Significance
Systemic lupus erythematosus (SLE) is characterized by compromised IL-2 production and regulatory T-cell function. Studies in human SLE and in murine lupus models report that IL-2 replenishment ameliorates clinical lupus manifestations. Here we show that engagement of signaling lymphocytic activation molecule family 3 (SLAMF3), a coregulatory receptor of T cells, restores the sensitivity of SLE CD4+ T cells to IL-2, increasing their response to exogenous IL-2 via up-regulation of the IL-2Rα subunit. Moreover, activation of naïve CD4+ T cells with a monoclonal antibody directed against SLAMF3 promotes T helper cell differentiation toward a suppressive phenotype. These data suggest that the SLAMF3 receptor may be a promising therapeutic target in SLE.
Keywords: SLE, autoimmunity, IL-2, Treg, SLAMF3
Abstract
Signaling lymphocytic activation molecule family 3 (SLAMF3/Ly9) is a coregulatory molecule implicated in T-cell activation and differentiation. Systemic lupus erythematosus (SLE) is characterized by aberrant T-cell activation and compromised IL-2 production, leading to abnormal regulatory T-cell (Treg) development/function. Here we show that SLAMF3 functions as a costimulator on CD4+ T cells and influences IL-2 response and T helper cell differentiation. SLAMF3 ligation promotes T-cell responses to IL-2 via up-regulation of CD25 in a small mothers against decapentaplegic homolog 3 (Smad3)-dependent mechanism. This augments the activation of the IL-2/IL-2R/STAT5 pathway and enhances cell proliferation in response to exogenous IL-2. SLAMF3 costimulation promotes Treg differentiation from naïve CD4+ T cells. Ligation of SLAMF3 receptors on SLE CD4+ T cells restores IL-2 responses to levels comparable to those seen in healthy controls and promotes functional Treg generation. Taken together, our results suggest that SLAMF3 acts as potential therapeutic target in SLE patients by augmenting sensitivity to IL-2.
Signaling lymphocytic activation molecule family members (SLAMF1-9) are type I transmembrane glycoprotein cell surface receptors that deliver downstream signals on their engagement (1). SLAMF3 (CD229/Ly9) is expressed on T cells, B cells, NK cells, macrophages, and dendritic cells (2). SLAMF3 acts as a self-ligand through the binding of the N-terminal Ig domain of SLAMF3 (3).
SLAMF3 has been proposed to be involved in the immunopathogenesis of systemic lupus erythematosus (SLE), a multisystem autoimmune disease characterized by a loss of tolerance to endogenous antigen, leading to autoantibody production and to a wide range of clinical manifestations (4). The SLAMF3 encoding gene is located on chromosome 1 within 1q23, a region known to be associated with increased susceptibility for SLE development (5, 6), and polymorphisms of SLAMF3 have been described in SLE families (7, 8). SLAMF3-deficient mice spontaneously develop SLE-associated autoantibodies (9) and display impaired T-cell proliferation and compromised antigen-driven IL-2 production (10).
T cells are key players in the pathogenesis of SLE. Impaired IL-2 production by activated T cells is a hallmark of both murine and human SLE (4, 11–13). Previous studies have shown that CD4+ T cells from SLE patients display lower levels of CD25, the α chain of the IL-2 receptor (IL-2R), compared with healthy subjects (14–16). IL-2 helps maintain peripheral immune self-tolerance and plays a critical role in the differentiation, survival, and function of regulatory T cells (Tregs) (4, 12). Tregs express the nuclear transcription factor forkhead box protein 3 (FoxP3) and the IL-2Rα chain CD25. IL-2–deficient mice develop a severe lupus-like phenotype, characterized by diminished number of Tregs (17). Most reports on human SLE have shown abnormal numbers/function of Tregs in the periphery (18–22). Replenishment of IL-2 in lupus-prone mice and in human SLE patients ameliorates disease manifestations through the expansion of Tregs (16, 23–27). The beneficial effect of low-dose IL-2 treatment has been characterized in graft-vs.-host disease (GVHD) and hepatitis C-associated vasculitis, where it augments the numbers of functional Tregs in the periphery after treatment (28–30).
Here we demonstrate that SLAMF3 costimulation enhances CD4+ T-cell sensitivity to IL-2 by up-regulating the IL-2Rα chain in a small mothers against decapentaplegic homolog 3 (Smad3)-dependent manner, resulting in the generation of functional Tregs. In CD4+ T cells isolated from SLE patients, engagement of SLAMF3 restores the sensitivity of IL-2R to IL-2 and the generation of competent Tregs.
Results
Anti-SLAMF3 Acts as a Coactivator of CD4+ T Cells.
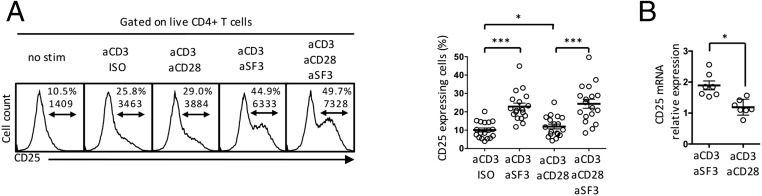
We examined the effect of SLAMF3 ligation on the IL-2/IL-2R/STAT5 pathway in naïve CD4+ T cells. IL-2 signals through the IL-2R, which is composed of three chains: CD25 (IL-2Rα), CD122 (IL-2Rβ), and CD132 (IL-2R common γ-chain). Binding of IL-2 to its receptor leads to phosphorylation of the STAT5A/B transcription factors (31). Compared with CD28 costimulation or to CD3/isotype control, SLAMF3 costimulation increased the expression of CD25 on the cell surface after 18 h of stimulation (Fig. 1A and Fig. S1).
Fig. 1.
SLAMF3 costimulation promotes the expression of the IL-2 receptor CD25 on human naïve CD4+ T cells. (A) Naïve CD4+ T cells were stimulated for 18 h, after which the expression of CD25 was assessed by flow cytometry. Shown are representative flow panels [top number, frequency (%); bottom number, mean fluorescence intensity (MFI)] and cumulative data (mean ± SEM); n = 18. (B) Naïve CD4+ T cells were stimulated for 6 h. CD25 mRNA was assessed by real-time quantitative RT-PCR (qRT-PCR) and is expressed as CD3/SLAMF3 or CD3/CD28 normalized to CD3/IgG isotype control. Data are mean ± SEM; n = 7.
Fig. S1.
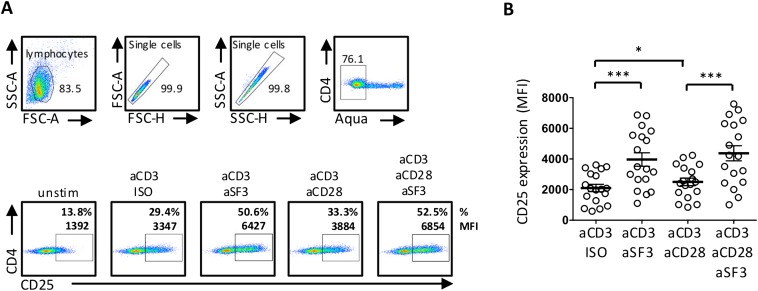
Human naïve CD4+ T cells were stimulated with aCD3, aCD28, aSF3, and/or IgG Isotype control (ISO) for 18 h, after which CD25 expression was assessed by flow cytometry. (A) Gating strategy and representative dot plot showing CD25 frequency (%, top number) and MFI (bottom number). (B) Cumulative data showing CD25 MFI expression. Data are mean ± SEM; n = 18.
SLAMF3-induced expression of CD25 is regulated at the transcriptional level, as shown by greater levels of CD25 mRNA after anti-CD3/anti-SLAMF3 stimulation compared with anti-CD3/anti-CD28 treatment (Fig. 1B). Examination of the promoter region of CD25 for transcription factor binding sites identified Smad3 as a candidate capable of controlling CD25 transcription (31).
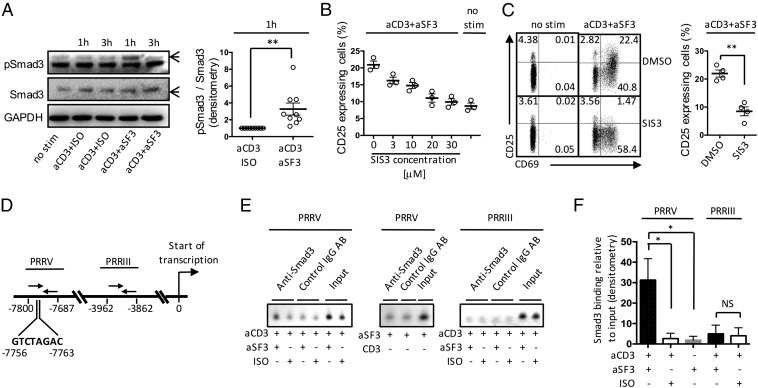
We examined phosphorylation of Smad3 after stimulation of naïve CD4+ T cells with anti-CD3 and anti-SLAMF3 monoclonal antibodies (mAbs) and found increased pSmad3 after 1 h (Fig. 2A), with no differences in the amount of total Smad3 protein. A specific inhibitor of Smad3 (SIS3) phosphorylation and Smad3-mediated cellular signaling, with no effect on Smad2, p38 MAPK, ERK, or PI-3K signaling (32), reduced SLAMF3-driven CD25 expression on naïve CD4+ T cells in a dose-dependent manner (Fig. 2B). This occurred without abolishing cell activation, with expression of CD69 barely affected by SIS3 (Fig. 2C).
Fig. 2.
CD25 up-regulation following SLAMF3 coengagement is Smad3-dependent. Expression of pSmad3, total Smad3, and GAPDH by stimulated naïve CD4+ T cells was assessed by Western blot analysis. (A) Representative blot showing the expression of pSmad3 and total Smad3 at 1 h and 3 h of stimulation, and cumulative data showing the densitometry ratio of pSmad3 over total Smad3 after 1 h of stimulation. Data are mean ± SEM; n = 9. (B) Naïve CD4+ T cells were stimulated for 18 h in the presence of increasing concentrations (3–30 μM) of Smad3 inhibitor SIS3, after which CD25 expression was assessed by flow cytometry. Data are mean ± SEM; n = 3. (C) CD25 and CD69 expression was assessed after 18 h of stimulation in the presence of SIS3 (30 μM). Data are mean ± SEM; n = 4. (D) Schematic representation of the PRRIII and PRRV of the CD25 gene. Numbers indicate forward (right arrows) and reverse (left arrows) primer positions relative to the start of CD25 gene transcription. The Smad3 putative binding site (GTCTAGAC) position is depicted as well. (E) ChIP data showing the binding of Smad3 to PRRV (Left and Middle) and PRRIII (Right) regions of CD25 in response to indicated stimulations. (F) Cumulative data of Smad3 binding relative to input (mean ± SEM); n = 3–4.
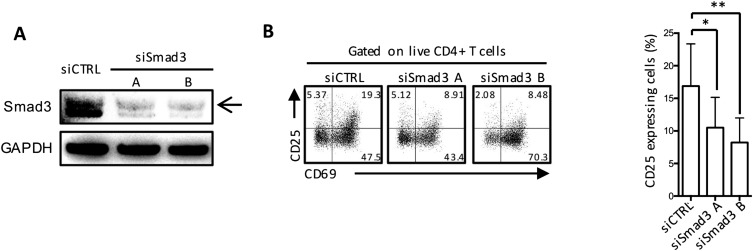
To confirm these findings, we used two small interfering RNAs (siRNAs) to knock down Smad3 in human CD4+ T cells, which resulted in an 80–90% reduction in Smad3 expression (Fig. S2A). Compared with cells transfected with control siRNA, Smad3 siRNA-transduced cells displayed a significant decrease in CD25 expression after 18 h of stimulation with anti-SLAMF3 mAb (Fig. S2B), suggesting that CD25 expression in response to SLAMF3 activation depends on Smad3. In accordance with the previous experiment, Smad3 siRNA CD4+ T-cell activation was not completely abrogated, with CD69 expression still increased after stimulation (Fig. S2B).
Fig. S2.
Human naïve CD4+ T cells transfected with Smad3 siRNA (siSmad3 A and siSmad3 B) or control siRNA (siCTRL) were stimulated with aCD3 and aSF3. CD25 and CD69 expression were assessed by flow cytometry after 18 h of stimulation. (A) Representative blot demonstrating maximal Smad3 silencing at 48 h after transfection. (B) Representative flow and cumulative data showing the expression of CD25 after 18 h of SLAMF3 coengagement. Data are mean ± SEM; n = 4.
To confirm that SLAMF3 favors CD25 gene expression in an Smad3-dependent manner, we performed chromatin immunoprecipitation (ChIP) assays in Jurkat cells to examine the binding of Smad3 to CD25 gene regulatory regions. Previous studies described six upstream positive regulatory regions (PRRs) that control the transcription of CD25 gene (31, 33). Among these six PRRs, Smad3 is known to be able to bind to PRRV in response to TGFβ1 and TCR engagement (34). Accordingly, we examined the binding of Smad3 to the regulatory PRRV region of CD25 gene in response to SLAMF3 costimulation. PRRIII was used as a negative control, because this region displays no Smad3-binding element (Fig. 2D). Compared with CD3/isotype control stimulation, SLAMF3 coengagement increased the binding of Smad3 to PRRV without binding to PRRIII (Fig. 2 E and F). Moreover, stimulation of cells with anti-SLAMF3 mAb alone (i.e., without CD3 engagement) did not increase the binding of Smad3 to PRRV (Fig. 2 E and F). Taken together, these data demonstrate that SLAMF3 coengagement promotes the phosphorylation of Smad3, which can then bind to the regulatory region of CD25 gene and promote its transcription.
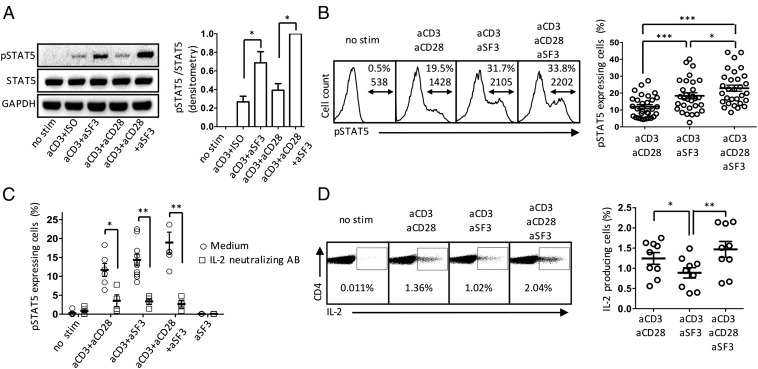
After IL-2 binds to its receptor, the IL-2/IL-2R/STAT5 pathway is activated (31). We measured the level of phosphorylated STAT5 (pSTAT5) to assess the intrinsic capacity of cells to produce IL-2 and activate the IL-2R/STAT5 signaling pathway in an autocrine fashion after TCR activation. We observed that SLAMF3 costimulation promoted the phosphorylation of STAT5 to a greater extent than CD28 (Fig. 3 A and B). Activation of the pathway was not a direct effect of SLAMF3 cross-linking, as demonstrated by the fact that phosphorylation of STAT5 was almost entirely abrogated in the presence of an IL-2–neutralizing antibody (Fig. 3C).
Fig. 3.
SLAMF3 coengagement promotes the activation of the IL-2/IL-2R/pSTAT5 pathway. Human naïve CD4+ T cells were stimulated for 18 h, after which expression of pSTAT5 was assessed by Western blot analysis and flow cytometry. (A) Representative blot and cumulative densitometry ratio of pSTAT5 over total STAT5. Data are mean ± SEM; n = 4. (B) Representative flow panels of pSTAT5 [top number, frequency (%); bottom number, MFI] and cumulative data (mean ± SEM); n = 17. (C) Naïve CD4+ T cells were stimulated for 18 h in the presence of an anti–IL-2 neutralizing antibody (2 μg/mL). Data are representative of 6–10 subjects per group (mean ± SEM). (D) Intracellular cytokine staining showing IL-2 production by naïve CD4+ T cells after 18 h of stimulation. Shown are representative flow panels and cumulative data (mean ± SEM); n = 9.
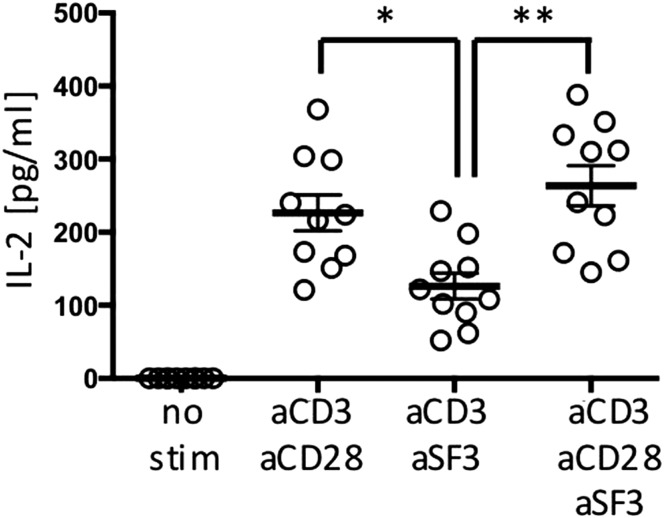
These results also may be explained by endogenous IL-2 production. To address this possibility, we assessed the frequency of IL-2–producing cells by intracellular cytokine staining after 18 h of stimulation of naïve CD4+ T cells. Contrary to CD28 coengagement, SLAMF3 did not increase IL-2 production significantly (Fig. 3D). Measurement of IL-2 in the supernatant of sorted naïve CD4+ T cells stimulated for 18 h with anti-CD3 and costimulatory molecules by ELISA showed that CD28 was more potent than SLAMF3 in inducing IL-2 production (Fig. S3).
Fig. S3.
IL-2 concentration was measured by ELISA in supernatants of isolated naïve CD4+ T cells after 18 h of stimulation. Data are mean ± SEM; n = 10.
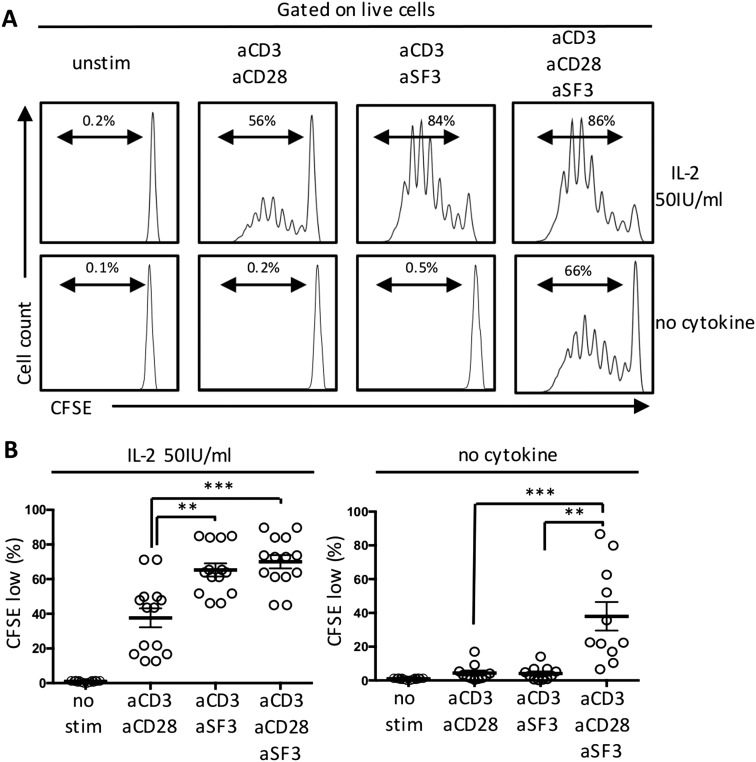
Compared with CD28 costimulation, SLAMF3-costimulated CD4+ T cells displayed significantly increased proliferation in response to exogenous IL-2 (Fig. S4 A and B). Proliferation of naïve CD4+ T cells was limited in the absence of exogenous IL-2. Of note, proliferation of naïve CD4+ T cells in response to both SLAMF3 and CD28 revealed a synergistic effect even in the absence of IL-2 (Fig. S4 A and B).
Fig. S4.
CFSE-labeled naïve CD4+ T cells were cultured for 6 d in the presence of recombinant IL-2 (50 IU/mL) or without cytokine. Shown are representative flow panels showing CFSE dilution (A) and cumulative data (B). Data are mean ± SEM; n = 7.
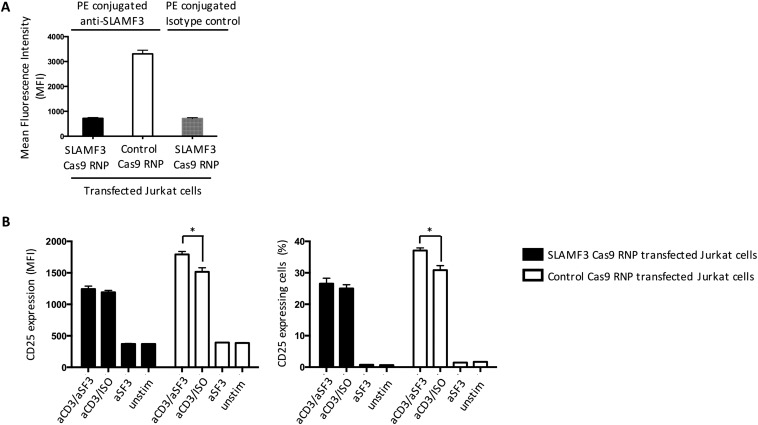
To ascertain whether anti-SLAMF3 antibody binds through specific interaction with SLAMF3 molecules on the cell surface, we silenced SLAMF3 in Jurkat cells using CRISPR/Cas9 (Fig. S5A) (35, 36). SLAMF3 knockout cells showed no difference in the expression of CD25 after 18 h of stimulation with anti-CD3 alone or with anti-CD3/anti-SLAMF3 (Fig. S5B), proving the specificity of the monoclonal anti-SLAMF3 antibody.
Fig. S5.
Anti-SLAMF3 antibody specifically binds to SLAMF3 on the cell surface. The CRISPR/Cas9 system was used to knock out SLAMF3 expression in Jurkat cells. (A) The expression of SLAMF3 (MFI) was assessed by flow cytometry after SLAMF3 Cas9 RNP treatment or control Cas9 RNP treatment. (B) SLAMF3/control Cas9 RNP-treated cells were stimulated with anti-SLAMF3 and/or isotype control (ISO) and/or anti-CD3 mAbs. After 18 h, CD25 expression (MFI and % of CD25-expressing cells) was measured by flow cytometry. Shown are the cumulative results of four independent experiments. Data are mean ± SEM.
In summary, SLAMF3 costimulation promotes cell proliferation by increasing the response of naïve CD4+ T cells to IL-2, not by increasing IL-2 cytokine production itself, but rather by up-regulating IL-2R and increasing activation of the IL-2/IL-2R/STAT5 pathway.
Costimulation by Anti-SLAMF3 Favors Treg Differentiation of Naïve CD4+ T Cells.
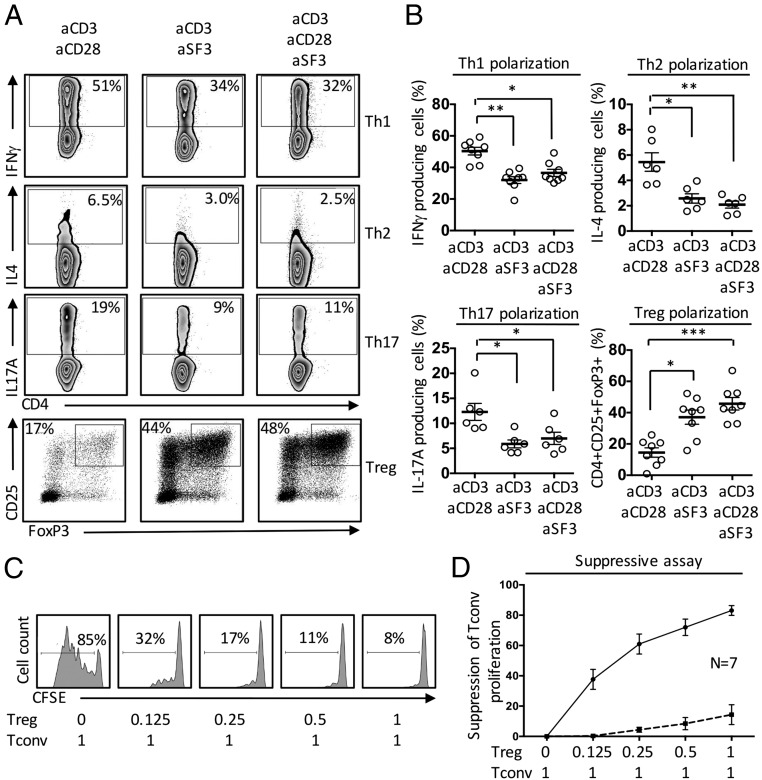
Because IL-2 is a key cytokine in T-cell homeostasis and activation, we examined the effect of SLAMF3 costimulation on CD4+ T-cell differentiation. When naïve CD4+ T cells were differentiated in Th1, Th2, and Th17 polarizing conditions, we observed decreased frequencies of IFNγ-, IL-4–, and IL-17A–producing cells, respectively, in the presence of SLAMF3 ligation compared with CD28 ligation (Fig. 4 A and B). In contrast, when naive CD4+ T cells were cultured under Treg polarizing conditions, SLAMF3 costimulation increased the frequency of CD25 and FoxP3 double-positive cells (Fig. 4 A and B).
Fig. 4.
SLAMF3 costimulation promotes the differentiation of human naïve CD4+ T cells into functional Tregs. Isolated naïve CD4+ T cells were cultured under Th1, Th2, Th17, and Treg polarizing conditions for 6 d. Intracellular cytokine staining was performed after 6 h of stimulation with PMA and ionomycin. Foxp3 was assessed after nuclear permeabilization for Treg differentiation. (A) Representative flow plots. (B) Cumulative data (mean ± SEM); n = 5–8. Human naïve CD4+ T cells were coactivated in the presence of anti-SLAMF3 mAb (solid line) or an isotype control (dashed line) and polarized under Treg conditions. After 6 d, increasing ratios of induced Tregs were cocultured in the presence of CFSE-labeled Tconv cells. Tconv proliferation was assessed after 5 d of coculture. (C) Representative flow plots. (D) Cumulative data (mean ± SEM); n = 7.
CD25 and FoxP3 expression, as well as STAT5 activation, are not unique to functional Tregs, and are also expressed by activated human effector T cells (37, 38). To clarify their function, we assessed the suppressive capacity of Tregs induced in the presence of SLAMF3 costimulation. Tregs induced in the presence of SLAMF3 ligation displayed a potent suppressive effect on the proliferation of autologous conventional T cells (Tconv) (Fig. 4 C and D), thus proving that these are functional Tregs.
SLAMF3 Costimulation Restores the Sensitivity of SLE Naïve CD4+ T Cells to IL-2 and Induces Treg Differentiation.
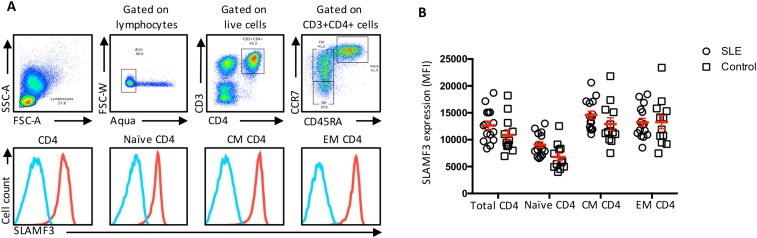
Compromised IL-2 production by T cells isolated from SLE patients is an important feature driving SLE immune dysregulation (12). In light of this abnormality and our findings regarding SLAMF3 costimulation, we hypothesized that treatment of SLE CD4+ T cells with anti-SLAMF3 should restore IL-2 sensitivity. We found SLAMF3 to be highly expressed on all CD4+ T-cell differentiated subsets in both SLE patients and controls (Fig. S6). SLAMF3 expression was slightly increased on SLE naïve CD4+ T cells compared with controls. We found no correlation between SLAMF3 expression and disease activity.
Fig. S6.
SLAMF3 expression by peripheral CD4+ T-cell subsets from patients with SLE and healthy subjects was measured by flow cytometry. (A) Representative flow panel and gating strategy (red, anti-SLAMF3 mAb; blue, isotype control). (B) Cumulative analysis of SLAMF3 expression by differentiated CD4+ T-cell subsets (SLE, n = 15; controls, n = 12). CM central memory, CD45RA-CCR7+; EM effector memory, CD45RA-CCR7+. Data are mean ± SEM.
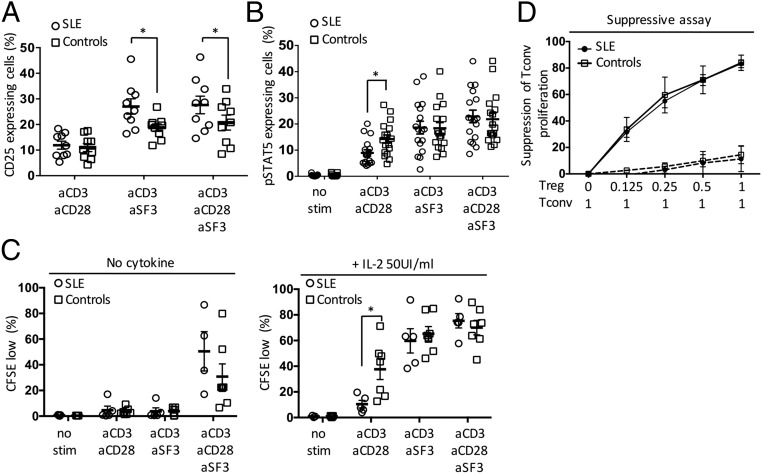
SLAMF3 coengagement increased expression of the IL-2Rα chain (CD25) on the surface of SLE naïve CD4+ T cells compared with T cells from control subjects (Fig. 5A). On CD3/CD28 costimulation, pSTAT5 was decreased in SLE patients compared with healthy controls (Fig. 5B), yet when we stimulated SLE naïve CD4+ T cells with anti-SLAMF3, STAT5 phosphorylation was restored to a level comparable to that seen in controls (Fig. 5B).
Fig. 5.
SLAMF3 costimulation restores the IL-2 response in human naïve CD4+ T cells and promotes Treg differentiation. (A and B) Naïve CD4+ T cells from SLE and healthy controls were stimulated for 18 h, after which expression of CD25 (SLE, n = 9; controls, n = 9) (A) and pSTAT5 (SLE, n = 17; controls, n = 17) (B) was evaluated by flow cytometry. (C) Naïve CD4+ T-cell proliferation was expressed as the percentage of CFSE-low cells after 6 d of stimulation (SLE, n = 4–5; controls, n = 5–7) without exogenous cytokine (Left) or in the presence of recombinant IL-2 (50 IU/mL) (Right). (D) Naïve CD4+ T cells from SLE and controls were coactivated with anti-SLAMF3 mAb (solid line) or an isotype control (dashed line) and polarized under Treg conditions for 6 d. Their suppressive capacity was evaluated by measuring inhibition of Tconv proliferation (SLE, n = 3; controls, n = 3). Data are mean ± SEM.
As mentioned above, when cells were activated with anti-CD3 and CD28, the proliferation of SLE naïve CD4+ T cells in response to exogenous IL-2 was decreased compared with that in healthy controls. However, when naïve CD4+ T cells were coactivated with anti-SLAMF3, the proliferation was restored to normal levels (Fig. 5C). Finally, SLE naïve CD4+ T cells were activated with anti-SLAMF3 in Treg polarizing conditions and found to display a suppressive function comparable to that of similarly treated cells from healthy subjects (Fig. 5D).
Discussion
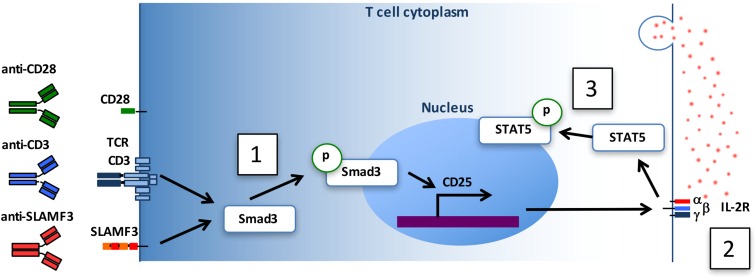
In this study, we observed that activation of CD4+ T cells by an mAb specifically directed against SLAMF3 exerts a costimulatory signal that increases cell sensitivity to IL-2. The mechanism involves increased expression of the IL-2R α chain CD25 (Fig. S7). The regulation of CD25 occurs at the transcriptional level and involves the transcription factor Smad3. Smad3 is phosphorylated on SLAMF3 costimulation, but not when cells are stimulated with anti-SLAMF3 in the absence of TCR engagement, and binds to the regulatory region of CD25 gene.
Fig. S7.
SLAMF3 costimulation promotes CD4+ T-cell responses to IL-2. Engagement of SLAMF3/CD3 with mAb promotes Smad3 phosphorylation (1), which in turn migrates to the nucleus and activates CD25 transcription, leading to its expression on the cell surface (2). This results in an enhanced response to endogenous IL-2, which promotes the activation of STAT5 transcription factor (3).
SLAMF3 costimulation does not significantly alter the production of IL-2 itself. When naïve CD4+ T cells are stimulated with both anti-SLAMF3 and anti-CD28 antibodies, a marked increase in cell proliferation occurs compared with the coengagement of either SLAMF3 or CD28 alone. This suggests that SLAMF3 and CD28 may act synergistically to activate CD4+ T cells through distinct pathways. SLAMF3 thereby promotes the expression of IL-2R on the cell surface, whereas CD28 preferentially enhances IL-2 production.
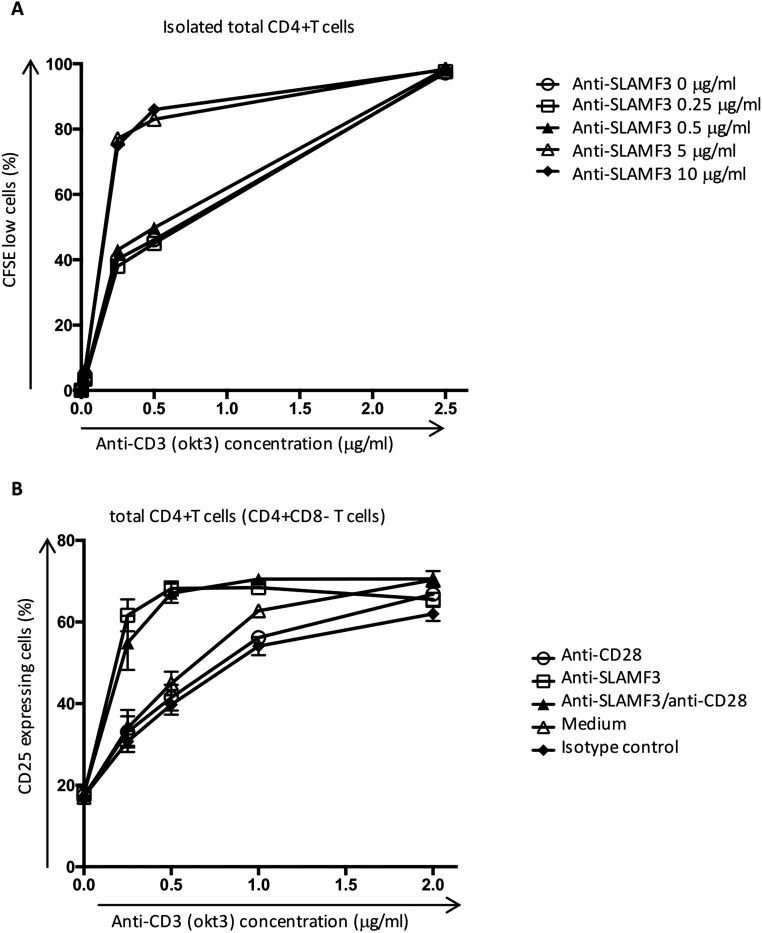
Differentiation and survival of Tregs in the periphery are dependent on the presence of IL-2 and on the activation of IL-2R–mediated phosphorylation of STAT5 (38, 39). Our experiments emphasize that SLAMF3 augments pSTAT5 expression, while promoting the differentiation of peripheral naïve CD4+ T cells into polarized Tregs and inhibiting cytokine production by other differentiated T helper subsets. In a previous study, SLAMF3 costimulation was suggested to increase IL-17 production by CD4+ T cells (40); however, in that study the anti-SLAMF3 antibody was used at a dose of 0.5 μg/mL, which was not sufficient to increase cell proliferation (Fig. S8 A and B).
Fig. S8.
(A) The optimal concentration of anti-SLAMF3 and anti-CD3 was determined by assessing the proliferation of isolated CD4+ T cells, cultured for 6 d in the presence of increasing concentrations of anti-SLAMF3 (expressed in μg/mL) and anti-CD3 mAb. Data are representative of three independent experiments. (B) T cells were stimulated with increasing concentrations of anti-CD3 mAb in the presence of the indicated costimulatory molecules (each at a concentration of 5 μg/mL). The expression of CD25 by total CD4+ T cells was assessed by flow cytometry after 18 h of stimulation. Data are mean ± SEM; n = 4.
Our results suggest that SLAMF3 is involved in immune tolerance. A recent study has shown that Ly9 (SLAMF3)-deficient mice develop spontaneous autoimmunity, and that in vitro anti-CD3– and anti-CD28–stimulated CD4+ T cells from Ly9-deficient mice exhibit greater IFNγ production compared with their wild type counterparts, suggesting SLAMF3 involvement in protection against autoimmunity (9).
Studies in SLE suggest that compromised IL-2 production is an important feature of the immune dysregulation involved in the pathogenesis of the disease. An impaired IL-2 level is associated with a reduced suppressive function of Tregs (4, 12, 16, 25). Moreover, IL-2 plays a role in mounting adequate cytotoxic responses. This process is compromised in SLE patients, contributing to the increased rate of infection, one of the leading causes of morbidity and mortality in SLE (41, 42). IL-2 also helps maintain peripheral immune self-tolerance by promoting activation-induced cell death (43), an important apoptotic process that contributes to the elimination of potentially autoreactive T cells. Here we show that SLAMF3 costimulation of SLE naïve CD4+ T cells restores the response to IL-2 to the level observed in healthy controls. This is illustrated by (i) the normal frequency of pSTAT5-expressing cells after 18 h of stimulation, (ii) the normal proliferation of SLE naïve CD4+ T cells in response to recombinant IL-2, and (iii) the generation of functional suppressive Tregs in the presence of SLAMF3 coengagement.
Recent case reports have emphasized that low-dose IL-2 administration could be beneficial in SLE (16, 27). Low-dose IL-2 treatment has been reported to have clinical value in patients with GVHD and hepatitis C-associated vasculitis by augmenting the numbers of functional Tregs in the periphery following treatment (28–30). In this context, our findings suggest that treatment-targeting SLAMF3 may represent a promising therapeutic option in SLE as well as in other conditions in which IL-2 availability/response is compromised, such as GVHD. SLAMF3 stimulation could be beneficial in patients who exhibit resistance to IL-2 therapy or who cannot tolerate recombinant IL-2 treatment. Similarly, engagement of SLAMF3 could offer significant adjuvant therapeutic value to clinical trials in which in vitro expanded Tregs are transfused to patients with SLE, diabetes, and GVHD (44).
Some emerging questions merit further investigation. Foremost, our results suggest that anti-SLAMF3 antibody will favor the generation of Tregs from CD4+ T cells, while inhibiting the development of proinflammatory CD4+ T helper subsets. However, our study focused on CD4+ T cells. SLAMF3 is also expressed on other hematopoietic cells (e.g., B cells and CD8+ T cells) and plays an important role in cell–cell interaction; thus, examining the effect and the safety of the administration of an mAb directed against SLAMF3 in in vivo settings is warranted.
Materials and Methods
More detailed information is provided in SI Materials and Methods.
SLE Patients and Controls.
SLE patients (n = 42) were diagnosed according to the American College of Rheumatology’s classification criteria (45) and recruited from the Division of Rheumatology, Beth Israel Deaconess Medical Center. Age-, sex-, and ethnicity-matched healthy individuals served as controls. Disease activity was measured using the SLE Disease Activity Index scoring system (Table S1). This study was approved by the medical center’s Institutional Review Board. Informed consent was obtained from each subject after the nature and possible consequences of the studies were explained.
Table S1.
Characteristics of the SLE patients included in the study (n = 42)
| Characteristic | Value |
| Age, y | |
| Median | 40.9 |
| Range | 21.0–71.0 |
| Sex, n (%) | |
| Female | 36 (85.7) |
| Male | 6 (14.3) |
| Ethnicity, n (%) | |
| African American | 11 (26.2) |
| Asian | 4 (9.5) |
| Hispanic | 7 (16.7) |
| Caucasian | 20 (47.6) |
| SLE disease activity index (SLEDAI) | |
| Median | 4.3 |
| Range | 0–21 |
| Treatments, n (%) | |
| Prednisone | 28 (66.7) |
| Hydroxychloroquine | 33 (78.6) |
| Mycophenolate mofetil | 15 (35.7) |
| Azathioprine | 7 (16.7) |
| Methotrextate | 2 (4.8) |
| Belimumab | 2 (4.8) |
| Intravenous immunoglobulin | 3 (7.2) |
Cell Isolation.
Peripheral blood mononuclear cells were enriched by density gradient centrifugation (Lymphocyte Separation Medium; Corning Life Sciences). T cells were isolated by negative selection (RosetteSep; Stem Cell Technologies). Naïve CD4+ T cells were isolated using the Cell Isolation Kit II (Miltenyi Biotec) or by FACS sorting (CD4+CD8-CD45RO-CD25-; >97% purity) using a FACSAria II (BD Bioscience).
Flow Cytometry.
Cells were stained for dead cells (Zombie Aqua/UV Fixable Viability Kit; Biolegend), and then labeled for surface antibodies (Table S2). Cells were permeabilized (Cytofix/Cytoperm; BD Biosciences) and stained for cytokines. For phosphoproteins, cells were permeabilized using the PerFix EXPOSE Kit (Beckman Coulter). For FoxP3, cells were permeabilized using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Data were acquired on an LSR II SORP digital cell analyzer (BD Biosciences) and analyzed using FlowJo version 10.0.8.
Table S2.
Antibodies list
| Antibody | Format/source | Clone | Company |
| Flow cytometry | |||
| Anti-CD3 | PE/Cy7 | UCHT1 | Biolegend |
| Anti-CD4 | APC | OKT4 | Biolegend |
| Anti-CD4 | FITC | OKT4 | Biolegend |
| Anti-CD8 | PerCP | RPA-T8 | Biolegend |
| Anti-CD45RA | APC | HI100 | Biolegend |
| Anti-CD45RA | APC/Cy7 | HI100 | Biolegend |
| Anti-CD45RO | APC/Cy7 | UCHL1 | Biolegend |
| Anti-CCR7 | Alexa Fluor 488 | G043H7 | Biolegend |
| Anti-CD25 | Brilliant Violet 421 | BC96 | Biolegend |
| Anti-CD122 | APC | TU27 | Biolegend |
| Anti-CD19 | PE/Dazzle 594 | HIB19 | Biolegend |
| Anti-CD14 | Alexa Fluor 700 | HCD14 | Biolegend |
| Anti-SLAMF3/CD229 | PE | HLy-9.1.25 | Biolegend |
| Anti-SLAMF3/CD229 | APC | HLy-9.1.25 | Biolegend |
| Anti-CD69 | APC | FN50 | Biolegend |
| Anti-CD69 | APC/Cy7 | FN50 | Biolegend |
| Anti–IL-2 | PE/Cy7 | MQ1-17H12 | Biolegend |
| Anti–IFNγ | Pacific Blue | 4S.B3 | Biolegend |
| Anti-TNFα | Alexa Fluor 488 | MAb11 | Biolegend |
| Anti–IL-17A | Alexa Fluor 647 | BL168 | Biolegend |
| Anti–IL-4 | Alexa Fluor 488 | 8D4-8 | Biolegend |
| Mouse IgG1 k isotype control | APC | MOPC-21 | Biolegend |
| Mouse IgG1 k isotype control | Brilliant Violet 421 | MOPC-21 | Biolegend |
| Mouse IgG1 k isotype control | PE | MOPC-21 | Biolegend |
| Anti-CD4 | PerCP eFLuor 710 | SK3 | eBioscience |
| Anti-FoxP3 | PE | 236A/E7 | eBioscience |
| Anti–IL-10 | eFluor 660 | JES3-9D7 | eBioscience |
| Mouse IgG1 k isotype control | PE | P3.6.2.8.1 | eBioscience |
| Anti–phospho-STAT5 (Tyr694) | Alexa Fluor 488 | C71E5 | Cell Signaling Technology |
| Rabbit IgG isotype control | Alexa Fluor 488 | DA1E | Cell Signaling Technology |
| Purified antibodies | |||
| Anti-CD3 | Purified | OKT3 | BioXcell |
| Anti-CD28 | LEAF purified | 28.2 | Biolegend |
| Anti-SLAMF/CD150 | LEAF purified | A12(7D4) | Biolegend |
| Anti-SLAMF2/CD48 | Purified | BJ40 | Biolegend |
| Anti-SLAMF3/CD229 | LEAF purified | HLy-9.1.25 | Biolegend |
| Anti-SLAMF4/CD244/2B4 | LEAF purified | C1.7 | Biolegend |
| Anti-SLAMF5/CD84 | Purified | CD84.1.21 | Biolegend |
| Anti-SLAMF6/CD352/NTBA | LEAF purified | NT-7 | Biolegend |
| Anti-SLAMF7/CD319/CRACC | Purified | 162.1 | Biolegend |
| Western blot/ChIP antibodies | |||
| Anti–phospho-Smad3(Ser423/425) | Rabbit polyclonal | Millipore | |
| Anti-Smad3 | Rabbit polyclonal | Cell Signaling Technology | |
| Anti–phospho-STAT5 (Tyr694) | Rabbit polyclonal | Santa Cruz Biotechnology | |
| Anti-STAT5 | Goat polyclonal | Santa Cruz Biotechnology | |
| Anti-GAPDH HRP conjugated | Santa Cruz Biotechnology | ||
| Secondary HRP-conjugated | Santa Cruz Biotechnology |
T-Cell Stimulation.
Total or naïve CD4+ T cells were stimulated in complete RPMI (supplemented with 10% FBS, 100 mg/mL streptomycin, and 100 U/mL penicillin) with precoated antibodies (anti-CD3, 0.5 μg/mL; anti-CD28 and anti-SLAMF3, 5 μg/mL) for 18 h in the presence of Brefeldin A (GolgiPlug 1 μL/mL; BD Biosciences). Alternatively, cells were stimulated with phorbol 12-myristate 13-acetate (PMA; 25 ng/mL) and ionomycin (0.5 μg/mL) for 6 h in the presence of Brefeldin A. In IL-2 neutralizing experiments, anti–IL-2 neutralizing antibody (R&D Systems) was added to the culture at a concentration of 2 μg/mL. Smad3 inhibitor SIS3 (EMD Millipore) was added to the cell culture at the indicated concentrations.
Suppressive Assay.
Sorted naïve CD4+ T cells were costimulated with anti-CD3 and anti-SLAMF3 in Treg polarizing conditions (i.e., in the presence of IL-2 and TGFβ) for 6 d. Differentiated cells were then cocultured with autologous carboxyfluorescein succinimidyl ester (CFSE)-labeled conventional T cells and stimulated with anti-CD3 and anti-CD28 antibodies in the presence of IL-2. Autologous T-cell proliferation was assessed on day 5 by flow cytometry.
Th1, Th2, Th17, and Treg Differentiation.
Freshly isolated naïve CD4+ T cells were stimulated in complete RPMI with precoated (anti-CD3, 0.5 μg/mL; anti-CD28 and SLAMF3, 5 μg/mL) antibodies. Th1, Th2, Th17, and Treg polarizing conditions are described in Table S3. Cytokines were replenished after 72 h. On day 6, cells were stimulated with PMA and ionomycin in the presence of Brefeldin A for the last 6 h of culture. Cytokines were purchased from Peprotech; neutralizing antibodies, from Biolegend.
Table S3.
CD4+ T helper polarizing conditions
| Polarization | Cytokines | Neutralizing antibodies | ||
| Concentration | Concentration | |||
| Th1 | IL-12 | 20 ng/mL | Anti–IL-4 | 5 μg/mL |
| Th2 | IL-4 | 50 ng/mL | Anti-IFNγ | 5 μg/mL |
| Th17 | IL-1β | 25 ng/mL | Anti–IL-4 | 5 μg/mL |
| IL-6 | 50 ng/mL | Anti-IFNγ | 5 μg/mL | |
| IL-23 | 50 ng/mL | |||
| TGFβ1 | 2 ng/mL | |||
| Treg | IL-2 | 50 UI/mL | Anti–IL-4 | 5 μg/mL |
| TGFβ1 | 5 ng/mL | Anti-IFNγ | 5 μg/mL | |
Statistics.
Statistical analyses were performed using the Mann–Whitney test (unpaired) or Wilcoxon matched-pair signed-rank test (paired). For multiple comparisons, analyses were performed using the Kruskal–Wallis test followed by Turkey’s test (unpaired) or Friedman’s test followed by Dunn’s test (paired). All analyses were performed with GraphPad Prism version 6. Statistical significance was reported as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
SI Materials and Methods
SLAMF3 Silencing Using CRISPR-Cas 9.
Jurkat cells were transduced with lentivirus constructed with pSPAX2, pVSVg, and lentiCas9-Blast (a gift from Feng Zhang, Broad Institute, Cambridge, MA; Addgene plasmid 52962). Following 3 d of selection with 20 μg/mL blasticidin, cells were expanded for 5 d, after which 2 × 106 cells were electroporated by the Amaxa system with 1 μg of plasmids expressing a guide RNA targeting the first common exon of slamf3 (pSPgRNA, a gift from Charles Gersbach, Duke University, Durham, NC; Addgene plasmid 47108). Guide primers were as follows: geLy9aF, caccgGATCCCTGACACCACTGTTG; geLy9aR, aaacCAACAGTGGTGTCAGGGATCc; geLy9bF, caccgGTGGTGTCAGGGATCCTAGG; geLy9bR, aaacCCTAGGATCCCTGACACCACc; geLy9cF, caccgGCCGACTAGACATCACCAAG; and geLy9cR, aaacGTGGTGTCAGGGATCCTAGGc. Two days later, cells were sorted based on SLAMF3 expression, and another 2 d later, sorting was repeated to yield the pure SLAMF3-negative population. Guide A was most efficacious, but all had activity.
Smad3 Silencing.
Transient transfections of purified human naïve CD4+ T cells were performed using the Amaxa Nucleofector human T-cell system (Lonza) according to the manufacturer’s instructions. In brief, 5–10 × 106 naïve CD4+ T cells were resuspended in 100 μL of Nucleofector solution in the presence of one of two different small interfering silencer RNAs against Smad3 (siSmad3 A or siSmad3 B) or a nonspecific control siRNA (siCTRL) at a final concentration of 250 nM. (siRNAs against Smad3 and siCTRL were purchased from Origene.) Maximal Smad3 down-regulation was achieved by 48 h after transfection and was evaluated by Western blot analysis.
ChIP Assays.
Jurkat cells were stimulated overnight with anti-CD3 and/or anti-SLAMF3 mAbs and/or IgG isotype control. At the end of stimulation, cells were collected, and the assay was performed using the MAGnify ChIP Kit (Life Technologies) according to the manufacturer’s instructions. In brief, cells were fixed in 1% formaldehyde to cross-link DNA–protein and protein–protein complexes. Glycine was subsequently added to stop the cross-linking. Cells were resuspended in lysis buffer containing protease inhibitors, sonicated to shear DNA, and then sedimented, after which diluted supernatants were immunoprecipitated with either rabbit anti-Smad3 antibody or a rabbit IgG isotype control. Ten percent of the diluted supernatants was kept as “input” for normalization.
Protein was digested with proteinase K, and the cross-linking was reversed at 65 °C. DNA was purified and amplified by qRT-PCR using specific primers close to PRRIII (PRRIII region position relative to start of transcription: -3780, -3703) (forward primer, 5′-GTGGGCCTTTCCTGATCACA-3′; reverse primer, 5′-TGACCTAGACTGCCTTCCCT-3′) or flanking PRRV (PRRV region position: -7664, -7556) (forward primer, 5′- TTTGAGTGAGGGAAGCCAGC-3′; reverse primer, 5′- AAACCCCCTTTGGAGCTCAG-3′) regions proximal of the IL-2Rα promoter (33). PCR fragments were quantified by densitometry (using ImageJ software) after migration on 1% agarose gel. Smad3 binding density was corrected for isotype control density and expressed relative to input density.
T-Cell Proliferation.
CFSE-labeled cells were stimulated for 6 d with precoated antibodies (anti-CD3, 0.25 μg/mL; anti-CD28 and anti-SLAMF3, 5 μg/mL) in complete RPMI with or without recombinant IL-2 (50 IU/mL; Peprotech). CFSE dilution was assessed by flow cytometry.
IL-2 ELISA.
IL-2 was measured in supernatants following the manufacturer's instructions (BioLegend).
Real-Time qRT-PCR.
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). Reverse transcription was performed with 100 ng of total RNA using the High-Capacity cDNA Archive Kit (Applied Biosystems). qRT-PCR (Light Cycler 480; Roche) was performed with 40 cycles at 94 °C for 12 s and 60 °C for 60 s using Taqman assays (Applied Biosystems). The comparative Ct method was used to quantify transcripts relative to the endogenous control gene large ribosomal protein.
Western Blot Analysis.
Cells were incubated in Triton 1% lysis buffer. Proteins were separated in 4–12% gradient Bis-Tris gels (Life Technologies) and transferred on PVDF membrane (Millipore). Membranes were blocked for 1 h with Tris-buffered saline solution containing 0.05% Tween and 5% nonfat dry milk. Membranes were incubated overnight at 4 °C with the indicated antibody, followed by incubation with an HRP-conjugated antibody. Detection was performed with Clarity Western ECL blotting reagents (Bio-Rad), and visualization was done with the ChemiDoc XRS+ Molecular Imager (Bio-Rad). Densitometric analysis was performed using ImageJ software.
Acknowledgments
This work was supported by National Institutes of Health Grants P01 AI065687, R01 AI42269, and R37 AI49954, and by a SICPA Foundation grant (to D.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605081113/-/DCSupplemental.
References
- 1.Romero X, et al. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J Immunol. 2005;174(11):7033–7042. doi: 10.4049/jimmunol.174.11.7033. [DOI] [PubMed] [Google Scholar]
- 2.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 3.Romero X, Sintes J, Engel P. Role of SLAM family receptors and specific adapter SAP in innate-like lymphocytes. Crit Rev Immunol. 2014;34(4):263–299. doi: 10.1615/critrevimmunol.2014010538. [DOI] [PubMed] [Google Scholar]
- 4.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 5.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wandstrat AE, et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Cunninghame Graham DS, et al. CaNIOS GenES Investigators Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9(2):93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 8.Margraf S, Garner LI, Wilson TJ, Brown MH. A polymorphism in a phosphotyrosine signalling motif of CD229 (Ly9, SLAMF3) alters SH2 domain binding and T-cell activation. Immunology. 2015;146(3):392–400. doi: 10.1111/imm.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Salort J, Cuenca M, Terhorst C, Engel P, Romero X. Ly9 (CD229) cell-surface receptor is crucial for the development of spontaneous autoantibody production to nuclear antigens. Front Immunol. 2013;4:225. doi: 10.3389/fimmu.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham DB, et al. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J Immunol. 2006;176(1):291–300. doi: 10.4049/jimmunol.176.1.291. [DOI] [PubMed] [Google Scholar]
- 11.Comte D, Karampetsou MP, Tsokos GC. T cells as a therapeutic target in SLE. Lupus. 2015;24(4-5):351–363. doi: 10.1177/0961203314556139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman LA, Tsokos GC. The IL-2 defect in systemic lupus erythematosus disease has an expansive effect on host immunity. J Biomed Biotechnol. 2010;2010:740619. doi: 10.1155/2010/740619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulton VR, Grammatikos AP, Fitzgerald LM, Tsokos GC. Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription. Proc Natl Acad Sci USA. 2013;110(5):1845–1850. doi: 10.1073/pnas.1214207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonelli M, et al. Phenotypic and functional analysis of CD4+ CD25- Foxp3+ T cells in patients with systemic lupus erythematosus. J Immunol. 2009;182(3):1689–1695. doi: 10.4049/jimmunol.182.3.1689. [DOI] [PubMed] [Google Scholar]
- 15.Solomou EE, Juang YT, Gourley MF, Kammer GM, Tsokos GC. Molecular basis of deficient IL-2 production in T cells from patients with systemic lupus erythematosus. J Immunol. 2001;166(6):4216–4222. doi: 10.4049/jimmunol.166.6.4216. [DOI] [PubMed] [Google Scholar]
- 16.von Spee-Mayer C, et al. Low-dose interleukin-2 selectively corrects regulatory T cell defects in patients with systemic lupus erythematosus. Ann Rheum Dis. 2016;75(7):1407–1415. doi: 10.1136/annrheumdis-2015-207776. [DOI] [PubMed] [Google Scholar]
- 17.Sadlack B, et al. Generalized autoimmune disease in interleukin-2–deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol. 1995;25(11):3053–3059. doi: 10.1002/eji.1830251111. [DOI] [PubMed] [Google Scholar]
- 18.Lyssuk EY, Torgashina AV, Soloviev SK, Nassonov EL, Bykovskaia SN. Reduced number and function of CD4+CD25highFoxP3+ regulatory T cells in patients with systemic lupus erythematosus. Adv Exp Med Biol. 2007;601:113–119. [PubMed] [Google Scholar]
- 19.Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007;178(4):2579–2588. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- 20.Bonelli M, et al. Quantitative and qualitative deficiencies of regulatory T cells in patients with systemic lupus erythematosus (SLE) Int Immunol. 2008;20(7):861–868. doi: 10.1093/intimm/dxn044. [DOI] [PubMed] [Google Scholar]
- 21.Miyara M, et al. Global natural regulatory T cell depletion in active systemic lupus erythematosus. J Immunol. 2005;175(12):8392–8400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- 22.Venigalla RK, et al. Reduced CD4+,CD25- T cell sensitivity to the suppressive function of CD4+,CD25high,CD127-/low regulatory T cells in patients with active systemic lupus erythematosus. Arthritis Rheum. 2008;58(7):2120–2130. doi: 10.1002/art.23556. [DOI] [PubMed] [Google Scholar]
- 23.Humrich JY, et al. Homeostatic imbalance of regulatory and effector T cells due to IL-2 deprivation amplifies murine lupus. Proc Natl Acad Sci USA. 2010;107(1):204–209. doi: 10.1073/pnas.0903158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koga T, et al. KN-93, an inhibitor of calcium/calmodulin-dependent protein kinase IV, promotes generation and function of Foxp3⁺ regulatory T cells in MRL/lpr mice. Autoimmunity. 2014;47(7):445–450. doi: 10.3109/08916934.2014.915954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizui M, et al. IL-2 protects lupus-prone mice from multiple end-organ damage by limiting CD4-CD8- IL-17–producing T cells. J Immunol. 2014;193(5):2168–2177. doi: 10.4049/jimmunol.1400977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koga T, et al. CaMK4-dependent activation of AKT/mTOR and CREM-α underlies autoimmunity-associated Th17 imbalance. J Clin Invest. 2014;124(5):2234–2245. doi: 10.1172/JCI73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humrich JY, et al. Rapid induction of clinical remission by low-dose interleukin-2 in a patient with refractory SLE. Ann Rheum Dis. 2015;74(4):791–792. doi: 10.1136/annrheumdis-2014-206506. [DOI] [PubMed] [Google Scholar]
- 28.Saadoun D, et al. Regulatory T-cell responses to low-dose interleukin-2 in HCV-induced vasculitis. N Engl J Med. 2011;365(22):2067–2077. doi: 10.1056/NEJMoa1105143. [DOI] [PubMed] [Google Scholar]
- 29.Koreth J, et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N Engl J Med. 2011;365(22):2055–2066. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klatzmann D, Abbas AK. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat Rev Immunol. 2015;15(5):283–294. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 31.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38(1):13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1–induced extracellular matrix expression. Mol Pharmacol. 2006;69(2):597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 33.Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17(5):349–366. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Kim HP, Kim BG, Letterio J, Leonard WJ. Smad-dependent cooperative regulation of interleukin 2 receptor alpha chain gene expression by T cell receptor and transforming growth factor-beta. J Biol Chem. 2005;280(40):34042–34047. doi: 10.1074/jbc.M505833200. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110(8):2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Passerini L, et al. STAT5-signaling cytokines regulate the expression of FOXP3 in CD4+CD25+ regulatory T cells and CD4+CD25- effector T cells. Int Immunol. 2008;20(3):421–431. doi: 10.1093/intimm/dxn002. [DOI] [PubMed] [Google Scholar]
- 39.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol. 2012;30:733–758. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee M, et al. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol. 2012;188(3):1206–1212. doi: 10.4049/jimmunol.1102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman LA, Tsokos GC. Lupus-prone mice fail to raise antigen-specific T cell responses to intracellular infection. PLoS One. 2014;9(10):e111382. doi: 10.1371/journal.pone.0111382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kis-Toth K, et al. Selective loss of signaling lymphocytic activation molecule family member 4 CD8 T cells contributes to the decreased cytotoxic cell activity in systemic lupus erythematosus. Arthritis Rheum. 2015;68(1):164–173. doi: 10.1002/art.39410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenardo MJ. Interleukin-2 programs mouse alpha beta T lymphocytes for apoptosis. Nature. 1991;353(6347):858–861. doi: 10.1038/353858a0. [DOI] [PubMed] [Google Scholar]
- 44.Singer BD, King LS, D’Alessio FR. Regulatory T cells as immunotherapy. Front Immunol. 2014;5:46. doi: 10.3389/fimmu.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]