the immunoprecipitates (Flag IP) are shown. Ser-188, Ser-205, and Ser-241 are the phospho-serine residues identified from mass spectrometry analyses in Fig. S3. Other serine residues conserved in human and mouse Pyrin were also included in the assay. Endogenous 14-3-3ε was examined. The experiments were performed three times; representative results are shown.
Ser-205/241 Are Dephosphorylated on Toxin Stimulation.
To examine Pyrin phosphorylation on Ser-205 and Ser-241 directly, we developed two rabbit monoclonal phospho-specific antibodies, Ab_pS241 and Ab_pS205, using mouse Pyrin-derived phosphopeptides as the antigens. Both antibodies could readily recognize mouse Pyrin expressed in 293T and DC2.4 cells but not the corresponding phosphorylation-site mutants (S241A and S205A) (Fig. 4C). Treatment with λ phosphatase diminished the immunoblotting signals of Ab_pS241 and Ab_pS205 (Fig. 4C). The phospho-specific immunoblotting signals also disappeared upon inflammasome activation by TcdA, TcdB, and Ibp_Fic1, as is consistent with the absence of pPyrin on the phos-tag gel (Fig. 4D).
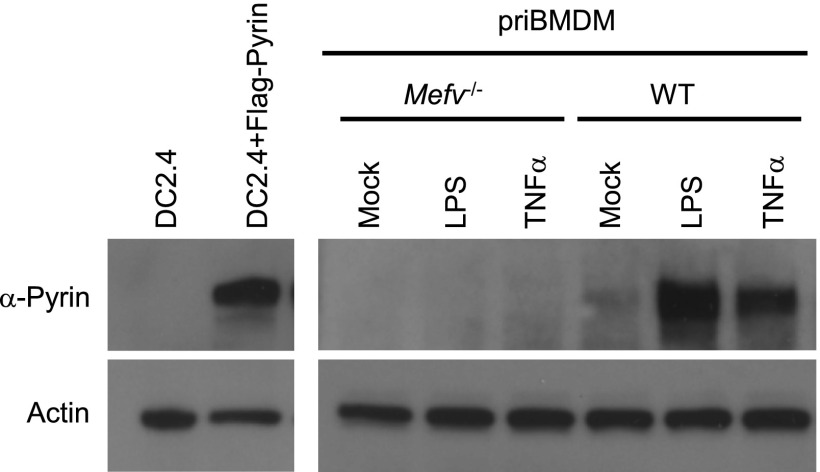
We also developed a monoclonal antibody that could detect endogenous Pyrin in primary BMDMs. As expected, the immunoblotting signal markedly increased upon LPS or TNF-α priming (Fig. S5). Both Ab_pS241 and Ab_pS205 produced a robust and specific immunoblotting signal on endogenous Pyrin precipitated using the monoclonal anti-Pyrin antibody from BMDM lysates (Fig. 4E). These phospho-specific signals were abolished by TcdA or TcdB, which correlated with the dissociation of 14-3-3 from endogenous Pyrin (Fig. 4E). Interestingly, individual mutation of Ser-205 and Ser-241 affected the phosphorylation status of one another, but theS188A mutation did not affect Ser-205 and Ser-241 phosphorylation (Fig. S6A). This effect might result from structural changes in the single serine mutation that can affect phosphorylation of the other serine; alternatively, phosphorylation or maintenance of phosphorylation on the two serine residues may have a synergistic effect. Moreover, S205D/E and S241D/E-mutant Pyrin resisted recognition by the phospho-specific antibodies (Fig. S6A), suggesting that the aspartate/glutamate substitutions cannot properly mimic phosphorylated Pyrin. Supporting this notion, S205D and S241D mutations also abolished the association between Pyrin and 14-3-3ε in 293T and DC2.4 cells (Fig. S6 A and B). These results provide direct evidence that Pyrin is phosphorylated on Ser-205 and Ser-241 and that Rho-modifying toxins trigger their dephosphorylation.
Fig. S5.
Validation of the rabbit monoclonal anti-Pyrin antibody. Lysates of DC2.4 cells or DC2.4 cells expressing Flag-Pyrin (Left) and primary mouse BMDMs (priBMDM) (Right) were subjected to immunoblotting analyses using the rabbit monoclonal antibody developed using recombinant mouse Pyrin (89–410) as the antigen and anti-actin antibody as the loading control. The BMDMs were derived from wild-type or Mefv−/− mice and were either left untreated (Mock) or stimulated with LPS or TNF-α.
Fig. S6.
The aspartate/glutamate substitutions of Ser-205 and Ser-241 do not functionally mimic the phosphorylated serine. (A and B) Flag-Pyrin_ΔN or the indicated single mutation stably expressed in DC2.4 cells (A) or transiently expressed in 293T cells (B) was subjected to anti-Flag immunoprecipitation. Cells were either left untreated or were stimulated with TcdB. Immunoblots of the precipitates (Flag IP) and the total cell lysates (Input) using the indicated antibodies are shown. Phosphorylation of Pyrin was also examined by the phos-tag gel immunoblotting (A). (C and D) Effects of aspartate substitution of Ser-205 and Ser-241 in Pyrin on TcdB-induced activation of the Pyrin inflammasome. DC2.4 cells stably expressing wild-type Flag-Pyrin or its S205D or S241D mutant were left untreated or were stimulated with TcdB. Cell supernatants were analyzed by anti–caspase-1 immunoblotting; Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. (C) Total cell lysates were blotted with anti-actin, anti-ASC, or anti-Pyrin antibody or were subjected to crosslinking before anti-ASC immunoblotting. (D) LDH release-based cell death is expressed as mean values ± SD from three technical replicates. Data shown are representative of three independent experiments.
Human Pyrin Adopts a Similar Phosphorylation/Dephosphorylation Pattern.
All the studies described above were performed with mouse Pyrin. We then examined whether phosphorylation-mediated control of Pyrin inflammasome activation also applies to human protein. Human Pyrin expressed in 293T cells also interacted with 14-3-3ε, and the interaction was disrupted by TcdB stimulation (Fig. 4 F and G). Alanine substitution of Ser-208 or Ser-242 in human Pyrin, equivalent to Ser-205 and Ser-241 in its mouse counterpart, abolished the association between Pyrin and 14-3-3ε. Notably, the two phospho-antibodies could also detect human Pyrin, which was abolished by TcdB stimulation (Fig. 4 F and G). S208C and S242R mutations have been observed in FMF patients (fmf.igh.cnrs.fr/ISSAID/infevers/search.php?n=1), further supporting the importance of the two phosphorylation sites. The two disease mutants, in contrast to several other nonrelated FMF disease mutants, resisted detection by the corresponding phospho-specific antibodies (Fig. 4G).The two phospho-specific antibodies might be of potential use in clinical diagnosis of FMF.
Ser-205/241 Dephosphorylation Is Important for Pyrin Activation.
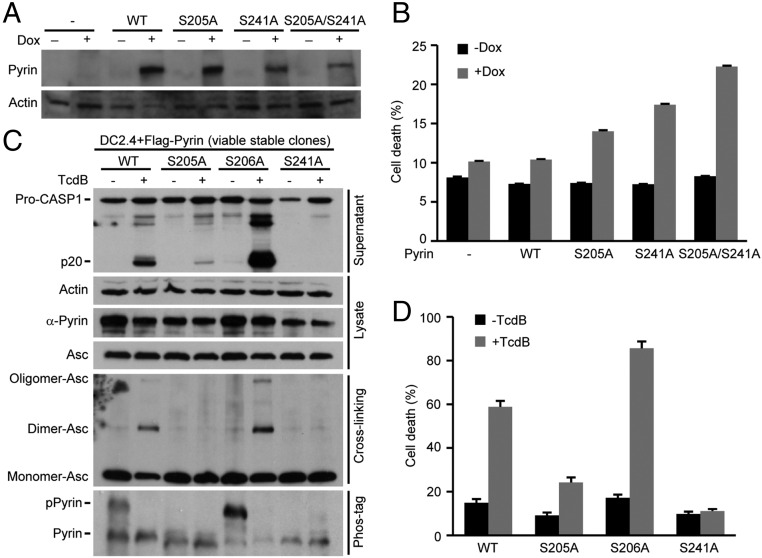
To investigate whether the change in the phosphorylation status of Pyrin on Ser-205 and Ser-241 plays any role in the activation of the Pyrin inflammasome, we generated tetracycline-inducible expression of Pyrin S205A, S241A, or the S205A/S241A double mutant in DC2.4 cells. When the drug was added to trigger the expression of the phosphorylation-negative mutants (Fig. 5A), a fraction of cells underwent pyroptotic cell death in the absence of inflammasome agonist stimulation (Fig. 5B). A higher percentage of pyroptosis was observed in cells expressing the S205A/S241A double mutant than in cells expressing the single S/A mutant. Because the inducible expression of the alanine mutants of Pyrin does not exactly mimic signal-induced dephosphorylation of the serine, these data do support the idea that S205 and S241 dephosphorylation plays a positive role in toxin-stimulated activation of the Pyrin inflammasome. Moreover, because ectopic expression of the phosphorylation-site mutant did not result in 100% killing, we were able to generate DC2.4 cells that stably expressed Pyrin S105A or S241A after selecting viable transfected cell clones. Phos-tag gel analyses confirmed the absence of pPyrin in cells expressing either mutant (Fig. 5C). When the cells were stimulated with TcdB, both the S205A and S241A mutations showed a complete inhibition on caspase-1 activation (Fig. 5C). Accordingly, ASC oligomerization and pyroptosis were also inhibited (Fig. 5 C and D). S105D and S241D mutant Pyrin showed similar inhibition on TcdB-induced activation of the Pyrin inflammasome (Fig. S6 C and D). We also noticed that the S206A mutation resulted in increased Pyrin phosphorylation (Fig. 5C) and association with 14-3-3ε (Fig. S4). The S206A mutation rendered more robust caspase-1 activation and inflammasome responses upon TcdB stimulation (Fig. 5 C and D). Phos-tag gel analyses revealed rapid dephosphorylation in the S206A mutant (Fig. 5C). Taken together, these findings indicate that both the signal-induced dephosphorylation on Ser-205/Ser-241 and phosphorylation of the resting-state Pyrin are important for activation of the Pyrin inflammasome by Rho-modifying toxins.
Fig. 5.
Phosphorylation and dephosphorylation of Pyrin on Ser-205/241 are important for toxin-induced activation of the Pyrin inflammasome. (A and B) Tet-on expression of Pyrin phosphorylation-negative mutants induces pyroptotic cell death. Wild-type Pyrin or the Pyrin S205A or S241A mutant was stably transfected into DC2.4 cells under a tetracycline-inducible promoter. Doxycycline (Dox) was added to induce Pyrin expression. Immunoblotting analyses of Pyrin expression are shown in A. (C and D) Assays of phosphorylation-site mutants on TcdB-induced Pyrin inflammasome activation. Viable DC2.4 cells stably expressing wild-type or the indicated serine mutants of Pyrin were subjected to TcdB stimulation. Cell supernatants were analyzed by anti–caspase-1 immunoblotting. Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. Total cell lysates were subjected to immunoblotting analyses as indicated (C), in which cross-linking and phos-tag gel electrophoresis were performed to examine ASC oligomerization and the phosphorylation status of Pyrin, respectively. LDH release-based cell death shown in B and D is expressed as mean values ± SD from three technical replicates. All data shown are representative of three independent experiments.
Microtubule Dynamics Controls Pyrin Inflammasome Activation Downstream of Its Dephosphorylation.
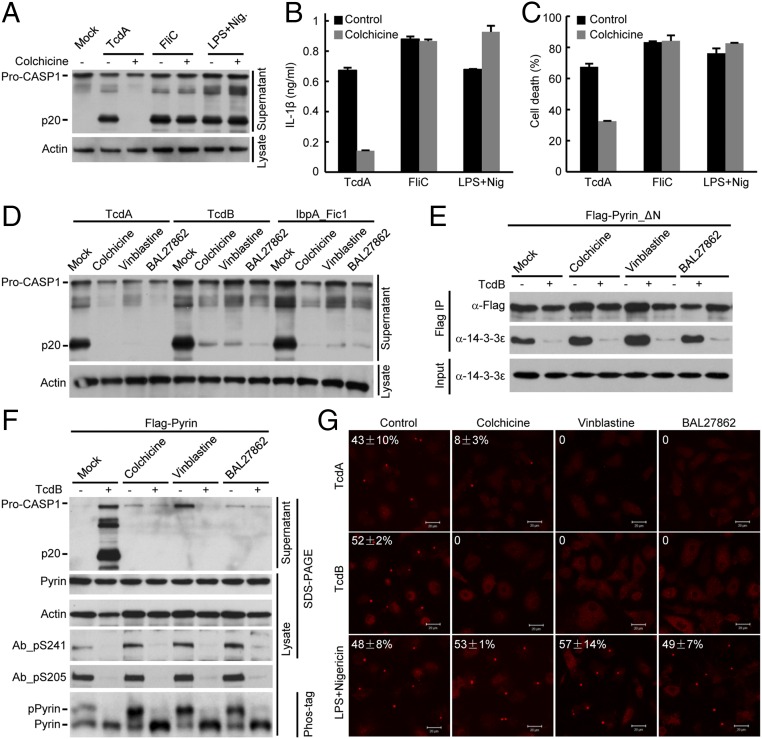
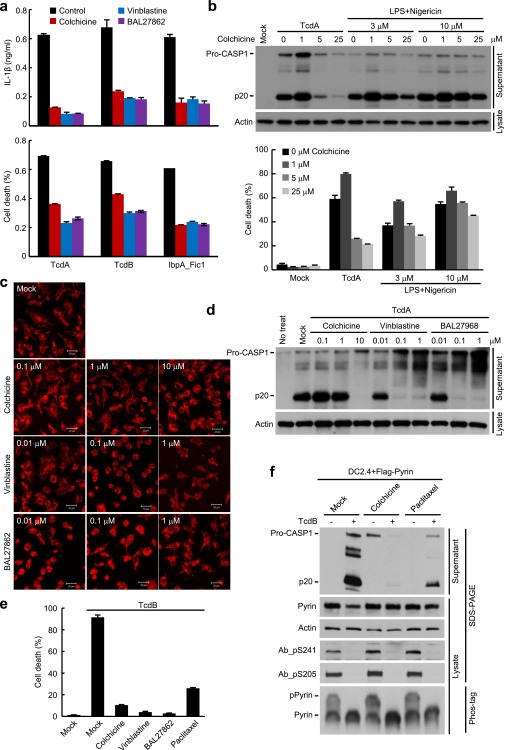
Pyrin has been shown to be associated with the microtubules (38). Colchicine, the microtubule-disrupting compound, is most effective in clinical treatment of FMF, but the exact mechanism is not clear (19). We found that colchicine could effectively block TcdA-induced caspase-1 activation in mouse BMDMs (Fig. 6A). Accordingly, IL-1β release and pyroptosis were both diminished (Fig. 6 B and C). Similarly, colchicine could block Pyrin inflammasome activation by TcdB and IbpA_Fic1 (Fig. 6D and Fig. S7A) but did not inhibit flagellin-stimulated NAIP5-NLRC4 inflammasome (Fig. 6 A–C). Consistent with the previous report (39), colchicine weakly inhibited the activation of the NLRP3 inflammasome induced by LPS plus 3 μM nigericin (Fig. S7B). The inhibition was not observed when the higher standard concentration of nigericin (10 μM) was used (Fig. 6 A–C and Fig. S7B). These results highlight a specific inhibition of the Pyrin inflammasome by colchicine.
Fig. 6.
Microtubule dynamics controls the activation of the Pyrin inflammasome downstream of Pyrin dephosphorylation. (A–D) Specific inhibition of Pyrin inflammasome activation by the microtubule-targeting drugs colchicine, vinblastine, and BAL27862. Primary BMDMs were preincubated with the indicated drugs and then were stimulated with TcdA, TcdB, IbpA_Fic1, or other inflammasome agonists. IbpA_Fic1 and FliC were delivered into the cells by the LFn-PA system. LPS plus nigericin (LPS+Nig) was used to stimulate NLRP3 inflammasome activation. (E and F) Effects of microtubule-targeting drugs on TcdB-induced 14-3-3 dissociation from Pyrin and Pyrin dephosphorylation. DC2.4 cells stably expressing Pyrin_ΔN (E) or full-length Pyrin (F) were pretreated with the indicated compounds and then were stimulated with TcdB. The interaction between Pyrin_ΔN and endogenous 14-3-3ε was assayed by anti-Flag immunoprecipitation (E), and the phosphorylation status of Pyrin was examined by the phospho-specific antibody blotting (Ab_pS241 and Ab_pS205) as well as by the phos-tag gel immunoblotting analysis (F). (G) Effects of microtubule-targeting drugs on TcdB-induced ASC aggregation downstream of Pyrin activation. Primary BMDM cells pretreated with colchicine, vinblastine, or BAL27862 were stimulated with TcdA, TcdB, or LPS plus nigericin. Shown are the anti-ASC (endogenous) immunofluorescence images of stimulated cells. The percentages of cells developing the ASC foci structure are marked on the images (mean values ± SD). (Scale bars: 20 μm.) Cell supernatants were analyzed by anti–caspase-1 immunoblotting in A, D, and F; Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. ELISA of IL-1β release (B) and LDH release-based cell death (C) are expressed as mean values ± SD from three technical replicates. All data shown are representative of three independent experiments.
Fig. S7.
Microtubule-disrupting or -stabilizing drugs inhibit the activation of the Pyrin inflammasome. (A and B) Effects of the microtubule-targeting drugs colchicine, vinblastine, and BAL27862 on activation of the Pyrin inflammasome. Primary BMDM cells were preincubated with indicated drugs and then were stimulated with TcdA, TcdB, or IbpA_Fic1 (A and B) or with LPS plus the indicated concentration of nigericin (B). IbpA_Fic1 was delivered into the cells by the LFn-PA system. (C and D) Comparison of the activity of colchicine, vinblastine, and BAL27862 on microtubule disruption and their inhibition of Pyrin inflammasome activation. Primary BMDMs were pretreated with the indicated compounds at different concentrations and then were stimulated with TcdA. Anti-tubulin fluorescence images are shown in C. (E and F) Assays of the effect of the microtubule-stabilizing drug paclitaxel on TcdB-induced activation of the Pyrin inflammasome. DC2.4 cells stably expressing full-length Pyrin were pretreated with paclitaxel or the indicated microtubule-disrupting compounds. ELISA of IL-1β release (A) and LDH release-based cell death (A, B, and E) are expressed as mean values ± SD from three technical replicates. Cell supernatants (B, D, and F) were analyzed by anti–caspase-1 immunoblotting. Pro-CASP1, the caspase-1 precursor; p20, the mature caspase-1. Total cell lysates were blotted with anti-actin (B, D, and F) or with anti-Pyrin antibody and the rabbit phospho-S241– and phospho-S205–specific antibodies (Ab_pS241 and Ab_pS205) (F). Pyrin phosphorylation was also examined by phos-tag gel immunoblotting. Data shown are representative of three independent experiments.
Microtubule-targeting drugs are classified into two classes that cause either a stabilizing or a destabilizing effect. The destabilizing drugs usually bind to one of two sites. The vinblastine-binding site is located between two longitudinally aligned tubulin dimers, and colchicine targets the intrasubunit interface within a tubulin dimer (40, 41). BAL27862, a new microtubule-destabilizing drug currently in clinical trials for treating advanced solid tumor, targets the same site in the unassembled tubulin as colchicine (42). BAL27862 binding to tubulin is reversible and has a higher affinity (in the nanomolar range) and different kinetics than colchicine. We found that both vinblastine and BAL27862 could inhibit activation of the Pyrin inflammasome (Fig. 6D and Fig. S7A). Dose titration performed in primary BMDMs showed that colchicine, vinblastine, and BAL27862 disrupted the microtubule network and caused cell rounding at the concentrations of 10 μM, 0.1 μM, and 0.1 μM, respectively; these dosages correlate with those required for inhibiting TcdA-induced capase-1 activation (Fig. S7 C and D). These findings support the idea that the three compounds block Pyrin activation by inhibiting microtubule polymerization. Paclitaxel, a classical microtubule-stabilizing drug, also was capable of inhibiting TcdB-induced caspase-1 activation and pyroptosis (Fig. S7 E and F). Thus, normal microtubule dynamics is likely important for the activation of the Pyrin inflammasome by toxin-mediated Rho modification.
We further investigated the step at which the tubulin drugs blocked the activation of the Pyrin inflammasome. Notably, TcdB-induced 14-3-3ε dissociation from Pyrin was not affected by colchicine, vinblastine, or BAL27862 (Fig. 6E). Consistently, the three drugs did not affect TcdB-triggered Pyrin dephosphorylation on Ser-241 and Ser-205 (Fig. 6F). Similar results were obtained with paclitaxel (Fig. S7F). Downstream of Pyrin dephosphorylation and 14-3-3 disassociation are the recruitment of the ASC adaptor and stimulation of ASC aggregation. We observed that TcdB-triggered endogenous ASC aggregation was completely inhibited by colchicine, vinblastine, and BAL27862 in primary BMDMs (Fig. 6G). Identical results were obtained with TcdA. The three compounds did not affect the formation of ASC foci induced by LPA plus nigericin (Fig. 6G). These data indicate that microtubule dynamics critically controls the activation of the Pyrin inflammasome, possibly by affecting the conformational change of dephosphorylated Pyrin and subsequent recruitment of ASC.
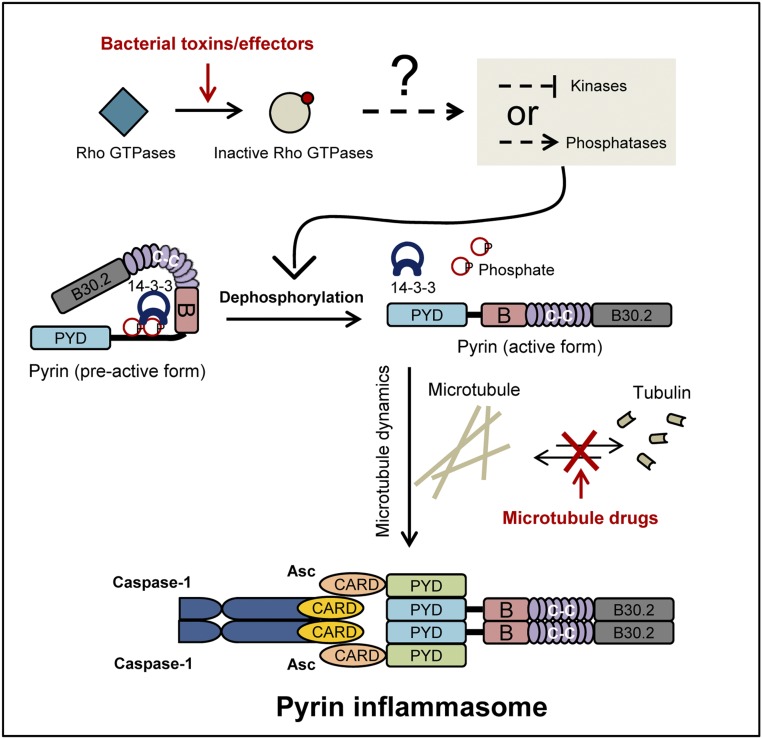
In summary (Fig. 7), we discovered that Pyrin is phosphorylated on Ser-205 and Ser-241 and therefore is inhibited by 14-3-3 binding in resting cells. Upon toxin stimulation or bacterial infection that leads to Rho modification and inactivation, Ser-205 and Ser-241 are dephosphorylated. The dephosphorylation triggers 14-3-3 dissociation (and possibly a series of conformational changes in Pyrin) that ultimately results in Pyrin activation and the formation of an oligomeric Pyrin–ASC inflammasome complex. Moreover, microtubule dynamics plays an important role in Pyrin activation, and microtubule-targeting drugs such as colchicine inhibit Pyrin-mediated ASC aggregation without affecting Pyrin dephosphorylation.
Fig. 7.
A model of Pyrin inflammasome activation by Rho-modifying toxins.
Discussion
Pyrin has two functional phosphorylation sites, Ser-205 and Ser-241 (Ser-208 and Ser-242 in human Pyrin), both of which are required for 14-3-3 binding and which undergo dephosphorylation upon toxin stimulation. There are two possible explanations for the requirement of both phosphorylation sites. Given that 14-3-3 is known to form a dimer in binding to its phosphorylated partner, one intuitive hypothesis is that the two 14-3-3 monomers simultaneously bind to the two phosphorylated serines in one Pyrin. An alternative explanation is that only one phosphorylated serine is directly engaged in 14-3-3 binding, but phosphorylation/dephosphorylation of the two serine residues is a highly coordinated event. In support of the latter model, mutation of Ser-205 could affect the phosphorylation of Ser-241. We speculate that the two serine residues are spatially close to each other and possibly interact, perhaps explaining why aspartate/glutamate substitution of only one of the two serine residues cannot functionally mimic the phosphorylated serine.
We observed that colchicine and other microtubule-targeting drugs could block the activation of the Pyrin inflammasome. This result provides a plausible mechanistic explanation for the clinical use of colchicine in FMF therapy. These drugs do not affect toxin-induced Pyrin dephosphorylation and dissociation of the Pyrin–14-3-3 complex but block the oligomerization of Pyrin with ASC. Thus, the microtubule network possibly serves as a platform for a conformational change in Pyrin and subsequent ASC recruitment following toxin-induced dephosphorylation. The Pyrin inflammasome responds to Rho-inactivating toxins (22, 23) and also to an inactivating mutation in WDR1 (25), both of which cause depolymerization of the actin filaments. Thus, Pyrin may function to monitor the proper dynamics of the actin cytoskeleton and particularly pathological disruptions of the actin dynamics. These notions suggest a paradigm in which the innate immune system is highly engaged with the cytoskeleton structure in mammalian cells. Because 14-3-3 has a rigid structure and generally does not function as a signal transducer (33), the upstream stimulus likely signals Pyrin to modulate its phosphorylation status. A kinase or a phosphatase associated with the actin cytoskeleton downstream of Rho GTPase likely regulates Pyrin phosphorylation and controls the activation of the Pyrin inflammasome.
Materials and Methods
Plasmids, Antibodies, and Reagents.
cDNA of mouse Mefv isoform1 (NM_001161790.1) was amplified from reverse-transcribed cDNA from primary BMDMs. cDNA of human MEFV isoform 1 (NM_000243.2) was synthesized by the in-house gene synthesis facility as previously described (22). The two genes were inserted into the pWPI-IRES-eGFP vector with an N-terminal Flag tag for generating the cell line stably expressing the pCS2 vector with an N-terminal triple HA tag for immunoprecipitation, the pLenti-NIrD vector with no tag for inducible expression, and the pGAD-GH for yeast two-hybrid assay. Truncations were constructed by the standard PCR cloning strategy. For 14-3-3 isoforms, cDNAs of 14-3-3ε (Ywhae, NM_009536) and 14-3-3γ (Ywhag, NM_018871) were amplified from reverse-transcribed cDNA from primary BMDM cells. CDNA of 14-3-3σ (Ywhas, NM_018754) was synthesized by the in-house facility. cDNAs of 14-3-3β (YWHAB, NM_003404), 14-3-3ζ (YWHAZ, NM_001135700), 14-3-3τ (YWHAQ, NM_006826), and 14-3-3η (YWHAH, NM_003405) were amplified from a commercial human cDNA library (Invitrogen). All the 14-3-3 genes were inserted into the pCS2 vector with an N-terminal 3xHA or 3xFlag tag for immunoprecipitation and into the pGBK-T7 vector for the yeast two-hybrid assay. cDNAs of λ phosphatase-like phosphatase from Yersinia enterocolitica (YE3016, NC_008800) and Escherichia coli (ECs0813, NC_002695) were amplified from the corresponding bacterial genomic DNA and cloned into the pET28a vector for recombinant expression in E. coli. pHIS1522-TcdB and pHIS1522-TcdA for expression in Bacillus megaterium were kindly provided by Hanping Feng, University of Maryland, Baltimore. pHIS1522-TcdBm (W102A/D288N), pET28a-LFn-C3, pET28a-LFn-IbpA-Fic1/Fic2, and pET28a-LFn-Flagellin were described in our previous publication (22). The PA (protective antigen)-expressing plasmid was obtained from Addgene. All point mutation constructs were generated by the overlapping PCR method or using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All plasmids were verified by DNA sequencing.
The rabbit polyclonal antibody against mouse Pyrin was generated using recombinant mouse Pyrin (residues 89–410) as the immunogen on which the rabbit monoclonal antibody (ab195975) was further developed by Abcam as a collaborative project. The rabbit monoclonal antibodies (Ab_pS241, ab200420 and Ab_pS205, ab201784) against phosphorylated mouse Pyrin were also developed by Abcam using the synthetic phospho-Ser-241/205–containing peptides as the antigens. Hybridoma lines were screened by Western blotting and immunoprecipitation analysis. Antibody for caspase 1(p20) was kindly provided by Vishva Dixit, Genentech, Inc., San Francisco. Home-made rabbit polyclonal anti-ASC antibody was described previously (43). Other antibodies used in this study include HA (MMS-101P; Covance), 14-3-3ε (Santa Cruz Biotechnology), rabbit anti-actin antibody (A2066; Sigma-Aldrich), and rabbit anti-Flag M2 antibody (F4049; Sigma-Aldrich).
Phos-tag Acrylamide AAL-107 aqueous solution (catalog no. 304-93526) was purchased from Wako. LPS (catalog no. L4524) and colchicine (catalog no. C9154) were purchased from Sigma-Aldrich. Vinblastine sulfate (catalog no. 11762) and paclitaxel (catalog no.10461) were obtained from Cayman Chemical Company. BAL27862 was synthesized by our in-house chemical facility (42). Anti-Flag M2 affinity beads were from Sigma-Aldrich, and Protein A agarose beads were from Thermo Fisher. The cross-linking agent disuccinimidyl suberate (DSS) was also purchased from Thermo Fisher (catalog no. 21555). Purified λ phosphatase was purchased from New England Biolabs. Cell-culture products are from Life Technologies, Inc., and all other chemicals used are Sigma-Aldrich products unless noted.
Cell Culture and Transgene Stable Expression.
iBMDM cells derived from HEK 293T and C57BL/6 mice were grown in DMEM. Primary BMDMs and the mouse DC2.4 dendritic cell line were grown in Roswell Park Memorial Institute (RPMI) medium 1640. All cell cultural media were supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, 100 U/L penicillin, and 100 mg/mL streptomycin. All cell lines were obtained from ATCC except for the iBMDM and DC2.4 cells, which have been described in our previous publications (11, 22). Cells were cultivated in a humidified atmosphere containing 5% CO2 at 37 °C. The commonly used PCR-based method was used to confirm that the cells were mycoplasma-negative.
Wild-type C57BL/6 mice were obtained from Vital River Laboratory Animal Technology Co. Asc−/− mice were provided by Vishva Dixit, and Mefv−/− mice were generated as described previously (22). Animal experiments were conducted following Ministry of Health national guidelines for housing and care of laboratory animals and were performed in accordance with institutional regulations after review and approval by the Institutional Animal Care and Use Committee at the National Institute of Biological Sciences. Primary BMDM cells were prepared from 6-wk-old male mice (C57BL/6 background) by following a standard procedure as previously described (43). For each relevant experiment, at least two mice were killed to prepare the BMDMs for assaying inflammasome activation.
Vigofect (Vigorous) was used according to the manufacturer’s instructions to transfect plasmid DNA into HEK 293T cells. For lentivirus packaging, 2.5 μg of pWPI-IRES-eGFP or pLenti-NIrD containing the indicated cDNA inserts, 1.0 μg of pMD2G, and 1.5 μg of psPAX2 packing plasmids (Addgene) were cotransfected into 293T cells in the six-well plate format. Four hours after transfection the cultural medium was replaced with 2 mM Hepes-buffered DMEM supplemented with 20% FBS and 2 mM l-glutamine. Forty-eight hours after transfection supernatants containing the lentivirus particles were collected and used to infect DC2.4 cells. Seventy-two hours postinfection cells carrying pWPI-IRES-eGFP insertions were isolated by FACS on the FACSAria II flow cytometer (BD Biosciences), and cells carrying pLenti-NIrD insertions were kept in medium containing Blasticidin (10 μg/mL) for 1 wk to generate the inducible Pyrin expression cell lines.
Phos-Tag Gel Detection of Pyrin Phosphorylation.
The samples to be analyzed were boiled in the SDS loading buffer. To examine the phosphorylation of full-length Pyrin, boiled samples were loaded onto the 6% SDS/PAGE gel containing 50 μmol/L Phos-tag AAL-107 and 100 μmol/L MnCl2. To examine the phosphorylation of Pyrin (89–410), 8% Phos-tag gel was used. Gels were run under a constant voltage condition of 150 V on ice. After the electrophoresis, gels were soaked twice (10 min each soaking) in the transfer buffer containing 10 mM EDTA to eliminate Mn2+ and then were soaked in the transfer buffer without EDTA for 10 min. Proteins were transferred to a PVDF membrane followed by immunoblotting analyses.
Cross-Linking Analysis of ASC Oligomerization.
DC2.4 cells stably expressing Pyrin and the indicated point mutations were harvested after TcdA stimulation. The cells were washed three times with PBS and were resuspended in PBS containing 1 mM DSS. The reaction mixtures were incubated at room temperature for 30 min and were stopped by Tris⋅HCl. The pellets were washed three times with PBS and were boiled in the SDS loading buffer for immunoblotting analyses.
Inflammasome Activation Assays.
Purified recombinant TcdA or TcdB was added to serum-free cell-culture medium at a final concentration of 1 μg/mL and 0.1 μg/mL, respectively. LFn-tagged proteins together with the PA protein were added into the culture medium at a final concentration of 1 μg/mL for cytosolic delivery of the recombinant protein. Cells were further incubated for 2 h (primary BMDMs) or 1 h (DC2.4 cells) before being subjected to the indicated assays. Supernatants of stimulated BMDM or DC2.4 cells were subjected to trichloroacetic acid precipitation, and the precipitates were analyzed by immunoblotting to detect caspase-1 activation. To measure IL-1β secretion, primary BMDMs were primed with LPS at a concentration of 100 ng/L for 3 h to induce IL-1β expression before being stimulated as indicated ; the released mature IL-1β was determined using the IL-1β ELISA kit (Neobioscience Technology Company). Percentages of pyroptotic cell death were measured by the lactate dehydrogenase assay using the CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega). For the microtubule drug inhibition experiment, each drug was added into cell-culture medium and incubated with the cells for 15 min at 37 °C before toxin stimulation.
Mass Spectrometric Analysis.
Protein bands on the silver-stained SDS/PAGE gel were destained and reduced in 10 mM DTT at 56 °C for 30 min followed by alkylation in 55 mM iodoacetamide in the dark for 1 h. The protein bands were then in-gel digested with sequencing-grade trypsin (10 ng/μL trypsin, 50 mM ammonium bicarbonate, pH 8.0) overnight at 37 °C. Peptides were extracted sequentially with 5% formic acid/50% acetonitrile and 0.1% formic acid/75% acetonitrile and then were concentrated to ∼20 μL. The extracted peptides were separated by an analytical capillary column (50 μm × 15 cm) packed with 5-μm spherical C18 reversed-phase material (YMC). A Waters nanoACQUITY UPLC system (Waters) was used to generate the following HPLC gradient: 0–30% B in 40 min and 30–70% B in 15 min (A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile). The eluted peptides were sprayed into an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nano-electrospray ionization ion source. The mass spectrometer was operated in data-dependent mode with one mass spectroscopy scan followed by four collision-induced dissociation and four high-energy collisional dissociation MS/MS scans for each cycle. For protein identification, database searches were performed on an in-house Mascot server (Matrix Science Ltd.) against the International Protein Index (IPI) mouse protein database. The search parameters were 10-ppm mass tolerance for precursor ions and 0.7-Da mass tolerance for product ions; two missed cleavage sites were allowed for trypsin digestion. Methionine oxidation was set as variable modification. The search results were filtered with both peptide significance threshold and expectation value below 0.05.
To identify the phosphorylation sites, database searches were performed against the mouse Pyrin isoform 1 protein sequence. The search parameters were 7-ppm mass tolerance for precursor ions and 0.5-Da mass tolerance for product ions; three missed cleavage sites were allowed for trypsin digestion, and the following variable modifications were included: oxidation on methionine, carbamidomethylation on cysteine, and phosphorylation on serine, threonine, and tyrosine. The tandem mass spectra of matched phosphorylated peptides were manually checked for their validity.
Bacterial Strains and Infection.
Wild-type B. cenocepacia J2315 strain obtained from the Belgian Coordinated Collection of Microorganisms/Ghent University Microbiology Laboratory (BCCM/LMG) bacteria collection and the previously described B. cenocepacia Δhcp mutant (22) were cultured overnight at 37 °C in 2xYT (16 g/l tryptone, 10 g/l yeast extract and 5 g/l NaCl) broth with shaking at 220 rpm. Bacterial cultures were diluted by 1:100 in fresh 2× YT broth and further grown to an OD600 of 0.8–1.0. The bacteria then were diluted in serum-free RPMI-1640 medium and added to the cell supernatant at a multiplicity of infection of 10. Cells were incubated at 37 °C after being centrifuged at 800 × g for 10 min, and the infection was allowed to proceed for 1 h before being subjected to the indicated assays.
SI Materials and Methods
Immunoprecipitation and Immunoblotting.
For immunoprecipitation of Flag-Pyrin in DC2.4, iBMDM, or HEK 293T cells, cells were harvested, washed with PBS, and lysed for 20 min on ice in the lysis buffer containing 50 mM Tris⋅HCl (pH 7.6), 150 mM NaCl, and 1% Triton X-100 supplemented with a protease inhibitor mixture (Roche Molecular Biochemicals). Cell lysates were centrifuged at 15,000 rpm for 15 min at 4 °C. The Flag M2 beads were then added to the soluble fraction and incubated for 2 h at 4 °C. After incubation, the immunoprecipitates were washed three times with the lysis buffer; then the beads were eluted with 0.5 mg/mL Flag peptide or were directly boiled in the SDS loading buffer. For phosphatase treatment, the indicated phosphatase and MnCl2 were added into the centrifuged soluble fraction and incubated at 30 °C for 20 min before the Flag M2 beads were added. For immunoprecipitation of endogenous Pyrin, primary Asc−/− BMDMs were primed with LPS (500 ng/mL) for 3 h to increase the expression of Pyrin before toxin stimulation. The anti-Pyrin monoclonal antibody was mixed with Protein A agarose beads and was incubated at 4 °C for 1 h. The antibody-attached beads were added to the above soluble fraction of centrifuged cell lysates. Following antigen–antibody binding, the beads were washed three times with PBS and boiled in the SDS loading buffer. For immunoblotting, Pyrin stably expressed in DC2.4 cells was detected with the rabbit polyclonal anti-Pyrin serum (1:1,000 dilution); immunoprecipitation and immunoblotting of endogenous Pyrin in BMDM cells were performed using the rabbit monoclonal anti-Pyrin antibody.
Immunofluorescence Staining.
Primary BMDMs were cultured on a coverslip in a 24-well plate. To visualize the formation of ASC foci, control cells or cells stimulated by the indicated toxin were washed with PBS followed by fixation in 4% paraformaldehyde in PBS for 10 min at room temperature. The fixed cells were washed three times with PBS and then were permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking with bovine serum, endogenous ASC was stained by the anti-ASC antibody and Alexa Fluor 546 goat anti-mouse IgG secondary antibody. For microtubule staining, drug-treated primary BMDMs were fixed with methanol at −20 °C for 10 min. The cells then were rinsed twice with cold acetone and washed further with PBS. Microtubules were directly stained with Cy3-conjugated monoclonal anti–β-tubulin antibody. Fluorescence images were captured with Zeiss Meta Confocal Microscope and analyzed using Zeiss LSM software.
Purification of Recombinant Proteins.
To express His-tagged TcdA and TcdB, B. megaterium harboring pHIS1522-TcdA, pHIS1522-TcdB, or HIS1522-TcdBm (W102A/D288N) expression plasmid was induced with 0.5% xylose. The toxins were purified from the lysed bacteria as previously described (22). To express LFn-tagged Flagellin, IbpA-Fic1, IbpA-Fic2, C. botulinum C3 toxin, and His-tagged phosphatase of Y. enterocolitica and E. coli, E. coli BL21 (DE3) strains transformed with the corresponding expression plasmid were grown in the Luria-Bertani (LB) medium at 37 °C. The bacteria were induced overnight at 22 °C with 0.5 mM isopropyl-B-d-thiogalactopyranoside (IPTG) after OD600 reached 0.6–0.8. Bacteria were lysed in a buffer containing 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, and 25 mM imidazole. His-tagged proteins were purified by affinity chromatography using Ni-NTA beads (Qiagen). Proteins were eluted with 250 mM imidazole in 50 mM Tris⋅HCl (pH 8.0) and 300 mM NaCl. Eluted samples were purified further using the HiTrap Q HP ion exchange column (GE Healthcare Life Sciences) and then were dialyzed against a buffer containing 50 mM Tris⋅HCl (pH 7.6) and 150 mM NaCl.
Acknowledgments
We thank X. Qi for help with chemical synthesis of BAL27862 and AAL-107 compounds; H. Chen and Abcam for generating rabbit monoclonal antibody; X. Liu and S. Chen for mass spectrometry analyses; and members of the F.S. laboratory for helpful discussions and technical assistance. This work was supported by China National Science Foundation Program for Distinguished Young Scholars Grant 31225002 and Program for International Collaborations Grant 31461143006; Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08020202; and National Basic Research Program of China 973 Program Grants 2012CB518700 and 2014CB849602 (to F.S.). The research also was supported in part by an International Early Career Scientist Grant from the Howard Hughes Medical Institute (to F.S.) and by the Beijing Scholar Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601700113/-/DCSupplemental.
References
- 1.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Shao F. Diverse mechanisms for inflammasome sensing of cytosolic bacteria and bacterial virulence. Curr Opin Microbiol. 2016;29:37–42. doi: 10.1016/j.mib.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Broderick L, De Nardo D, Franklin BS, Hoffman HM, Latz E. The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol. 2015;10:395–424. doi: 10.1146/annurev-pathol-012414-040431. [DOI] [PubMed] [Google Scholar]
- 4.Henao-Mejia J, Elinav E, Thaiss CA, Flavell RA. Inflammasomes and metabolic disease. Annu Rev Physiol. 2014;76:57–78. doi: 10.1146/annurev-physiol-021113-170324. [DOI] [PubMed] [Google Scholar]
- 5.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 6.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 7.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Zhao Y, Shao F. Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr Opin Immunol. 2015;32:78–83. doi: 10.1016/j.coi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 10.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477(7366):592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477(7366):596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 12.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J Immunol. 2013;191(8):3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci USA. 2013;110(35):14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol Cell. 2014;54(1):17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Shao F. The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol Rev. 2015;265(1):85–102. doi: 10.1111/imr.12293. [DOI] [PubMed] [Google Scholar]
- 16.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243(1):109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 17.Anonymous French FMF Consortium A candidate gene for familial Mediterranean fever. Nat Genet. 1997;17(1):25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 18.Anonymous The International FMF Consortium Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell. 1997;90(4):797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 19.Chae JJ, Aksentijevich I, Kastner DL. Advances in the understanding of familial Mediterranean fever and possibilities for targeted therapy. Br J Haematol. 2009;146(5):467–478. doi: 10.1111/j.1365-2141.2009.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13(2):236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 21.Chae JJ, et al. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 2011;34(5):755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513(7517):237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 23.Aubert DF, et al. A Burkholderia Type VI Effector Deamidates Rho GTPases to Activate the Pyrin Inflammasome and Trigger Inflammation. Cell Host Microbe. 2016;19(5):664–674. doi: 10.1016/j.chom.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Waite AL, et al. Pyrin and ASC co-localize to cellular sites that are rich in polymerizing actin. Exp Biol Med (Maywood) 2009;234(1):40–52. doi: 10.3181/0806-RM-184. [DOI] [PubMed] [Google Scholar]
- 25.Kim ML, et al. Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J Exp Med. 2015;212(6):927–938. doi: 10.1084/jem.20142384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47(2):349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Just I, et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270(23):13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 28.Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: The ABCD model. Trends Microbiol. 2008;16(5):222–229. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Lyras D, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458(7242):1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuehne SA, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467(7316):711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 31.Ng J, et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 2010;139(2):542–552. doi: 10.1053/j.gastro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Jéru I, et al. Interaction of pyrin with 14.3.3 in an isoform-specific and phosphorylation-dependent manner regulates its translocation to the nucleus. Arthritis Rheum. 2005;52(6):1848–1857. doi: 10.1002/art.21050. [DOI] [PubMed] [Google Scholar]
- 33.Obsil T, Obsilova V. Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol. 2011;22(7):663–672. doi: 10.1016/j.semcdb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci USA. 2006;103(46):17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petosa C, et al. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem. 1998;273(26):16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 36.Liu D, et al. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376(6536):191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4(10):1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 38.Mansfield E, et al. The familial Mediterranean fever protein, pyrin, associates with microtubules and colocalizes with actin filaments. Blood. 2001;98(3):851–859. doi: 10.1182/blood.v98.3.851. [DOI] [PubMed] [Google Scholar]
- 39.Misawa T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14(5):454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 40.Gigant B, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435(7041):519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 41.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 42.Prota AE, et al. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J Mol Biol. 2014;426(8):1848–1860. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Gong Y-N, et al. Chemical probing reveals insights into the signaling mechanism of inflammasome activation. Cell Res. 2010;20(12):1289–1305. doi: 10.1038/cr.2010.135. [DOI] [PubMed] [Google Scholar]