Blending molecular developmental biology and quantitative genetics, Hlusko et al. (1) present in PNAS a framework for identifying phenotypic traits whose genetics are suitable for testing hypotheses about adaptive evolution in the fossil record. In recent years, startling advances have been made in our understanding of the molecular mechanisms of development and the genomic structures that underpin them. Laboratory manipulations are able to determine the patterns of gene expression that produce phenotypic structures such as brains, bones, and teeth (2, 3); computational models of the spatial dynamics of development are capable of predicting the phenotypic outcomes of up- or down-regulation of gene expression (4); and genomic analyses reveal the regulatory mechanisms that govern cascades of gene expression (5). These tools have mapped the causal links between genotype and phenotype that were largely missing from 20th century evolutionary biology.
However, this new understanding is based on a small number of model organisms. Translating molecular genetics from them to the fossil record, which provides the only direct evidence for most evolutionary transitions, can be problematic because many aspects of morphogenesis depend on the organism’s genetic and environmental context (6, 7). In the fossil record, genetic and nongenetic components of phenotypic variation can rarely be separated, and many important fossil taxa are phylogenetically distant from model organisms like mice and chickens. Hlusko et al. (1) combine the predictive power of gene expression and quantitative genetics to identify fossilizable morphological traits that are both heritable and uncomplicated by pleiotropy (genetic correlations) with other traits. Traits that meet these criteria are ones from which the history of natural selection, genetic drift, and taxonomic differentiation can most accurately be reconstructed from the fossil record.
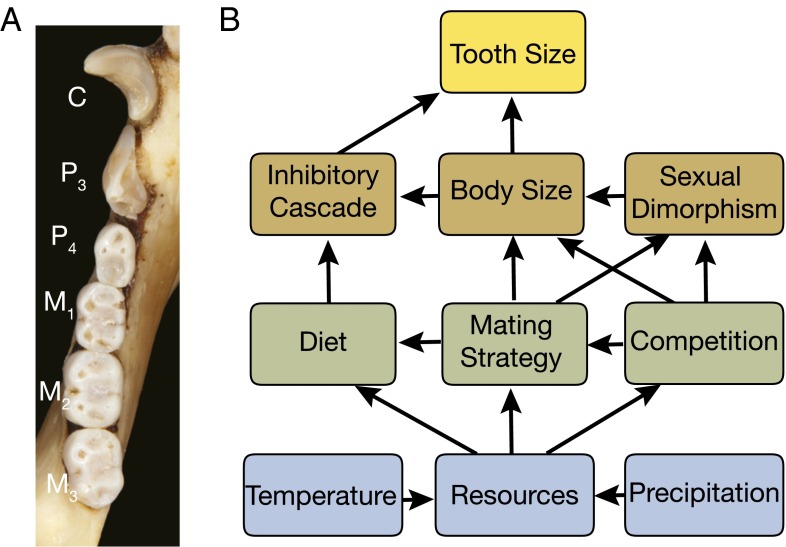
Teeth best exemplify the problem addressed by Hlusko et al. (1). For mammals, compact, dense, and durable teeth are the most commonly preserved objects in the fossil record (8, 9), and because they have complex, quickly evolving phenotypes, even an isolated mammal tooth can usually be identified at the level of species (10). Tooth traits provide a rich source of evidence for genetic differentiation and dietary function in the geological past (11–13). Teeth are frequently used to measure rates and modes of evolution (14, 15) and to study which selective factors were important in the evolutionary history of clades such as our own (16, 17). However, the genetic and developmental processes that produce teeth are often linked to confounding factors, links that differ from one clade to another. Hlusko et al. (1) offer the example of the complex relationship between teeth and body size. Sexual dimorphism in size, including the dentition, is common in primates (18, 19), but proportional tooth size is also linked to molecular genetic cascades of activators and inhibitors that produce sequences of molar teeth, and those proportions are linked to dietary function across a wide range of mammals, including primates (20–24). Selection for body size, sexual dimorphism, and dietary specialization can thus all have competing effects on primate dental traits (Fig. 1). A change in primate dental proportions observed in the fossil record could therefore be selectively linked to temperature’s effect on body size according to Bergmann’s rule (25), to precipitation’s effect on vegetation and diet (26), to resource competition from a newly evolved species (27), or to interspecific competition for mates and sexual dimorphism resulting from changes in habitat availability (28). Reconstructing historical patterns of selection from the fossil record can easily become a multivariate, multifactorial quagmire. Model organisms like mice do not provide easy solutions because, unlike primates, murid rodents are not sexually dimorphic in size, so their nexus of selective factors is necessarily different (29).
Fig. 1.
(A) Lower dentition of a De Brazza’s monkey. C, canine; M, molar; P, premolar (Michigan State University Museum 29074). Reprinted from Animal Diversity Web, with permission from Phil Myers (University of Michigan, Ann Arbor, MI). (B) Links between a few of the factors that influence tooth size in primates.
Although the interaction between selective factors and pleiotropic traits is of intrinsic interest (30), interpretations of paleontological data would be easier if better predictive models were available for traits that are likely to evolve independently. Hlusko et al. (1) marshal the predictive power of quantitative genetics to identify traits that are highly evolvable and unlikely to be linked in potentially confounding ways. Phenotypes are a complex interaction between genetics and environment, and any trait has components of both (31). The genetic proportion of phenotypic variation is known as narrow sense heritability (h2). Because natural selection only works on heritable variation, traits with higher heritabilities and more genetic variance are therefore more evolvable (32). Covariation between traits also has heritable and nonheritable components, all of which can be estimated using data from parents and offspring or individuals connected by a larger pedigree (31). Multivariate heritability estimates (also known as G matrices) are therefore a complement to molecular developmental interactions in defining the genotype-to-phenotype map.
Using a baboon colony, Hlusko et al. (1) assessed heritability and pleiotropy of traits that had previously been identified as being correlated based on molecular developmental work in mice. Their goal was to verify which mouse model traits have the desirable properties of high heritability and low pleiotropy in primates. They found that h2 in primates ranged from about 0.4 to nearly 0.9. Two composite traits were classified as “genetically patterned” (GP) because of their high heritability, developmental linkage by gene expression cascades, and relevance to dental function: the proportional length of the third molar to the first molar and the proportional length of the fourth premolar to the second molar. The authors then tested whether these traits have indeed evolved independently in response to selection in Old World monkeys. Once phylogenetic covariances were removed, these traits showed no correlation with one another, with sexual dimorphism, or with age. Furthermore, using the primate fossil record, they showed that rates of evolution and disparity in these traits increased as vegetation changed during the global climatic transition in the Late Miocene (∼15–5 million y ago). Species richness and clade diversity changed along with the GP traits as the primate fauna turned over from one dominated by extinct Miocene apes to today's monkey-dominated fauna that includes baboons. Likewise, many of the dental specializations possessed by apes during the early Miocene are now found in living Old World monkeys. The processes that have shaped evolution are putatively clearer in GP traits because they are independently evolvable, uncluttered by pleiotropic associations with multiple factors.
Hlusko et al. (1) argue that heritability and pleiotropy can be estimated in new taxa more quickly and cheaply than data on molecular developmental interactions, especially with pedigrees derived from molecular markers. Quantitative genetics therefore allow phenotypic traits to be triaged in almost any clade that has extant members, thus providing a first check on assumptions about how traits will evolve that are based on developmental genetics in model organisms. Furthermore, they argue that quantitative genetics provide critical information about nonheritable variation that should be taken into account in evaluating alternative hypotheses about the causes of phenotypic change observed in the fossil record.
The fusion of quantitative genetics with molecular developmental biology is not a panacea for evolutionary paleontology, of course. The genetic structure of traits can evolve so quickly that even local estimates from extant members of a clade may not be applicable even to closely related extinct subclades (33). In addition, the methods used to estimate heritability and pleiotropy, like morphometrics, have limits because they typically require that the same set of variables be present in all taxa, and therefore cope poorly with evolutionary novelty (34). Nevertheless, the work of Hlusko et al. (1) sets a new, higher standard for how to study selection and morphological evolution in the fossil record.
Acknowledgments
This research is supported by the National Science Foundation (Grant EAR 1338298).
Footnotes
The author declares no conflict of interest.
See companion article on page 9262.
References
- 1.Hlusko LJ, Schmitt CA, Monson TA, Brasil MF, Mahaney MC. The integration of quantitative genetics, paleontology, and neontology reveals genetic underpinnings of primate dental evolution. Proc Natl Acad Sci USA. 2016;113:9262–9267. doi: 10.1073/pnas.1605901113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thesleff I, Juuri E. Tooth development. In: McCauley LK, Somerman MJ, editors. Mineralized Tissues in Oral and Craniofacial Science. Wiley–Blackwell; Oxford: 2012. pp. 119–128. [Google Scholar]
- 3.Hall BK, Gillis JA. Incremental evolution of the neural crest, neural crest cells and neural crest-derived skeletal tissues. J Anat. 2013;222(1):19–31. doi: 10.1111/j.1469-7580.2012.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salazar-Ciudad I, Marín-Riera M. Adaptive dynamics under development-based genotype-phenotype maps. Nature. 2013;497(7449):361–364. doi: 10.1038/nature12142. [DOI] [PubMed] [Google Scholar]
- 5.Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14(3):168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- 6.Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat Rev Genet. 2011;12(3):204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 7.Drown DM, Wade MJ. Runaway coevolution: Adaptation to heritable and nonheritable environments. Evolution. 2014;68(10):3039–3046. doi: 10.1111/evo.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badgley C. Taphonomy of mammalian fossil remains from Siwalik rocks of Pakistan. Paleobiology. 1986;12(2):119–142. [Google Scholar]
- 9.Briggs DEG. The role of decay and mineralization in the preservation of soft-bodied fossils. Annu Rev Earth Planet Sci. 2003;31:275–301. [Google Scholar]
- 10.Polly PD, Le Comber SC, Burland TM. On the occlusal fit of tribosphenic molars: Are we underestimating species diversity in the Mesozoic? J Mamm Evol. 2005;12(1):285–301. [Google Scholar]
- 11.Szuma E. Geography of dental polymorphism in the red fox Vulpes vulpes and its evolutionary implications. Biol J Linn Soc Lond. 2007;90(1):61–84. [Google Scholar]
- 12.Santana SE, Strait S, Dumont ER. The better to eat you with: Functional correlates of tooth structure in bats. Funct Ecol. 2011;25(4):839–847. [Google Scholar]
- 13.Schultz JA, Martin T. Function of pretribosphenic and tribosphenic mammalian molars inferred from 3D animation. Naturwissenschaften. 2014;101(10):771–781. doi: 10.1007/s00114-014-1214-y. [DOI] [PubMed] [Google Scholar]
- 14.Meloro C, Raia P. Cats and dogs down the tree: The tempo and mode of evolution in the lower carnassial of fossil and living Carnivora. Evol Biol. 2010;37(4):177–186. [Google Scholar]
- 15.Gingerich PD. Rates of evolution. Annu Rev Ecol Evol Syst. 2009;40:657–675. [Google Scholar]
- 16.Bobe R, Behrensmeyer AK. The expansion of grassland ecosystems in Africa in relation to mammalian evolution and the origin of the genus Homo. Palaeogeogr Palaeoclimatol Palaeoecol. 2004;207(3-4):399–420. [Google Scholar]
- 17.Ungar PS. Reproductive fitness and tooth wear: Milking as much as possible out of dental topographic analysis. Proc Natl Acad Sci USA. 2005;102(46):16533–16534. doi: 10.1073/pnas.0508642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingerich PD, Smith BH, Rosenberg K. Allometric scaling in the dentition of primates and prediction of body weight from tooth size in fossils. Am J Phys Anthropol. 1982;58(1):81–100. doi: 10.1002/ajpa.1330580110. [DOI] [PubMed] [Google Scholar]
- 19.Ungar PS. Dental allometry in mammals: A retrospective. Ann Zool Fenn. 2014;51(1-2):177–187. [Google Scholar]
- 20.Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449(7161):427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 21.Renvoisé E, et al. Evolution of mammal tooth patterns: New insights from a developmental prediction model. Evolution. 2009;63(5):1327–1340. doi: 10.1111/j.1558-5646.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- 22.Halliday TJ, Goswami A. Testing the inhibitory cascade model in Mesozoic and Cenozoic mammaliaforms. BMC Evol Biol. 2013;13(79):79. doi: 10.1186/1471-2148-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroer K, Wood B. Modeling the dental development of fossil hominins through the inhibitory cascade. J Anat. 2015;226(2):150–162. doi: 10.1111/joa.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans AR, et al. A simple rule governs the evolution and development of hominin tooth size. Nature. 2016;530(7591):477–480. doi: 10.1038/nature16972. [DOI] [PubMed] [Google Scholar]
- 25.Meiri S. Bergmann’s Rule–what’s in a name? Glob Ecol Biogeogr. 2011;20(1):203–207. [Google Scholar]
- 26.Godfrey LR, Winchester JM, King SJ, Boyer DM, Jernvall J. Dental topography indicates ecological contraction of lemur communities. Am J Phys Anthropol. 2012;148(2):215–227. doi: 10.1002/ajpa.21615. [DOI] [PubMed] [Google Scholar]
- 27.Plavcan JM, van Schaik CP. Intrasexual competition and canine dimorphism in anthropoid primates. Am J Phys Anthropol. 1992;87(4):461–477. doi: 10.1002/ajpa.1330870407. [DOI] [PubMed] [Google Scholar]
- 28.Sauther ML, Sussman RW. A new interpretation of the social organization and mating system of the Ringtailed Lemur (Lemur catta) In: Kappeler PM, Ganzhorn JU, editors. Lemur Social Systems and Their Ecological Basis. Springer; New York: 1993. pp. 111–121. [Google Scholar]
- 29.Lindenfors P, Gittleman JL, Jones KE. Sexual size dimorphism in mammals. In: Fairbairn DJ, Blanckenhorn WU, Székely T, editors. Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism. Oxford Univ Press; Oxford: 2007. pp. 16–26. [Google Scholar]
- 30.Kingsolver JG, Schemske DW. Path analyses of selection. Trends Ecol Evol. 1991;6(9):276–280. doi: 10.1016/0169-5347(91)90004-H. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 32.Hansen TF, Houle D. Measuring and comparing evolvability and constraint in multivariate characters. J Evol Biol. 2008;21(5):1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. [DOI] [PubMed] [Google Scholar]
- 33.Eroukhmanoff F. Just how much is the G-matrix actually constraining adaptation? Evol Biol. 2009;36(3):323–326. [Google Scholar]
- 34.Polly PD. Developmental dynamics and G-matrices: Can morphometric spaces be used to model evolution and development? Evol Biol. 2008;35:83–96. [Google Scholar]