Significance
Proteins often exhibit large-scale collective motions that are essential for biological macromolecules to perform their functions. To understand the nature of the large-scale conformational dynamics, we applied transition path theory to analyze the Markovian microstates that are obtained from extensive unrestrained molecular dynamics simulations, using the activating conformational changes in the kinase domain of c-Src nonreceptor tyrosine kinase as an example. These results elucidate the fine details of the conformational transition and offer a perspective for the interpretation of the conformational transition. Microstates that are not crucial to the thermodynamics but are important for the kinetics can be identified by transition path theory, explaining why extensive conformational sampling is needed to reproduce the accurate kinetics.
Keywords: transition path theory, conformational transition, Markov state models
Abstract
Nonreceptor tyrosine kinases of the Src family are large multidomain allosteric proteins that are crucial to cellular signaling pathways. In a previous study, we generated a Markov state model (MSM) to simulate the activation of c-Src catalytic domain, used as a prototypical tyrosine kinase. The long-time kinetics of transition predicted by the MSM was in agreement with experimental observations. In the present study, we apply the framework of transition path theory (TPT) to the previously constructed MSM to characterize the main features of the activation pathway. The analysis indicates that the activating transition, in which the activation loop first opens up followed by an inward rotation of the αC-helix, takes place via a dense set of intermediate microstates distributed within a fairly broad “transition tube” in a multidimensional conformational subspace connecting the two end-point conformations. Multiple microstates with negligible equilibrium probabilities carry a large transition flux associated with the activating transition, which explains why extensive conformational sampling is necessary to accurately determine the kinetics of activation. Our results suggest that the combination of MSM with TPT provides an effective framework to represent conformational transitions in complex biomolecular systems.
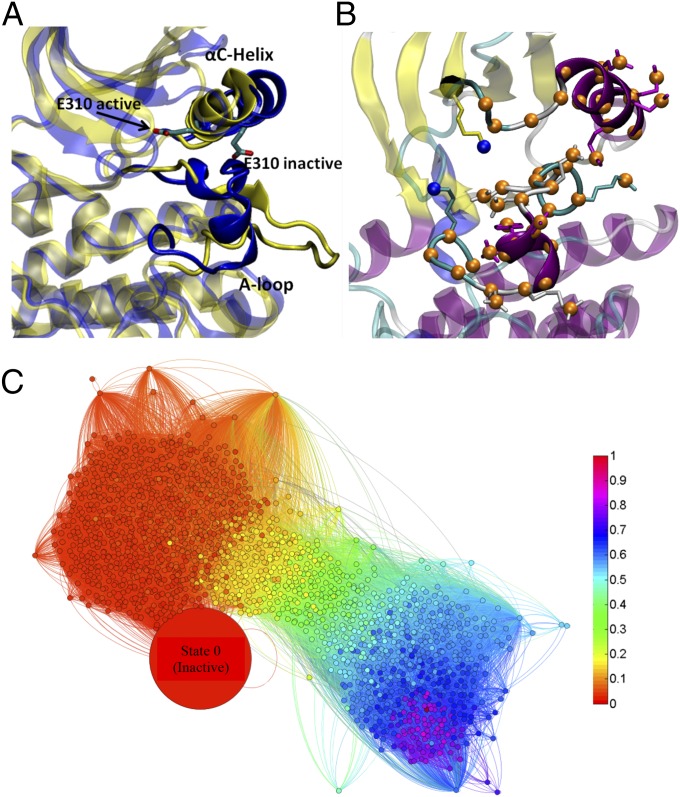
Proteins, rather than being static molecular structures, often exhibit large-scale collective motions that are biologically essential for their function (1, 2). Dynamics and flexibility form the foundation of both the conformational selection and induced-fit mechanisms of protein–protein or protein–ligand binding (3). An energy landscape theory was proposed in the early 1990s to conceptualize protein dynamics (4) and was extensively used to characterize the protein folding problem (5) and allostery (6). According to the energy landscape theory, the complex topography of the landscape that underlies the dynamics can give rise to multiple and significantly populated conformational states (metastable states). The study of protein dynamics is not only interested in obtaining the thermodynamic properties of those metastable states but also pays significant attention to the transition pathways linking them and the kinetic information. A noteworthy example of biological significance is presented by the nonreceptor tyrosine kinases of the Src family. A well-characterized prototypical tyrosine kinase is c-Src, which plays vital roles in cellular signaling pathways (7); overactivation is key to tumorgenesis and metastasis and obesity (8). Although the configurations of the regulatory domains are important in controlling kinase activity, the conformational changes occurring within the catalytic domain itself (Fig.1A and SI Methods for more details) is of special interest. Intramolecular motions, both at short and large length scales, control the activation of c-Src. Therefore, a better understanding of the internal motions displayed within the catalytic domain of c-Src during activation will clarify how protein kinases act as molecular switches, as well as help identify novel metastable states that are potentially suitable for the design of potent inhibitors.
Fig. 1.
(A) Cartoon representation of the conformational transition in c-Src catalytic domain. The inactive conformation is colored in blue, and the active conformation is colored in yellow. The main structural changes include the rotation of the αC-helix and the movement of the activation (A-) loop. E310 (chicken c-Src numbering) in the αC-helix is explicitly shown to illustrate the movement of the αC-helix in the transition. E310 is pointing outward in the inactive conformation, whereas it is pointing inward in the active-like conformation so that a catalytically important salt bridge can be formed (36). The A-loop is partially folded in the inactive kinase, but it becomes fully extended in the active conformation. (B) Atoms that define the space of CVs. The Cartesian coordinates of the selected atoms are used as CVs. A total of 50 atoms are selected. The carbon atoms are colored in orange, and the nitrogen atoms are colored in blue. (C) Network representation of the conformational transition. Each node represents a microstate from MSM, and edges represent nonzero elements of the transition probability matrix. The size of each node is proportional to the Boltzmann weight of the corresponding microstate. The largest node size is set to be 150, whereas the smallest node size is 10. Microstates are colored by their committor probability values. The program Gephi (37) was used to generate this network representation.
Molecular dynamics (MD) simulations offer an attractive approach to examine the conformational dynamics in biological macromolecules at the atomic level. One possibility is to capture spontaneous transitions during long unbiased MD simulations (9). However, this strategy becomes inefficient for very slow transitions, because the system spends a large fraction of its time returning to regions that were previously visited due to the high barriers in the energy landscape. To accelerate the sampling of the conformational space, a biasing potential can be introduced in an MD simulation to overcome the high energy barriers; umbrella sampling (10) and metadynamics (11) are popular methods in studies of conformational transitions. An alternative strategy is to combine the information from a large number of short simulations to construct Markov state models (MSMs) representative of the process of interest (12–15). MSMs use discrete-time Markov chain to describe the conformational dynamics of proteins in terms of jumps between the microstates extracted from the simulation data, providing structural, thermodynamic, and long-timescale kinetic information (16, 17). For example, this approach was used to examine the activation of PKA regulatory subunit RIα in response to cAMP binding (18). Yang et al. (19) carried out a MSM analysis of a simplified coarse-grained model of the catalytic domain of Hck, a member of the Src family of kinases. More recently, Shukla et al. (20) constructed an MSM for the activation process in c-Src catalytic domain using ∼500 μs of aggregated sampling generated from Folding@Home.
A fundamental issue with conformational transitions occurring in complex macromolecular structures is that they are difficult to comprehend because of their inherent multidimensional nature. An elegant framework, called transition pathway theory (TPT), developed to facilitate the analysis of MSMs can help overcome this challenge (21, 22). TPT has previously been applied to the study of protein folding (17, 23) and the dynamics of HIV-1 protease flaps (24) and has offered atomistic insights into the pathways and kinetics. The availability of a meaningful MSM analysis based on extensive MD simulation data provides a great opportunity to further examine the nature of the conformational transition pathways associated with the activation process within the c-Src kinase domain. The present study uses TPT to help elucidate the fine details of the activation transition pathway in an important signaling protein like c-Src.
SI Methods
Atomic Models.
Chicken c-Src numbering is used throughout this paper. Only the isolated kinase domain (residues W260 to T521) in the absence of the regulatory domains SH2 and SH3 was considered in the present study. Hydrogen atoms were built to the crystallographic structures using the HBUILD module of CHARMM (38). The all-atom protein was solvated in a truncated octahedral simulation cell constructed from an equilibrated 80- × 80- × 80-Å3 box. The final water box comprises 11,216 TIP3P (39) water molecules. The size of the simulation cell was chosen to extend at least 10 Å away from the surface of the protein. One ATP molecule and two Mg2+ ions were placed in the active site, based on the high-resolution structure of PKA [Protein Data Bank ID code 1ATP (40)]. In addition, 22 Na+ ions and 19 Cl− ions were inserted into the system to neutralize the total charge and to model a salt concentration of ∼150 mM. Y416 in the A-loop is not phosphorylated at all times.
MSM.
MSM employs discrete-time Markov chain to describe the conformational dynamics of proteins in terms of jumps between the microstates obtained from the clustering of the simulation data into discrete states. It offers a way to combine many short trajectories and yield information at longer timescales. The transition probability matrix T includes the probability of transitions from state i to state j at a certain time interval τ. An element in the transition probability matrix (e.g., Tij) is estimated by counting the number of transitions nij observed between time t and t + τ. The transition probability matrix is then row-normalized. To enforce detailed balance (which ensures that population of states is conserved), a maximum likelihood estimate of the transition probability matrix that follows the detailed balance is obtained. The transition probability matrix can be used to obtain the population of the system at any time using the Chapman–Kolmogrov equation. The eigenvalues/eigenvectors spectrum of the transition matrix gives information about the aggregate transitions between subsets of states in the model and at what timescales these transitions occur. The equilibrium populations of the individual states are estimated from the first eigenvector of the transition probability matrix. The eigenvalues µ of the transition probability matrix are related to the implied timescales 1/k of transitions via the following expression:
To characterize the conformational changes in c-Src kinase domain with MSM, 500 μs of unbiased MD simulations had been accumulated using Folding@Home. The details of the simulation and the protocol for building MSM are given elsewhere (20). In summary, 1,798 Markov states (microstates) were generated out of 500 μs of simulations, using a lag time of 5 ns. A network representation of the MSM is given in Fig. 1C.
TPT.
MSM employs discrete-time Markov chain to describe the conformational dynamics of proteins in terms of jumps between the microstates obtained from cluster analysis of the simulation data. It offers a way to combine many short trajectories and yield information at longer timescales. However, when dealing with complex dynamical processes such as the activating conformational transition in c-Src kinase domain, MSMs themselves can be complicated and require advanced tools to facilitate the analysis. TPT was developed for such a purpose. It aims to describe the reactive trajectories that connect the end states and their probability density function, probability current and flux, and the transition rate. In the framework of TPT, two subsets of the state space (e.g., states from MSM), A and B, are defined to represent the two stable conformations that one wants to study. Without loss of generality, A can be assigned to denote the reactant and B is the product. Reactive trajectories are pathways leaving A and entering B, through intermediates states (I), which are among the remaining states. Central to the TPT, the committor probability, , is defined as the probability that the system will first enter B rather than A when being at state i. It can be interpreted as the fraction of trajectories initiated with Boltzmann-distributed momenta from state i that commits to B first. The committor probability of each state can be computed by the following equation:
in which Tij is the element in the transition probability matrix T and is the complement of the union of A and B. The first term is the probability that the trajectory goes to B in one step after leaving state i, and the second term represents the system entering another intermediate j and then reaching B first. By definition, the committor probability of any state in A has the value of zero, whereas it is one for any state in B. One quantity that relates to committor probability is the backward committor probability, , that is the probability that the system was in A previously rather than in B when being at state i. When studying a system at equilibrium, due to time reversibility. Another useful quantity is the effective flux fij, which is the probability of a reactive trajectory visiting state i and j consecutively and can be computed as follows:
in which πi is the equilibrium population of state i. After anti symmetrizing fij by adopting the following,
the net flux from A to B in the reactive trajectory can be obtained. Once the net fluxes are at hand, one could compute the total flux from A to B using the following equation:
The total flux represents the expected number of observed A → B transitions per unit time (it is usually the lag time τ in the analysis of MSM). Therefore, the transition rate from A to B is related with F as follows:
Another useful concept in the TPT is the reactive trajectory, which is an equilibrium continuous trajectory that transits from A to B. Suppose there is a very long equilibrium trajectory and it travels back and forth between A and B. Reactive trajectories are pieces of the long trajectory that has last left A before entering B. In practical applications, one is often interested in knowing the probability of observing a reactive trajectory at a microstate. This probability can be calculated as πiR = qi─πiqi+, which is a multiplication of three factors. They are the forward and the backward committor probabilities and the equilibrium probability of finding microstate i. By definition, the reactant and the product states have πAR = πBR = 0. After adding πiR for all states, one could obtain the equilibrium probability for the trajectory to be reactive, that is, the fraction of time a trajectory spends on the pieces that leave A and enter B. One could refer to E and Vanden-Eijnden (22), Metzner et al. (41), and Noé et al. (17) for the details of TPT.
Markovian Milestoning Calculation.
Markovian milestoning simulations were also carried out to characterize the activating conformational transition. Like MSM, this method aims to use the transition rate among different Voronoi cells to restore the statistical properties of the longtime dynamics of the system. Theoretically, a Markovian milestoning simulation establishes a continuous-time Markov model, whereas MSM relies on a discrete-time Markov model. It was shown previously that Markovian milestoning formulation can reconstruct the statistical properties of the long-time dynamics of the system from independent simulations, each confined inside one of the K cells (33, 34). The key feature required for this result is that the confined dynamics must be equivalent to that from a long unbiased trajectory passing through the same set of cells. More specifically, the confinement must leave unperturbed the dynamical properties of the systems when it is in the interior of the cell as well as the probability flux in and out of the cells. Both the hard-wall method (a strict reflection rule) (34) and soft-wall potential (planar half-pseudoharmonic restraining potential) (33) are able to confine the system within each cell. The conservation of probability flux through the boundaries of the cell gives a way to compute, πR, the equilibrium probability of the system to be in cell R, and the associated free energy, . For a Markovian milestoning simulation restrained to cell α, the rate of escape from cell α to cell γ can be calculated as ναγ = Nα/Tα, where Nα is the number of collisions with the boundary between α and γ, and Tα is the time spent in cell α. The conservation of probability flux at steady state yields the following linear equation:
with a constrain that .
Each Voronoi cell was centered at an image along an optimized string which was obtained previously (20), resulting in a total of 51 Voronoi cells. Principal component analysis on the strings identified that the top 15 principal components covered ∼90% of the variation in the full CV space. A soft-wall restraining potential as described in Maragliano et al. (33) was applied to the 15-dimensional CV space. Fifty-nanosecond MD trajectories were accumulated for each Voronoi cell. All Markovian milestoning simulations were performed using version 2.9 of the molecular simulation package NAMD (42) and the CHARMM27 force field (43). The isobaric–isothermal (NPT) ensemble was used in all MD calculations. The pressure and temperature were controlled by Langevin piston method (44) and Langevin dynamics and kept at 1 atmosphere pressure and 300 K, respectively. Periodic boundary conditions were applied and particle-mesh Ewald (45) was used to treat long-range nonbonded interactions. The short-range nonbonded interactions were truncated at 12 Å, with a switching function turned on from 10 to 12 Å. The nonbonded list was updated at every MD step with a cutoff distance of 16 Å. Covalent bonds involving hydrogen atoms were constrained at their equilibrium distances by SHAKE algorithm (46) so that a 2-fs time step can be used in all milestoning simulations. The force constant of the restraining potential is 500 kcal/(mol⋅rad2).
Results and Discussion
Selection of the Reactant and the Product States from MSM.
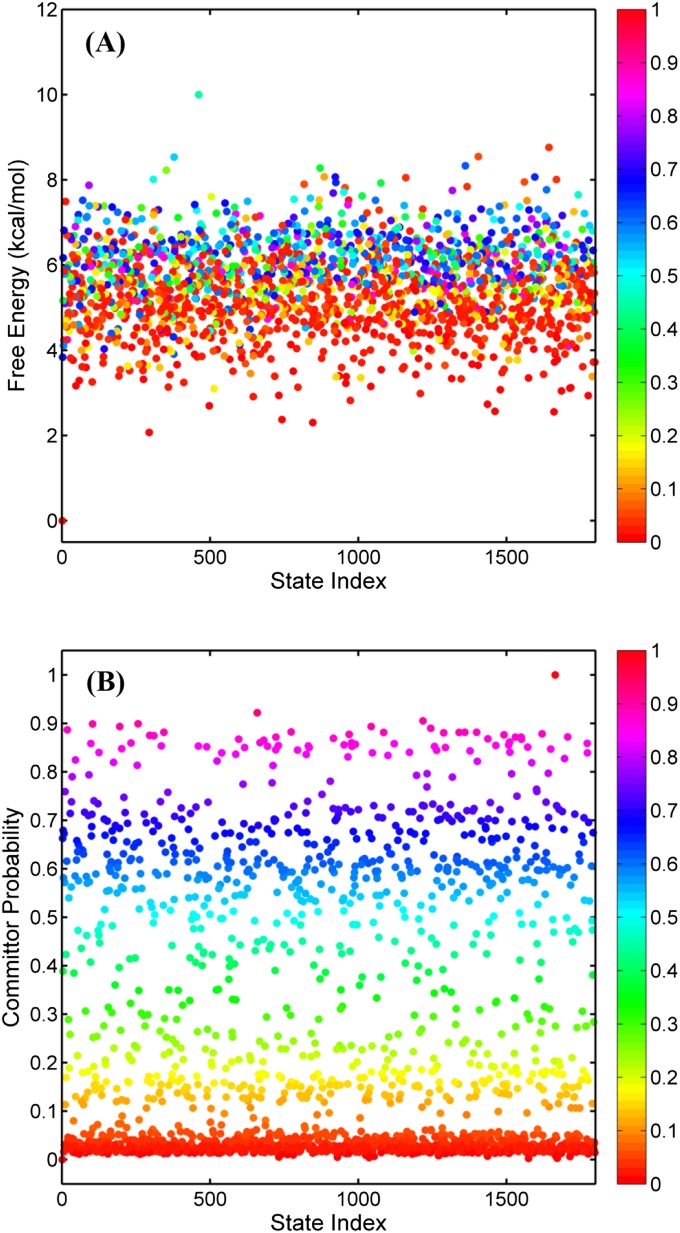
In a previous study, an MSM comprising 1,798 microstates with a 5-ns lag time was constructed from ∼550 μs of aggregate MD trajectories. A network (1,798 nodes and 46,089 edges) representation of the MSM is displayed in Fig. 1C. In this network, each node represents a microstate and the edges denote a nonzero transition probability between a pair of microstates i and j. Such a large number of nodes and edges indicates that advanced analysis scheme such as TPT is necessary to reveal the inherent features of the conformational transition. The first step in applying TPT is to define the reactant and the product states. To simplify the present analysis, a single MSM microstate was assigned to be the reactant and the product, respectively. The reactant state (inactive kinase) is selected based both on structural similarity with the crystallographic structure of the inactive c-Src kinase 2SRC (25), and on the free energy of the microstates in the MSM analysis. This leads to microstate 0: It displays the lowest free energy (Fig. S1A) and the rmsd of all Cα atoms relative to the kinase domain of the crystal structure 2SRC (25) is 1.0 Å. Furthermore, inspection of the structure shows that its αC-helix adopts an outward-rotated conformation and the A-loop is in the partially folded and closed conformation, which are two key features of the inactive conformation. This indicates that microstate 0 is indeed a good representation of the inactive state. Structurally, the product state is expected to resemble the kinase domain from the crystal structure of active-like c-Src 1Y57 (26). Several microstates display an rmsd for Cα atoms that is fairly small (less than 1∼2 Å). In principle, a broad basin of microstates could have been ascribed to the product state. For the sake of simplicity, a single microstate state was chosen, the one yielding the smallest possible rmsd together with a distribution of committor probabilities covering the full range [0,1] with no gaps within bins of 0.1 (see below for more discussion about committor probabilities). The microstate (1663) chosen to stand for the product state has an rmsd value of all Cα atoms of 1.5 Å relative to the kinase domain of 1Y57 (26) and yields a range of committor probabilities that is well distributed (Fig. S1B). The free energy difference between the reactant and the product, which can be computed with the equilibrium probabilities of the two end states, is about 3.9 kcal/mol. This value is coincidentally very close to the ∆G between the inactive and active basins after integrating the 2D potential of the mean force (2D-PMF; Fig. S2A). One advantage of using TPT to investigate conformational transition is that all kinetic information can be incorporated in closed form from the MSM. Using the current definition of the reactant and product states, the rate of reaction A → B (kAB; see SI Methods for the calculation of kAB) is estimated to be 1/95 μs−1, based on TPT. This is consistent with the mean first passage time of 110 μs previously determined from the slowest eigenvalue from the MSM, further validating our choice of the two end states.
Fig. S1.
(A) Free energy of MSM microstates as a function of state index. (B) Committor probability of microstates as a function of state index. Microstates in both plots are colored by their committor probability values.
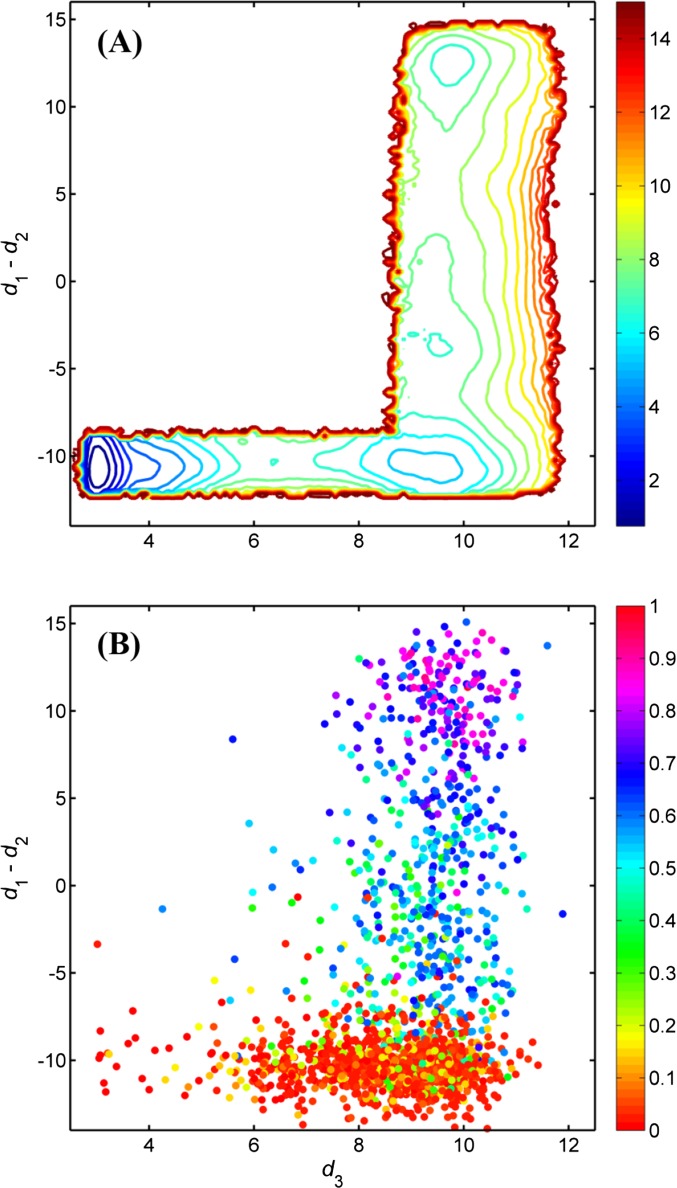
Fig. S2.
(A) Two-dimensional PMF from replica-exchange molecular dynamics/umbrella sampling calculation. The inactive conformation of kinase domain occupies the lowest free energy region, which is located at the lower left corner. The active conformation occupies the upper right corner. A free energy basin which associates with an intermediate state can be seen at the lower right corner. d1 is the distance between E310 Cδ atom and R409 Cζ atom. d2 is the distance between E310 Cδ atom and K295 Nζ atom. d3 is the average of distances between O-Asp413 and N-Thr417, O-Asn414 and N-Ala418, and O-Glu415 and N-Arg419. The 2D-PMF reflects the free energy landscape projected onto the αC-helix and the A-loop motions. (B) Projection of MSM microstates onto 2D-PMF coordinates. MSM microstates are colored by their committor probability values. Although the microstates were obtained from unbiased MD simulations, projection of the microstates onto 2D-PMF still helps understand features of the conformational changes and the energy landscape.
Analysis of All Markov Microstates.
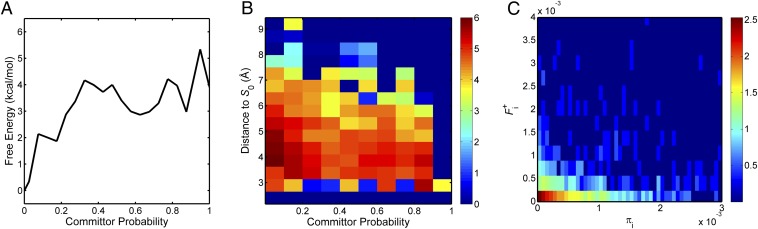
When studying conformational transitions, the reaction coordinate is a single variable that quantifies progress along a pathway. The committor probability (i.e., the probability, starting from any microstate, to first reach state A rather than state B), serves as an ideal reaction coordinate (27, 28). By constructing a histogram of the microstates based on the committor probabilities and accumulating the equilibrium probabilities in each bin, the free energy profile along the “reaction coordinate”–committor probability can be obtained. The resulting 1D free energy profile is plotted in Fig. 2A. One can observe the existence of multiple metastable intermediate states after lumping microstates along the reaction coordinate. The 1D free energy profile demonstrates that the basin with lowest free energy contains the inactive state of the kinase domain. The shallow free energy well centered at q ≈ 0.2 corresponds to the intermediate state observed in the 2D-PMF calculated from replica exchange molecular dynamics/umbrella sampling calculation (the lower-right corner of Fig. S2A). A new free energy well centered at q ≈ 0.6 can be found along the 1D free energy profile, indicating a second intermediate state. This intermediate state appears in the later stage of the transition and was not captured by the umbrella sampling simulations. However, this intermediate state was identified by distributed MD simulations with ∼500 μs of aggregated sampling, highlighting the importance of conformational sampling in investigating dynamical behavior of biological macromolecules. One possible reason is that umbrella sampling simulations used structural properties (linear combination of distances) as order parameters. Different regions in the 2D-PMF correspond to degenerate values of the committor probabilities (Fig. S2B). A new metastable state appears when projecting the 2D-PMF onto the reaction coordinate. A piece of evidence to support this reasoning is that visual inspection of the structural ensemble of the intermediate state centered at q ≈ 0.6 reveals large structural flexibility, especially for the αC-helix. This can also be observed in Fig. S2B: Microstates having q ≈ 0.6 are scattered in a large region of the 2D-PMF coordinates. The present analysis confirms that structural features indeed tend to be oversimplified when characterizing conformational transitions on the basis of a few chosen order parameters. Several widely used enhanced sampling strategies such as umbrella sampling and metadynamics rely on choosing a few (usually one to three) predefined structural collective variables (CVs) to coarse-grain a conformational transition (i.e., mapping a high-dimensional transition to a much lower dimensional structural CV space) and to bias the sampling in the CV space. They could potentially suffer from this oversimplified mapping and fail to capture crucial aspects of the transition. Similar issues might also be encountered with the string method, which also depends on a set of predefined CVs, although this is somewhat alleviated because the subspace of CVs is typically of a fairly large dimension.
Fig. 2.
TPT analysis of all MSM microstates. (A) Free energy profile as a function of the committor probability. (B) The heat map of F+(qi, di). The heat map is plotted on a log10 scale of the net effective flux. Regions having F+ to be zero are assigned a value of 10−7 when taking the logarithm. (C) Two-dimensional histogram of (πi, ). It is zoomed-in so that nonzero counts are highlighted.
Although the microstates were obtained from unbiased MD simulations, projection of the microstates onto 2D-PMF still helps understand features of the conformational changes and the energy landscape. According to Fig. S2B, opening the A-loop occurs at the very early stage of the activation, because most of the metastable states in the horizontal band have qi ≤0.3. Microstates with a committor probability around 0.5 mostly scatter in the region between the intermediate and the active states, suggesting the kinetic bottleneck (the transition states) lies between the intermediate and the active states. As expected, the free energy basin in the 2D-PMF that includes the kinase domain in an active conformation also contains metastable microstates with committor probabilities close to 1.0. However, one can still observe microstates with q ≈ 0.5 within a basin close to the active conformation, suggesting that some slow degrees of freedom exist despite the structural similarities of those microstates. This further confirms that there is a need to go beyond a structural perspective to fully understand the inherent complexity of conformational transition in biological macromolecular systems.
The Transition Tube.
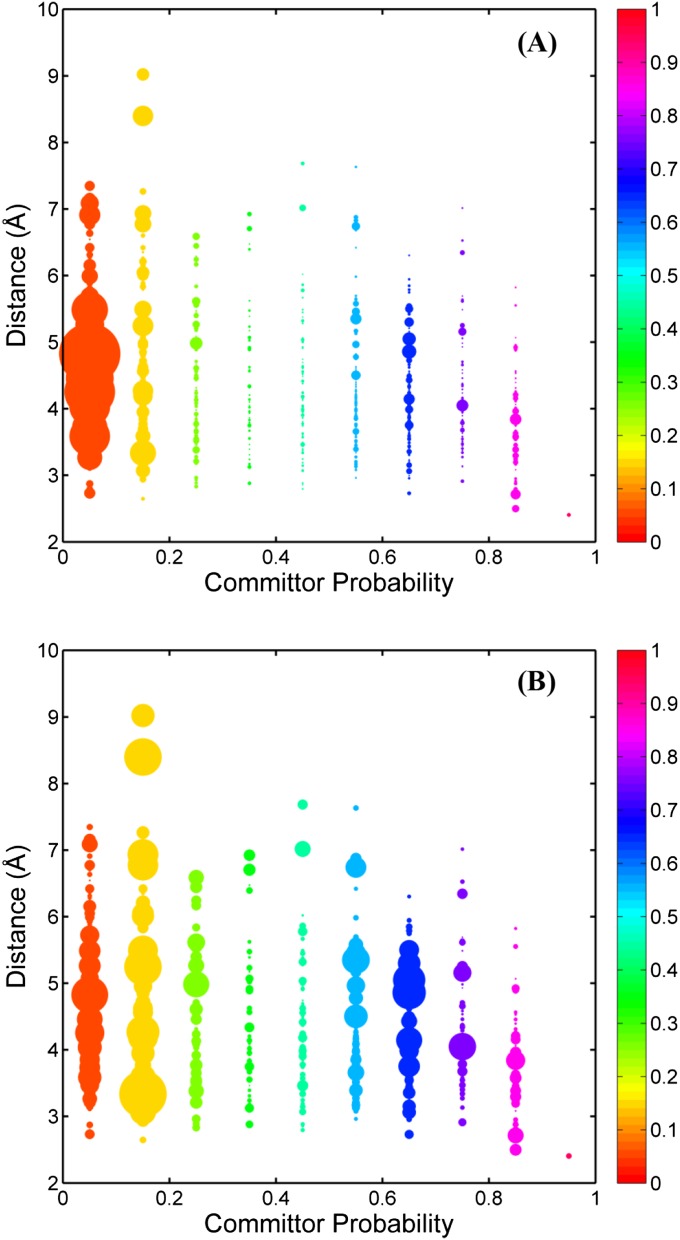
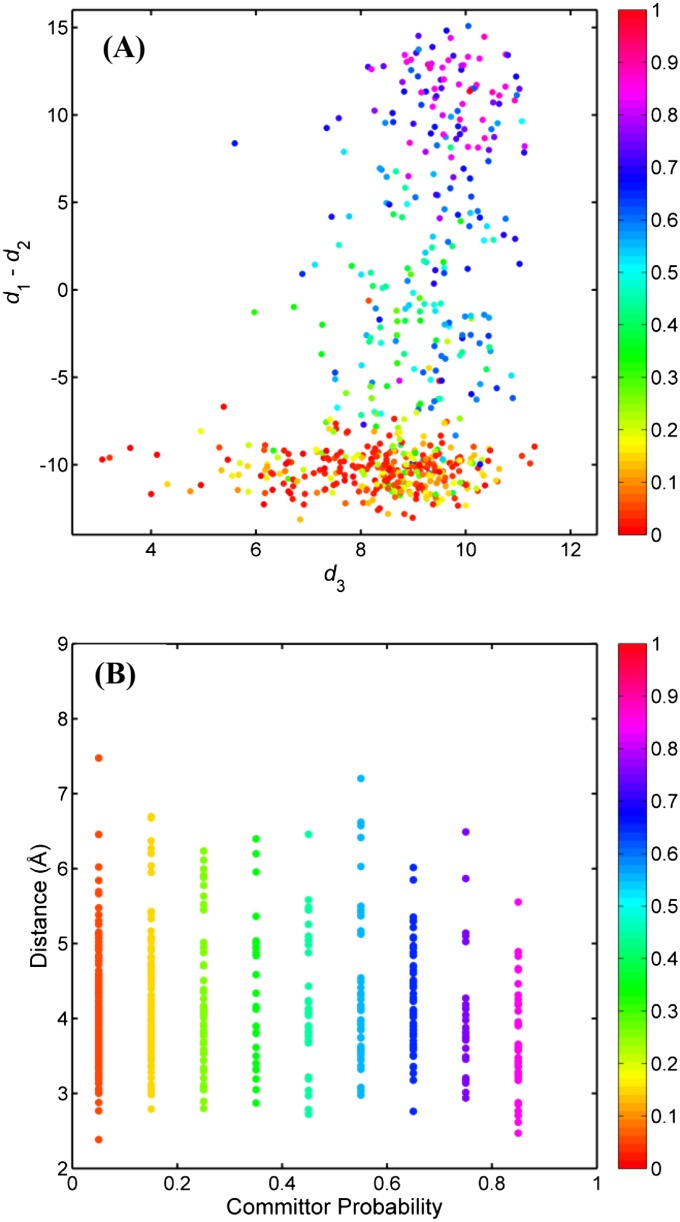
When the committor probability is used as the reaction coordinate, a “reaction tube” connecting the two end states can be defined by grouping microstates according to their committor probabilities. We are particularly interested in knowing the spatial distribution of the equilibrium probability (πi), net flux (), and the equilibrium probability of observing a reactive trajectory () within this transition tube because those are key quantities characterizing a conformational transition. Together, they define the shape of the transition tube. To progress in our analysis, a histogram of committor probability was constructed (bin width of 0.1). Through this discretization process, the transition tube was separated into 12 slices or cross-sections. Microstates that fall into each slice along the transition tube were then collected, and their geometric center in the CV space was computed. The geometric center corresponds to a line at the center of the tube. In the present analysis, the Cartesian coordinates of 50 atoms (Fig. 1B) are used to define the CV space; Euclidean distances between each microstates and the geometric center were calculated in the CV space and those Euclidean distances were normalized by the square root of the number of CV atoms (i.e., 50) to correspond more closely to the commonly used rmsd value. The effective normalized distance, referred to as di, will be used throughout the remainder of the analysis. Fig. S3A displays di as a function of committor probability. According to Fig. S3A, the microstates distributed around the corresponding geometric center display a lower bound of distance value of ∼2 Å, and an upper bound that is between 6 and 9 Å. Microstates are scattered in a large region of the CV space, indicating that the transition tube is quite broad. Linear regression of the di values in response to committor probability resulted in a straight line with a slope of −0.38 and intercept of 4.5. The shape of the tube on the basis of the committor probability seems to resemble a spindle. The transition tube becomes wider as it leaves the reactant state and reduces its width as it approaches the product state. Eventually, all fluxes enter the one-point product state.
Fig. S3.
(A) Distance from a microstate to the geometric center as a function of the committor probability. The size of each circle is proportional to the Boltzmann weight (equilibrium probability of a microstate, πi). According to our selection of the reactant and the product, there is only one microstate for each, so that the reactant and the product are not included in generating A. Furthermore, only two microstates were found to fall into the bin centered at q = 0.9. Hence, only one distance value (one point in this figure) of 2.4 Å could be obtained. (B) Distance from a microstate to the geometric center as a function of the committor probability. The size of each circle is proportional to the equilibrium probability of the reactive trajectory ().
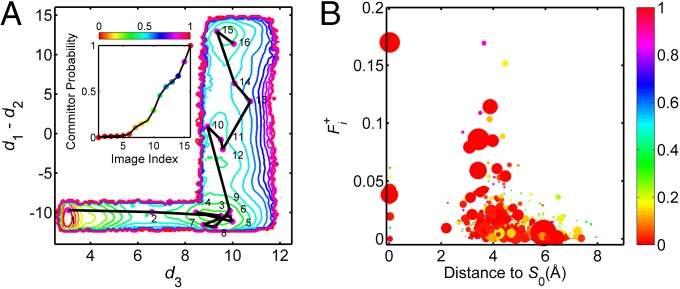
To probe the distribution of the net reactive flux within the transition tube, we used the row-sum of the net-flux matrix , first introduced by Vanden-Eijnden (21). The quantity represents the flux leaving microstate i and eventually entering the product state. In TPT, such “productive flux elements” play an important role in determining the kinetics of the conformational transition. The total flux leaving the reactant state () is the total transition flux. As expected, the total transition flux that leaves microstate 0 (the reactant) equals the total flux entering microstate 1663 (the product). As shown in Fig. 2B, large fluxes tend to populate microstates located closer to the center of the transition tube; large probability currents flow through the central region of the transition tube. To clarify the relationship between the equilibrium probabilities πi (important for thermodynamics) and the productive flux elements (important for kinetics), a 2D histogram of (πi, ) was constructed. The 2D histogram, shown in Fig. 2C, illustrates that states with large productive flux elements are likely to have small equilibrium probabilities πi, and vice versa. The TPT analysis of c-Src activation reveals that microstates that are not highly probable at equilibrium can nonetheless carry large productive fluxes to act as hubs for the transitions. Those states are important for understanding kinetics. However, microstates that possess relatively large equilibrium probability (and carry a small productive flux as shown in Fig. 2B) can be found far away from the geometric center of the transition tube. This simply shows that microstates that are important for thermodynamics are not necessarily important for kinetics. Transition fluxes and equilibrium probabilities need both to be taken into account to characterize conformational changes.
Reactive trajectories generated within the TPT framework, in which the system transits from the state A (inactive) to the state B (active), also provide important information on the activation mechanism. In practical applications, one is often interested in knowing the probability if a specific microstate is (or is not) along the path of a reactive trajectory (). By definition, the reactant and the product states have . Among the remaining 1,796 microstates, state 513 has the largest probability to be part of a reactive trajectory ( ≈ 0.022). Fig. S3B shows di with respect to qi, with the size of each point proportional to . Microstates having qi between 0.1 and 0.2 are more likely to belong to a reactive trajectory than other committor probabilities. Similar to the distribution of πi in the transition tube, microstates that are located at the outer region of the transition tube can still belong to reactive trajectories, even though those reactive trajectories do not carry a large flux.
Constructing Pathways from TPT.
The string method, by which a transition pathway is discretized into a chain of states called images, is a powerful technique to study rare event in complex systems (29, 30). Once the optimized string has been obtained, thermodynamic and kinetic information can be extracted from milestoning simulations. Recent successes (31, 32) in applying the string method to study transition events prompted us to incorporate this concept in our TPT analysis of the MSM for Src kinase activation. Here we adopt the following prescription to generate path strings connecting the reactant and the product based on a steepest descent/ascent type of strategy to the net flux. Starting from the reactant (microstate 0), microstate j, which has the largest net flux , is selected. Then, microstate j is set to be the starting point and the previous criterion is applied. This leads to microstate k because is maximal among net fluxes leaving microstate j. The process is repeated until the product state (here microstate 1663) is reached. The chain of discrete microstates can be viewed as the discretized form of a string, with increasing image index as the microstates move toward the product state. Following the above protocol, a discretized string (S0) with 16 images can be constructed. Projection of S0 onto the 2D-PMF is shown in Fig. 3A. Fig. 3A illustrates that S0 mostly travels in the valley of the free energy landscape. This observation is consistent with the assumption that the optimized string corresponds to a minimum free energy pathway. Further, the intermediate state traps a large fraction of S0 (seven images, ∼44% of the images in S0), indicating the complex nature of the transitions in the intermediate state region. Fig. 3A also displays that the committor probability increases monotonically along S0. Images 11 and 12 approximately correspond to the transition state in S0.
Fig. 3.
Analysis of string S0. (A) Projection of string S0 onto 2D-PMF coordinates. (Inset) The committor probability as a function of the index of string images. (B) Scatter plot of the net flux of a microstate vs. its distance to S0 in the CV space. The size of each circle is proportional to the equilibrium probability of the microstate. Microstates are colored by their committor probability values. The reactant state is excluded when making this plot because of its large Boltzmann weight and net flux.
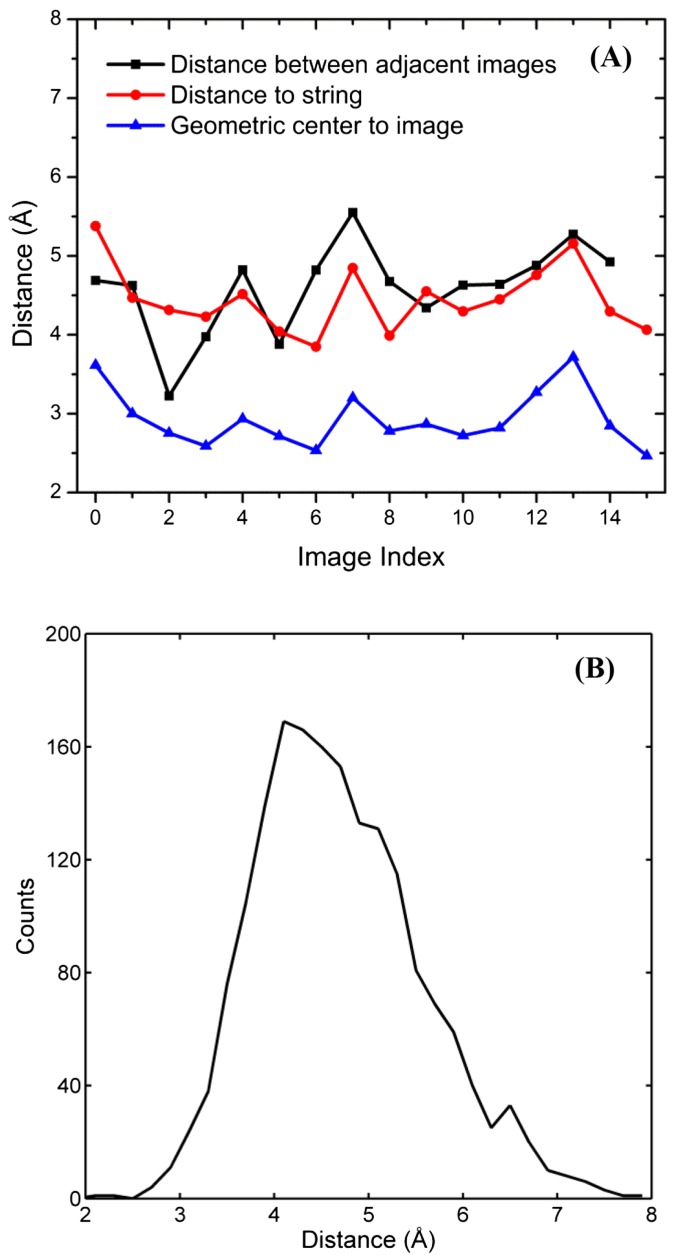
To analyze how the microstates distribute around S0, a Voronoi tessellation of the CV space is used, using the 16 images of S0 as the center of the cells. The distance between neighboring images, and the average distance from a microstate to S0 are computed and plotted in Fig. S4A. The mean value of distances between adjacent cells center-to-center is 4.6 Å, with an SD of 0.6 Å. Remarkably, the sum of each segment, which is approximately the total length of the string, is ∼70 Å whereas the distance between the two end-points is only ∼10 Å. The large ratio between the length of the string and the distance between two end points suggests that the optimal productive path S0 in the CV space is highly “crumpled.” The system must navigate on the rugged free energy landscape, thereby traveling a much longer total distance in the CV space than the actual distance between the end states. When viewing S0 in the 2D-PMF coordinates, S0 zigzags with loops in the free energy landscape. We also examined the relative position between an image (defined as the cell center) and the true geometric center of all microstates in that cell, for all cells. The distances between two types of center are also plotted in Fig. S4A. Comparing the average distance to S0 (the red curve in Fig. S4A) and the distance between two types of centers (the blue curve in Fig. S4A), the latter is shorter than the former, suggesting that S0 travels near the center of the data cloud. Moreover, the curve showing the average distance to S0 is more or less flat, suggesting that microstates surround S0 with equal thickness on average. A histogram of the distance from a microstate to S0 () is given in Fig. S4B. A single peak is observed and occurs at ∼4.1 Å with small counts at both short and large distances.
Fig. S4.
(A) Analysis of microstates around S0. (B) Histogram of the distance from a microstate to S0. A statistical test of the normality is performed on the distribution of values, using the null hypothesis (H0) that the probability density is Gaussian. The P value yielded from the Shapiro–Wilk test is smaller than 0.01. The values are hence not normally distributed, although the shape of the histogram resembles a Gaussian.
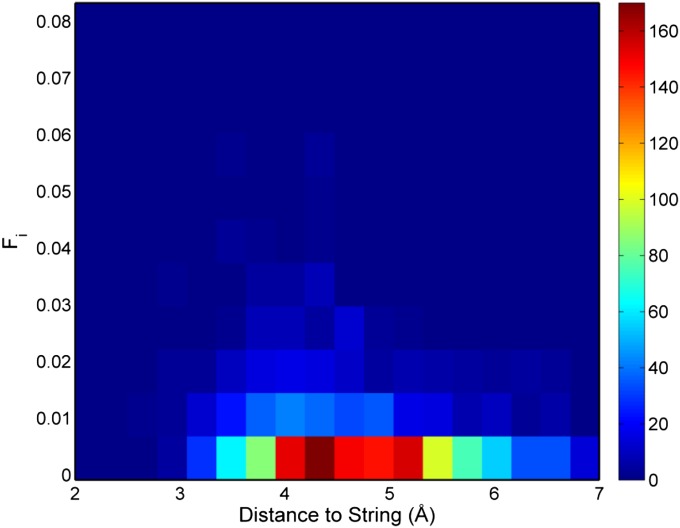
In addition to the geometrical distribution of microstates around S0 we are also interested in knowing the distribution of the net fluxes and equilibrium probability around S0. To achieve this, we plot Fi+ vs. and use the size of a point to reflect the πi (Fig. 3B). From this figure, one can see that microstates with large are close to S0 (upper left of Fig. 3B), whereas microstates that are far away from S0 have small (lower right of Fig. 3B). A 2D histogram of and is also constructed (Fig. S5) to augment the scatter plot of vs. . The pattern demonstrated in Fig. 3B can also be observed from the 2D histogram. Furthermore, the 2D histogram shows that microstates that are 4–5.25 Å away from S0 and have very small net fluxes are more probable than the rest. We further examine how πi distribute around S0. As demonstrated previously, large correlates with relatively small πi, whereas large πi correlates with small .
Fig. S5.
Two-dimensional histogram of the distance to S0 and the net effective flux.
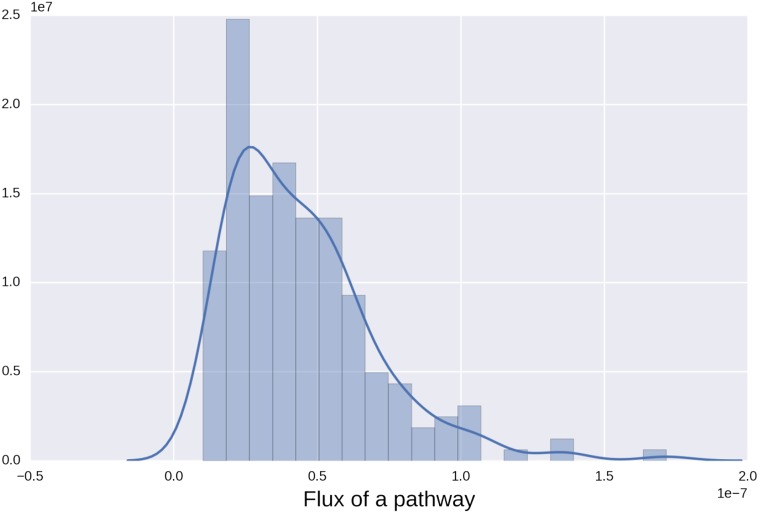
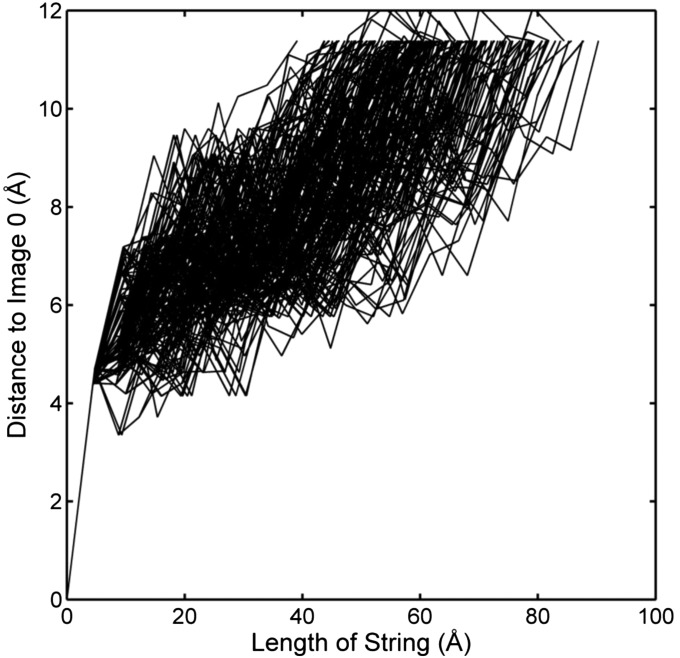
Multiple strings (transition pathways) are possible for a complex system like the c-Src kinase domain. Each string carries a certain amount of transition flux. In TPT, the flux of a pathway can be calculated as the minimal flux of an edge in the pathway (17). In the case of c-Src kinase domain, the transition network contains 1,798 nodes and 46,094 edges and an edge can be used in more than one pathway. Therefore, a complete scanning of all possible pathways sorted by their fluxes is extremely time-consuming. Instead, a two-step protocol is repeated to generate a number of paths. (i) The steepest descent/ascent type of strategy is adopted to generate a pathway, as described above. (ii) Immediately following the generation of a pathway, the flux of the pathway was subtracted by all edges in that pathway. Thus, although an edge in that selected pathway could be used in other pathways, the flux is modified every time the edge contributes to a pathway. We selected the first 200 pathways that resulted from the above procedure and used them in our analysis. Although the combined reactive flux carried by the set of 200 pathways accounts for approximately 18% of the total reactive flux from the reactant state, the set includes all of the largest flux paths. The distribution of the reactive fluxes among those 200 pathways can be found in Fig. S6. The spread of the 200 strings is similar to that of all microstates, as shown in Fig. S7, suggesting that those 200 strings are not confined in a particular region of the transition tube. The accumulated length of segments (approximated length of the string) in a string is plotted against the distance between the corresponding image and microstate 0 which is the starting image (Fig. S8) to reflect the straightness of the strings. Fig. S8 further supports that the transition tube has to travel a long distance to connect the two end points. In polymer theory, the length of a fully stretched chain L can be determined as N⋅b, in which N is the number of segments and b is the length of each segment. Assuming the chain follows a random walk model, in which the growth of segments is independent of each other, the mean of the square of the end-to-end distance R satisfies the following relation: . Therefore, the length of the chain can be expressed as . In the present study, each one of the 200 pathways can be viewed as a random chain. The end-to-end distance is the distance between the first image and the last one and is a constant (11.4 Å) because it represents the rmsd in the CV space between the reactant and the product. The average of L and N was determined to be 63.6 Å and 14, respectively, using the 200 pathways, which yield R to be ∼16 Å. One may picture as if the pathways fill the transition tube, much like a random coil in the multidimensional subspace of the CVs. Therefore, the transition pathways for the activation of c-Src correspond to random walks in the subspace of CV, confined within a reaction tube of about 4.5-Å radius rmsd. Although the activation of c-Src involves large-scale motions, the finite and relatively constant width of the reaction tube that encompasses the transition pathways does not seem to be consistent with the partial unfolding protein-quake model of conformational transition previously proposed by Miyashita et al. (6).
Fig. S6.
Histogram of the reactive fluxes among those 200 pathways. An estimated kernel density is also displayed as a solid line.
Fig. S7.
(A) Projection of microstates onto 2D-PMF coordinates. (B) Distance from a microstate to the geometric center as a function of the committor probability. In both plots, the microstates are collected from the 200 pathways. Microstates are colored by their committor probability values. The spatial distribution of microstates from 200 pathways resembles that from using all microstates.
Fig. S8.
Comparison of the distance between an image to the starting point of the string and the length of the string up to that image, along each transition pathway for the 200 pathways.
Milestoning Simulations Reveal Similar Structural Features.
The Markovian milestoning method (33, 34) is another way to reveal both the thermodynamic and kinetic information of conformational transitions. In a Markovian milestoning simulation, partitioning the CV space into Voronoi cells is used a priori. Because the milestoning method also relies on a predefined CV subspace of low dimensionality, it too could potentially fail to capture important features of the landscape of accessible conformations.
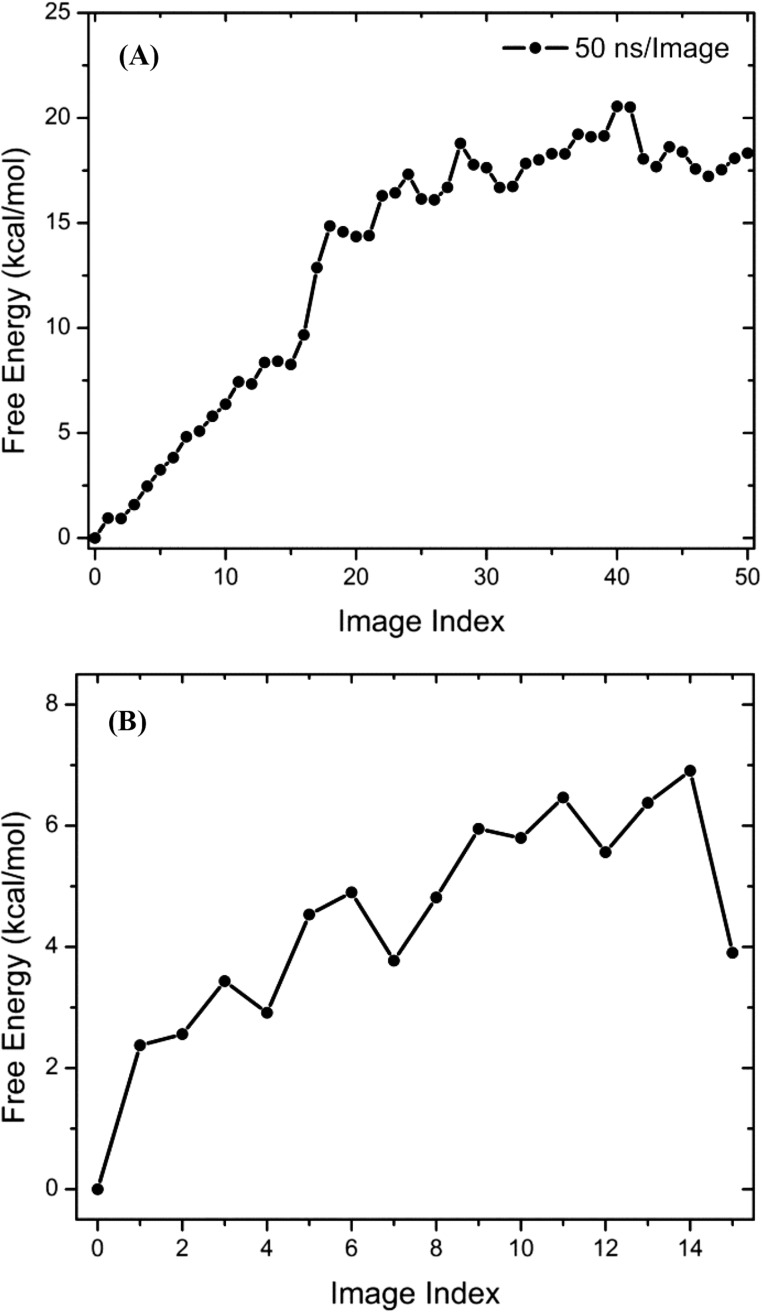
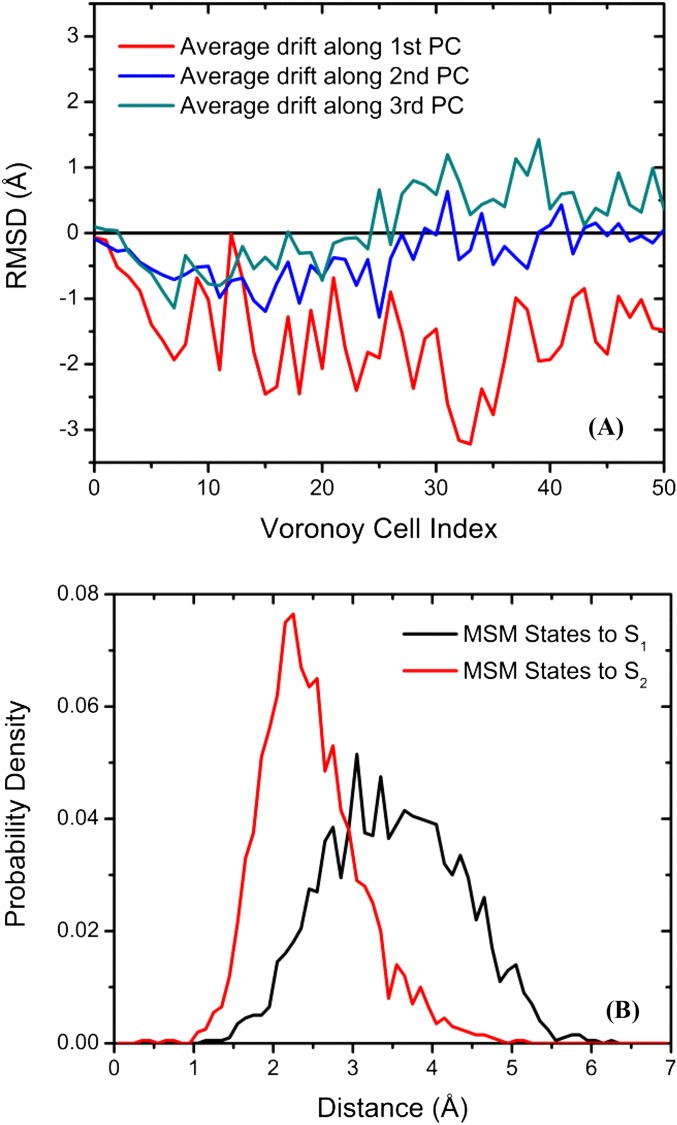
One strategy to partition the CV space is to use a perfectly converged string representing the minimum free energy pathway. Here, the milestoning simulations started from an optimized string S1 that was also used to seed the Folding@Home simulations. Fifty-nanosecond MD trajectories were accumulated for each Voronoi cell, and a total of 2.55 μs of MD sampling was eventually achieved. Using the 50-ns milestoning trajectories, a 1D-PMF as a function of image index (also the index of Voronoi cell centers) is computed and plotted in Fig. S9A, whereas the free energy profile along S0 is shown in Fig. S9B. One should note here that the 1D-PMF in Fig. S9B is based on ∼500 μs of MD sampling. Comparing the two 1D-PMFs, one could see that the PMF from milestoning simulations yields two metastable states (one for the inactive kinase domain and the other for the active kinase domain) separated by an energy barrier. The same behavior is also seen from the TPT analysis. However, both the relative free energy between inactive and active metastable states and the free energy barrier are too high for the milestoning simulations, when quantitatively compared with the PMF yielded from MSM and TPT. To understand the difference, we recomputed the geometric center from the milestoning simulation in each Voronoi cell. A new string S2 can be constructed by connecting adjacent geometric center. A drift from S1 can be observed when comparing S1 and S2, as demonstrated in Fig. S10. Further, projecting the MSM microstates onto S1 and S2 also demonstrates that the microstates are closer to S2. Our work suggests that an optimized string may seem to be a minimum free energy pathway during the optimization process of string methods (i.e., plateaus can be seen when examining the rmsd with respect to the initial and/or final strings). However, the string can still be drifting very slowly toward the true minimum free energy pathway: This drift becomes clear in a timescale of 50 ns per image in our application of the milestoning method to the Src activation. Our work indicates that obtaining a minimum free energy pathway using the string method is a slow process and requires large-scale conformational sampling. In addition, choosing/designing a metric that is able to indicate the convergence of the string method better than simply comparing the rmsd with respect to the initial and/or final strings is also an important aspect of practical application of the string method.
Fig. S9.
Free energy profiles along strings. (A) Free energy profile along S1 from Markovian milestoning simulations. (B) Free energy profile along S0 from the MSM microstates.
Fig. S10.
Analysis of the markovian milestoning simulations. (A) Average drift along principal components as a function of image index. For the sake of clarity, only the drifts along the first three principal components are displayed. (B) Histogram of distances from MSM microstates to strings S1 and S2.
Conclusion
Large-scale collective motions are essential to biological macromolecules such as proteins. Independent all-atom MD simulations with explicit solvent models can be aggregated to elucidate the molecular mechanisms of such motions and to quantitatively characterize the thermodynamic and kinetic properties, based on MSMs. When the conformational transition becomes complex, more powerful theoretical frameworks such as TPT and the transition reweighted analysis method (35) can be combined with MSMs to interpret the dynamical process.
In the present study, we aimed to understand the nature of conformational transitions in c-Src tyrosine kinase, which is an example of allosteric biological macromolecules. The conformational transition that activates the c-Src kinase domain was analyzed based on aggregated data from unbiased MD simulations within a theoretical framework provided by MSM and TPT: TPT was applied to the Markov microstates from the massively distributed MD simulations. The TPT analysis indicates that the activating transition takes place via a dense set of intermediate microstates distributed within a broad and curved multidimensional “reaction tube” connecting the two end-point conformations. Large transition fluxes tend to appear near the central axis of the tube and are likely to pass through metastable microstates that are not highly populated at equilibrium. Thus, microstates with low Boltzmann weight cannot be ignored in the study of conformational transition because they may be needed to correctly represent the kinetics. Therefore, it is understandable that conformational sampling is crucial in estimating kinetic quantities accurately. However, metastable states with large equilibrium probability and with large probability to observe a reactive trajectory can be found at the outer region of the transition tube and do not necessarily contribute to the flow of transition probability current significantly. Although the pathways display considerable variability, they are nonetheless confined within a broad “reaction tube” consistent with the view that the inactive-to-active transition in c-Src kinase proceeds first by an opening of the A-loop, followed by an inward rotation of the αC-helix (19, 20).
More generally, the present analysis offers a perspective for the interpretation of a conformational transition in an important signaling allosteric protein. Our findings point out that both transition flux and equilibrium probability are essential to understand conformational changes and they need to be taken into account simultaneously. Furthermore, transition pathways populate a broad and curly reaction tube. This could explain the difficulty in representing the conformational transition via a unique and well-defined minimum free energy pathway in practice. Also, we show that metastable states with low Boltzmann weight are able to affect kinetics and should not be ignored in the study of conformational transition. Finally, our previous modeling has suggested that compounds that stabilize the inactive and the intermediate conformations of the kinase domain might be used as c-Src inhibitors, because they can prevent activating c-Src kinase domain (20). Therefore, an understanding of the dynamic process can have a broader impact.
Methods
TPT.
A brief description of TPT is given in SI Methods, and one could refer to Noé et al. (17) and Vanden-Eijnden (21) for more details.
Markovian Milestoning Calculation.
Markovian milestoning simulations were also carried out to characterize the activating conformational transition in the catalytic domain of c-Src (see SI Methods for an introduction to the methodology and the computational details).
Acknowledgments
Y.M. thanks Drs. Avisek Das, Mikolai Fajer, and Luca Maragliano for insightful discussions. This work was supported by National Cancer Institute, NIH Grant CAO93577 (to Y.M. and B.R.) and the Simulation of Biological Structures NIH National Center for Biomedical Computation through NIH Roadmap for Medical Research Grant U54 GM07297 (to D.S. and V.P). The computations were supported by Extreme Science and Engineering Discovery Environment Grant OCI-1053575, by NIH through resources provided by the Computation Institute and the Biological Sciences Division of the University of Chicago and Argonne National Laboratory under Grant S10 RR029030-01, and by a Biomedical Data Science Initiative Postdoctoral Fellowship from Stanford School of Medicine (D.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602790113/-/DCSupplemental.
References
- 1.Grant BJ, Gorfe AA, McCammon JA. Large conformational changes in proteins: Signaling and other functions. Curr Opin Struct Biol. 2010;20(2):142–147. doi: 10.1016/j.sbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JC, Roux B. Eppur si muove! The 2013 Nobel Prize in Chemistry. Structure. 2013;21(12):2102–2105. doi: 10.1016/j.str.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 5.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338(6110):1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 6.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100(22):12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 8.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22(4):337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 9.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 10.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comput Phys. 1977;23(2):187–199. [Google Scholar]
- 11.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99(20):12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chodera JD, Noé F. Markov state models of biomolecular conformational dynamics. Curr Opin Struct Biol. 2014;25:135–144. doi: 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan AC, Roux B. Building Markov state models along pathways to determine free energies and rates of transitions. J Chem Phys. 2008;129(6):064107. doi: 10.1063/1.2959573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pande VS. Understanding protein folding using Markov state models. Adv Exp Med Biol. 2014;797:101–106. doi: 10.1007/978-94-007-7606-7_8. [DOI] [PubMed] [Google Scholar]
- 15.Noé F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18(2):154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Lane TJ, Shukla D, Beauchamp KA, Pande VS. To milliseconds and beyond: challenges in the simulation of protein folding. Curr Opin Struct Biol. 2013;23(1):58–65. doi: 10.1016/j.sbi.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci USA. 2009;106(45):19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boras BW, Kornev A, Taylor SS, McCulloch AD. Using Markov state models to develop a mechanistic understanding of protein kinase A regulatory subunit RIα activation in response to cAMP binding. J Biol Chem. 2014;289(43):30040–30051. doi: 10.1074/jbc.M114.568907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Banavali NK, Roux B. Mapping the conformational transition in Src activation by cumulating the information from multiple molecular dynamics trajectories. Proc Natl Acad Sci USA. 2009;106(10):3776–3781. doi: 10.1073/pnas.0808261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla D, Meng Y, Roux B, Pande VS. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat Commun. 2014;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanden-Eijnden E. Transition path theory. Adv Exp Med Biol. 2014;797:91–100. doi: 10.1007/978-94-007-7606-7_7. [DOI] [PubMed] [Google Scholar]
- 22.E W, Vanden-Eijnden E. Transition-path theory and path-finding algorithms for the study of rare events. Annu Rev Phys Chem. 2010;61:391–420. doi: 10.1146/annurev.physchem.040808.090412. [DOI] [PubMed] [Google Scholar]
- 23.Voelz VA, Bowman GR, Beauchamp K, Pande VS. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1-39) J Am Chem Soc. 2010;132(5):1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng NJ, Zheng W, Gallicchio E, Levy RM. Insights into the dynamics of HIV-1 protease: A kinetic network model constructed from atomistic simulations. J Am Chem Soc. 2011;133(24):9387–9394. doi: 10.1021/ja2008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3(5):629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 26.Cowan-Jacob SW, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13(6):861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Peters B, Trout BL. Obtaining reaction coordinates by likelihood maximization. J Chem Phys. 2006;125(5):054108. doi: 10.1063/1.2234477. [DOI] [PubMed] [Google Scholar]
- 28.Best RB, Hummer G. Reaction coordinates and rates from transition paths. Proc Natl Acad Sci USA. 2005;102(19):6732–6737. doi: 10.1073/pnas.0408098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G. String method in collective variables: Minimum free energy paths and isocommittor surfaces. J Chem Phys. 2006;125(2):24106. doi: 10.1063/1.2212942. [DOI] [PubMed] [Google Scholar]
- 30.Pan AC, Sezer D, Roux B. Finding transition pathways using the string method with swarms of trajectories. J Phys Chem B. 2008;112(11):3432–3440. doi: 10.1021/jp0777059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovchinnikov V, Cecchini M, Vanden-Eijnden E, Karplus M. A conformational transition in the myosin VI converter contributes to the variable step size. Biophys J. 2011;101(10):2436–2444. doi: 10.1016/j.bpj.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ovchinnikov V, Karplus M, Vanden-Eijnden E. Free energy of conformational transition paths in biomolecules: The string method and its application to myosin VI. J Chem Phys. 2011;134(8):085103. doi: 10.1063/1.3544209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maragliano L, Vanden-Eijnden E, Roux B. Free energy and kinetics of conformational transitions from Voronoi tessellated milestoning with restraining potentials. J Chem Theory Comput. 2009;5(10):2589–2594. doi: 10.1021/ct900279z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanden-Eijnden E, Venturoli M. Markovian milestoning with Voronoi tessellations. J Chem Phys. 2009;130(19):194101. doi: 10.1063/1.3129843. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Mey AS, Rosta E, Noé F. Statistically optimal analysis of state-discretized trajectory data from multiple thermodynamic states. J Chem Phys. 2014;141(21):214106. doi: 10.1063/1.4902240. [DOI] [PubMed] [Google Scholar]
- 36.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastian M, Heymann S, Jacomy M. 2009. Gephi: An open source software for exploring and manipulating networks. Proceedings of the Third International AAAI Conference on Weblogs and Social Media (Assoc for the Advancement of Artificial Intelligence, Palo Alto, CA), pp 361–362. [DOI] [PMC free article] [PubMed]
- 38.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 40.Zheng J, et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49(Pt 3):362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 41.Metzner P, Schutte C, Vanden-Eijnden E. Transition path theory for Markov jump processes. Multiscale Model Simul. 2009;7(3):1192–1219. [Google Scholar]
- 42.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 44.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 45.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 46.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]